Abstract
The fusion proteins of the alphaviruses and flaviviruses have a similar native structure and convert to a highly stable homotrimer conformation during the fusion of the viral and target membranes. The properties of the alpha- and flavivirus fusion proteins distinguish them from the class I viral fusion proteins, such as influenza virus hemagglutinin, and establish them as the first members of the class II fusion proteins. Understanding how this new class carries out membrane fusion will require analysis of the structural basis for both the interaction of the protein subunits within the homotrimer and their interaction with the viral and target membranes. To this end we report a purification method for the E1 ectodomain homotrimer from the alphavirus Semliki Forest virus. The purified protein is trimeric, detergent soluble, retains the characteristic stability of the starting homotrimer, and is free of lipid and other contaminants. In contrast to the postfusion structures that have been determined for the class I proteins, the E1 homotrimer contains the fusion peptide region responsible for interaction with target membranes. This E1 trimer preparation is an excellent candidate for structural studies of the class II viral fusion proteins, and we report conditions that generate three-dimensional crystals suitable for analysis by X-ray diffraction. Determination of the structure will provide our first high-resolution views of both the low-pH-induced trimeric conformation and the target membrane-interacting region of the alphavirus fusion protein.
Enveloped viruses must have a mechanism to bypass the cellular and viral membranes that block entry of the viral genome into the cytoplasm. Fusion of the virus membrane with a cell membrane, either at the cell surface or from within the endocytic pathway, allows access to the cytoplasm and initiates infection. Reagents that inhibit virus fusion block virus infection and are an important current focus of antiviral therapies (10, 35).
One of the best-understood viruses to use the endocytic entry pathway is the alphavirus Semliki Forest virus (SFV) (21, 42, 46). SFV is a small enveloped RNA virus with a relatively simple structure. The nucleocapsid is composed of the positive-stranded RNA genome and associated capsid protein, and the enveloping membrane contains two type I glycoproteins, E1 and E2, each about 50 kDa. E1 and E2 form stable heterodimers that associate into trimers and form an icosahedral lattice on the surface of the virus (30, 38, 53). E1 is the membrane fusion protein and contains the virus fusion peptide, while E2 binds the virus receptor and regulates the fusion activity of E1. Upon exposure to the low pH of endosomes, the E1-E2 heterodimer dissociates, allowing the E1 proteins to undergo a series of conformational changes, including the interaction of the fusion peptide region with target membranes and the formation of an extremely stable E1 homotrimer. The E1 homotrimer has been shown to be essential in viral membrane fusion (23, 47), but the molecular mechanism of the fusion reaction is not fully understood.
Extensive biochemical analysis has revealed a number of key features of the SFV fusion reaction (see reference 21 for review). Rapid and efficient fusion with either cell membranes or protein-free liposomes is triggered at a pH of about 6.2 or below (4). Both in vitro fusion and virus infection of host cells specifically require the presence of cholesterol in the target membrane (37, 51). A monomeric ectodomain form of E1 termed E1* can be produced by proteolytic cleavage (22). E1* lacks the transmembrane domain and membrane-proximal stem regions and is water soluble. Treatment of E1* at low pH in the presence of cholesterol-containing target membranes leads to insertion of the fusion peptide into the target membrane and formation of a very stable homotrimer (1, 26). Similar to the full-length homotrimer, this ectodomain homotrimer (E1*HT) is resistant to proteases, detergents, and chemical or heat denaturation (16). Thus, E1* retains biological activity. At low pH, the ectodomain recapitulates the conformational changes and membrane association that take place in E1 during fusion but is nonfusogenic due to the absence of the transmembrane domain.
A similar water-soluble ectodomain form of E1, E1ΔS, was purified, crystallized, and used to determine the X-ray structure of the neutral-pH form of the protein (30, 50). The native E1 molecule is an elongated three-domain structure composed mainly of β-sheets, with the fusion peptide located in a loop at the most membrane distal tip and the transmembrane domain located at the opposite end of the molecule. Comparison of the structures of the native fusion glycoproteins of the alpha- and flaviviruses demonstrates that, despite the lack of any identifiable sequence similarity, they have a strikingly similar molecular architecture, and they have thus been grouped together as class II virus fusion proteins (30). The alphavirus and flavivirus fusion proteins share a number of interesting characteristics (reviewed in references 17 and 21). Both are regulated by their interaction with a companion subunit, which is cleaved by furin during virus biosynthesis to activate subsequent virus fusion at the pH of the endosome. During fusion both proteins convert from a native homo- or heterodimer to a stable homotrimer. Similar to SFV, the ectodomain of the fusion protein of the flavivirus tick-borne encephalitis virus forms a membrane-bound homotrimer when treated at acid pH in the presence of cholesterol-containing target membranes (45). The structures of alpha- and flavivirus particles have been characterized by cryoelectron microscopy (cryo-EM) and reconstruction. Strikingly, although the symmetry of their arrangement on the virus surface differs, both fusion proteins lie tangential to the virus membrane and define the regular protein lattice covering the virus particle (11, 28, 30, 38, 53). Therefore, in addition to their well-documented fusion activities, these class II proteins contribute significantly to both virus particle formation during budding and virus disassembly during membrane fusion (12). However, while much is known about the native structures and their fusogenic conformational changes, the structure of the class II homotrimer, the rearrangements and motifs that mediate its high stability, and the nature of the trimer-target membrane interaction are unknown.
The class I virus fusion proteins include those of orthomyxoviruses, paramyxoviruses, retroviruses, coronaviruses, and filoviruses. The stable postfusion conformation of the class I fusion proteins has been structurally characterized for a number of these viruses (reviewed in references 8, 10, and 49). The rearrangements of the protein during fusion are best understood in the case of the influenza virus hemagglutinin (HA) (reviewed in reference 43), which typifies the general features shared by the class I viral fusion proteins. HA is synthesized as an inactive precursor, HA0, which is activated by proteolytic cleavage to produce a trimer of two disulfide-linked subunits, the receptor-binding HA1 and the transmembrane HA2 containing the virus fusion peptide. Upon triggering of the fusion reaction at low pH, a region of extended structure of HA2 refolds to form a continuation of the central trimeric α-helical coiled coil. This translocates the N-terminal HA2 fusion peptide to the top of the molecule, where it can insert into the target membrane, producing an intermediate with the HA2 protein anchored in both the viral and target membranes. A C-terminal region adjacent to the virus membrane then folds back to pack in the grooves of the core coiled coil, thus producing a highly stable α-helical bundle or hairpin with the HA2 transmembrane domain and fusion peptide at the same end of the molecule (5). The transition to this stable hairpin conformation drives the class I membrane fusion reaction (33, 41).
