Abstract
Stem and progenitor cells maintain a robust DNA replication program during the tissue expansion phase of embryogenesis. The unique mechanism that protects them from the increased risk of replication-induced DNA damage, and hence permits self-renewal, remains unclear. To determine whether the genome integrity of stem/progenitor cells is safeguarded by mechanisms involving molecules beyond the core DNA repair machinery, we created a nucleostemin (a stem and cancer cell-enriched protein) conditional-null allele and showed that neural-specific knockout of nucleostemin predisposes embryos to spontaneous DNA damage that leads to severe brain defects in vivo. In cultured neural stem cells, depletion of nucleostemin triggers replication-dependent DNA damage and perturbs self-renewal, whereas overexpression of nucleostemin shows a protective effect against hydroxyurea-induced DNA damage. Mechanistic studies performed in mouse embryonic fibroblast cells showed that loss of nucleostemin triggers DNA damage and growth arrest independently of the p53 status or rRNA synthesis. Instead, nucleostemin is directly recruited to DNA damage sites and regulates the recruitment of the core repair protein, RAD51, to hydroxyurea-induced foci. This work establishes the primary function of nucleostemin in maintaining the genomic stability of actively dividing stem/progenitor cells by promoting the recruitment of RAD51 to stalled replication-induced DNA damage foci.
Keywords: DNA damage repair, homologous recombination, conditional knockout, replication fork stalling, neural development
Stem and progenitor cells play critical roles in embryonic organogenesis, adult tissue regeneration, and tumor development. To maintain self-renewing proliferation, they must be protected from replication-induced DNA damage that limits the proliferative lifespan of most dividing cells. Replication-induced DNA damage may occur spontaneously as a result of stalled and collapsed replication forks, caused by the slowing of the DNA replication machinery over replication “trouble” zones or previously unrepaired damage sites (1–3). Alternatively, replication stalling can be triggered by drugs that deplete the endogenous nucleotide pool (e.g., hydroxyurea) or inhibit the activity of DNA replication (e.g., camptothecin). Prolonged replication stalling will lead to the collapse of replication machinery and double-strand DNA breaks (DSBs), causing cell cycle arrest and genomic instability (4, 5). To date, it remains unclear how stem and progenitor cells survive the increased risk of replication-induced DNA damage during this hyperactive mitotic window of embryogenesis.
Nucleostemin (NS) is a stem cell-enriched nucleolar protein (6). Its biological significance has been illustrated by the early embryonic lethal phenotype of NS germ-line knockout mice (7), which ineluctably hinders further analyses of its in vivo function beyond the blastula stage. To date, the mechanism of NS action in vivo remains unclear. Although some suggested a connection to the p53 pathway (6, 8–10), others have shown that NS is functionally indispensable even in p53-null cells (11, 12). We recently discovered a function of NS in preventing DNA damage on interstitial and telomeric chromosomes, which, by far, best recapitulates its obligatory role in biology (13). Although the telomere-protecting function of NS is mediated by telomeric repeat binding factor 1 (TRF1) SUMOylation and promyelocytic leukemia protein isoform IV (PML-IV) recruitment (13), the role of NS in protecting nontelomeric chromosomes is yet unknown. We hypothesize that NS may formulate a unique mechanism that protects stem/progenitor cells from spontaneous DNA damage as a result of DNA replication. To investigate this idea, we created the NS-flox (NSf) allele and two NS conditional-knockout (NScko) mouse models. Analyses of these mouse models revealed that neural-specific deletion of NS increases the frequency of spontaneous DNA damage and eliminates neural stem cells (NSCs) in the developing neuroepithelium. DNA damage triggered by NS deletion occurs in a replication-dependent manner and independently of the p53 status or ribosomal RNA (rRNA) synthesis. More importantly, NS is capable of being recruited to DNA damage sites and is required for the efficient recruitment of RAD51 to stalled replication-induced damage foci. This work signifies a primary role of NS in maintaining the genomic stability of stem and progenitor cells.
Results
Creation of a NS Conditional-Knockout Allele.
