Abstract
The human mixed-lineage leukemia 5 (MLL5) protein mediates hematopoietic cell homeostasis, cell cycle, and survival; however, the molecular basis underlying MLL5 activities remains unknown. Here, we show that MLL5 is recruited to gene-rich euchromatic regions via the interaction of its plant homeodomain finger with the histone mark H3K4me3. The 1.48-Å resolution crystal structure of MLL5 plant homeodomain in complex with the H3K4me3 peptide reveals a noncanonical binding mechanism, whereby K4me3 is recognized through a single aromatic residue and an aspartate. The binding induces a unique His–Asp swapping rearrangement mediated by a C-terminal α-helix. Phosphorylation of H3T3 and H3T6 abrogates the association with H3K4me3 in vitro and in vivo, releasing MLL5 from chromatin in mitosis. This regulatory switch is conserved in the Drosophila ortholog of MLL5, UpSET, and suggests the developmental control for targeting of H3K4me3. Together, our findings provide first insights into the molecular basis for the recruitment, exclusion, and regulation of MLL5 at chromatin.
The mixed-lineage leukemia 5 (MLL5) protein was initially identified as a candidate tumor suppressor and more recently has been shown to play a critical role in hematopoiesis and lymphopoiesis (1). The MLL5 gene-containing region of chromosome 7 is frequently deleted in patients with hematological disorders, including acute myeloid and therapy-induced leukemias and myeloid dysplastic syndrome (MDS). Depletion of Mll5 in mice causes mild postnatal lethality, with some of the surviving animals showing retarded growth, male sterility, and decreased size of thymus, spleen, and lymph nodes (2–4). Genetic analyses of these Mll5 deficiencies reveals a 30% decrease in the number of hematopoietic stem cells (HSC) and progenitors, defects in HSC self-renewal mechanisms, and impaired myeloid differentiation (2–4). In addition to being an essential mediator of HSC homeostasis, MLL5 has been implicated in cytokinesis, the DNA damage response, and genome maintenance (5–7). Overexpression and knockdown of MLL5 both induce cell cycle arrest at various phases, suggesting a versatile function of MLL5 throughout the cell cycle (5).
MLL5 belongs to the MLL family of methyltransferases that regulate gene expression during developmental processes. These enzymes catalyze the addition of methyl groups to the ε-amino moiety of lysine and are highly specific for lysine 4 of histone H3. Along with MLL5, also known as a lysine methyltransferase 2E (KMT2E), the MLL family contains MLL1-4, SET1A, and SET1B (KMT2A-KMT2D, KMT2F, and KMT2G, respectively) (8). Full-length MLL5 is ∼200 kDa and is evolutionarily distant from the more canonical and better-characterized members of this family. Unlike the other multimodular MLL proteins, MLL5 consists of only two conserved motifs near the N terminus, a plant homeodomain (PHD) finger, followed by a catalytic Su(var)3–9, enhancer of zeste, trithorax (SET) domain. A long, ∼1,000-residue C-terminal region of MLL5 displays no apparent homology to any known structural domain and is truncated in a shorter ∼75-kDa isoform, MLL5 short (MLL5s). Furthermore, MLL5 has no intrinsic histone lysine methyltransferase (HKMT) activity (7); however, once posttranslationally modified through glycosylation at Thr440 in the SET domain, the short isoform MLL5s is capable of generating mono- and dimethylated H3K4 marks (9). The HKMT activity of MLL5s stimulates expression of retinoic acid receptor α target genes and facilitates retinoic acid-dependent granulopoiesis in human promyelocytes (9). The glycosylation-induced catalytic function of MLL5s appears to be unique to mammals. The yeast and drosophila orthologs of MLL5 (Set3/4 and UpSET, respectively) lack the HKMT activity and instead are found in histone deacetylase complexes (10). One of the most discernible features of MLL5 is that, in contrast to other MLLs that harbor multiple PHD finger cassettes, it contains a single PHD module, the biological role of which remains unclear.
In this study, we show that MLL5 associates with the chromatin regions downstream of transcriptional start sites (TSSs) of active genes. The PHD finger of MLL5 binds strongly and specifically to histone H3K4me3 through a noncanonical mechanism. Phosphorylation of H3T3 and H3T6 disrupts binding of MLL5 to H3K4me3, and this regulatory mechanism is conserved in the Drosophila ortholog of MLL5, UpSET. Our results suggest that the phospho-methyl switch plays an essential role in the regulation of MLL5/UpSET during mitosis and development.