An enormous amount of information on viral fusion and fusion protein conformational changes has been derived from the work on the class I fusion proteins, but much is still unknown about how viral proteins carry out membrane fusion. Since the characterization of the hairpin structure has been performed with ectodomain fragments lacking both the fusion peptide region and the transmembrane domains, there are presently no postfusion structures of the fusion protein regions mediating membrane interaction. With the exception of influenza virus (52) and partial information on Newcastle disease virus (7), the native fusion protein structures have not been determined, therefore limiting our views of the overall protein rearrangements that lead to the final stable hairpin. Additionally, since the class I virus particles studied do not display regular structures, it has not yet been possible to analyze them by high-resolution EM. Therefore the existing protein crystal structures cannot be placed within the context of the overall macromolecular assembly to explain how individual trimers might work together during the process of fusion.
To address these and other questions about the class II virus fusion mechanism, we set out to obtain material suitable for high-resolution structural studies of the SFV E1 homotrimer. To this end, we developed a method to purify the stable membrane-inserted ectodomain homotrimer from target membranes. The purified homotrimer is detergent soluble, retains the stability of the starting homotrimer, and is free of lipid and protein contaminants. Screening of solution and crystallization conditions demonstrated substantial changes from the water-soluble native ectodomain monomer. Several habits of three-dimensional (3D) crystals were identified, at least one of which was suitable for analysis by X-ray diffraction. This is among the first viral fusion proteins to be crystallized in the postfusion conformation with the fusion peptide present and thus provides a new approach to understanding virus fusion machines and their interaction with membranes.
(Portions of the data in this paper are from a thesis submitted by D. L. Gibbons in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Sue Golding Graduate Division of Medical Sciences, Albert Einstein College of Medicine, Yeshiva University.)
MATERIALS AND METHODS
Virus and cells.
BHK-21 cells were cultured at 37°C in Dulbecco's modified Eagle's medium containing 5% fetal calf serum, 10% tryptose phosphate broth, and 100 U of penicillin and 100 μg of streptomycin per ml (37). The SFV used for these experiments was produced in BHK-21 cells from a well-characterized plaque-purified isolate (25) and was purified by banding on tartrate gradients (20).
Preparation of SFV glycoprotein ectodomains.
Soluble forms of the E1 and E2 glycoprotein subunits (E1* and E2*) were prepared as previously described (16, 22). In brief, purified unlabeled SFV (1 to 2 mg of protein) was digested with subtilisin for 60 to 90 min on ice at a protein-to-protease ratio of ∼3:1 (wt/wt) in phosphate-buffered saline buffer containing 0.9 mM CaCl2, 0.5 mM MgCl2, and 0.5% Triton X-114 (TX-114). The ectodomains were then purified by TX-114 phase separation (3) and concanavalin A chromatography, followed by dialysis against morpholineethanesulfonic acid (MES) buffer (20 mM MES, pH 7.0, and 130 mM NaCl). The purified proteins were stored at −140°C until use.
Liposomes.
Liposomes were prepared by extrusion as previously described (6), by using equimolar amounts of cholesterol and phospholipids and a molar ratio of 1:1:1:3 phosphatidylcholine (from egg yolk)-phosphatidylethanolamine (from egg phosphatidylcholine by transphosphatidylation)-sphingomyelin (bovine brain)-cholesterol (24). Phospholipids were from Avanti Polar Lipids (Alabaster, Ala.), and cholesterol was from Steraloids (Wilton, N.H.). All liposome stocks contained either [3H]cholesterol or 125I-labeled phosphatidylcholine as a tracer.
Generation and assay of the E1* homotrimer.
Purified ectodomains (∼0.25 mg of total protein/ml) were mixed with liposomes at a final concentration of 1.0 mM lipid, a lipid-to-protein ratio of ∼12.5 nmol per μg of E1*, or ∼830:1 on a molar basis. The mixture was preincubated for 5 min at 37°C, adjusted to pH 5.5 by addition of a precalibrated volume of 0.5 N acetic acid, incubated for 10 min at 37°C, and then neutralized by addition of 0.5 N NaOH. The E1*HT was assayed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 11% polyacrylamide gels after treating samples in SDS-sample buffer (200 mM Tris, pH 8.8, 4% SDS, 10% glycerol, and 0.02% bromophenol blue) at either 30°C to preserve the homotrimer (47) or at 95°C with reduction and alkylation to separate E1* and E2* (22). Proteins were visualized by Coomassie blue (R-250) or silver staining. Alternatively, the E1*HT was assayed by digestion for 1 h at 37°C with 125 μg of trypsin (type XIII; Sigma Chemical Co., St. Louis, Mo.)/ml in phosphate-buffered saline containing 0.5% TX-100. The digestion was stopped by addition of phenylmethylsulfonyl fluoride to a final concentration of 5 mM, and samples were analyzed by SDS-PAGE as described above.
Sucrose flotation gradients.
To assay E1*-liposome binding, low-pH-treated and neutralized samples were adjusted to a final concentration of 40% (wt/vol) sucrose, layered into the bottom of TLS55 ultracentrifuge tubes, and then overlaid with 1.4 ml of 25% (wt/vol) sucrose and 0.2 ml of 5% (wt/vol) sucrose in TN buffer (100 mM NaCl, 50 mM Tris-HCl, pH 7.4) (26). For samples solubilized with detergent, the detergent was included throughout the gradient at a final concentration of 1.5%, except where indicated. Samples containing cholate or deoxycholate were buffered at pH 8.5 instead of pH 7.4 to prevent precipitation of the detergent during centrifugation and handling. Gradients were centrifuged in the TLS55 rotor for 3 h at 54,000 rpm at 4°C, fractionated into seven 300-μl fractions, and analyzed for the presence of protein and [3H]cholesterol. Preparative samples used an SW60 rotor with a 40% sucrose layer of 1.8 ml, a 25% sucrose layer of 2 ml, and a 5% sucrose layer of 0.2 ml. These samples were centrifuged for 3 h at 54,000 rpm, and the top three 400-μl fractions containing the membranes were collected. Following flotation, membrane-bound E1* homotrimer was stored on ice until further purification.
Purification of the E1* homotrimer by sucrose sedimentation gradients.
Samples of the membrane-bound E1* homotrimer from sucrose flotation gradients were solubilized in 1.5% (wt/vol) octyl-β-d-glucopyranoside (OG; Fluka Chemical Corp., Milwaukee, Wis.) in TN buffer for 1 h at room temperature. The mixture was concentrated to a final volume of 0.5 ml by using a Centricon microconcentrator with a 30-kDa molecular mass cutoff. The sample was layered on top of an 11.5-ml, 7 to 15% (wt/wt) sucrose gradient in TN buffer containing 1% OG and 0.1 mM phenylmethylsulfonyl fluoride and was centrifuged in an SW41 rotor at 4°C for 22 h at 35,000 rpm.
Purification of the E1* homotrimer by centrifugal concentrator.
