Abstract
Persistent infection with hepatitis C virus (HCV) is a leading cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. It has recently been shown that HCV RNA replication is susceptible to small interfering RNAs (siRNAs), but the antiviral activity of siRNAs depends very much on their complementarity to the target sequence. Thus, the high degree of sequence diversity between different HCV genotypes and the rapid evolution of new quasispecies is a major problem in the development of siRNA-based gene therapies. For this study, we developed two alternative strategies to overcome these obstacles. In one approach, we used endoribonuclease-prepared siRNAs (esiRNAs) to simultaneously target multiple sites of the viral genome. We show that esiRNAs directed against various regions of the HCV coding sequence as well as the 5′ nontranslated region (5′ NTR) efficiently block the replication of subgenomic and genomic HCV replicons. In an alternative approach, we generated pseudotyped retroviruses encoding short hairpin RNAs (shRNAs). A total of 12 shRNAs, most of them targeting highly conserved sequence motifs within the 5′ NTR or the early core coding region, were analyzed for their antiviral activities. After the transduction of Huh-7 cells containing a subgenomic HCV replicon, we found that all shRNAs targeting sequences in domain IV or nearby coding sequences blocked viral replication. In contrast, only one of seven shRNAs targeting sequences in domain II or III had a similar degree of antiviral activity, indicating that large sections of the NTRs are resistant to RNA interference. Moreover, we show that naive Huh-7 cells that stably expressed certain 5′ NTR-specific shRNAs were largely resistant to a challenge with HCV replicons. These results demonstrate that the retroviral transduction of HCV-specific shRNAs provides a new possibility for antiviral intervention.
Hepatitis C virus (HCV) is an enveloped virus with a single-stranded 9.6-kb RNA genome of positive polarity (reviewed in reference 4). The 5′ nontranslated region (NTR) of the genome contains an internal ribosome entry site (IRES) that directs the translation of a single long open reading frame. The encoded polyprotein is co- and posttranslationally cleaved into 10 viral proteins (core, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B). Recently, the production of an additional viral protein by a ribosomal frame shift has been reported (66, 69). HCV has been classified as a member of the genus Hepacivirus within the family Flaviviridae (62). Based on nucleotide sequence comparisons, HCV genomes can be grouped into at least six genotypes, or clades, that differ from each other by 31 to 34%. Furthermore, several subtypes have been defined, with a nucleotide sequence diversity of about 20%. In Western Europe and the United States, infections caused by genotypes 1a and 1b are the most frequent, followed by infections with genotype 2 and 3 viruses. The other genotypes are rare and can only be found in distinct geographical regions, such as Egypt (genotype 4), South Africa (genotype 5), and Southeast Asia (genotype 6). The genomic variability of HCV is due to the high error rate of the viral RNA-dependent RNA polymerase, which has been calculated to be in the range of about 10−4 (42). This number is in line with the mutation rates of 1.44 × 10−3 and 1.92 × 10−3 base substitutions per site per year that were found in a chronically infected human and chimpanzee, respectively (47, 48).
HCV has infected an estimated 170 million people worldwide (68). In most cases, the virus has established a persistent infection, frequently associated with chronic hepatitis and liver fibrosis. Chronic hepatitis C often progresses to cirrhosis and eventually to hepatocellular carcinoma (26, 60). Currently, hepatitis C patients are treated with alpha interferon (IFN-α) alone or in combination with ribavirin. However, there is still no cure for a large proportion of patients, even with the most advanced therapy regimens (43). Thus, alternative therapeutic approaches for chronic hepatitis C are needed.
RNA interference (RNAi) is an ancient mechanism of sequence-specific gene regulation. RNAi pathways are known to regulate heterochromatin formation, protein translation, and RNA degradation (reviewed in reference 23). The latter is initiated by Dicer, a member of the RNase III family that chops double-stranded RNA (an intermediate in the replication of many, if not all, viruses) into so-called small interfering RNAs (siRNAs) that have a length of 21 to 25 nucleotides, 3′ overhangs of 2 or 3 nucleotides, and phosphorylated 5′ ends. Dicer is also known to process the precursor of a recently discovered species of noncoding RNAs, named micro-RNAs (miRNAs) or small temporal RNAs. These transcripts are about 60 to 80 nucleotides long and form stem-loop hairpins with asymmetric bulges within the stem. Dicer cuts within the stem, thereby producing double-stranded RNAs of approximately 21 nucleotides. Both siRNAs and mature miRNAs induce posttranscriptional gene silencing by binding to a multiprotein complex that has been termed the RNA-induced silencing complex (RISC) (20, 22). In the case of siRNAs, the incorporation results in RNA unwinding. The complex is then guided to specific substrates by Watson-Crick base pairing between the now single-stranded siRNA and complementary nucleotides of the target molecule. Hereupon, the target is cleaved within the recognition sequence, which in most cases initiates its rapid degradation (46). This kind of RNAi is highly sequence specific. Even a single mismatch between the siRNA and its target sequence can dramatically decrease the efficiency of RNA degradation. A similar mechanism of degradation has been proposed for RNAs that encounter an miRNA-loaded RISC complex if the miRNA shows complete complementarity to the target, whereas imprecise base pairing seems to induce translational arrest rather than degradation (reviewed in references 3 and 11).
In plants, RNAi serves as a potent antiviral defense mechanism which has provoked the evolution of counteracting strategies by many viruses (reviewed in reference 36). For example, the p19 protein of tomato bushy stunt virus and related tombusviruses binds siRNAs, thereby blocking systemic silencing in plants (57). Recently, it was shown that flock house virus, an insect pathogen, also encodes a protein that inhibits RNAi (35). Whether mammalian viruses encounter RNAi-based defense mechanisms is not clear. Although mammalian cells express Dicer homologs, they normally do not produce siRNAs from exogenous double-stranded RNAs (discussed in reference 9). Instead, they activate the IFN system, which leads to the enhanced expression of numerous genes, including those that encode proteins with strong antiviral activities (19, 52). Nevertheless, RNAi can be used to inhibit virus replication in mammalian cells. It has been demonstrated that the transfection of virus-specific siRNAs efficiently inhibits the multiplication of human immunodeficiency virus type 1 (HIV-1) and of several other viruses (reviewed in reference 9). Most recently, it was shown that HCV RNA replication is also sensitive to RNAi (29, 51, 67, 73; M. Y. Seo, S. Abrignani, M. Houghton, and J. H. Han, Letter, J. Virol. 77:810-812, 2003). However, the notoriously error-prone replication of RNA viruses is a problem for the development of siRNA-based gene therapies. Here we report the use of endonuclease-prepared siRNAs (esiRNAs) to simultaneously target multiple sites of the HCV genome for degradation, a strategy that should prevent the evolution of escape mutants. Furthermore, we show that HCV replication can be blocked by means of retrovirally transduced short hairpin RNAs (shRNAs) directed against the 5′ NTR or nearby coding sequences.
MATERIALS AND METHODS
Cells.
The Huh-7 cell clones 9-13, 20-1, and 9B (containing the HCV 1b replicons I377/NS3-3′, I389/Core-3′/5.1, and I389/NS3-3′/LucUbiNeo-ET, respectively) have been described previously (18, 40, 50). The newly established Huh-7 cell clone A-3/3 contains the chimeric HCV 1a/1b replicon I389/NS3-3′/H77/DR (the construction of this replicon is described below). Cells were maintained in Dulbecco's modified Eagle's medium (Life Technologies, Gaithersburg, Md.) supplemented with 10% fetal calf serum, 200 U of penicillin G per ml, and 200 μg of streptomycin per ml. For cell clones carrying HCV replicons, various amounts of G418 (Life Technologies) were added to the culture medium (0.1 mg per ml for clone 20-1, 0.25 mg per ml for clone 9B, 0.5 mg per ml for clone A-3/3, and 1 mg per ml for clone 9-13). For the selection of retrovirally transduced cells, the culture medium was supplemented with 10 μg of zeocin (Invitrogen, Carlsbad, Calif.) per ml.