Results
MLL5 Is Recruited to Actively Transcribed Genes.
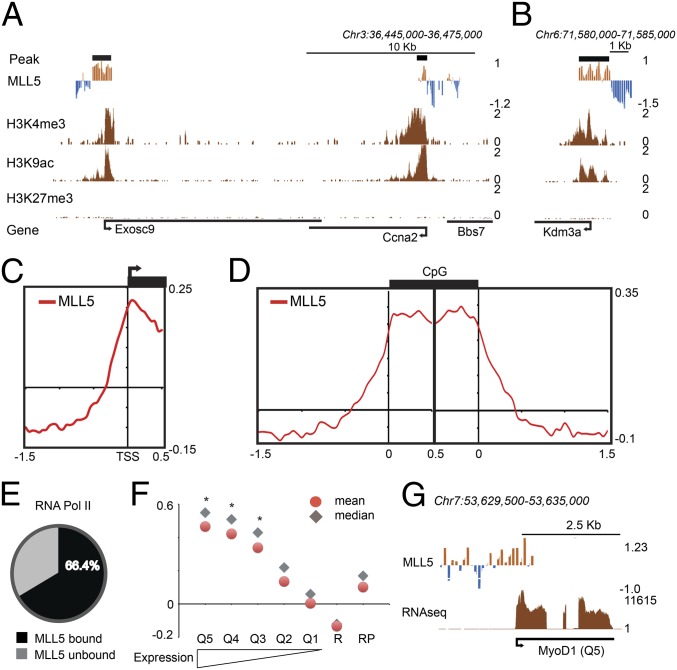
To determine the genome-wide distribution of MLL5 in mammals, we performed a DNA adenine methyltransferase identification (DamID)-based chromatin analysis of MLL5 in C2C12 cells using promoter arrays covering ∼32,000 regions plus CpG islands (11). The DamID profiling revealed that MLL5 is recruited to ∼15,000 genomic regions, including ∼13,000 genes in myoblasts. Consistent with Sebastian et al. (7), we found that MLL5 binds to the genomic regions downstream of TSSs of Ccna2 and Exosc9 (Fig. 1A). To better characterize the global recruitment of MLL5, we carried out a meta-analysis around TSSs using the MLL5 DamID signal. This analysis revealed that MLL5 preferentially targets the regions downstream of TSSs, and this result was well reproduced in experimental replicates (Fig. 1 A–C; Fig. S1 A and B). In mammals, ∼60% of promoter regions overlap with CpG islands (12); therefore, we tested whether MLL5 occupies CpG islands or flanking sequences. In agreement with the initial meta-analysis, MLL5 DamID signal was enriched at CpG islands but not at the flanking regions (Fig. 1D).
Fig. 1.
MLL5 preferentially binds actively transcribed genes. (A and B) MLL5 chromatin profile over a 30-kb region of chromosome 3 (A) or a 5-kb region of chromosome 6 (B). MLL5 DamID signal (a log2 ratio), peaks (a thick black line), and normalized ChIP-seq data for H3K4me3, H3K9ac, and H3K27me3 are shown. The mean signal was smoothed + whiskers. (C) Meta-analysis of MLL5 binding around transcriptional start sites (−1.5 kb upstream; –0.5 bp downstream). (D) Meta-analysis of MLL5 binding around CpG islands (1.5-kb upstream and downstream regions flanking CpG islands). (E) Percentage of MLL5-bound promoter regions overlapping with RNA polymerase II (Table S1). (F) Graph showing the mean and median of MLL5 DamID signal around gene promoters clustered by expression quintiles. High (Q5)- to low (Q1)-expressing gene groups are shown. R, ∼4,000 random sequences; RP, promoter regions randomly selected. Asterisk (*) indicates statistically significant groups in comparison with RP. (G) MLL5 binding over a genomic region containing the MyoD1 gene (quintile 5). Coding regions are shown in black (Fig. S1).
To establish whether MLL5 recruitment correlates with gene expression, we examined the overlap between MLL5-bound and RNA polymerase-bound promoter regions in C2C12 cells (13). We found that MLL5 co-occupies ∼66% (7,195 of 10,834) of RNA polymerase-bound promoters (Fig. 1E; Table S1). To confirm this finding, we measured the mean and median of the MLL5 DamID signal at the gene promoters arranged by expression-based quintiles (Materials and Methods). As shown in Fig. 1 F and G, MLL5 is bound preferentially at the promoters of the three most highly expressed gene groups compared with the low- or nonexpressed genes, 4,000 randomly selected genes, or a cohort of 4,000 random sequences (P = 2.2 × 10−16; Fig. S1 C–F). Together, our data suggest that MLL5 associates with the transcribed regions in mammalian cells.