Samples of the membrane-bound E1* homotrimer from sucrose flotation gradients were solubilized in 1.5% OG in TN or 1 M NaCl-50 mM Tris-HCl (pH 7.4) (HSTN buffer) for 1 h at room temperature. The sample was then concentrated to a final volume of 0.5 ml in a filtration device with a 100-kDa molecular mass cutoff (Millipore or Vivascience). The sample was repeatedly washed with fresh buffer by dilution and reconcentration to 0.5 ml. After removal of the majority of the contaminating lipid as monitored by [3H]cholesterol, the protein was exchanged into TN or HSTN buffer containing 0.7% OG and 0.02% NaN3 by several sequential washes. The final sample volume was 0.5 ml, with a typical protein concentration of ∼0.5 mg/ml. The samples were stored on ice or at 4°C until use.
Lipid and detergent analysis.
E1*HT samples were analyzed for lipid and detergent content by thin-layer chromatography (29). Merck Silicagel 60 F254 plates were activated just prior to use by prerunning in ethyl ether and allowing them to air dry. Samples were applied in multiple small volumes with drying between applications to minimize spot size. Plates were developed in a preequilibrated tank containing a 65:25:4 mixture of CHCl3:methanol:H2O and were stained with iodine vapor to visualize the positions of OG and phospholipids. The amount of OG in the samples was estimated by comparison to standards on the same plate.
For lipid phosphate analysis, aqueous samples were extracted with methanol:CHCl3 (2) and were analyzed for total phospholipid content (36).
Dynamic light-scattering experiments.
Samples of the purified E1* protein in the indicated buffers were analyzed in a DynaPro-801 DLS instrument by using the manufacturer's software for analyses (Protein Solutions, Charlottesville, Va.). This instrument uses the dynamic scattering of laser light, detected at a 90° angle, to determine the diffusion constant of the molecules in solution by photon correlation spectroscopy. The hydrodynamic radius (Rh) is calculated from this measurement and is dependent upon the viscosity and the temperature of the solution. All measurements were carried out at 19°C.
Crystal screening and X-ray diffraction.
Purified E1*HT in 0.7% OG and HSTN was concentrated to 5 to 10 mg of protein/ml and a volume of about 25 to 50 μl by using a Vivaspin 500 concentrator with a molecular mass cutoff of 100 kDa (Vivascience, Inc.). Protein concentration was determined by absorbance spectroscopy at 276 nm and a calculated extinction coefficient for the E1* ectodomain of 47,810 M−1 cm−1. It was found that either 1 or 0.5 M salt maintained HT solubility during concentration and detergent exchange (see Fig. 4 and data not shown). Detergent exchange was therefore performed by diluting the 25- to 50-μl concentrated sample with 0.5 ml of 0.5 M salt (either NaCl or NaBr), 50 mM Tris-HCl, pH 7.5, 0.02% NaN3, and detergent above the critical micelle concentration (CMC) (most often 15 mM N,N-dimethyldecylamine-N-oxide [DDAO; Fluka] or 3.0 mM lauryldimethylamine-N-oxide [LDAO, N,N-dimethyldodecylamine-N-oxide; Fluka]), and reconcentrating the sample to 25 to 50 μl. This step was repeated several times to ensure proper detergent exchange. Where indicated, the E1*HT was exchanged into different detergents at this step. For screening of crystallization conditions, 1-μl aliquots of concentrated protein in the above buffer (or other test buffers) were mixed with equal volumes of a solution containing the same detergent at a concentration above the CMC, along with test additives as indicated, and the indicated concentration of polyethylene glycol (PEG) (Hampton or Merck). This 2-μl drop was then placed over a well containing the indicated PEG concentration in 0.5 M salt with or without 50 mM Tris-HCl, pH 7.5. Importantly, it was found that the PEG solutions had to be aged until the pH was at least 3.5 to 4.0, producing a final pH in the drop of approximately 4.0. Using a fresh PEG solution and adjusting the pH to 4 did not yield crystals. For the crystals described in the Fig. 6 legend, the protein was concentrated in buffer containing 15 mM DDAO, 0.5 M NaBr, 50 mM Tris-HCl, pH 7.5, and 0.02% NaN3 and was then mixed 1:1 with a solution of 15 mM DDAO and aged PEG 400, at starting PEG concentrations in the drop ranging from 2.5 to 12.5% (wt/vol). These drops were supplemented with HoCl3, to a starting concentration of 10 mM. At 4°C, crystals with the shape of a hexagonal rod appeared within 1 week and grew for several weeks to approximately 200 μm in length and 60 to 80 μm in thickness as shown in Fig. 6. Preliminary diffraction data were collected at beam-line X06SA at the Swiss Light Source in Viligen, Switzerland.
FIG. 4.
Dynamic light-scattering analysis of the purified E1* homotrimer. Each panel shows a single experiment containing the cumulative data from 50 to 100 measurements graphed as intensity versus the Rh (in nanometers on a log scale). The data are not corrected for the changes in solution viscosity due to detergent and salt differences between samples; therefore, the Rh values shown are intended only for relative qualitative comparisons. (A) Analysis of purified E1* homotrimer (∼0.5 mg/ml) in buffer containing 1 M NaCl, 50 mM Tris-HCl, pH 7.5, and the detergent OG at (0.7%) (A1) or below (0.08%) (A2) the CMC. (B) Analysis of purified E1* homotrimer in buffer containing 0.7% OG, 50 mM Tris-HCl, pH 7.5, and 100 mM NaCl (B1). Panel B2 gives the analysis of the sample from panel B1 after addition of NaCl to a final concentration of 1 M and incubation for 5 min.
FIG. 6.
3D crystals and X-ray diffraction of the E1* homotrimer. The left panel shows a crystal grown from 6% PEG 400, 0.25 M NaBr, 10 mM HoCl3, 25 mM Tris-HCl, pH 7.5, and 15 mM DDAO, by use of the hanging-drop vapor diffusion technique as explained in Materials and Methods. The crystals are hexagonal prisms that grow to approximately 200 μm in length and to approximately 80 μm in thickness. The right panel shows the diffraction pattern from a similar crystal, a 0.2° oscillation image taken at beam-line X06SA at the Swiss Light Source in Viligen, Switzerland. The detector was a charge-coupled device made by MAR Research (Hamburg, Germany). The wavelength of the X-ray beam was 1.1808 Å, the crystal-to-detector distance was 200 mm, and the exposure time was 2 s. The resolution at the edge of the detector was 3.18 Å.
RESULTS
Solubilization and purification.
Low-pH treatment of the SFV membrane protein ectodomains in the presence of cholesterol and sphingolipid-containing target membranes leads to E1* membrane insertion and conversion of ∼50 to 60% of E1* to the E1*HT (1, 26). As a first step in E1*HT purification, sucrose gradient flotation was used to separate the membrane-bound homotrimer from the soluble ectodomains (E1*, E2*, and p62*). After centrifugation E1*HT cofloated in the top three or four fractions of the gradient (Fig. 1A) with the [3H]cholesterol-labeled liposomes (Fig. 1B), while the bulk of the E2* and the remaining monomeric E1* remained in the bottom fractions of the gradient. The fractions containing membrane-bound E1*HT could be stored for extended periods on ice without aggregation, denaturation, or proteolytic degradation (see also reference 16).