Construction of HCV replicons.
The chimeric HCV 1a/1b replicon I389/NS3-3′/H77/DR was constructed by using plasmid pCV-H77 (70), which contains a full-length clone of the HCV genotype 1a strain H77 (kindly provided by J. Bukh, National Institutes of Health, Bethesda, Md.). Briefly, an ApaLI-AflII fragment of pCV-H77 (containing the NS3 to NS5B coding region) was amplified by using the PCR primers S3418-NcoI (CGCCATGGCGCCCATCACGGCGTACGC) and A3998-SpeI (GGCCACTAGTGGGCCACCTGGAAGCTCTGGGG). After restriction with ApaLI and NcoI, the amplification product was inserted between the AflII and NcoI sites of pFK-EMCVΔluc8991-9605, thereby generating the intermediate construct pFK-EMCVΔluc/H77. Next, the PmeI-SpeI portion of plasmid pFK-I389/NS3-3′/wt (40) was replaced by an analogous fragment isolated from pFK-EMCVΔluc/H77. Cell culture-adaptive mutations were introduced by a PCR-based approach into the resulting plasmid, pFK-I389/NS3-3′/H77/wt (24). First, an arginine substitution for serine at amino acid position 2204 was generated by using the primers S4351 (GGATGAACCGGCTAATAGCC), A-S2204R (GCGGACAGCTGCCTAGCCGAGG), S-S2204R (CCTCGGCTAGGCAGCTGTCCGC), and A7558 (CGGCCCCACTACTGACCGTCG). Second, an aspartic acid substitution for alanine at amino acid position 1226 was introduced by using the primers S2066 (GCTGGCCCGCTCCTCAAGG), A-A1226D (GCATGCAGGTGGTCCACCTGGAAGC), S-A1226D (GCTTCCAGGTGGACCACCTGCATGC), and A2979 (GGCATTGATGCCCAATGCG). The presence of both mutations in the resulting plasmid, pFK-I389/NS3-3′/H77/DR, was confirmed by sequence analysis. Note that given amino acid positions refer to the Con1 consensus genome. The replicon construct I389/NS3-3′/Luc-ET (used in transient transfection experiments) was described previously under the name of Luc-ET (18, 41).
Generation and transfection of HCV RNAs.
The generation of HCV RNAs by in vitro transcription and the electroporation of Huh-7 cells with these RNAs have been described previously (39, 41).
Generation and transfection of esiRNAs.
PCR standard protocols and primers with 5′-terminal T7 or T3 promoter sequences were used to amplify fragments of the HCV genotypes 1a (strain H77), 1b (strain Con1), and 2a (strain HC-J6CH, clone pJ6CF; kindly provided by J. Bukh). In addition, fragments of genes encoding firefly luciferase (FLuc), enhanced green fluorescent protein (EGFP), and β-galactosidase (LacZ) were amplified (for more detailed information, see Table 1 and Fig. 1A). PCR products were transcribed in vitro with a MEGAscript kit (Ambion, Austin, Tex.). Complementary transcripts were annealed, and the resulting double-stranded RNAs were digested with highly purified, recombinant RNase III essentially as described previously (8, 72). Cleavage products were purified by using QIAquick columns (Qiagen, Hilden, Germany), the flowthrough was collected, and a heterogenous population of double-stranded RNAs of approximately 15 to 40 bp was precipitated with 2-propanol. The pellet was washed with 70% ethanol, purified esiRNAs were dissolved in double-distilled water, and the concentration was determined photometrically.
TABLE 1.
Templates, PCR primers, and amplicons used for the generation of esiRNAs
| Template (reference or source) | Primer name | Primer sequence (5′ → 3′) | Amplicon name | Amplicon size (nt) |
|---|---|---|---|---|
| pFK-I377neo/NS3-3′/wt (40) | T7-5′NTR/1b_F | ATTAATACGACTCACTATAGGGGCGACACTCCACCATAGAT | 5′NTR/1b | 337 |
| T7-5′NTR/1b_R | CGTAATACGACTCACTATAGGACTCGCAAGCACCCTATCAG | |||
| T7-ORF-1/1b_F | TTAATACGACTCACTATAGGTGCTTCTGCTTTCGTAGGC | ORF1/1b | 1,126 | |
| T3-ORF-1/1b_R | TAATTAACCCTCACTAAAGGTTAGCCGTCTCCGCCGTAAT | |||
| T7-ORF-2/1b_F | TTAATACGACTCACTATAGGTCCTTGGCCAGCTCATCAG | ORF2/1b | 1,132 | |
| T3-ORF-2/1b_R | TAATTAACCCTCACTAAAGGTTTTGCCATGATGGTGGTGT | |||
| T7-ORF-3/1b_F | TTAATACGACTCACTATAGGAATGAGGTTTTCTGCGTCCA | ORF3/1b | 1,125 | |
| T3-ORF-3/1b_R | TAATTAACCCTCACTAAAGGCGATGTCTCCAGACTCGCAA | |||
| pCV-H77C (70) | T7-ORF-1/1a_F | ATTAATACGACTCACTATAGGCGGGAGCTCTTGTAGCATTC | ORF1/1a | 999 |
| T7-ORF-1/1a_R | CGTAATACGACTCACTATAGGAGGGATCAGTGAGCATGGAC | |||
| T7-ORF-2/1a_F | ATTAATACGACTCACTATAGGGCTCCATCTCTCAAGGCAAC | ORF2/1a | 1,046 | |
| T7-ORF-2/1a_R | CGTAATACGACTCACTATAGGACGGAGTTGATGTGGGCTAC | |||
| T7-ORF-3/1a_F | ATTAATACGACTCACTATAGGCCAAGAACGAGGTTTTCTGC | ORF3/1a | 1,081 | |
| T7-ORF-3/1a_R | CGTAATACGACTCACTATAGGGCCACCCTATTGATTTCACC | |||
| pJ6CF-pGEM (71) | T7-ORF-1/2a_F | ATTAATACGACTCACTATAGGGCAGCATAGGCTTGGGTAAG | ORF1/2a | 1,061 |
| T7-ORF-1/2a_R | CGTAATACGACTCACTATAGGGGATGGGGATGGGTCTGTTAGCATGG | |||
| T7-ORF-2/2a_F | ATTAATACGACTCACTATAGGGCAAGGCCTATGATGTGGAC | ORF2/2a | 1,125 | |
| T7-ORF-2/2a_R | CGTAATACGACTCACTATAGGTTTTGGCCATGATGGTTGTA | |||
| T7-ORF-3/2a_F | ATTAATACGACTCACTATAGGCGCCTTATCGTTTACCCTGA | ORF3/2a | 1,049 | |
| T7-ORF-3/2a_R | CGTAATACGACTCACTATAGGCCAAGTTTTCTGAGGGCTGA | |||
| pEGFPLuc (Clontech) | T7-FLuc_F | TAATACGACTCACTATAGGGAGAGCAACTGCATAAGG | FLuc | 737 |
| T3-FLuc_R | AATTACCCTCACTAAAGGGAGAATCTGACGCAGGCAGT | |||
| pEGFPLuc (Clontech) | T7-EGFP_F | TTAATACGACTCACTATAGGTGAGCAAGGGCGAGGA | EGFP | 711 |
| T3-EGFP_R | TAATTAACCCTCACTAAAGGGTACAGCTCGTCCATGCCGA | |||
| pSV-paX (7) | T7-LacZ_F | TAATACGACTCACTATAGGGAGAATCGTAATCACCCGAGTGTGA | LacZ | 1,363 |
| T7-LacZ_R | AATTACCCTCACTAAAGGGAGCCCTAATCCGAGCCAGTTTA |
FIG. 1.