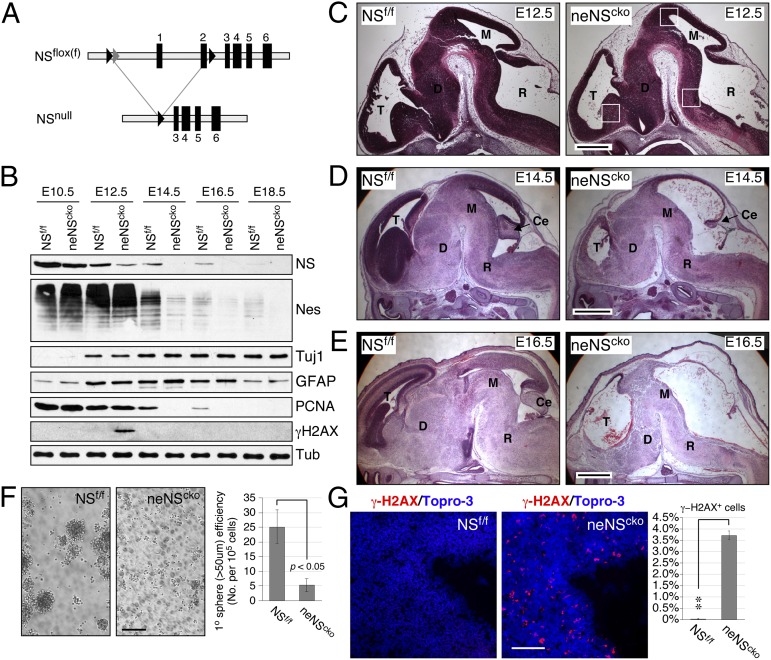
The gene-targeting strategy was used to generate ES cells with a NSfloxneo (NSfn) allele, which contains a loxP site linked to an frt-flanked phosphoglycerate kinase-neomycin (pgk-neo) cassette at 820 bp upstream of the first exon and a second loxP site in the second intron (Fig. 1A and Fig. S1A). After germ-line transmission, the pgk-neo cassette was deleted from the NSfn allele to generate the NSflox (NSf) allele (Fig. S1 B–D). NSf/f homozygotes manifest normal developmental, growth, and reproductive activities, and homozygous deletion of the floxed sequence (NSnull) results in early embryonic lethality at embryonic day (E) 3.5 (Fig. S1E). These findings confirm that the NSf allele is a bona fide conditional-null allele.
Fig. 1.
Neural-specific deletion of nucleostemin (NS) induces DNA damage, decreases embryonic neural stem cells (NSCs), and causes severe brain defects in vivo. (A) Diagrams of the NS-flox (NSf) and null (NSnull) alleles. Black arrowheads, loxP; gray arrowheads, frt. (B) Crenestin-driven NS deletion (neNScko) decreases its protein in the developing forebrain from embryonic day 12.5 (E12.5). The levels of nestin and PCNA proteins are reduced from E14.5. An increase of γ-H2AX proteins is noted in the neNScko forebrain at E12.5. Tuj1, neuron-specific β-tubulin; GFAP, glial fibrillary acid protein. (C) Compared with control NSf/f embryos, neNScko embryos begin to show a decreased cellularity in the telencephalic (T), diencephalic (D), mesencephalic (M), and rhombencephalic (R) neuroepithelium at E12.5. Squares indicate regions shown in G and Fig. S2B. At E14.5 (D) and E16.5 (E), neNScko embryos exhibit severe brain defects throughout the neural axis. Ce, cerebellar primordium. (F) At E12.5, the forebrains of neNScko embryos contain much less sphere-forming NSCs than that of NSf/f embryos. (G) A notable phenotype of NS deletion is the increase of γ-H2AX+ cells in the ventricular zone of the E12.5 neuroepithelium. The region shown here is close to the midbrain roof. (Scale bars: 500 µm in C, 1 mm in D and E, and 100 µm in F and G.) Bar graphs, mean ± SEM; *P < 0.01; **P < 0.001; ***P < 0.0001.
Neural-Specific Deletion of NS Triggers DNA Damage and Decreases NSCs in Vivo.
To study the NS function in the developing brain, a nestin promoter-driven Cre (Crenestin) (14) was used to delete NS in the neuroepithelial stem/progenitor cells (neNScko). Homozygous neNScko mice die immediately after birth due to respiratory failure. Western blots showed that Crenestin-driven NS deletion reduces NS proteins in the developing forebrain most significantly at E12.5 and older, followed by the decrease of nestin and proliferative cell nuclear antigen (PCNA) proteins in E14.5 and older embryos (Fig. 1B). The delayed onset of PCNA down-regulation compared with that of NS suggests a delayed cell cycle exit following the loss of NS. To investigate the neural defects of neNScko mice, embryos were collected at consecutive developmental stages. Histological analyses showed that the neuroepithelial morphology of neNScko embryos appears the same as that of NSf/f embryos at E10.5 (Fig. S2A) but begins to show reduced cellularity throughout the neural axis at E12.5 (Fig. 1C). At E14.5 and E16.5, neNScko embryos show major defects in the telencephalic cortex, ganglionic eminence, cerebellar primordium, midbrain tectum, smaller diencephalon, midbrain and hindbrain tegmentum, and spinal cord compared with their NSf/f littermates (Fig. 1 D and E). Associated with those defects is a decrease of NSCs in neNScko neuroepithelium at E12.5 (Fig. 1F). We also noticed that E12.5 neNScko embryos show a significant increase of γ-H2AX proteins and γ-H2AX+ cells in their neuroepithelium (Fig. 1 B and G and Fig. S2B).
NS Protects NSCs from DNA Damage and Maintains Their Self-Renewal.