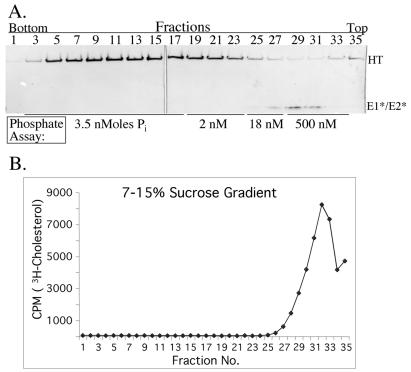
FIG. 1.
Sucrose gradient flotation analysis of ectodomains and liposomes. (A) Purified ectodomains were mixed with liposomes containing [3H]cholesterol as a tracer, treated at pH 5.5 for 10 min at 37°C, returned to neutral pH, and analyzed without detergent treatment. The sample was loaded in the bottom of a sucrose step gradient, centrifuged in a TLS55 rotor at 55,000 rpm for 3 h at 4°C, and fractionated into seven 300-μl fractions. The fractions were precipitated by acid, solubilized in sample buffer at 95°C, reduced and alkylated, and analyzed by SDS-PAGE and Coomassie blue staining. An aliquot of the starting ectodomain prep is shown (ectos). Fraction 1 is the top of the gradient. The positions of E1*, E2*, p62*, and BSA are indicated. (B) Samples were analyzed by flotation as done for panel A, either without detergent pretreatment (open squares) or after treatment for 1 h at room temperature with the indicated detergent at a final concentration of 1.5%. The sucrose gradients also contained the indicated detergent except for the OG sample marked ± (filled diamonds). The amount of [3H]cholesterol in aliquots of each fraction was measured by liquid scintillation counting. CHAPS, 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate.
It has previously been shown that the E1*HT associates with liposomes via an interaction of the fusion peptide with cholesterol-rich regions of the membrane that are resistant to solubilization with TX-100 (1). [3H]cholesterol thus serves as a useful marker to monitor the complete solubilization of the liposomes and the removal of lipid from the E1*HT preparation. Several detergents were found to solubilize the liposomes so that both the [3H]cholesterol marker (Fig. 1B) and the E1*HT (data not shown) no longer floated to the top fractions of sucrose step gradients. Importantly, however, at least for OG the solubilization was partially reversible. In the absence of OG in the gradient, the solubilized E1*HT and lipid could reassociate to form buoyant liposomes (Fig. 1B) (see also reference 1).
Together these data and concurrent EM studies (15) suggested that OG solubilized the E1*HT without significantly affecting its structure or biochemical properties. In addition, OG is available in highly purified form, is relatively invisible in spectroscopic measurements, is easily removed due to its high CMC, and has been extensively used for a variety of biochemical and structural studies of membrane proteins, including nuclear magnetic resonance and X-ray crystallography (13, 14, 18, 19, 27, 32). The homotrimer-liposome complexes from sucrose flotation gradients were therefore solubilized with 1.5% OG at room temperature for 1 h and were sedimented on 7 to 15% sucrose gradients containing 1.5% OG. SDS-PAGE analysis showed that the E1*HT migrated as a broad peak in the middle of the gradient (Fig. 2A). The broadness of this peak might reflect different folding states, further delipidation during gradient centifugation, or other factors. This homotrimer peak was essentially lipid free as assayed by [3H]cholesterol quantitation (Fig. 2B), thin-layer chromatography, and lipid phosphate analysis of pooled gradient fractions (Fig. 2A). The top of the gradient (fractions 27 to 35) contained monomeric E1* and E2* and essentially all of the [3H]cholesterol and phospholipids. Thus, both neutral lipids and phospholipids were efficiently removed from the homotrimer during purification.
FIG. 2.
Sucrose sedimentation gradient purification of the E1* homotrimer. Liposome-bound homotrimer was separated by flotation in the absence of detergent as done for Fig. 1A, and a sample (∼100 μg of protein) was solubilized in 1.5% OG for 1 h at room temperature and was layered on top of a 7 to 15% sucrose gradient containing 1% OG. The gradient was centrifuged for 22 h at 35,000 rpm in an SW41 rotor, fractionated from the bottom, and assayed by SDS-PAGE and silver staining after solubilization in SDS-sample buffer at 30°C to preserve the homotrimer (A). The [3H]cholesterol marker was assayed by liquid scintillation counting (B). Fractions were pooled as indicated beneath the gel depicted in panel A, lipid extracts were prepared, and lipid phosphate was quantitated as outlined in Materials and Methods. Background in this assay was ∼3 nmol of Pi.
Although purification by sucrose sedimentation gradients first demonstrated that the E1*HT could be solubilized by OG and separated from contaminating lipids, this method proved impractical for the generation of large amounts of purified homotrimer. Instead, an alternative method was developed based on centrifugal filtration devices with semipermeable membranes. Microconcentrators with a molecular mass cutoff of 100 kDa retained the detergent-solubilized homotrimer, presumably as a homotrimer-detergent micelle complex. Since pure OG micelles and OG-lipid micelles are much smaller, they were readily removed by repeated washing of the sample with fresh buffer containing OG at concentrations above the CMC (∼0.7%). Table 1 summarizes the efficiency of lipid removal as a function of wash volume. Four sequential washes with buffer containing 1.5% OG removed ∼98.7% of the [3H]cholesterol. Two further washes with 0.7% OG yielded E1*HT that was ∼99% free of lipid, as monitored by [3H]cholesterol (Table 1), 125I-labeled phosphatidylcholine, lipid phosphate analysis, or thin-layer chromatography (data not shown). Together these analyses demonstrated that microconcentrator purification removed the bulk lipids from the HT without concentrating the OG.
TABLE 1.
Lipid removal from the E1*HT versus wash volume
| Purification step | Wash vol (ml) | [3H]cholesterol removed per wash (%)a,b,c | [3H]cholesterol remaining (% total) |
|---|---|---|---|
| Starting material (16 ml) | 100 | ||
| Concentrate (0.5 ml) | 71.8 | 28.2 | |
| 1st washd,e | 7.5 | 14.4 | 13.8 |
| 2nd washd,e | 7.5 | 6.1 | 7.7 |
| 3rd washd,e | 7.5 | 4.0 | 3.7 |
| 4th washd,e | 7.5 | 2.4 | 1.3 |
| Buffer exchangee,f | (2×) 3.0 | 2.0 | |
| Final concentrate (0.5 ml) | 1.3g | ||
| Total filtrate | 51.5 | ≥99 |
Measured by liquid scintillation counting of the filtrates.