The PHD Finger of MLL5 Recognizes H3K4me3.
To define the basis for MLL5 recruitment to active chromatin, we examined the epigenetic features of MLL5-bound promoter regions. We analyzed ChIP-sequencing (ChIP-seq) datasets for histone posttranslational modifications (PTMs) in C2C12 cells and mapped them to the ∼32,000 promoter regions of the mouse genome. Consistent with the above results, we found that MLL5 occupies promoters that contain marks of actively transcribed regions, including H3K4me3, H3K4me2, and H3K9ac (Fig. S2A; Table S1). To corroborate these data, we examined colocalization of the long isoform MLL5 and the short isoform MLL5s with H3K4me3 and the repression-associated H3K9me3 mark in C2C12 cells by fluorescence microscopy (Fig. S2B). Both MLL5 isoforms overlapped well with H3K4me3 but were excluded from the regions enriched in H3K9me3, confirming that MLL5 binds chromatin with the expression-linked PTMs (i.e., H3K4me3) in vivo. Furthermore, these results revealed that the N-terminal region of MLL5, present in both isoforms and encompassing the PHD finger and SET domain, is responsible for this association.
The sharp MLL5 enrichment around TSSs and its overlap with H3K4me3 strongly suggested that MLL5 can be recruited by this histone modification. Because the PHD domain of MLL5 is the best candidate to mediate this recruitment, we analyzed its interaction with H3K4me3 by NMR. We collected 1H,15N heteronuclear single-quantum coherence (HSQC) spectra of the 15N-labeled MLL5 PHD finger as unlabeled H3K4me3 peptide (amino acids 1–12 of H3) was titrated in (Fig. S2C). Addition of the peptide induced large changes in the protein amide resonances. The binding was in the intermediate-to-fast exchange regime on the NMR time scale, which is characterized by progressive shifting and broadening of cross-peaks and is indicative of the strong interaction between MLL5 and H3K4me3.
The Crystal Structure of MLL5 PHD in Complex with H3K4me3.
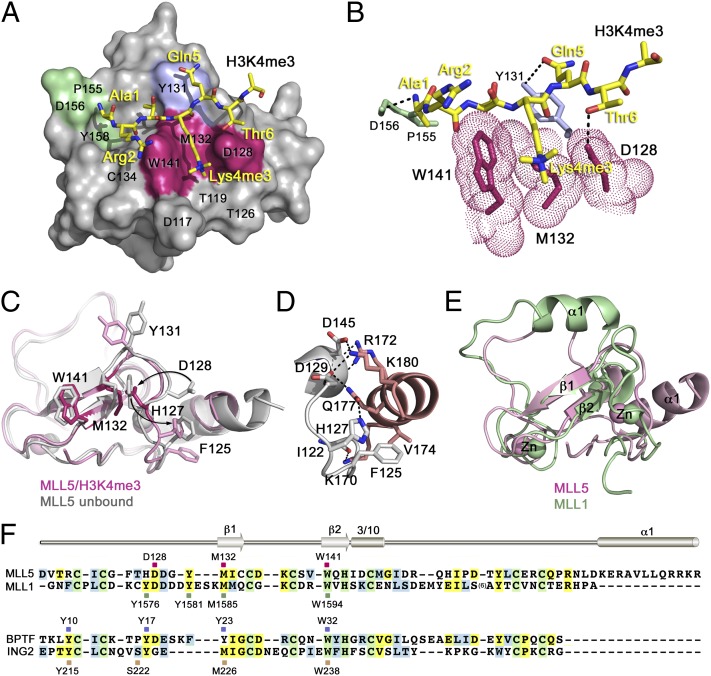
To determine the molecular mechanism of the histone recognition, we obtained a 1.48-Å resolution crystal structure of the MLL5 PHD finger bound to the H3K4me3 peptide (Fig. 2 A and B; Table S2). The structure shows a characteristic really interesting new gene (RING)-finger fold, consisting of long loops and a short antiparallel β-sheet stabilized by two zinc-binding clusters. A 310-helical turn follows the second β-strand, and an additional α-helix is present at the C terminus. The H3K4me3 peptide adopts an extended conformation and pairs with the existing β-sheet, forming the third antiparallel β-strand. The intermolecular backbone–backbone hydrogen bonds are formed between the carbonyl groups (COs) of Arg2 and Lys4me3 of the peptide and the amino groups (NHs) of C134 and M132 of the protein, as well as between NHs of Lys4me3 and Thr6 and COs of M132 and G130.