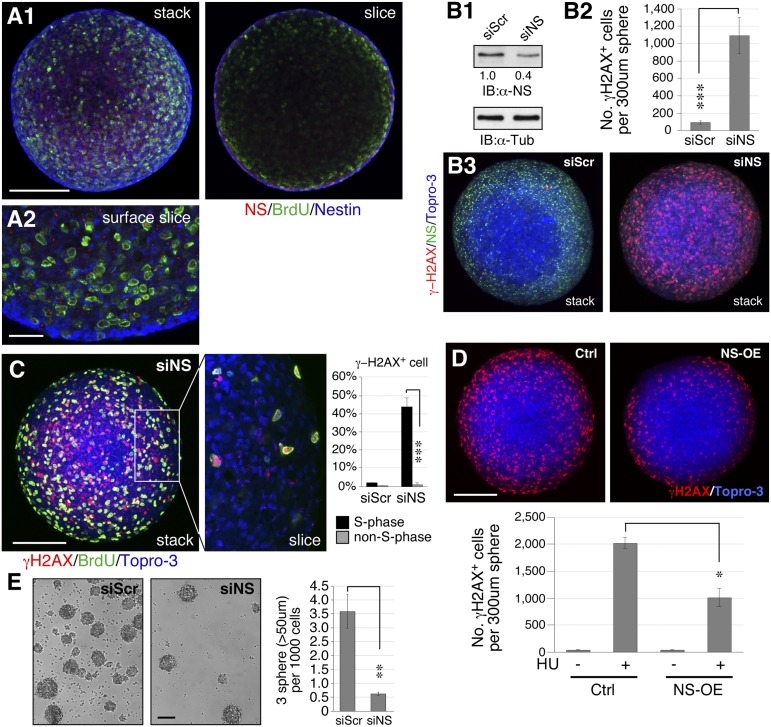
NSCs were isolated from the E12.5 forebrain cortex and grown as neurospheres in suspension culture. Confocal analyses showed that most nestin+, NS+, and BrdU-labeled cells are found in the peripheral region of the sphere (Fig. 2A). The siRNA-based knockdown approach allows a 60% reduction of NS proteins in neurospheres (Fig. 2B1) and produces a striking increase of γ-H2AX+ cells spontaneously (P < 0.0001, Fig. 2 B2 and B3). This NS RNAi (siNS)-mediated knockdown (NSKD) efficiency for the whole neurosphere is moderate compared with what we’ve seen in monolayer cultures (80–90%), which may be due to the three-dimensional structure of neurospheres. DNA replication is one major source of spontaneous DNA damage that occurs in the S-phase cells. We noted that NSKD causes more DNA damage on S-phase (BrdU+) neurosphere cells compared with non–S-phase (BrdU−) cells (Fig. 2C). Conversely, overexpression of NS can reduce the amount of DNA damage caused by hydroxyurea (HU) in NSCs (Fig. 2D). Along with its DNA damage effect, NS depletion also impairs the self-renewal of NSCs, as indicated by the decreased tertiary sphere formation from secondary neurosphere cells (358.3 versus 61.3 spheres per 105 plated cells) (Fig. 2E). These results demonstrate that NS plays an important role in maintaining the self-renewal of NSCs by protecting them from replication-dependent DNA damage.
Fig. 2.
Loss of NS induces DNA damage and perturbs self-renewal in NSCs. (A) NSCs were isolated from the E12.5 cortex of NSf/f embryos and cultured as neurospheres. Dividing neuroepithelial precursors (BrdU+ [green] and nestin+ [blue]) are found in the peripheral zone of the sphere and coincide mostly with the NS signal (red). (A1) shows a stacked image (Left) and a single slice through the center (Right). (A2) shows an enlarged view of a single slice close to the surface. (B1) RNAi-based NS knockdown (siNS) gives a 60% knockdown efficiency in the whole neurosphere. (B2 and B3) Loss of NS significantly increases the number of γ-H2AX+ cells in neurospheres. (C) The S-phase (BrdU+) cells are much more susceptible to the DNA damage effect of NSKD than the non–S-phase (BrdU−) cells. (D) NS overexpression protects neurosphere cells from hydroxyurea (HU)-induced DNA damage (γ-H2AX+). (E) NSKD by siNS significantly diminishes the ability of secondary neurosphere cells to form tertiary neurospheres. y axis indicates the number of tertiary neurospheres formed for every 1,000 secondary neurosphere cells plated. (Scale bars: 100 µm in A1 and B–E, 25 µm in A2.) Bar graphs (see description in Fig. 1).
NSKO-Induced DNA Damage Does Not Depend on p53 or Affect rRNA Synthesis.