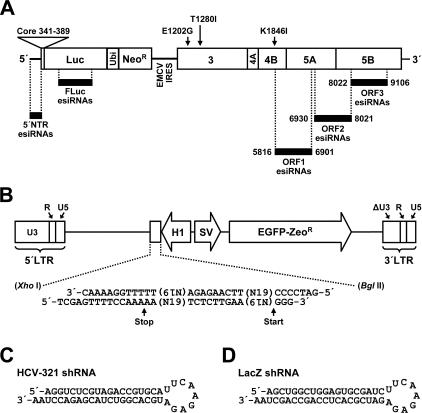
Target sequences of esiRNAs and design of a retroviral vector system for the delivery of shRNAs. (A) Schematic representation of the subgenomic HCV replicon I389/NS3-3′/LucUbiNeo-ET that persistently replicates in cells of the Huh-7 cell clone 9B. The replicon is composed of the HCV 5′ NTR; nucleotides 342 to 389 of the core coding region (core) fused to the coding sequences of the firefly luciferase gene (Luc), the ubiquitin gene (Ubi), and the neomycin phosphotransferase gene (Neor); the IRES of the encephalomyocarditis virus (EMCV IRES); the coding region of the HCV nonstructural proteins NS3 to NS5B; and the HCV 3′ NTR. Black bars indicate viral and replicon nonviral sequences targeted by shRNAs. Note that numbers refer to nucleotide positions of the Con1 consensus genome. (B) Schematic representation of pBABE/H1/SV40/EGZ/ΔU3. The vector contains two heterologous promoter elements that operate in a bidirectional manner. The polymerase III H1-RNA promoter (H1) is used to direct the transcription of shRNAs, whereas an SV40 promoter (SV) directs the transcription of an mRNA which encodes an EGFP-zeocin resistance fusion protein (EGFP-Zeor). Note that the 3′ LTR carries a large deletion in the U3 region in order to inactivate U3-dependent gene expression of the integrated provirus. Labeled arrows indicate the start and stop sites of the RNA polymerase III. (C) Predicted secondary structure of the HCV-specific shRNA HCV-321. (D) Predicted secondary structure of the β-galactosidase-specific shRNA LacZ.
For the transfection of esiRNAs, cells were seeded into 6-well plates (105 cells per well). About 24 h later, cells were transfected with esiRNAs or control RNAs [yeast tRNA or poly(IC)] by the use of Optimem (Life Technologies) and the Oligofectamine transfection reagent (Invitrogen) according to the manufacturer's recommendations.
Construction of retroviral vectors encoding shRNAs.
The vector pBABE/H1/SV40/EGZ/ΔU3 was constructed by modifying the Moloney murine leukemia virus (Mo-MuLV)-based vector pBABE/puro (44). The puromycin coding sequence of pBABE/puro was replaced by a fragment of pcz-CFG5-IEGZ containing an EGFP-zeocin fusion protein coding sequence (5) (kindly provided by D. Lindemann, Institute of Virology, TU Dresden, Germany). The resulting construct was digested with XbaI and NheI and religated to delete a large portion of the U3 region within the 3′ long terminal repeat (LTR). Next, a BglII restriction site within the EGFP-zeocin coding sequence was destroyed by silent mutagenesis. Finally, the H1-RNA promoter element of pSUPER (kindly provided by R. Agami, The Netherlands Cancer Institute, Amsterdam, The Netherlands) (6) was excised by using EcoRI and EcoO109 restriction enzymes. The resulting fragment was treated with the Klenow enzyme and inserted between the EcoRI and BamHI restriction sites of the intermediate vector pBABE/SV40/EGZ/ΔU3, thereby generating pBABE/H1/SV40/EGZ/ΔU3 (Fig. 1B).
Chemically synthesized DNA oligonucleotides were used to generate shRNA coding sequences (6). After being annealed, the oligonucleotides were inserted between the XhoI and BglII sites of pBABE/H1/SV40/EGZ/ΔU3 (for details, see Fig. 1B). Finally, the integrity of the shRNA coding region was verified by sequencing. Note that the sequences of all HCV-specific shRNAs can be deduced from their names, which refer to corresponding nucleotide positions of the Con1 genome (41). As an example of an HCV-specific shRNA, the sequence and secondary structure of HCV-321 is shown in Fig. 1C. The p53-specific shRNA, as used in our experiments, has been described previously (6), and the β-galactosidase-specific shRNA is shown in Fig. 1D.
Production of pseudotyped retroviruses.
The stock virus was generated essentially as described previously (58). Briefly, 293T cells (14) were cotransfected with the parental vector pBABE/H1/SV40/EGZ/ΔU3 (or constructs that encode shRNAs), pHIT60 (encoding the Gag-Pol proteins of Mo-MuLV) (58), and pcz-VSV-Gwt (encoding the G protein of vesicular stomatitis virus) (28) by the calcium phosphate precipitation method. The next day, cells were treated with 10 mM sodium butyrate for 9 h. After an additional incubation period of 16 h in the absence of sodium butyrate, supernatants were collected and the cell debris was removed by filtration (pore size, 0.45 μm). The stock virus was either used directly after filtration or stored at 4°C.
Luciferase assays and HCV RNA quantification.
The firefly luciferase activity and the HCV RNA concentration were measured essentially as described previously by Krieger and coworkers (33) and Vrolijk and coworkers (64), respectively. Note that both assays were performed in parallel by using aliquots of the same cell lysates.
Immunofluorescence analysis.
Cells grown on glass coverslips were fixed with 3% paraformaldehyde and permeabilized with 0.5% Triton X-100. Immunostaining was performed according to standard protocols, with goat antibodies conjugated to the cyanine dye Cy3 (Dianova, Hamburg, Germany) as secondary antibodies. NS5A and EGFP were detected by using the mouse monoclonal antibody 3924-4940-8858 (Biogenesis, Poole, United Kingdom) and the immunoglobulin G (IgG) fraction of a rabbit serum from an animal that was immunized with wild-type GFP (Molecular Probes, Eugene, Oreg.), respectively. Cellular DNA was stained with 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI) (Molecular Probes).
Sequence analysis and database accession numbers.
Nucleotide sequences were retrieved and analyzed by using either tools of the Hepatitis C Virus DataBase (http://hepatitis.ibcp.fr) or the VECTOR NTI ADVANCE program (INFORMAX, Bethesda, Md.). HCV sequences with the following database accession numbers were used: AJ238799 (strain Con1), AF009606 (strain H77), AF177036 (strain HC-J6CH), D17763 (strain NZL1), D45193 (strain HEMA51), Y13184 (strain EUH1480), and D84262 (strain Th580).
RESULTS
Inhibition of HCV replicons by esiRNAs.
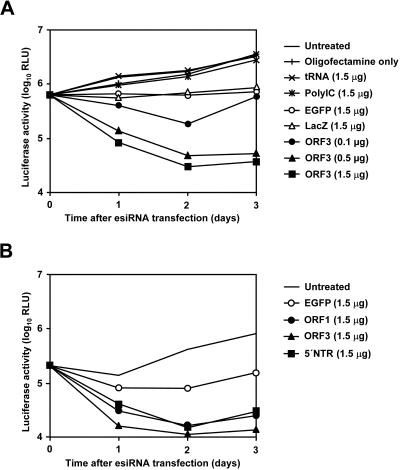
A sensitive and precise quantification of HCV RNA replication can be achieved by using replicons encoding firefly luciferase as a reporter (33). In an initial set of experiments, we transfected cells of the Huh-7 cell clone 9B that contained the subgenomic HCV genotype 1b replicon I389/NS3-3′/LucUbiNeo-ET with HCV-specific esiRNAs and various control RNAs (for a schematic representation of the replicon and the positions of targeted sequences, see Fig. 1A). As shown in Fig. 2A, transfection of the ORF3 esiRNA preparation inhibited luciferase reporter gene expression in a dose-dependent manner. The effect was detectable as early as 24 h after transfection and became even more pronounced at later time points. We calculated that the transfection of 1.5 μg of esiRNAs per 2 × 105 cells suppressed the luciferase activity within 72 h to approximately 1% of the levels in untreated control cells. Interestingly, we observed that the transfection of unrelated control esiRNAs targeting coding sequences of EGFP or LacZ also reduced the luciferase activity, albeit to a much lesser extent (for unknown reasons, this phenomenon was observed only in some experiments). However, incubation with the transfection reagent alone or with mixtures of the transfection reagent and longer double-stranded RNAs such as poly(IC) had no effect on reporter gene expression. In additional experiments, we compared the antiviral activity of different HCV-specific esiRNAs. The results shown in Fig. 2B demonstrate that the luciferase reporter activity in 9B cells is inhibited to almost the same extent and with similar kinetics by esiRNAs targeting HCV coding sequences and by the highly structured 5′ NTR. Similar results were obtained for esiRNAs targeting the firefly luciferase coding sequence (data not shown). Next, we analyzed whether the complexity of our esiRNA preparations was a limiting factor in the inhibition of HCV RNA replication. To that end, we transfected 9B cells with a mixture of different esiRNAs (ORF1, ORF2, and ORF3) and compared the antiviral effect with that of the individual preparations. However, no enhanced antiviral activity was observed with mixed preparations (data not shown), indicating that the individual preparations contained enough different esiRNAs to cover a redundant number of target sites.