Fig. 2.
The crystal structure of the PHD finger of MLL5 in complex with the H3K4me3 peptide. (A) The protein residues involved in the interaction with Lys4me3, Ala1, and Gln5 of the peptide are colored magenta, green, and blue, respectively. (B) A zoom-in view of the H3K4me3 binding pocket. Dashed lines represent intermolecular hydrogen bonds. (C) An overlay of the structure of the H3K4me3-bound MLL5 PHD finger (pink) with the structure of MLL5 PHD in the apo-state (gray; PDB ID code 2LV9). The H3K4me3 peptide is omitted for clarity. (D) A zoom-in view of the MLL5 PHD structure showing close contacts between the N-terminal loop and the C-terminal α-helix. (E) An overlay of the structure of the H3K4me3-bound MLL5 PHD finger (pink) with the structure of the H3K4me3-bound MLL1 PHD3 (green; PDB ID code 3LQJ). (F) Alignment of PHD finger sequences: absolutely, moderately, and weakly conserved residues are colored green, yellow, and blue, respectively. The Lys4me3 binding residues of MLL5, MLL1, BPTF, and ING2 are indicated by magenta, green, blue, and wheat, respectively, squares, and labeled. Secondary structure elements of MLL5 PHD are shown.
The side chain of trimethylated Lys4 occupies an elongated groove formed by the W141, M132, and D128 residues of the PHD finger (Fig. 2 A and B, magenta). The side chains of W141 and D128 create the groove’s walls, whereas M132 resides at the bottom. The trimethylammonium group of Lys4 is centrally positioned, with the distance to W141 and D128 being 4.4 Å and 4.0 Å, respectively. The N-terminal amino moiety of Ala1 donates hydrogen bonds to the backbone COs of P155 and D156 (Fig. 2 A and B, green). The methyl group of Ala1 fits in a small hydrophobic cavity formed by Y128, I133, and I154. The other two intermolecular hydrogen bonds restrain Gln5 and Thr6 of the peptide. The hydroxyl group of Y131 is hydrogen-bonded to the side chain carbonyl group of Gln5, and the hydroxyl group of Thr6 makes a hydrogen bond with the carboxyl group of D128.
H3K4me3 Binding Induces a His–Asp Swapping Rearrangement in MLL5.
The structure of MLL5 PHD highlights three significant features that have not been observed previously for the association of PHD fingers with H3K4me3. An overlay of the structures of the MLL5 PHD finger in complex with the H3K4me3 peptide and in the apo-state [Protein Data Bank (PDB) ID code 2LV9; NSGC] reveals that the binding is accompanied by a large conformational change in the protein N-terminal loop (residues G124–Y131; Fig. 2C). In the unbound state (gray), D128 points toward the C-terminal α-helix and is engaged in hydrogen bonding and ionic contacts with K180. Unexpectedly, the imidazole ring of the preceding residue, H127, occupies exactly the same position as the carboxyl group of D128 would occupy in the complex, thereby creating a wall of the groove even in the absence of the ligand. However, upon binding to H3K4me3, H127 and D128 swap their positions (pink). The side chains of H127 and D128 swing in opposite directions, each by almost 180 degrees. H127 moves away from the groove and interacts with the α-helix through a newly formed hydrogen bond with Q177 (Fig. 2 C and D). Concomitantly, D128 dissociates from the α-helix, breaking contacts with K180, and flips inward to complete the Lys4me3 binding pocket. The H127–D128 swapping rearrangement causes conformational changes in other residues of this loop, including F125 and Y131 (Fig. 2C).
The involvement of the C-terminal α-helix of MLL5 underlines the second unique feature of this binding mechanism. Typically, PHD fingers have a short α-helix or an α-helical turn at the C terminus that contains a zinc-coordinating Cys/His residue but protrudes away from the globular core. In contrast, the unusually long α-helix of MLL5 is in direct contact with and stabilizes the core of the PHD finger; it packs against the N-terminal loop and the 310 helix, forming an intricate network of hydrogen bonding, electrostatic, and hydrophobic interactions at the interface (Fig. 2D). A long chain of hydrogen bonds and salt bridges is seen between K170, R172, Q177, and K180 of the α-helix, and I122 (backbone contact), H127, D129, and D145 (both side chain and backbone contacts) of the protein core. Furthermore, the α-helix in MLL5 is not involved in coordination of a zinc ion, being located further downstream in sequence compared with a typical C-terminal helix of PHD fingers or an α-helix in the PHD3 finger of MLL1 (Fig. 2E).