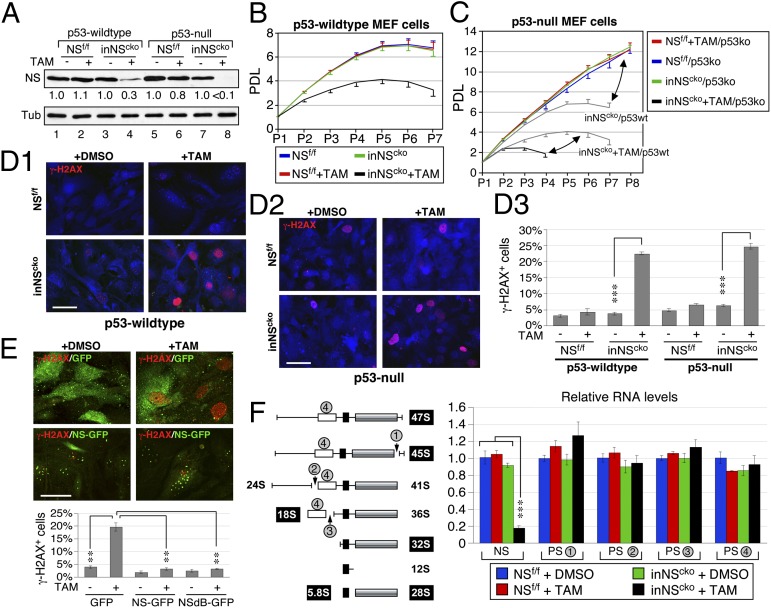
To define the role of p53 in the NSKO event, a TAM-inducible NScko mouse model (inNScko) was created by introducing the CreER transgene (15) into NSf/f mice, followed by breeding of inNScko and NSf/f mice into the p53-null background (16). Mouse embryonic fibroblast (MEF) cells were prepared from E13.5 embryos. Western blots showed that TAM treatment (0.1 µM) decreases the NS protein level by 70% in p53-wild type inNScko cells (lanes 3 and 4) and by >90% in p53-null inNScko cells (lanes 7 and 8), but has no effect on NSf/f cells (Fig. 3A). Cell growth studies showed that loss of NS impairs the long-term proliferative potential of MEF cells, as shown by the reduced population doubling level (PDL) and early growth plateau (Fig. 3B), and that p53 deletion allows NS wild-type MEF cells to escape senescence but significantly shortens the lifespan of NSKO cells (Fig. 3C). DNA damage analyses showed that NS deletion increases the percentage of γ-H2AX+ MEF cells regardless of their p53 status (P < 0.0001) (Fig. 3D), indicating that the p53 activity is not required for the NSKO-induced DNA damage. To verify that the DNA damage phenotype of NSKO cells is caused specifically by the loss of NS, we restored the NS expression by introducing a recombinant NS-GFP into NSKO MEF cells, and confirmed that the expression of NS-GFP can reduce the γ-H2AX+ percentage of NSKO MEF cells (3.1%) compared with the expression of GFP (19.6%, P < 0.0001) (Fig. 3E). In addition, the NSdB mutant of NS, which lacks the nucleolar localization signal and hence is distributed in the nucleoplasm, retains the same ability as wild-type NS to rescue the DNA damage effect of NSKO, suggesting that nucleolar localization is not a prerequisite for the genome-protecting function of NS. To address the issue of whether NSKO may cause DNA damage secondarily to the perturbation of ribosomal synthesis, we measured the NSKO effect on the levels of pre-rRNA transcripts containing the processing site-1 (PS-1), PS-2, PS-3, or 18S rRNA sequences by real-time RT-PCR (Fig. 3F, Left). The different PS-containing products represent precursor species that exist before the splicing events occurring at different stages of pre-rRNA processing. Although NSKO reduces NS transcripts and elicits a clear DNA damage response, it has no effect on the splicing events occurring on the 47S pre-rRNA or at the junctions of 5′ETS-18S (PS-2) or 18S-ITS1 (PS-3). Neither does it reduce the total amount of pre-rRNAs and rRNAs, as measured by the PS-4 qRT-PCR (Fig. 3F, Right). These results prove that the DNA damage effect of NSKO does not depend on the p53 activity and nor is it caused by the impairment of rRNA synthesis.
Fig. 3.
NSKO-induced DNA damage does not depend on p53 or rRNA synthesis. (A) A tamoxifen (TAM)-inducible NS conditional-KO model (inNScko) was generated. TAM treatment (0.1 µM, 4 d) reduces NS proteins in p53 wild-type and null inNScko MEF cells. (B) The population doubling level (PDL) of p53-wildtype MEF cells is decreased by NSKO. (C) Deletion of p53 increases the PDL of NS wild-type cells (red, green, blue) and decreases the PDL of NS-null cells (black) compared with their respective p53wt controls (gray). (D) TAM-induced NSKO increases the percentage of γ-H2AX+ cells in p53 wild-type and null MEF cells. (E) Transfection of NS-GFP or NSdB-GFP reduces the γ-H2AX+ percentage in transfected NSKO MEF cells compared with GFP-transfected NSKO cells. (Scale bars, 50 µm.) (F Left) Diagram of the pre-rRNA–processing step and the primer-targeting sites for the processing site-1 (PS-1), PS-2, PS-3, and the 18S-containing transcripts (PS-4). (Right) Quantitative RT-PCR shows that NSKO reduces the NS transcript but has no effect on the PS-1, PS-2, and PS-3 events. Neither does it reduce the total amount of pre-rRNAs and rRNAs (PS-4). The transcript levels are quantified by referencing to high mobility group protein 14 (HMG-14) and compared with the DMSO-treated NSf/f samples.