Similar results were obtained with radiolabeled phosphatidylcholine.
The data shown are an average obtained from 55 preparations.
OG (1.5%) in HSTN buffer, 15-fold dilution.
Buffer was added to 0.5 ml of concentrated sample, mixed, and reconcentrated to 0.5 ml.
OG in TN buffer, sixfold dilution.
Measured by direct liquid scintillation counting of the final retained material.
Characterization of the purified homotrimer.
We tested the SDS and trypsin resistance of the purified homotrimer to confirm that its structure and properties were preserved during purification. Before purification the ectodomain preparation contained primarily of p62*, E1*, E2*, and bovine serum albumin (BSA) as visualized by SDS-PAGE (Fig. 3A, lane 1). After gradient flotation and microconcentrator purification only the E1* homotrimer was observed, as judged by its migration at ∼130 kDa following SDS solubilization at 30°C (Fig. 3A, lane 2). SDS-PAGE analysis of the microconcentrator filtrate showed that contaminating E1* monomer, BSA, and E2* were removed by filtration, similar to the results of the gradient purification (data not shown). Electrophoresis of increasing amounts of the purified HT under reducing conditions showed a single band with the same electrophoretic mobility as the starting E1* (Fig. 3A, lanes 3 through 5). Digestion of the purified E1*HT for 1 h with trypsin at 37°C showed that, similar to the starting trimer, the purified protein was highly resistant to proteolysis, maintaining its trimeric structure (Fig. 3B, lane 1) and showing no evidence of cleavage products (Fig. 3B, lane 2). The purified HT migrated as a single peak (fractions 13 to 19) when analyzed by sucrose gradient sedimentation as in Fig. 2 and thus by this criterion was less heterogeneous than the gradient-purified HT (data not shown).
FIG. 3.
Analysis of microconcentrator-purified E1* homotrimer. (A) Samples of the starting ectodomains (lane 1, Ectos) or the final microconcentrator-purified homotrimer (Purified HT) were incubated in SDS-containing sample buffer for 3 min at 30°C or were reduced at 95°C and alkylated before analysis by SDS-PAGE and Coomassie blue staining. The last three lanes show increasing amounts of the purified material (10, 20, and 30 μl). The positions of the homotrimer (HT) and the monomeric proteins E1*, E2*, p62*, and BSA are indicated. (B) Samples of the concentrator-purified homotrimer were incubated for 1 h at 37°C with 0.5% TX-100 and 125 μg of trypsin/ml (lanes 1 and 2). For comparison, samples of the stock trypsin (lane 3) and of the purified E1*HT (lane 4) are shown. The samples were solubilized in SDS-sample buffer at 30°C to retain the HT (lane 1) or at 95°C with reduction and alkylation (lanes 2 to 4) and were analyzed by SDS-PAGE and Coomassie blue staining. The two arrows indicate the positions of reduced and nonreduced trypsin.
Conversion of the E1* monomer to the E1*HT involves insertion of the fusion peptide into the target membrane and thus conversion from a soluble protein to a membrane protein (1, 26). In keeping with this conversion, the properties of the E1*HT were highly sensitive to detergent and buffer conditions. Dynamic light-scattering analysis of purified E1*HT in high-salt buffer (1.0 M NaCl) containing OG at or above the CMC showed that the protein was monodisperse even after storage at 4°C for months at a concentration of ∼0.5 mg/ml (Fig. 4A1, 0.7% OG). Preliminary small-angle X-ray-scattering experiments confirmed this observation (data not shown). Upon dilution of the detergent below the CMC, the protein became markedly heterogeneous and progressively aggregated over a time scale of minutes, as indicated by the presence of multiple peaks with much larger Rh values (Fig. 4A2, 0.08% OG). These results are consistent with EM analysis of similar samples that showed that purified E1*HT formed rosettes upon detergent dilution or dialysis (15). The aggregation of the E1*HT was partially reversible by adding back OG to a final concentration above the CMC (data not shown). Although monomeric E1* is stable in buffers containing 100 to 130 mM NaCl (22, 26), the purified E1*HT aggregated in low-salt buffer (100 mM NaCl) in spite of the presence of OG at concentrations at or above the CMC (0.7 to 1.0%) (Fig. 4B1). This behavior was partially reversible by adjusting the salt concentration back to 1 M (Fig. 4B2). Results from the literature and independent measurements of the detergent monomer-to-micelle transition (not shown) both indicated that the effective CMC of OG is decreased at high salt concentrations (48). This enhanced detergent effect may explain the more uniform behavior of the protein in high salt. In addition, the purified E1*HT may have both strongly polar and hydrophobic surfaces, leading to a requirement for both high salt and detergent to maintain optimal solubility (see Discussion). Based on these results the purification protocol was modified to include high salt (1.0 M) in the buffer washes, resulting in easier removal of the lipids (Table 1) and significantly improved protein recovery.
3D crystal screening.
Initial crystallization trials were based on the E1*HT in OG solution using the hanging-drop vapor diffusion method (31). Extensive screening of standard matrices generally produced disappointing results, yielding only amorphous precipitate or extensive phase separation in the drops. Attempts to lower the initial OG concentration in order to avoid phase separation were unsuccessful and usually produced protein precipitation, as discussed above. The purified protein was therefore exchanged into other detergents, including dodecyl-β-d-maltoside, LDAO, and DDAO. Initial crystals were obtained in 3.0 mM LDAO, 25 mM Tris-HCl, pH 7.5, 0.25 M NaCl, 18 to 23% PEG 400, and 2.5 mg of protein/ml (initial concentrations) (Fig. 5A). At 19°C the crystals developed in 24 to 72 h and grew for 10 to 14 days. DDAO produced crystals under similar conditions (15 mM DDAO, 25 mM Tris-HCl, pH 7.5, 0.25 M NaCl, and 9 to 11.5% PEG 400). Both detergents were effective over a range of concentrations at or above the CMC, and the crystals appeared as trigonal or hexagonal rods. These crystals never grew thicker than about 40 to 50 μm and yielded diffraction data to about 5 to 7 Å.
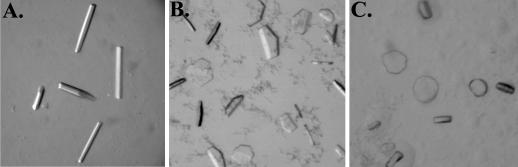
FIG. 5.
3D crystals of the E1* homotrimer grown at 19°C. (A) An example of the rod-shaped crystals grown by hanging-drop vapor diffusion in 11% PEG 400, 3.0 mM LDAO, 0.25 M NaCl, 25 mM Tris-HCl, pH 7.5, and 2.5 mg of protein/ml (initial concentrations in the drop). (B) Hexagonal plate-shaped crystals grown in 7.5% PEG 1000, 1.5 mM LDAO, 0.25 M NaCl, 25 mM Tris-HCl, pH 7.5, and 2.5 mg of protein/ml. Some precipitate is also seen. (C) Rounded plate shaped-crystals grown in 5% PEG 1000, 3.0 mM LDAO, 0.25 M NaCl, 25 mM Tris-HCl, pH 7.5, and 2.5 mg of protein/ml.