The Noncanonical Lys4me3 Binding Mechanism of MLL5.
The most striking feature, however, is the mechanism by which the MLL5 PHD recognizes trimethylated Lys4. It has been well established that PHD fingers accommodate Lys4me3 in a binding pocket, composed of four, three, or two aromatic residues (14). For example, the PHD finger of bromodomain PHD finger transcription factor (BPTF) has three tyrosine residues and a tryptophan in the pocket (blue); the MLL1 PHD3 has two tyrosine residues, a tryptophan and a methionine (green); and the inhibitor of growth protein 2 (ING2) PHD has a tyrosine, a tryptophan, a methionine and a serine (wheat; Fig. 2F; Fig. S3). The aromatic rings, being arranged roughly perpendicular to each other or to the protein surface, create an aromatic cage (or a semiaromatic cage) around Lys4me3 and are engaged in cation-π, hydrophobic, and van der Waals contacts with the trimethylammonium group (Fig. S3). Surprisingly, the Lys4me3 binding groove in the MLL5 PHD contains only one aromatic residue, W141. Furthermore, the MLL5 PHD uses an acidic residue, D128, instead of an aromatic or a hydrophobic residue, to complete the opposite wall of the groove. The presence of an acidic residue in the aromatic pocket of Tudor domains or mutated PHD of BPTF has been shown to dictate their preference for di- or monomethylated lysine substrates over trimethylated lysine (reviewed in ref. 14). The carboxyl group of an aspartate can form additional favorable hydrogen bonding contacts with the di- or mono- but not trimethylammonium group of Lys4. Despite this, the MLL5 PHD finger selects for H3K4me3 (see below).
MLL5 PHD Is Specific for H3K4me3.
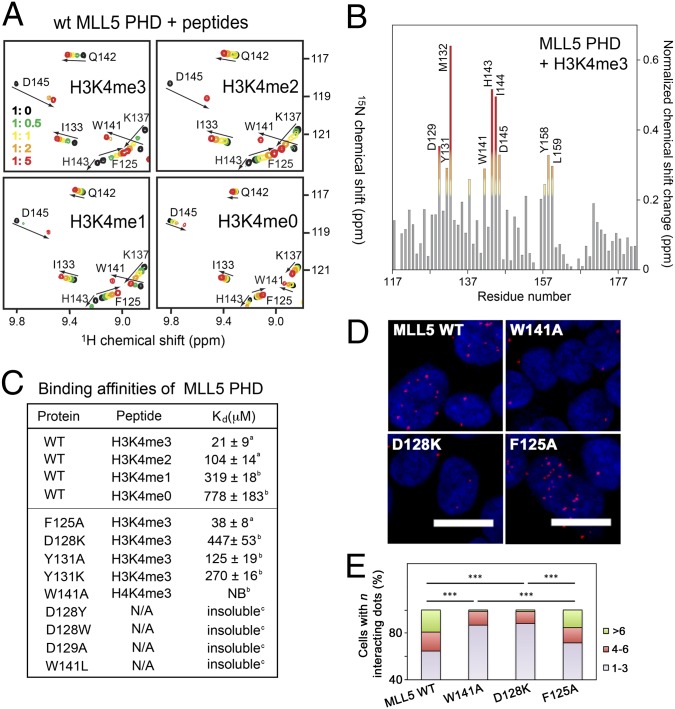
To establish the preference of the MLL5 PHD finger for the mono-, di-, or trimethylated state of H3K4, we investigated interactions with histone H3 peptides using NMR and fluorescence spectroscopy (Fig. 3). The similar pattern of chemical shift changes induced by H3K4me3, H3K4me2, and H3K4me1 in the 1H,15N HSQC spectra of PHD (Fig. 3 A and B) suggested that these peptides occupy the same binding pocket; however, H3K4me2 and H3K4me1 were bound five- and 16-fold more weakly than H3K4me3 (Kd of 21 μM; Fig. 3C). Unmodified H3K4me0 peptide caused very small changes, indicating a weak association with the PHD finger (Kd of 778 μM). Thus, the MLL5 PHD binds specifically and strongly to H3K4me3 and, despite the presence of an aspartate residue in the binding pocket, prefers the trimethylated state to any lower-methylated state of Lys4.