FIG. 2.
Effect of different esiRNAs on the replication of a subgenomic HCV replicon. (A) Dose-dependent inhibition of replicon-dependent luciferase reporter gene expression by HCV-specific esiRNAs. (B) Antiviral effect of esiRNAs targeting different regions of the HCV genome. About 2 × 105 cells of the Huh-7 clone 9B (containing replicon I389/NS3-3′/LucUbiNeo-ET) were transfected with the indicated amounts of HCV genotype 1b-specific esiRNAs (ORF1, ORF3, or 5′ NTR), unrelated esiRNAs (EGFP or LacZ), tRNA, or poly(IC). As additional controls, cells were treated with the transfection reagent only or left untreated. At the times indicated, cells were lysed and luciferase activities were determined, at least in duplicate. RLU, relative light units.
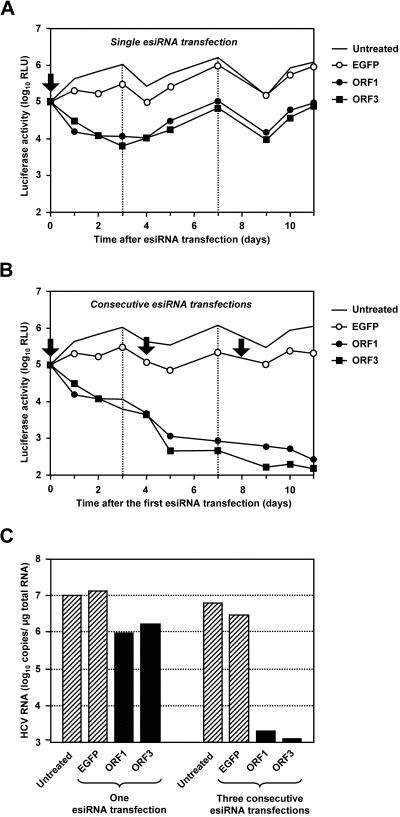
Previous time course experiments indicated that esiRNA-induced silencing is maximal after 6 days and levels off thereafter (72). To study whether HCV RNAs can be purged from transfected cells by the use of esiRNAs, we transfected 9B cells with HCV-specific or unrelated control esiRNAs and monitored the fates of the replicons by measuring the luciferase reporter gene activity up to 11 days posttransfection. Similar to the experiments described above, HCV-specific esiRNAs rapidly suppressed reporter gene expression by approximately 98% (compare Fig. 3A and 4A). Moreover, reporter gene expression did not recover in these cells (even 11 days after the transfection of HCV-specific esiRNAs). In contrast, the inhibition of reporter gene expression induced by nonrelated EGFP-specific esiRNAs was only transient, and at 7 days posttransfection, luciferase activities were comparable to those measured for untreated control cells (Fig. 3A). Based on experiments in which the expression of a host gene was silenced, we estimated that the efficiency by which we transfected esiRNAs into Huh-7 cells was in the range of 90 to 99% (data not shown). Thus, we inferred that consecutive transfections of HCV-specific esiRNA transfections might completely remove HCV replicons from 9B cells. The results shown in Fig. 3B demonstrate that this is indeed the case. Three days after the last of three consecutive transfections of HCV-specific esiRNAs, luciferase activities were close to the detection limit. We calculated the suppression compared to untreated control cells to be >99.97 and >99.98% for ORF1 and ORF3 esiRNAs, respectively. In contrast, additional transfections of EGFP-specific esiRNAs did not further reduce luciferase expression levels. To confirm these findings, we determined the replicon copy number by quantitative real-time reverse transcription (RT)-PCR. As shown in Fig. 3C, the reduction in reporter gene activity was always paralleled by an equivalent reduction in the number of replicon RNAs.
FIG. 3.
Long-term effect of esiRNAs on the replication of a subgenomic HCV replicon. The effects of single (A) and consecutive (B) esiRNA transfections on replicon-dependent luciferase reporter gene expression are shown. About 2 × 105 cells of the Huh-7 clone 9B (containing the HCV 1b replicon I389/NS3-3′/LucUbiNeo-ET) were transfected with 1 μg of HCV genotype 1b-specific esiRNAs (ORF1 and ORF3) or unrelated esiRNAs (EGFP) or were left untreated. At the times indicated, cells were lysed and luciferase activities were determined, at least in duplicate. Arrows and dotted lines indicate times of transfection and cell splitting (ratio of 1:3), respectively. RLU, relative light units. (C) Effect of genotype 1b-specific esiRNA transfections on the replicon copy number. RNAs were prepared from cells that had been lysed 11 days after the first esiRNA transfection, and the numbers of replicon molecules were determined by quantitative RT-PCR. The concentrations of viral RNAs are given in replicon copy numbers per microgram of total RNA. Hatched columns, viral RNA concentration in control cells; black columns, viral RNA concentration after transfection of HCV-specific esiRNAs. The figure shows the results from a single representative experiment.
FIG. 4.
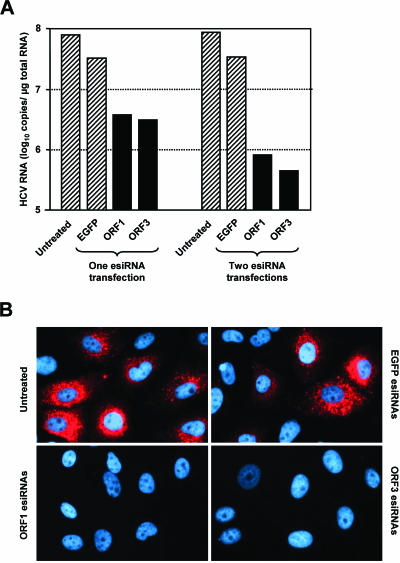
Effect of esiRNAs on the replication of a full-length HCV replicon. (A) Effect of esiRNAs on HCV RNA copy number. About 2 × 105 cells of the Huh-7 clone 20-1 (containing the replicon I389/Core-3′/5.1) were transfected with 1 μg of genotype 1b-specific esiRNAs (ORF1, ORF3, or EGFP) or were left untreated. Half of the cells were harvested 2 days later. The other half was passaged (split ratio, 1:4), seeded into new cell culture dishes, and transfected a second time or left untreated. Two days later, these cells were also harvested, RNAs were prepared, and the replicon copy numbers per microgram of total RNA were determined by quantitative RT-PCR. Hatched columns, viral RNA concentration in control cells; black columns, viral RNA concentration after the transfection of HCV-specific esiRNAs. The figure shows the results from a single representative experiment. (B) Effect of esiRNAs on the expression of NS5A. About 2 × 105 cells of clone 20-1 were transfected with 1.5 μg of esiRNAs (ORF1, ORF3, or EGFP) or were left untreated. Two days later, the cells were split in a ratio of 1:3 and seeded onto glass coverslips. After 2 days of further cultivation, cells were fixed, permeabilized, and immunostained for NS5A (red) and counterstained for DNA (blue) by using an NS5A-specific mouse monoclonal antibody and DAPI, respectively.