Optimization of crystal growth.
A number of different factors were manipulated in an effort to optimize these initial crystals, including the precipitant used (PEG 200 to 20,000), the temperature, the type and concentration of salt, and the presence of additives. PEG 200 had no effect, producing no precipitation or phase separation in the drops. PEG 400 and 550 worked equally well to produce rods with a trigonal or hexagonal appearance within a few days from drops that were uniphasic. Conditions that caused phase separation in the drops prevented crystal development. PEG 600 or larger produced thin, usually hexagonal plates (15 to 30 μm thick) that developed within 10 to 14 days and grew slowly over several weeks (Fig. 5B). These crystals were often associated with or growing within a detergent-rich phase and sometimes had a more rounded morphology (Fig. 5C). Occasionally, especially with the higher-molecular-weight PEGs, crystals would begin to appear as the drop equilibrated but would then disappear as the detergent concentration increased and phase separation occurred in the drop.
The final salt conditions in the crystallization drop had a pronounced effect on the crystal quality. At lower salt concentrations (e.g., 50 mM) the crystals were more rounded, while at increasing concentrations up to 0.5 M the crystals became more regular, with better overall morphology of both the rod and plate forms. At final salt concentrations greater than 0.75 M the solutions had a pronounced tendency to phase separate, yielding either no crystals or crystals of poor quality. Substitution of NaBr for NaCl produced crystals of the same form, but the NaBr crystals often grew more quickly, at lower PEG concentrations (4 to 12% PEG 400), and diffracted to higher resolution, although they were less stable over time (both in the mother drop and in the X-ray beam).
Crystallization of membrane proteins is often quite sensitive to the presence of additives (including impurities from commercial detergents and chemicals) (13, 27, 44). We therefore tested crystallization by using a primary detergent with a variety of additives, including other detergents at very low concentrations, the substances found in the Hampton Additive Screens 1, 2, and 3, and the Hampton heavy atom screen. The combinations of primary and additive detergents that were tried included the following: LDAO+OG, OG+LDAO, LDAO + dodecylmaltoside, LDAO + C12E8, and LDAO + SDS. The other additives that were used included chaotropes, polyamines, micelle-manipulating amphiphiles (e.g., 1,2,3-heptanetriol), and various nonvolatile and volatile organics. The most interesting results were found with the alcohols, especially isopropanol and ethanol. For both the plate- and rod-shaped crystals, crystal growth was aided by the presence of the alcohol (1.5 to 6%), producing fewer but larger crystals at lower PEG concentrations. The presence of the alcohol appeared to affect the nucleation event, possibly by modulating the micelle size, changing the way in which the protein interacted with the detergent, or lowering the overall protein solubility. An important variable was found to be the PEG solutions used for the crystallization, which needed to be aged until the pH reached ∼3.5 to 4.0, as described in Materials and Methods. This may reflect the reported generation of aldehydes in PEG solutions (39), which could be acting as an important additive during crystallization. To screen for heavy atom derivatives, we soaked preformed crystals in various solutions of heavy atoms. These tests revealed that holmium bound the protein in a fashion that stabilized the oligomer, producing better diffraction data. Similarly, crystal growth in the presence of 10 mM HoCl3 yielded better-diffracting crystals with a more regular morphology.
All of the crystal forms were extremely sensitive to temperature during both the nucleation and growth phases. At temperatures higher than ∼20°C crystals did not form at all or required higher precipitant concentrations (5 to 10% more PEG 400), while preformed crystals began to dissolve at temperatures higher than 20 to 22°C. The presence of alcohols as additives made the crystals more resistant to the effects of higher temperatures. Incubation at 4°C produced overall results similar to those found at 19°C with the same conditions, although the crystals appeared and grew more slowly and at lower PEG 400 concentrations. The 4°C crystals were generally larger and produced better diffraction data.
Optimal crystals for X-ray diffraction were grown at 4°C from hanging drops under starting conditions of 15 mM DDAO, 0.25 M NaBr, 10 mM HoCl3, 25 mM Tris-HCl pH 7.5, 2.5 to 12.5% PEG 400, and a final measured pH of 4.0, yielding hexagonal rods from 40 to 80 μm thick and of variable length, usually 200 to 300 μm. A representative example of these crystals is shown in the left panel in Fig. 6. The right panel in Fig. 6 shows a sample diffraction pattern of a crystal analyzed with synchrotron radiation, displaying diffraction extending equally well in all directions. The crystals were found to belong to the space group P3121, with cell parameters of a = b = 196.702 and c = 115.506, α = β = 90°, and γ = 120°. The crystals are very sensitive to radiation damage, which has so far limited the overall resolution of the diffraction data (3.18-Å resolution for the diffraction pattern shown in Fig. 6, right panel). We are currently in the process of determining the structure of the homotrimer by a combination of multiwavelength anomalous dispersion and isomorphous replacement.
DISCUSSION
The native fusion proteins of SFV, tick-borne encephalitis virus, and dengue virus have strikingly similar structures (30, 34, 40) and are general paradigms for the fusion proteins of the Togaviridae and Flaviviridae families. The class II fusion proteins represented by these viruses have important mechanistic and structural differences from the class I proteins. As the class II viruses include a number of significant global pathogens, understanding their fusion mechanism at the molecular level is important to the development of antiviral therapies. A key question is the structural basis of the low-pH-induced trimeric form of the fusion protein. Starting with the membrane-inserted SFV E1* homotrimer, we here describe a method to prepare milligram amounts of E1*HT as a detergent-protein complex. The first step of gradient flotation enriched for functional, membrane-bound HT and removed monomeric E1and E2 ectodomains. Subsequent detergent solubilization and filtration steps removed the lipid, resulting in a final E1*HT preparation that retained the characteristics of the starting homotrimer. This material was crystallized for structural analysis by X-ray diffraction.
Previous experiments demonstrated that low pH specifically triggers both virus fusion and E1* association with target membranes containing cholesterol and sphingolipid (9, 26). The fusion peptide of the membrane-inserted E1* is strongly associated with cholesterol-enriched membrane domains (“rafts”), and detergent conditions that preserve rafts also fail to solubilize E1* (1). Interestingly, not all detergents that maintained the purified E1*HT in solution allowed effective initial delipidation of the protein. For example, if the same microconcentrator protocol was used, LDAO did not efficiently separate the protein from the [3H]cholesterol marker (data not shown). This may suggest differences in the solubilization of cholesterol and/or rafts or in the detergent's ability to replace a protein-cholesterol interaction. While the OG-purified E1*HT was free of detectable lipid, given the limitations of the assays, it is possible that stoichiometric amounts of lipid could remain associated with the protein, perhaps specifically.