Fig. 3.
A unique H3K4me3 binding mechanism of MLL5 PHD. (A) Superimposed 1H,15N HSQC spectra of MLL5 PHD collected upon titration of indicated peptides. Spectra are color-coded according to the protein:peptide molar ratio. (B) The normalized chemical-shift changes observed in the PHD finger upon binding to H3K4me3 as a function of residue. (C) Binding affinities of WT and mutated MLL5 PHD for the indicated histone peptides measured by tryptophan fluorescence (a) or NMR (b). NB, no binding. (D and E) HEK293T cells expressing wild-type FLAG-MLL5 and mutants were fixed and stained with anti-FLAG and anti-H3K4me3 antibodies and with secondary antibodies conjugated with Duolink PLA probes and Duolink PLA detection reagents and imaged using a confocal microscope. Foci numbers in each cell were counted manually (400–500 cells were scored for each sample) and grouped into three categories: 1–3, 4–6, and >6. ***P < 0.001.
The critical role of the binding-site residues was supported by mutational analysis. Substitution of W141 with an alanine completely abolished the MLL5 PHD–H3K4me3 interaction, and replacement of D128 with a lysine severely decreased binding even as the structures of the mutants remained intact (Fig. 3C; Fig. S4). All attempts to replace D128 with an aromatic residue or an alanine resulted in an insoluble protein. As anticipated, binding of the Y131A and Y131K mutants was reduced approximately six- to 13-fold, whereas mutation of F125 had no effect on the interaction (Fig. 3C; Fig. S4).
Binding to H3K4me3 Stabilizes MLL5 at Chromatin.
To assess if the interaction of the PHD finger with H3K4me3 is necessary for localization of MLL5 in vivo, we performed a Duolink in situ proximity ligation assay (PLA; Fig. 3 D and E). HEK 293T cells were transfected with plasmids expressing FLAG-tagged wild-type MLL5 and MLL5 mutants, W141A and D128K, impaired in binding to H3K4me3. The FLAG-F125A mutant of MLL5 that retains the H3K4me3 binding ability was used as a positive control. After cells were stained with anti-FLAG antibody and anti-H3K4me3 antibody, secondary antibodies conjugated with PLA probes were applied to detect the interacting foci. Though all four MLL5 constructs displayed similar expression protein levels, the number of interacting foci in each cell, as specified by red dots in Fig. 3D, was different (Fig. 3E). A significantly higher number of cells expressing wild-type MLL5 showed six or more interacting foci, which are indicative of specific interactions, than the cells expressing loss-of-function W141A and D128K mutants (P < 0.001; Fig. 3E). As expected, the F125A mutant colocalized with H3K4me3-enriched chromatin essentially as wild-type MLL5 did (P = 0.866). These results are consistent with in vitro binding data and indicate that the PHD–H3K4me3 interaction stabilizes MLL5 at chromatin in the intact cells.
Phosphorylation of H3T3 and H3T6 Disrupts Binding of the MLL5 PHD.
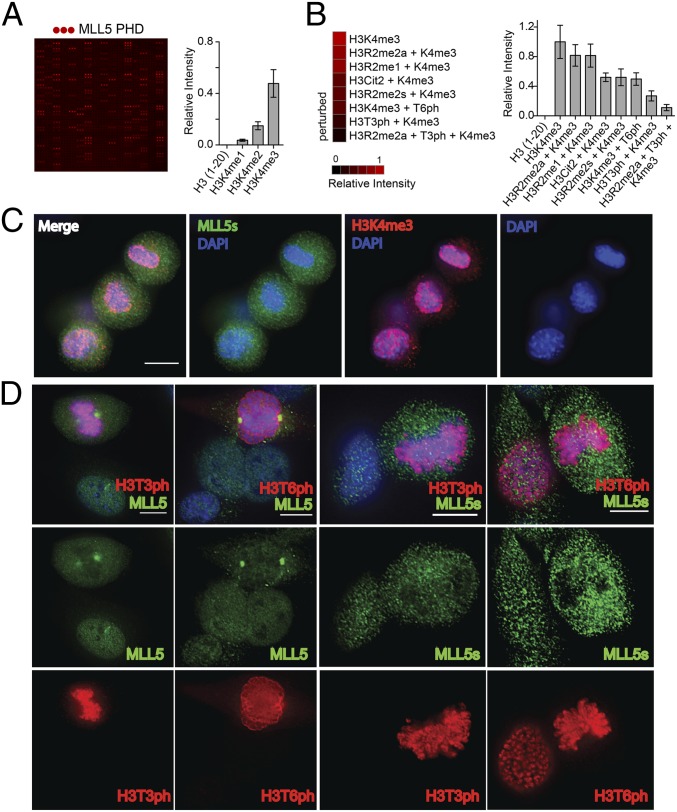
We next examined the effect of the neighboring PTMs on the interaction of MLL5 PHD with H3K4me3 using a high-throughput microarray (Fig. 4 A and B). The GST-tagged MLL5 PHD finger was screened against a library of ∼200 peptides corresponding to unmodified and posttranslationally modified histone tails (Dataset S1). In agreement with the above data, in the microarray, MLL5 PHD associated with H3K4me3; however, PTMs flanking Lys4me3 significantly affected this interaction. As shown in Fig. 4B, phosphorylation of Thr3 or Thr6 (H3T3ph or H3T6ph) inhibited binding, and methylation of Arg (H3R2me) or citrullination of Arg2 (H3Cit2) slightly reduced it.