All experiments described so far were performed with cells containing a subgenomic replicon that does not encode the viral proteins core, E1, E2, p7, and NS2. Consequently, we could not rule out that one or more of these proteins interfere with RNAi. Therefore, we transfected cells of the Huh-7 cell clone 20-1 that contained the selectable genomic HCV replicon I389/Core-3′/5.1 with ORF1 or ORF3 esiRNAs. Since this replicon does not carry a reporter gene, we used quantitative RT-PCR and indirect immunofluorescence to measure viral replication. As shown in Fig. 4, transfections with HCV-specific esiRNAs, but not with unrelated control RNAs, caused a decrease in both the HCV RNA copy number and HCV protein expression. Thus, HCV structural proteins and the NS2 protease do not counteract the effect of esiRNAs. To substantiate this conclusion, we quantified the antiviral effect of esiRNAs in additional cell clones containing various HCV replicons (cell clones 9-13, 5-15, and 21-5 containing the replicons I377/NS3-3′, I389/NS3-3′, and I389/Core-3′/5.1, respectively). The results of these experiments confirmed that subgenomic replicons are not more susceptible to esiRNAs than genomic replicons (data not shown).
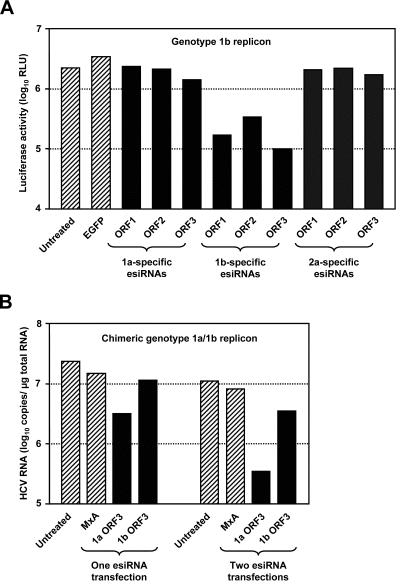
HCV genomes differ as much as 34% in their nucleotide sequences. Nevertheless, several genomic regions are highly conserved among all known HCV isolates (most notably, both NTRs, but also certain sequences within the core and NS5B coding regions). We speculated that these regions might well serve as potential binding sites for esiRNAs, which would allow the silencing of multiple genotypes with a single esiRNA preparation. To test this hypothesis, we transfected genotype 1a-, 1b-, or 2a-derived esiRNAs into 9B cells (containing the genotype 1b replicon I389/NS3-3′/LucUbiNeo-ET) and measured the luciferase reporter activity. As shown in Fig. 5A, we found that only genotype 1b-derived esiRNAs efficiently inhibited reporter gene expression. To corroborate this finding, we assayed the antiviral activities of different esiRNA preparations in cells with the chimeric replicon I389/NS3-3′/DR, which contains the NS3 to NS5B coding sequence of a genotype 1a virus and the NTRs of a genotype 1b virus. In this experimental setting, we found that genotype 1a- but not genotype 1b-derived esiRNAs efficiently blocked HCV replication (Fig. 5B). However, slightly lower HCV RNA levels were detected in cells that had been transfected with heterologous ORF3 esiRNAs. Thus, it is tempting to speculate that individual esiRNA molecules that target the highly conserved sequence motifs within the NS5B sequence account for this phenomenon. In summary, these data suggest that the efficient silencing of multiple HCV genotypes is only possible with esiRNAs that target highly conserved regions of the viral genome.
FIG. 5.
Sequence specificity of esiRNAs. (A) Effect of genotype 1a-, 1b-, and 2a-specific esiRNAs on the replication of an HCV genotype 1b replicon. About 2 × 105 cells of Huh-7 clone 9B (containing the replicon I389/NS3-3′/LucUbiNeo-ET) were transfected with 1.5 μg of esiRNAs, as indicated, or were left untreated. Two days later, cells were lysed and luciferase activities were determined, at least in duplicate. RLU, relative light units. (B) Effect of genotype 1a- and 1b-specific esiRNAs on the replication of a chimeric HCV replicon with genotype 1a coding sequences. About 2 × 105 cells of the Huh-7 clone A-3/3 (containing the replicon I389/NS3-3′/H77/DR) were transfected with 1 μg of esiRNAs, as indicated, or were left untreated. Half of the cells were harvested 2 days later. The other half was passaged (split ratio, 1:4), seeded into new cell culture dishes, and transfected a second time or left untreated. Two days later, these cells were also harvested, RNAs were prepared, and the replicon copy numbers per microgram of total RNA were determined by quantitative RT-PCR. Hatched columns, viral RNA concentration in control cells; black columns, viral RNA concentration after the transfection of HCV-specific esiRNAs. The figure shows the results from a single representative experiment.
Inhibition of HCV replicons by shRNAs.
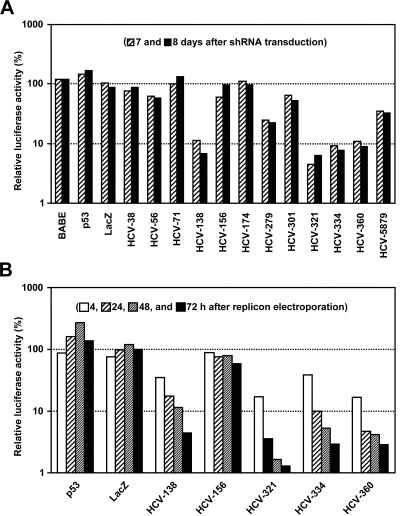
In an attempt to establish an alternative method to purge HCV RNAs from infected cells and to confer resistance against HCV infections, we explored the utility of shRNAs. These siRNA-like transcripts were first described by Brummelkamp and coworkers, who also demonstrated that the constitutive expression of shRNAs results in persistent silencing (6). These findings prompted us to design shRNAs to target highly conserved regions of the HCV genome. Since it is difficult to predict which sequence is targeted most efficiently, we tested a series of 12 HCV-specific shRNAs for the ability to inhibit HCV RNA replication. Eleven of these targeted various highly conserved sequence motifs within the 5′ NTR or the core coding region, and one was directed against a comparatively variable sequence within the NS4B coding region (Fig. 6 and 7). For the delivery and constitutive expression of shRNAs, we constructed a retroviral vector that we named pBABE/H1/SV40/EGZ/ΔU3 (for details, see Materials and Methods). The main characteristic features of this Mo-MuLV-based vector construct are (i) a large deletion within the U3 region of the 3′ LTR, (ii) a BglII/XhoI cloning site for the insertion of shRNA-encoding oligonucleotides downstream of an RNA polymerase III H1-RNA promoter, and (iii) an EGFP-zeocin cassette downstream of a simian virus 40 (SV40) promoter element (Fig. 1B). In a pilot experiment, we tested the ability of this vector to induce RNAi by using a human p53-specific shRNA design that was described previously (6). As expected, the transduction of naive Huh-7 cells with the recombinant retrovirus pBABE/H1/SV40/EGZ/ΔU3/p53 resulted in the expression of the EGFP-zeocin fusion protein and silenced the endogenous p53 gene, thereby demonstrating the functionality of our retroviral vector (data not shown). Next, we infected 9B cells (containing the replicon I389/NS3-3′/LucUbiNeo-ET) with different retroviruses that transduce HCV-specific shRNAs or unrelated control shRNAs. As additional controls, cells were infected with the parental vector or incubated with medium from untransfected 293T cells. Seven and 8 days later, cells were harvested and luciferase assays were performed. We found that neither the parental vector (BABE) nor vectors transducing unrelated control shRNAs (p53 and LacZ) inhibited the expression of the luciferase gene, whereas half of the HCV-specific shRNAs caused a reduction in reporter gene activity (Fig. 8A). Those that targeted sequences in close proximity to the start codon in domain IV were especially efficient at inducing RNAi (e.g., HCV-321, HCV-334, and HCV-360). Other regions of the 5′ NTR, however, appeared to be rather resistant towards shRNA-induced gene silencing. Nearly unhindered luciferase expression occurred in cells that expressed shRNAs targeting domain II of the 5′ NTR (namely HCV-38, HCV-56, and HCV-71), and only one of five shRNAs targeting domain III induced efficient silencing of luciferase expression (HCV-138). We also targeted by shRNAs a site in the NS4B coding sequence that was successfully targeted in a previous study by chemically synthesized siRNAs (51). HCV-5879, however, only moderately inhibited HCV RNA replication compared to 5′ NTR-specific shRNAs such as HCV-321. One could argue that the observed differences in the ability of HCV-specific shRNAs to induce RNAi are due to different transduction efficiencies. However, indirect immunofluorescence using GFP-specific antibodies revealed that, in all cases, >90% of the cells were transduced (data not shown).