Loss of NS Triggers Replication-Induced DNA Damage.
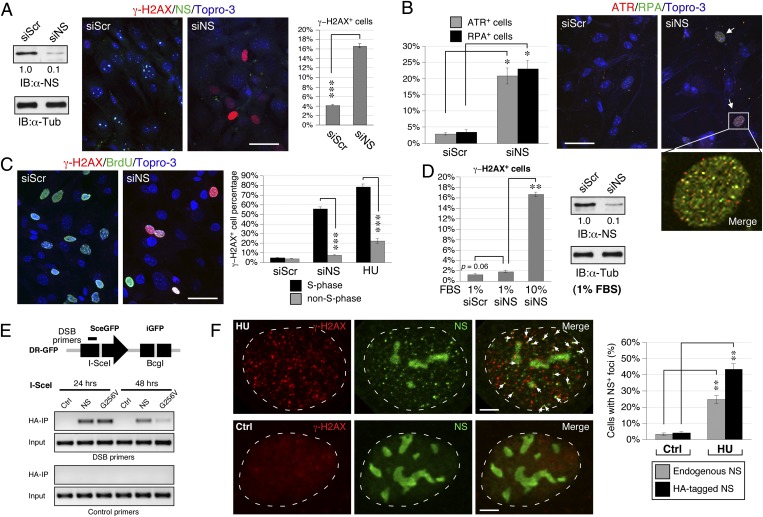
We noticed that NSKD in MEF cells increases not only the percentage of γ-H2AX+ cells (from 4.1% to 16.6%, P < 0.0001) but also the percentages of ataxia telangiectasia and Rad3-related (ATR)+ (from 3% to 21%, P < 0.01) and replication protein A-32 (RPA32)+ cells (from 4% to 23%, P < 0.01) (Fig. 4 A and B). Most of the ATR+ and RPA32+ foci colocalize with each other. NSKD also increases the number of DNA damage foci labeled by γ-H2AX, ATR, or breast cancer type 1 (BRCA1) in dysplastic oral keratinocytes cells (Fig. S3). We noted that 55.7% of the NS-depleted MEF cells in S-phase show γ-H2AX+ signals and only 7.8% of the non–S-phase cells are γ-H2AX+. This DNA damage profile resembles that of HU treatment (Fig. 4C). To bolster the link between NSKD-triggered damage and DNA replication, we performed propidium iodide-γ-H2AX–doubled labeled flow cytometry on control-KD (siScr) and NSKD (siNS) MEF cells. The flow cytometry results confirm that NSKD significantly increases the percentage of γ-H2AX+ cells compared with the control-KD samples (Fig. S4A). Cell cycle analyses on the γ-H2AX–labeled (Fig. S4B1) versus γ-H2AX–nonlabeled subpopulation (Fig. S4B2) of NSKD cells also prove that a significant portion of NSKD-induced DNA damage occurs in the S-phase cells. By pooling the S-phase cells from both the γ-H2AX–labeled and nonlabeled populations, we find that 80.1% of the S-phase cells show γ-H2AX+ signals, whereas only 25.5% of the non–S-phase cells show γ-H2AX+ signals (P < 0.01, Fig. S4C). To further support the idea, we demonstrated that the DNA damage effect of NSKD is greatly reduced in slowly dividing MEF cells grown under the serum deprivation condition (1.9%) compared with those grown in 10% (vol/vol) serum (16.6%), and shows no difference from the scrambled RNAi (siScr)-treated cells grown in low-serum medium (1.3%) (Fig. 4D, Left). This lack of siNS effect in slowly dividing MEF cells is not caused by a decreased NSKD efficiency (Fig. 4D, Right). To determine whether NS is directly involved in the response and/or repair of DNA damage, we performed the DSB-chromatin coimmunoprecipitation (ChIP) assay in DR-GFP-U2OS cells (17). The DR-GFP-U2OS cells were stably transfected with the SceGFP transgene, which contains an internal I-SceI site (depicted in Fig. 4E, Upper). DSBs were introduced specifically at the I-SceI site by the expression of I-SceI enzyme. Primers were designed to detect the DNA sequence localized at 500 bp upstream to the I-SceI site. The DSB-ChIP results showed that immunoprecipitating HA-tagged NS can physically pull down the I-SceI–cut DSB site on the SceGFP locus at the 24-h and 48-h time points following the expression of I-SceI. Compared with the wild-type NS, the non–GTP-binding mutant of NS (G256V) shows a faster clearance from the I-SceI–cut site (Fig. 4E, Lower). In consistence, confocal studies demonstrated that, following the HU treatment, the endogenous NS protein in 25% of the cells forms discrete foci in the nucleoplasm without losing its nucleolar signals, and 44% of the cells overexpressing HA-tagged NS shows foci formation by the recombinant protein (Fig. 4F). Some NS+ and NS-HA+ foci are colocalized with the γ-H2AX+ signals. These data indicate that NS may play a direct and primary role in reducing the amount of spontaneous DNA damage associated with the DNA replication process.