Fig. 4.
Phosphorylation of Thr3 and Thr6 inhibits binding of the MLL5 PHD finger with H3K4me3. (A and B) GST-MLL5 PHD was probed by a histone peptide microarray. Red spots indicate binding of MLL5 PHD to H3K4me3 peptides. The complete list of peptides is shown in Dataset S1. (C) H3K4me3 remains present during mitosis. C2C12 cells were stained with anti-MLL5s (green) and anti-H3K4me3 antibodies (red). Blue indicates DNA stained with DAPI. (D) MLL5 is displaced from chromatin during mitosis. C2C12 cells were stained with MLL5 and MLL5s antibodies (green) and with anti-H3T3ph and anti-H3T6ph antibodies (red). Scale bar, 10 μm.
The structure of the MLL5 PHD–H3K4me3 complex provides insight into the inhibitory effects of PTMs adjacent to Lys4me3. Phosphorylation of Thr3 is sterically and electrostatically unfavorable because this residue is bound in a narrow channel flanked by the hydrophobic residues I133, I154, and Y131 of the protein. Addition of a phosphate group to Thr6 would not only eliminate the hydrogen bond to the carboxylic group of D128 but also increase repulsion between the two negatively charged moieties. Although the guanidino group of Arg2 is not hydrogen-bonded, it can transiently interact with nearby D136, which may explain a mild effect of PTMs on Arg2.
H3T3ph and H3T6ph Exclude MLL5 from Chromatin in Mitosis.
MLL5 is involved in the regulation of cell cycle and G1/S transition (5); however, its functioning in mitosis remains unclear. Because Thr3 and Thr6 become phosphorylated during mammalian cell division (15), we tested the idea that the PHD finger mediates the MLL5 release from chromatin when cells enter mitosis. We monitored localization of MLL5 in C2C12 cells during interphase and metaphase using fluorescence microscopy (Fig. 4 C and D; Fig. S2B). MLL5 associated with chromatin in the interphase nucleus, however it dissociated from mitotic chromosomes, even though H3K4me3 was still detected on chromosomes (Fig. 4C). Costaining of the cells with antibodies against H3T3ph or H3T6ph confirmed that mitotic chromosomes are hyperphosphorylated on both threonine residues (Fig. 4D). Disengagement of MLL5 from chromatin was accompanied by its accumulation at centrosomes. In contrast, disengagement of the short isoform MLL5s led to the speckled distribution of the protein throughout the nucleus, indicating that the C-terminal region, but not its PHD finger, is responsible for MLL5 association with the mitotic spindle. Notably, these results suggest that the PHD finger mediates localization of MLL5 at chromatin as well as translocation of MLL5 in response to phosphorylation of H3T3 or H3T6 during cell cycle progression.
Recognition of H3K4me3 Is Conserved in the MLL5 Orthologs Set3 and UpSET.