In parallel with the experiments reported here, we also characterized the membrane-inserted E1 homotrimer by EM (15). Visualization of E1*HT on membranes by negative staining and cryo-EM revealed that the protein reorients perpendicular to the target membrane and inserts the tip of domain II into the membrane. The bulk of the trimer remains exposed to solution. These extramembranous trimer domains can associate in hexagonal arrays on the membrane surface, enabling a three-dimensional reconstruction of this portion of the protein to 25-Å resolution. Solubilization in OG disrupts these intertrimer interactions, but dilution or removal of the detergent in the presence of lipids allows the E1*HT to reassociate with membranes and reform hexagonal lattices. Such cooperative interactions of the HT may be important in the membrane fusion mechanism (15). These results were important to the experiments reported here, since the EM demonstrated that the E1*HT has both domains that mediate trimer-membrane interaction and domains that confer trimer-trimer interaction.
The requirements for purification of the E1*HT agree with the multifaceted nature of the E1 protein observed in the previous biochemical and EM studies. Release of the E1*HT from the target membrane required detergent. Both the EM and dynamic light-scattering results indicated that the purified homotrimer acts as a bona fide membrane protein, requiring micellar concentrations of detergent to remain soluble. The detergent-solubilized, purified molecule remains a highly stable trimer, in keeping with stability being conferred by protein-protein interactions within the trimer, rather than by the interaction of the protein with the membrane (16). The need for high ionic strength to prevent the formation of E1*HT aggregates or higher-order oligomers suggests that E1*HT has a tendency to interact via the water-soluble regions. In agreement with this biochemical property, the EM studies showed that formation of the hexagonal lattice was most efficient at lower salt concentrations (150 mM NaCl).
The factors governing the crystallization of membrane-associated proteins are not well understood or predictable. The type of membrane interaction displayed by the E1* homotrimer, with the bulk of the protein exposed to solution, is the most favorable for producing high-quality 3D crystals because the mechanism of crystallization is generally governed by the same principles as for soluble proteins (27, 44). Clearly the choice of detergent was a critical variable in our studies. OG was very effective for E1*HT purification and produced the protein used here for successful 3D crystallization. The OG-solubilized protein also generated 2D crystals upon detergent dialysis and membrane reconstitution (15). However, OG was not useful as a detergent for maintaining the purified E1*HT during the actual generation of 3D crystals. In contrast, both DDAO and LDAO, with a simpler alkyl (C10 or C12) amine structure, allowed crystallization under several different sets of conditions, even though LDAO was less efficient at the initial solubilization and delipidation of membrane-associated E1*HT, as discussed above. Similar to results with other detergent-soluble proteins, we found that the E1*HT could crystallize either from the aqueous phase or from a hydrophobic, detergent-rich phase. The latter crystals were platelike and tended to grow in the plane of the plate rather than in thickness. This is analogous to the 2D hexagonal lattice discussed above (15) and suggests that the hexagonal platelike crystals might be associated with detergent micelle surfaces, similar to the association of the 2D lattice with the membrane surface.
The SFV E1*HT crystallized with the intact fusion peptide, while all of the postfusion structures of the class I proteins to date have been determined by using ectodomain fragments missing the fusion peptide. The postfusion structures of the class I proteins also have the bulk of the protein exposed to solution, with the membrane interaction mediated by the fusion peptide. However, in general the class I fusion peptides are located at or near the N terminus, and their structures may not be comparable to those of the class II fusion peptides. In addition, the membrane-inserted fusion peptide of the influenza virus HA, unlike that of SFV, was not found to be associated with membrane rafts (1). Nonetheless, it is very possible that these apparent differences are not limiting and that the postfusion forms of some class I proteins can be crystallized with their fusion peptides. This would allow direct comparison of the structural basis by which the two classes anchor in target membranes. Perhaps most importantly, though, the solution of the E1*HT structure will answer many questions about the overall refolding reaction used by the class II proteins to mediate membrane fusion.
Acknowledgments
We thank Jean Lepault for many helpful discussions and insights and Stephane Duquerroy, Stephane Bressanelli, and Fasselli Coulibaly for help with the crystallization trials, crystal manipulation, and data collection. We also thank Alyson Urian for expert technical assistance with virus and protein purification and the members of the Kielian lab for their helpful comments on the manuscript.
This work was supported by a grant to M.K. from the Public Health Service (R01 GM52929) and by Cancer Center Core Support Grant NIH/NCI P30-CA13330. The work was also supported by grants to F.A.R. from the CNRS and INRA, the “SESAME Program of the Region Ile-de-France,” “the Foundation pour la Recherche Medicale,” “the Association pour la Recherche contre le cancer,” and the “CNRS programs Physique et Chimie du Vivant and Dynamique et reactivite des assemblages biologiques.” D.L.G. was supported through the Medical Scientist Training Program of the Albert Einstein College of Medicine (NIH T32 GM07288) and by the Albert Cass Traveling Fellowship Award and the CNRS.