Fig. 4.
NS depletion triggers replication-induced DNA damage. (A) The siNS treatment produces a 90% protein knockdown efficiency in MEF cells and a significant increase of γ-H2AX+ cells. (B) NSKD also increases the percentages of ATR+ and RPA32+ cells. Most of the ATR+ and RPA32+ foci are colocalized. (C) S-phase (BrdU+) cells are much more susceptible to the DNA damage effect of NSKD than non–S-phase cells, which resembles the DNA damage profile of HU treatment. (D) The siNS-induced DNA damage is significantly diminished in MEF cells grown in low serum-containing medium. The siNS-mediated KD efficiency for the low-serum culture condition is the same as that for the normal-serum culture condition. (E) DR-GFP U2OS cells (upper panel) were transfected with the empty vector or HA-tagged NS plasmid. After introducing DSB at the I-SceI site, ChIP analyses were performed by precipitation with anti-HA antibody and PCR amplification with DSB-specific or control primers. The DSB-ChIP assay demonstrates that NS is physically associated with DSB sites. (F) HU treatment (in U2OS cells) induces the foci formation of endogenous NS (detected by Ab2438) in the nucleoplasm without apparently diminishing its nucleolar signals. The percentages of cells showing ≥ 5 foci of colocalized NS (green) and γ-H2AX (red) signals are quantified by confocal analyses (gray bars), and independently confirmed by using the HA-tagged NS recombinant protein (black bars). (Scale bars: 50 µm in A–C; 5 µm in F.)
NS Depletion Perturbs the Recruitment of RAD51 to Stalled Replication-Induced Damage Foci.
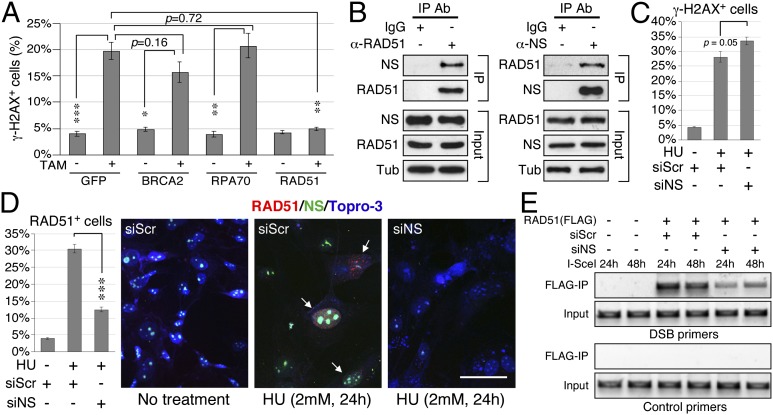
Increased DNA damage by NS depletion may be caused by an increased source of DNA damage, induced secondarily by stress, or by impaired DNA damage repair. In the latter case, one should be able to identify the point of perturbation in the DNA repair axis. As homologous recombination (HR) is the main mechanism responsible for repairing replication-induced DNA damage (18) and knockout of the core HR protein, RAD51, shows the same early embryonic lethal phenotype as does NSKO (19), we hypothesize that the recruitment of RAD51 to DNA damage sites may be a target of NS regulation. To date, only a handful of proteins (i.e., BRCA2, RPA70, Bloom’s syndrome protein, and RAD52) are implicated in the loading of RAD51 to DNA damage sites. To look for the potential targets of NS, we tested the ability of BRCA2, RPA70 and RAD51 in rescuing the DNA damage phenotype of NSKO MEF cells, and found that only RAD51, but not BRCA2 or RPA70, is capable of doing so (Fig. 5A), suggesting that RAD51 may be a direct target of NS. Indeed, coimmunoprecipitation assays confirm that the endogenous NS and RAD51 proteins can interact with each other in vivo (Fig. 5B). To determine the role of NS in regulating the recruitment of RAD51, control-KD and NSKD MEF cells were treated with HU (2 mM) for 24 h and assayed for their RAD51 recruitment efficiency. The results showed that, in control-KD MEF cells, HU treatment significantly increases the percentages of γ-H2AX+ (28.0%, P < 0.0001, Fig. 5C) and RAD51+ cells (30.4%, P < 0.0001, Fig. 5D) over the non–HU-treated MEF cells (4.1% for γ-H2AX+ cells and 3.8% for RAD51+ cells). In contrast, HU treatment of NSKD MEF cells increases γ-H2AX+ cells significantly (33.6%, Fig. 5C), but its effect on triggering RAD51+ foci is greatly reduced (12.4%) compared with its effect in control-KD cells (30.4%, P < 0.0001) (Fig. 5D). Finally, we performed DSB-ChIP experiments to confirm that NS is required for the physical recruitment of RAD51 to DSB sites (Fig. 5E). The ChIP results showed that the recruitment of RAD51 to I-SceI–induced DSB sites at the 24-h and 48-h time points is much attenuated by NS knockdown. These findings, in conjunction with the lack of RAD51 foci formation in NS-depleted cells, establish the importance of NS in promoting RAD51 recruitment to DSB sites and refute the possibility that the DNA damage effect of NS depletion is caused secondarily by increased cellular stress.