FIG. 6.
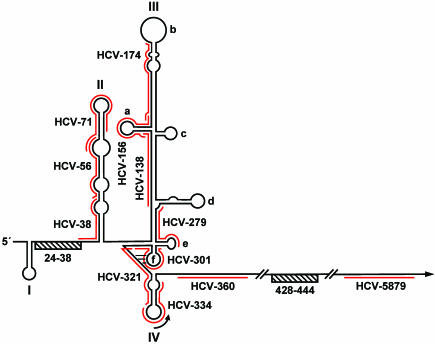
Target structures of HCV-specific shRNAs. A schematic representation of the secondary structure of the HCV 5′ NTR, including adjacent coding sequences (modified from the work of Honda et al. [25]), is shown. Domains and subdomains are labeled with roman numerals and lowercase letters, respectively. The position of the AUG start codon within domain IV is indicated by an arced arrow. A pseudoknot structure that is generated by base pairing between domain IIIf and the intervening sequence connecting domains IIIf and IV is indicated by three horizontal lines. Furthermore, the positions of two regions (ranging from nucleotides 24 to 38 and 428 to 444) that are involved in a long-range RNA-RNA interaction (31) are marked with striped bars. Red lines indicate sequences targeted by shRNAs. Note that numbers refer to nucleotide positions of the Con1 consensus genome.
FIG. 7.
Target sequences of HCV-specific shRNAs. The CLUSTAL W algorithm (61) was used to align the first 389 nucleotides of the HCV Con1 consensus genome (genotype 1b) with homologous sequences of six other genomes representing genotypes 1a (strain H77), 2a (strain HC-J6CH), 3a (strain NZL1), 4a (strain HEMA51), 5a (strain EUH1480), and 6b (strain Th580). Black lines indicate sequences targeted by shRNAs. Coding nucleotides are highlighted in gray.
FIG. 8.
Antiviral activity of HCV-specific shRNAs. (A) Effect of shRNAs on replicon-dependent luciferase reporter gene expression in cells persistently replicating a subgenomic HCV replicon. About 6 × 104 cells of clone 9B (containing replicon I389/NS3-3′/LucUbiNeo-ET) were transduced thrice with retroviral vectors encoding either an HCV-specific shRNA or an unrelated control shRNA (as indicated). Control cells were transduced with the parental retroviral vector (BABE) or incubated with medium from untransfected 293T cells. About 24 h after the last transduction, the cells were passaged (split ratio, 1:4) and further cultivated. About 96 and 120 h later, the cells were lysed and luciferase activities were determined, at least in duplicate. The mean luciferase activities of nontransduced control cells were set at 100% and used to normalize luciferase activities of transduced cells. The resulting relative luciferase values are shown. (B) Transduction of HCV-specific shRNAs confers resistance against a subgenomic HCV replicon. Naive Huh-7 cells were transduced twice with retroviral vectors that encode either an HCV-specific shRNA or an unrelated control shRNA (as indicated). After an incubation period of 13 days, during which the cells were cultured in the presence of zeocin and split at regular intervals, the cells were electroporated with the subgenomic genotype 1b replicon I389/NS3-3′/Luc-ET, seeded into multiple cell culture dishes, and harvested at given time points. The average luciferase activities of nontransduced control cells were set at 100% and used to normalize the corresponding activities of transduced cells. The resulting relative luciferase values are shown.
To find out whether shRNAs can also confer antiviral resistance, we infected naive Huh-7 cells with a number of retroviruses that encoded either HCV-specific or unrelated shRNAs and cultivated the cells for nearly 2 weeks in the presence of zeocin in order to select for transduced cells. The cells were then challenged with the cell culture-adapted subgenomic HCV replicon I389/NS3-3′/Luc-ET (18, 41), and viral replication was quantified by measuring luciferase reporter activities. We found that those shRNAs which inhibited HCV replication in 9B cells also blocked HCV replication in this experiment (Fig. 8B). This result suggests that the transduction of shRNAs may indeed protect cells from HCV infections.
Taken together, the results demonstrate that the 5′ NTR of the HCV genome contains sequences that are highly susceptible to esiRNAs and shRNAs. Note that some of these sequences are extremely conserved, which might allow the inhibition of all known genotypes by universal shRNAs such as HCV-321. Furthermore, the results suggest that it may be possible in the near future to confer resistance against HCV infections by the constitutive expression of shRNAs.
DISCUSSION
In contrast to plants and invertebrates, mammals have not yet been reported to use RNAi as an antiviral defense mechanism. Nevertheless, RNAi can be initiated in mammalian cells artificially by the delivery of siRNAs (15). This technique has recently been used in cultured cells to block the replication of important human pathogens, such as HIV (10, 13, 27, 34, 45, 49) and HCV (29, 51, 67, 73; Seo et al., letter). In all of these studies, RNAi was induced in a highly specific manner by the use of chemically synthesized oligonucleotides, with the exception of the work of Yokota and coworkers, for which eukaryotic expression vectors to deliver HCV-specific siRNAs and shRNAs were also used (73). A problem with siRNAs as well as shRNAs, however, is that the efficiency by which they trigger RNAi varies dramatically and is difficult to predict (16, 24a). Another problem is the specificity with which they bind to their target sequences. Even a single nucleotide mismatch can completely abolish their capacity to induce RNAi (2, 12, 21). Given these limitations, it is challenging to design siRNAs that efficiently target all quasispecies of a virus population. RNA viruses and retroviruses will be especially difficult to eradicate because of their notoriously error-prone mode of replication which allows the rapid evolution of escape mutants. Since esiRNAs are directed against multiple sites of a targeted sequence, they can be used to inhibit the replication of a heterogenous population of related viruses. Furthermore, it seems extremely unlikely that a viral genome can accumulate enough point mutations to escape the antiviral activity of esiRNAs. Our results demonstrate that esiRNAs can indeed be used efficiently to block viral replication. Since esiRNAs are much cheaper to produce than chemically synthesized oligonucleotides, they represent a powerful and rather inexpensive molecular tool for many virologists.
Many plant viruses and an insect virus are known to encode proteins that counteract RNAi (35, 36). Interestingly, it was recently noted that the double-stranded RNA-binding protein σ3 of the mammalian orthoreovirus also inhibits RNAi if it is expressed as a recombinant protein in transgenic plants (37). Since the HCV core proteins bind RNA in a rather promiscuous manner (17, 53, 56), it is conceivable that the core protein also binds to esiRNAs, which would interfere with RNAi. However, our finding that the replication of subgenomic replicons is as sensitive as that of full-length HCV RNAs to the transfection of esiRNAs indicates that the core protein cannot counteract RNAi. This observation is in line with a recent study showing that the replication of a full-length HCV RNA is sensitive to RNAi induced by chemically synthesized siRNAs (51). Nevertheless, the data do not exclude the possibility that the core protein protects the viral genome from Dicer, which would block the endogenous production of siRNAs. It is, however, unlikely that such an ability of core is of relevance to the replication of HCV in human cells, as most mammals do not seem to use RNAi as an antiviral defense mechanism.