REFERENCES
- 1.Ahn, A., D. L. Gibbons, and M. Kielian. 2002. The fusion peptide of Semliki Forest virus associates with sterol-rich membrane domains. J. Virol. 76:3267-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 3.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 4.Bron, R., J. M. Wahlberg, H. Garoff, and J. Wilschut. 1993. Membrane fusion of Semliki Forest virus in a model system: correlation between fusion kinetics and structural changes in the envelope glycoprotein. EMBO J. 12:693-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee, P. K., M. Vashishtha, and M. Kielian. 2000. Biochemical consequences of a mutation that controls the cholesterol dependence of Semliki Forest virus fusion. J. Virol. 74:1623-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, L., J. J. Gorman, J. McKimm-Breschkin, L. J. Lawrence, P. A. Tulloch, B. J. Smith, P. M. Colman, and M. C. Lawrence. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure 9:255-266. [DOI] [PubMed] [Google Scholar]
- 8.Colman, P. M., and M. C. Lawrence. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 4:309-319. [DOI] [PubMed] [Google Scholar]
- 9.Corver, J., L. Moesby, R. K. Erukulla, K. C. Reddy, R. Bittman, and J. Wilschut. 1995. Sphingolipid-dependent fusion of Semliki Forest virus with cholesterol-containing liposomes requires both the 3-hydroxyl group and the double bond of the sphingolipid backbone. J. Virol. 69:3220-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 11.Ferlenghi, I., M. Clarke, T. Ruttan, S. L. Allison, J. Schalich, F. X. Heinz, S. C. Harrison, F. A. Rey, and S. D. Fuller. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7:593-602. [DOI] [PubMed] [Google Scholar]
- 12.Forsell, K., L. Xing, T. Kozlovska, R. H. Cheng, and H. Garoff. 2000. Membrane proteins organize a symmetrical virus. EMBO J. 19:5081-5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garavito, R. M., D. Picot, and P. J. Loll. 1996. Strategies for crystallizing membrane proteins. J. Bioenerg. Biomembr. 28:13-27. [PubMed] [Google Scholar]
- 14.Garavito, R. M., and J. P. Rosenbusch. 1986. Isolation and crystallization of bacterial porin. Methods Enzymol. 125:309-328. [DOI] [PubMed] [Google Scholar]
- 15.Gibbons, D. L., I. Erk, B. Reilly, J. Navaza, M. Kielian, F. A. Rey, and J. Lepault. 2003. Visualization of the target-membrane-inserted fusion protein of Semliki Forest virus by combined electron microscopy and crystallography. Cell 114:573-583. [DOI] [PubMed] [Google Scholar]
- 16.Gibbons, D. L., and M. Kielian. 2002. Molecular dissection of the Semliki Forest virus homotrimer reveals two functionally distinct regions of the fusion protein. J. Virol. 76:1194-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinz, F. X., and S. L. Allison. 2000. Structures and mechanisms in flavivirus fusion. Adv. Virus Res. 55:231-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helenius, A., and J. Kartenbeck. 1980. The effects of octylglucoside on the Semliki Forest virus membrane. Eur. J. Biochem. 106:613-618. [DOI] [PubMed] [Google Scholar]
- 19.Henry, G. D., and B. D. Sykes. 1994. Methods to study membrane protein structure in solution. Methods Enzymol. 239:515-535. [DOI] [PubMed] [Google Scholar]
- 20.Kääriäinen, L., K. Simons, and C.-H. von Bonsdorff. 1969. Studies of Semliki Forest virus subviral components. Ann. Med. Exp. Biol. Fenn. 47:235-248. [PubMed] [Google Scholar]
- 21.Kielian, M., P. K. Chatterjee, D. L. Gibbons, and Y. E. Lu. 2000. Specific roles for lipids in virus fusion and exit: examples from the alphaviruses. Subcell. Biochem. 34:409-455. [DOI] [PubMed] [Google Scholar]
- 22.Kielian, M., and A. Helenius. 1985. pH-induced alterations in the fusogenic spike protein of Semliki Forest virus. J. Cell Biol. 101:2284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kielian, M., M. R. Klimjack, S. Ghosh, and W. A. Duffus. 1996. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J. Cell Biol. 134:863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kielian, M. C., and A. Helenius. 1984. The role of cholesterol in the fusion of Semliki Forest virus with membranes. J. Virol. 52:281-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kielian, M. C., S. Keränen, L. Kääriäinen, and A. Helenius. 1984. Membrane fusion mutants of Semliki Forest virus. J. Cell Biol. 98:139-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klimjack, M. R., S. Jeffrey, and M. Kielian. 1994. Membrane and protein interactions of a soluble form of the Semliki Forest virus fusion protein. J. Virol. 68:6940-6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhlbrandt, W. 1988. Three-dimensional crystallization of membrane proteins. Q. Rev. Biophys. 21:429-477. [DOI] [PubMed] [Google Scholar]
- 28.Kuhn, R. J., W. Zhang, M. G. Rossman, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kundu, S. K. 1981. Thin-layer chromatography of neutral glycosphingolipids and gangliosides. Methods Enzymol. 72:185-204. [DOI] [PubMed] [Google Scholar]
- 30.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 31.McPherson, A. 1982. Preparation and analysis of protein crystals. John Wiley & Sons, New York, N.Y.
- 32.McPherson, A., S. Koszelak, H. Axelrod, J. Day, R. Williams, L. Robinson, M. McGrath, and D. Cascio. 1986. An experiment regarding crystallization of soluble proteins in the presence of B-octyl glucoside. J. Biol. Chem. 261:1969-1975. [PubMed] [Google Scholar]
- 33.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 100:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore, J. P., and R. W. Doms. 2003. The entry of entry inhibitors: a fusion of science and medicine. Proc. Natl. Acad. Sci. USA 100:10598-10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morrison, W. R. 1964. A fast, simple, and reliable method for the microdetermination of phosphorus in biological materials. Anal. Biochem. 7:218-224. [DOI] [PubMed] [Google Scholar]
- 37.Phalen, T., and M. Kielian. 1991. Cholesterol is required for infection by Semliki Forest virus. J. Cell Biol. 112:615-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pletnev, S. V., W. Zhang, S. Mukhopadhyay, B. R. Fisher, R. Hernandez, D. T. Brown, T. S. Baker, M. G. Rossmann, and R. J. Kuhn. 2001. Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell 105:127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ray, W. J., Jr., and J. M. Puvathingal. 1985. A simple procedure for removing contaminating aldehydes and peroxides from aqueous solutions of polyethylene glycols and of nonionic detergents that are based on the polyoxyethylene linkage. Anal. Biochem. 146:307-312. [DOI] [PubMed] [Google Scholar]
- 40.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 41.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlesinger, S., and M. J. Schlesinger. 2001. Togaviridae: the viruses and their replication, p. 895-916. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott, Williams and Wilkins, Philadelphia, Pa.
- 43.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 44.Song, L., and J. Gouaux. 1997. Membrane protein crystallization: application of sparse matrix to alpha-hemolysin heptamer. Methods Enzymol. 277:60-74. [DOI] [PubMed] [Google Scholar]
- 45.Stiasny, K., S. L. Allison, J. Schalich, and F. X. Heinz. 2002. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J. Virol. 76:3784-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wahlberg, J. M., R. Bron, J. Wilschut, and H. Garoff. 1992. Membrane fusion of Semliki Forest virus involves homotrimers of the fusion protein. J. Virol. 66:7309-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walter, A., G. Kuehl, K. Barnes, and G. VanderWaerdt. 2000. The vesicle-to-micelle transition of phosphatidylcholine vesicles induced by nonionic detergents: effects of sodium chloride, sucrose and urea. Biochim. Biophys. Acta 1508:20-33. [DOI] [PubMed] [Google Scholar]
- 49.Weissenhorn, W., A. Dessen, L. J. Calder, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1999. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16:3-9. [DOI] [PubMed] [Google Scholar]
- 50.Wengler, G., G. Wengler, and F. A. Rey. 1999. The isolation of the ectodomain of the alphavirus E1 protein as a soluble hemagglutinin and its crystallization. Virology 257:472-482. [DOI] [PubMed] [Google Scholar]
- 51.White, J., and A. Helenius. 1980. pH-dependent fusion between the Semliki Forest virus membrane and liposomes. Proc. Natl. Acad. Sci. USA 77:3273-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the hemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289:366-378. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, W., S. Mukhopadhyay, S. V. Pletnev, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. 2002. Placement of the structural proteins in Sindbis virus. J. Virol. 76:11645-11658. [DOI] [PMC free article] [PubMed] [Google Scholar]