Fig. 5.
NS binds RAD51 and promotes its recruitment to DNA damage foci. (A) Overexpression of RAD51, but not that of BRCA2 or RPA70, rescues the γ-H2AX+ cell percentage of NSKO MEF cells. (B) CoIP of endogenous NS and RAD51 by anti-RAD51 (left) or anti-NS antibody (right) confirms that these two proteins interact with each other in vivo. (C) HU treatment increases the percentages of γ-H2AX+ in both siScr- and siNS-treated MEF cells. siNS-treated cells show more DNA damage events than siScr-treated cells. (D) The HU effect on triggering RAD51+ foci in siNS-treated MEF cells is significantly reduced compared that in siScr-treated cells. (Scale bar: 50 µm.) (E) DSB-ChIP assays show that the physical recruitment of RAD51 to I-SceI–induced DSBs is significantly attenuated by NSKD (siNS) at the 24 and 48-h time points following the induction of I-SceI expression.
Discussion
This study identifies NS as an essential player in protecting stem/progenitor cells from replication-induced DNA damage in vivo and demonstrates its ability to bind and promote the recruitment of RAD51 to stalled replication-induced damage foci. The neNScko mouse model shows that deleting NS in the neural stem/progenitor cell population increases the frequency of DNA damage, reduces the number of stem/progenitor cells, and causes major developmental defects in the embryonic brain. DNA damage triggered by neNScko becomes most prominent at E12.5. This window of appearance may be determined collectively by the activity of nestin promoter and the differentiation and dying of NS-deleted neural progenitor cells. As a result, the peak of the NSKO event should parallel or immediately follow the highest of nestin expression at E10.5 and E12.5 (6). The more DNA damage events in the E12.5 compared with the E10.5 neuroepithelium of neNScko embryos may be caused by the increased frequency of homozygous deletion of NS (evidenced by the amount of NS protein reduction) and the increased frequency of replication stalling events over additional rounds of cell division. The lack of γ-H2AX signals in the E14.5 or older forebrains may be due to the rapid loss of proliferative neural precursors. In support of these in vivo findings, NS depletion by siNS triggers massive DNA damage in NSCs and MEF cells and greatly reduces their ability to self-renew or propagate in culture. The self-renewing activity of NSCs is indicated by their ability to form spheres over multiple passages. Given its role in promoting the RAD51 recruitment during DNA damage repair, loss of NS is expected to reduce the protection of NSCs against spontaneous DNA damage caused by DNA replication, thereby introducing cell cycle arrest and perturbing the formation of tertiary neurospheres from secondary neurospheres. Notably, restoring the NS expression can reverse the DNA damage phenotype of NS-deleted MEF cells, which supports the specific effect of NS depletion in causing spontaneous DNA damage in vitro and in vivo. Finally, we establish the direct role of NS in the response/repair of DNA damage by showing that NS is physically recruited to the DSB sites and that the DNA damage effect of NSKO depends neither on the p53 status nor on the synthesis of rRNAs.
The two major sources of cell-intrinsic DNA damage are oxidative stress and replication stalling. We noted that the S-phase cells are much more susceptible to the NSKO effect than the non–S-phase cells, a feature also shared by the HU treatment. This idea that NS deletion leads to replication-dependent DNA damage is supported by the lack of DNA damage effect of NSKD on slowly dividing MEF cells and the protective effect of NS against HU-induced DNA damage in NSCs. Further analyses identify RAD51 as a direct molecular target of NS with the ability to rescue the NSKO-induced phenotype. More importantly, loss of NS perturbs the recruitment efficiency of RAD51 to stalled replication-induced damage foci and DSB sites, which refutes that the DNA damage effect of NS depletion arises secondarily due to increased cellular stress. Because the DNA damage phenotype of NSKO cells can be compensated by overexpressing RAD51 alone, we reason that NS more likely plays a promoting role than a determining role in the RAD51 recruitment and that the NS-promoted RAD51 recruitment should be inactive without the preexisting DNA damage event, which, otherwise, would incur nonspecific recombination and genomic instability. This NS function, although modulatory in nature, becomes indispensable for cells that undergo extended DNA replication, such that both NSKO and RAD51-KO mice show the same phenotype of early embryonic lethality (19). In addition, we noted that this obligatory function of NS does not require its nucleolar localization and that the damage-induced foci formation of NS occurs without the apparent translocation of its protein from the nucleolus to the nucleoplasm, suggesting that DNA damage may activate the nucleoplasmic NS via a posttranslational mechanism.