The 5′ NTR has long been a focus of antiviral research because this highly conserved region of the HCV genome is indispensable for both RNA translation and replication. In a number of studies, antisense oligodeoxynucleotides have been tested for the ability to block the activity of the HCV IRES (1, 63, 65). Furthermore, the 5′ NTR has been targeted by ribozymes containing complementary sequences (38). These studies indicated that sequences in close proximity to the start codon are most suitable for the efficient inhibition of HCV RNA translation in a number of surrogate assays. These observations were confirmed by our finding that all domain IV-specific shRNAs (HCV-321 and HCV-334) blocked the replication of HCV RNAs. In contrast, none of the domain II-specific shRNAs and only one of five domain III-specific shRNAs (HCV-138) showed a similar degree of antiviral activity. We do not know why most of the shRNAs that targeted domain II and III failed to induce RNAi. However, one could speculate that some of them target RNA sequences that are not accessible to the RISC because of unfavorable secondary structures (for a definition of target requirements, see reference 32). Furthermore, it is possible that the tight interaction of the HCV IRES with components of the cellular translation machinery protects large sections of the 5′ NTR from binding shRNAs. In this context, it is interesting that most of domain III is involved in the recruitment of the 40S ribosomal subunit and the subsequent binding of the eukaryotic initiation factor eIF3 (reviewed in reference 54). Furthermore, it was shown by Spahn and coworkers that domain II is heavily engaged in the induction and/or stabilization of extensive conformational changes that take place in the 40S subunit before the 60S subunit joins the complex (59). Moreover, several noncanonical translation factors and other yet uncharacterized cellular proteins have been identified which bind to the HCV IRES (reviewed in reference 4) and may thereby also interfere with the binding of shRNAs.
Recently, Yokota and coworkers tested five different 5′ NTR-specific chemically synthesized siRNAs for the ability to block the replication of a subgenomic HCV replicon (73). The findings of Yokota and coworkers are somewhat difficult to compare with ours because the target sequences of their siRNAs and those of our shRNAs do not match. Nevertheless, a few conclusions can be drawn. First, as with our shRNAs, the most effective siRNA targeted a sequence within domain IV which encompasses the start codon. Second, an siRNA that targeted the subdomain IIIe only slightly inhibited HCV replication (similar to shRNA HCV-279). Most interesting, however, is the observation that an siRNA that targeted domain II efficiently inhibited HCV RNA replication but failed to induce the degradation of mRNAs containing the HCV IRES. Notably, the 5′ ends of that siRNA possess a pronounced sequence asymmetry (the strand that is complementary to the negative strand starts with a UAAC sequence, whereas the 5′ sequence of the other strand is GCGU). Given the recent finding that the siRNA strand whose 5′ end is less tightly paired to its complement is preferentially incorporated into the RISC (30, 55), it seems likely that this particular siRNA targets the 3′ terminus of the HCV negative strand rather than the 5′ NTR of the viral genome. It is tempting to speculate that some of our HCV-specific shRNAs also targeted the HCV negative strand, a mode of action which might have contributed to the antiviral activity of HCV-138.
A main goal of this study was the identification of shRNAs that can be used to target all common HCV genotypes. With HCV-138 and HCV-321, we found two shRNAs that fulfill this criterion. Furthermore, we established a system that allows the efficient transduction of these shRNAs into potential host cells. This know-how may be used in future approaches to inhibit HCV replication in vivo.
Acknowledgments
We are grateful to Reuven Agami, Jens Bukh, and Dirk Lindemann for the kind provision of reagents. Furthermore, we thank Nicole Krieger, Artur Kaul, Ulf Zeuge, and Ulrike Herian for technical assistance and Kerry Mills for critical reading of the manuscript.
This work was supported by grants from the Bundesministerium für Bildung und Forschung (Kompetenznetz Hepatitis, projects 01KI0102 and D.10022130) and an Unrestricted Biomedical Research Grant in Infectious Diseases from the Bristol-Myers Squibb Foundation.
REFERENCES
- 1.Alt, M., R. Renz, P. H. Hofschneider, G. Paumgartner, and W. H. Caselmann. 1995. Specific inhibition of hepatitis C viral gene expression by antisense phosphorothioate oligodeoxynucleotides. Hepatology 22:707-717. [PubMed] [Google Scholar]
- 2.Amarzguioui, M., T. Holen, E. Babaie, and H. Prydz. 2003. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 31:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambros, V. 2003. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113:673-676. [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., M. Frese, and T. Pietschmann. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res., in press. [DOI] [PubMed]
- 5.Berberich-Siebelt, F., S. Klein-Hessling, N. Hepping, B. Santner-Nanan, D. Lindemann, A. Schimpl, I. Berberich, and E. Serfling. 2000. C/EBPbeta enhances IL-4 but impairs IL-2 and IFN-gamma induction in T cells. Eur. J. Immunol. 30:2576-2585. [DOI] [PubMed] [Google Scholar]
- 6.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 7.Buchholz, F., L. Ringrose, P. O. Angrand, F. Rossi, and A. F. Stewart. 1996. Different thermostabilities of FLP and Cre recombinases: implications for applied site-specific recombination. Nucleic Acids Res. 24:4256-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calegari, F., W. Haubensak, D. Yang, W. B. Huttner, and F. Buchholz. 2002. Tissue-specific RNA interference in postimplantation mouse embryos with endoribonuclease-prepared short interfering RNA. Proc. Natl. Acad. Sci. USA 99:14236-14240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caplen, N. J. 2003. RNAi as a gene therapy approach. Expert Opin. Biol. Ther. 3:575-586. [DOI] [PubMed] [Google Scholar]
- 10.Capodici, J., K. Kariko, and D. Weissman. 2002. Inhibition of HIV-1 infection by small interfering RNA-mediated RNA interference. J. Immunol. 169:5196-5201. [DOI] [PubMed] [Google Scholar]
- 11.Carrington, J. C., and V. Ambros. 2003. Role of microRNAs in plant and animal development. Science 301:336-338. [DOI] [PubMed] [Google Scholar]
- 12.Chiu, Y. L., and T. M. Rana. 2002. RNAi in human cells: basic structural and functional features of small interfering RNA. Mol. Cell 10:549-561. [DOI] [PubMed] [Google Scholar]
- 13.Coburn, G. A., and B. R. Cullen. 2002. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 76:9225-9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DuBridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 16.Elbashir, S. M., J. Harborth, K. Weber, and T. Tuschl. 2002. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26:199-213. [DOI] [PubMed] [Google Scholar]
- 17.Fan, Z., Q. R. Yang, J. S. Twu, and A. H. Sherker. 1999. Specific in vitro association between the hepatitis C viral genome and core protein. J. Med. Virol. 59:131-134. [PubMed] [Google Scholar]
- 18.Frese, M., K. Barth, A. Kaul, V. Lohmann, V. Schwärzle, and R. Bartenschlager. 2003. Hepatis C virus RNA replication is resistant to tumor necrosis factor-α. J. Gen. Virol. 84:1253-1259. [DOI] [PubMed] [Google Scholar]
- 19.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 20.Grishok, A., A. E. Pasquinelli, D. Conte, N. Li, S. Parrish, I. Ha, D. L. Baillie, A. Fire, G. Ruvkun, and C. C. Mello. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106:23-34. [DOI] [PubMed] [Google Scholar]
- 21.Hamada, M., T. Ohtsuka, R. Kawaida, M. Koizumi, K. Morita, H. Furukawa, T. Imanishi, M. Miyagishi, and K. Taira. 2002. Effects on RNA interference in gene expression (RNAi) in cultured mammalian cells of mismatches and the introduction of chemical modifications at the 3′-ends of siRNAs. Antisense Nucleic Acid Drug Dev. 12:301-309. [DOI] [PubMed] [Google Scholar]