This newly discovered function of NS parallels the one in which NS protects against DNA damage on the telomere by regulating telomeric repeat binding factor 1 (TRF1) SUMOylation and the recruitment of PML-IV (13). Together, these two mechanisms help protect the genome integrity of stem/progenitor cells throughout their proliferative lifespan. NS has also been noted to regulate the p53 pathway via a direct mouse double minute 2 homologue (MDM2) interaction (9, 10). The MDM2-regulatory mechanism of NS, however, cannot account for its essential role in cell proliferation and development, as these events do not require the presence of p53 (11). We have observed that this MDM2-stabilizing function of NS is mostly silent under the normal condition and becomes activated only when the nucleolar organization is disrupted by stress signals (10). Therefore, our current view is that the DNA/telomere-protecting function of NS is required for continuously dividing cells under normal growth conditions and is operated by the nucleoplasmic pool of NS in a damage-dependent manner, whereas its MDM2-stabilizing function is only turned on under nucleolar stress conditions and is operated by the NS protein released from the disassembled nucleolus.
In conclusion, both in vivo and in vitro studies show that loss of NS predisposes stem and progenitor cells to spontaneous DNA damage closely related to the DNA replication event. This essential function of NS operates independently of p53 or rRNA synthesis and acts by promoting the DSB recruitment of RAD51. These discoveries suggest that cells with a robust DNA replication program may need additional molecules to enhance the core HR-based repair activity to safeguard their genome integrity.
Materials and Methods
Animal Studies.
All animals were housed by the Program for Animal Resources at the Texas A&M Health Science Center (Houston, TX) and handled in accordance with the principles of the Guide for the Care and Use of Laboratory Animals and the procedures approved by the Institutional Animal Care and Use Committee.
ChIP at DSB Sites.
DR-GFP U2OS cells (17) were transfected with the indicated plasmid or treated with the siRNA duplex. After 24 h, DSBs were introduced at the I-SceI site on the SceGFP transgene by transfecting with the I-SceI plasmid, crosslinked with formaldehyde, and subjected to five rounds of sonication (8 s) with 1-min interval using the 60 Sonic Dismembrator. Immunoprecipitation was performed using the anti–HA- or FLAG-agarose beads. See SI Materials and Methods for primer sequences.
Other Methods.
See SI Materials and Methods for details.
Supplementary Material
Acknowledgments
This work was supported by National Cancer Institute–Public Health Service Grant R01 CA113750 and Texas A&M Research Development and Enhancement Award (to R.Y.L.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301672110/-/DCSupplemental.
References
- 1.Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412(6846):553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 2.Katou Y, et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424(6952):1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 3.Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell. 2006;21(4):581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 4.Saintigny Y, et al. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 2001;20(14):3861–3870. doi: 10.1093/emboj/20.14.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanada K, et al. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006;25(20):4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai RY, McKay RD. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16(23):2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Q, Yasumoto H, Tsai RY. Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability. Mol Cell Biol. 2006;26(24):9279–9290. doi: 10.1128/MCB.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma H, Pederson T. Depletion of the nucleolar protein nucleostemin causes G1 cell cycle arrest via the p53 pathway. Mol Biol Cell. 2007;18(7):2630–2635. doi: 10.1091/mbc.E07-03-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai MS, Sun XX, Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol Cell Biol. 2008;28(13):4365–4376. doi: 10.1128/MCB.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng L, Lin T, Tsai RY. Nucleoplasmic mobilization of nucleostemin stabilizes MDM2 and promotes G2-M progression and cell survival. J Cell Sci. 2008;121(Pt 24):4037–4046. doi: 10.1242/jcs.037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beekman C, et al. Evolutionarily conserved role of nucleostemin: Controlling proliferation of stem/progenitor cells during early vertebrate development. Mol Cell Biol. 2006;26(24):9291–9301. doi: 10.1128/MCB.01183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jafarnejad SM, Mowla SJ, Matin MM. Knocking-down the expression of nucleostemin significantly decreases rate of proliferation of rat bone marrow stromal stem cells in an apparently p53-independent manner. Cell Prolif. 2008;41(1):28–35. doi: 10.1111/j.1365-2184.2007.00505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu JK, Lin T, Tsai RY. Nucleostemin prevents telomere damage by promoting PML-IV recruitment to SUMOylated TRF1. J Cell Biol. 2012;197(5):613–624. doi: 10.1083/jcb.201109038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tronche F, et al. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat Genet. 1999;23(1):99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: A tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244(2):305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 16.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356(6366):215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 17.Peng G, et al. BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nat Cell Biol. 2009;11(7):865–872. doi: 10.1038/ncb1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helleday T. Amplifying tumour-specific replication lesions by DNA repair inhibitors - a new era in targeted cancer therapy. Eur J Cancer. 2008;44(7):921–927. doi: 10.1016/j.ejca.2008.02.044. [DOI] [PubMed] [Google Scholar]
- 19.Tsuzuki T, et al. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci USA. 1996;93(13):6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.