- 22.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 23.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 24.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 24a.Holen, T., M. Amarzguioui, M. T. Wiiger, E. Babaie, and H. Prydz. 2002. Positional effects of short interfering RNAs targeting the human coagulation trigger tissue factor. Nucleic Acids Res. 30:1757-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Honda, M., M. R. Beard, L. H. Ping, and S. M. Lemon. 1999. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 73:1165-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoofnagle, J. H. 1997. Hepatitis C: the clinical spectrum of disease. Hepatology 26(Suppl. 1):15S-20S. [DOI] [PubMed] [Google Scholar]
- 27.Jacque, J. M., K. Triques, and M. Stevenson. 2002. Modulation of HIV-1 replication by RNA interference. Nature 418:435-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalajzic, I., M. L. Stover, P. Liu, Z. Kalajzic, D. W. Rowe, and A. C. Lichtler. 2001. Use of VSV-G pseudotyped retroviral vectors to target murine osteoprogenitor cells. Virology 284:37-45. [DOI] [PubMed] [Google Scholar]
- 29.Kapadia, S. B., A. Brideau-Andersen, and F. V. Chisari. 2003. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl. Acad. Sci. USA 100:2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khvorova, A., A. Reynolds, and S. D. Jayasena. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115:209-216. [DOI] [PubMed] [Google Scholar]
- 31.Kim, Y. K., S. H. Lee, C. S. Kim, S. K. Seol, and S. K. Jang. 2003. Long-range RNA-RNA interaction between the 5′ nontranslated region and the core-coding sequences of hepatitis C virus modulates the IRES-dependent translation. RNA 9:599-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kretschmer-Kazemi, F. R., and G. Sczakiel. 2003. The activity of siRNA in mammalian cells is related to structural target accessibility: a comparison with antisense oligonucleotides. Nucleic Acids Res. 31:4417-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, N. S., T. Dohjima, G. Bauer, H. Li, M. J. Li, A. Ehsani, P. Salvaterra, and J. Rossi. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20:500-505. [DOI] [PubMed] [Google Scholar]
- 35.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 296:1319-1321. [DOI] [PubMed] [Google Scholar]
- 36.Li, W. X., and S. W. Ding. 2001. Viral suppressors of RNA silencing. Curr. Opin. Biotechnol. 12:150-154. [DOI] [PubMed] [Google Scholar]
- 37.Lichner, Z., D. Silhavy, and J. Burgyan. 2003. Double-stranded RNA-binding proteins could suppress RNA interference-mediated antiviral defences. J. Gen. Virol. 84:975-980. [DOI] [PubMed] [Google Scholar]
- 38.Lieber, A., C. Y. He, S. J. Polyak, D. R. Gretch, D. Barr, and M. A. Kay. 1996. Elimination of hepatitis C virus RNA in infected human hepatocytes by adenovirus-mediated expression of ribozymes. J. Virol. 70:8782-8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohmann, V., J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 41.Lohmann, V., F. Körner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lohmann, V., A. Roos, F. Körner, J. O. Koch, and R. Bartenschlager. 2000. Biochemical and structural analysis of the NS5B RNA-dependent RNA polymerase of the hepatitis C virus. J. Viral Hepat. 7:167-174. [DOI] [PubMed] [Google Scholar]
- 43.McHutchison, J. G., and M. W. Fried. 2003. Current therapy for hepatitis C: pegylated interferon and ribavirin. Clin. Liver Dis. 7:149-161. [DOI] [PubMed] [Google Scholar]
- 44.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Novina, C. D., M. F. Murray, D. M. Dykxhoorn, P. J. Beresford, J. Riess, S. K. Lee, R. G. Collman, J. Lieberman, P. Shankar, and P. A. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8:681-686. [DOI] [PubMed] [Google Scholar]
- 46.Nykanen, A., B. Haley, and P. D. Zamore. 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107:309-321. [DOI] [PubMed] [Google Scholar]
- 47.Ogata, N., H. J. Alter, R. H. Miller, and R. H. Purcell. 1991. Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:3392-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okamoto, H., M. Kojima, S. Okada, H. Yoshizawa, H. Iizuka, T. Tanaka, E. E. Muchmore, D. A. Peterson, Y. Ito, and S. Mishiro. 1992. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology 190:894-899. [DOI] [PubMed] [Google Scholar]
- 49.Park, W. S., N. Miyano-Kurosaki, M. Hayafune, E. Nakajima, T. Matsuzaki, F. Shimada, and H. Takaku. 2002. Prevention of HIV-1 infection in human peripheral blood mononuclear cells by specific RNA interference. Nucleic Acids Res. 30:4830-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Randall, G., A. Grakoui, and C. M. Rice. 2003. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc. Natl. Acad. Sci. USA 100:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Santolini, E., G. Migliaccio, and N. La Monica. 1994. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J. Virol. 68:3631-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarnow, P. 2003. Viral internal ribosome entry site elements: novel ribosome-RNA complexes and roles in viral pathogenesis. J. Virol. 77:2801-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwarz, D. S., G. Hutvagner, T. Du, Z. Xu, N. Aronin, and P. D. Zamore. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115:199-208. [DOI] [PubMed] [Google Scholar]
- 56.Shimoike, T., S. Mimori, H. Tani, Y. Matsuura, and T. Miyamura. 1999. Interaction of hepatitis C virus core protein with viral sense RNA and suppression of its translation. J. Virol. 73:9718-9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silhavy, D., A. Molnar, A. Lucioli, G. Szittya, C. Hornyik, M. Tavazza, and J. Burgyan. 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21:3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soneoka, Y., P. M. Cannon, E. E. Ramsdale, J. C. Griffiths, G. Romano, S. M. Kingsman, and A. J. Kingsman. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spahn, C. M., J. S. Kieft, R. A. Grassucci, P. A. Penczek, K. Zhou, J. A. Doudna, and J. Frank. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291:1959-1962. [DOI] [PubMed] [Google Scholar]
- 60.Theodore, D., and M. W. Fried. 2000. Natural history and disease manifestations of hepatitis C infection. Curr. Top. Microbiol. Immunol. 242:44-54. [DOI] [PubMed] [Google Scholar]
- 61.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Virus taxonomy: the VIIth report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif..
- 63.Vidalin, O., M. E. Major, B. Rayner, J. L. Imbach, C. Trepo, and G. Inchauspe. 1996. In vitro inhibition of hepatitis C virus gene expression by chemically modified antisense oligodeoxynucleotides. Antimicrob. Agents Chemother. 40:2337-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vrolijk, J. M., A. Kaul, B. E. Hansen, V. Lohmann, B. L. Haagmans, S. W. Schalm, and R. Bartenschlager. 2003. A replicon-based bioassay for the measurement of interferons in patients with chronic hepatitis C. J. Virol. Methods 110:201-209. [DOI] [PubMed] [Google Scholar]
- 65.Wakita, T., and J. R. Wands. 1994. Specific inhibition of hepatitis C virus expression by antisense oligodeoxynucleotides. In vitro model for selection of target sequence. J. Biol. Chem. 269:14205-14210. [PubMed] [Google Scholar]
- 66.Walewski, J. L., T. R. Keller, D. D. Stump, and A. D. Branch. 2001. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA 7:710-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wilson, J. A., S. Jayasena, A. Khvorova, S. Sabatinos, I. G. Rodrigue-Gervais, S. Arya, F. Sarangi, M. Harris-Brandts, S. Beaulieu, and C. D. Richardson. 2003. RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc. Natl. Acad. Sci. USA 100:2783-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.World Health Organization. 2000. Hepatitis C: global prevalence (update). Wkly. Epidemiol. Rec. 75:18-19. [PubMed] [Google Scholar]
- 69.Xu, Z., J. Choi, T. S. Yen, W. Lu, A. Strohecker, S. Govindarajan, D. Chien, M. J. Selby, and J. Ou. 2001. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 20:3840-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. USA 94:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1999. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262:250-263. [DOI] [PubMed] [Google Scholar]
- 72.Yang, D., F. Buchholz, Z. Huang, A. Goga, C. Y. Chen, F. M. Brodsky, and J. M. Bishop. 2002. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA 99:9942-9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yokota, T., N. Sakamoto, N. Enomoto, Y. Tanabe, M. Miyagishi, S. Maekawa, L. Yi, M. Kurosaki, K. Taira, M. Watanabe, and H. Mizusawa. 2003. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Rep. 4:602-608. [DOI] [PMC free article] [PubMed] [Google Scholar]