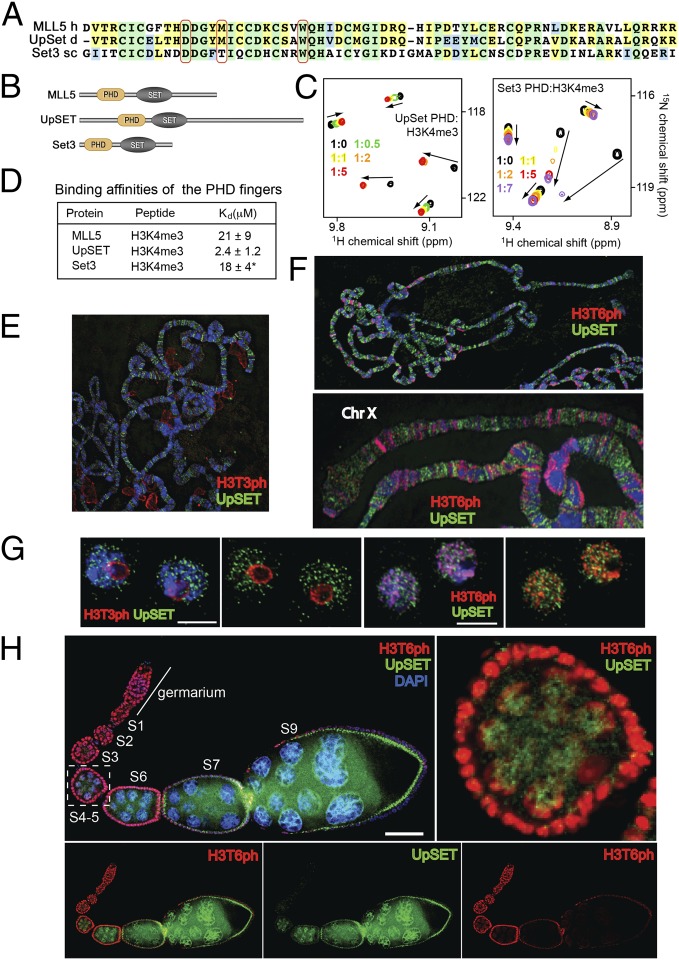
Set3 from Saccharomyces cerevisiae and UpSET from Drosophila melanogaster have the same architecture as MLL5, including the combination of a single PHD finger and a SET domain (Fig. 5 A and B). The alignment of PHD finger sequences of MLL5, Set3, and UpSET shows a high degree similarity, thus suggesting similar functions. Interestingly, the Set3 PHD finger contains a threonine in place of the K4me3-caging methionine of MLL5, but nevertheless, it recognizes H3K4me3 (Fig. 5C), supporting previous findings (16). Titration of the H3K4me3 peptide induced chemical shift perturbations in 1H,15N HSQC spectra of the UpSET PHD finger, indicating direct interaction with this histone modification, which was an order of magnitude stronger than binding of MLL5 or Set3 (Fig. 5 C and D).
Fig. 5.
Binding to H3K4me3, antagonized by H3T3ph and H3T6ph, is conserved in the MLL5 orthologs Set3 and UpSET. (A) Alignment of the PHD finger sequences. The Lys4me3 binding residues are indicated by red ovals. (B) Architecture of the MLL5/UpSET/Set3 family. (C) Superimposed 1H,15N HSQC spectra of the PHD fingers of UpSET and Set3 collected upon titration of H3K4me3. (D) Binding affinities of the PHD fingers measured by tryptophan fluorescence. (Data from ref. 16.) (E and F) Third larval instar polytene chromosomes (E) and polytene chromosomes (F) stained with a mix of antibodies to UpSET N- and C-terminus (green), and H3T3ph (red). DNA counterstained with DAPI (blue). See also Fig. S5 A and B. (G) Drosophila S2 cells stained with antibodies to UpSET N terminus (green) and H3T3ph or H3T6ph (red). Scale bar, 5 μm. (H) UpSET protein levels are differentially regulated during oogenesis. Wild-type ovarioles were stained with antibodies against UpSET (green), H3T6ph (red), and costained with DAPI (blue). (Upper Right) Zoom-in of the S4 egg chamber showing a reduced overlap between UpSET and H3T6ph. Scale bar, 100 μm.
The Phospho-methyl Regulatory Switch Is Conserved in Drosophila UpSET.
To determine whether the installation of the H3T3ph and H3T6ph marks is a conserved mechanism to eliminate binding to chromatin, we investigated UpSET colocalization with H3T3ph or H3T6ph on polytene chromosomes from wild-type salivary glands using fluorescence microscopy (Fig. 5). UpSET staining did not overlap with either phosphorylated mark on polytene chromosomes (Fig. 5 E and F; Fig. S5). In insect S2 cells, H3T3ph was highly abundant at the perinucleolar region, and low levels of this PTM can be detected in discrete regions along polytene chromosomes (Fig. 5 E and G; and Fig. S5A). In contrast, H3T6ph was mainly located around or close to heterochromatin regions in insect cells and polytene chromosomes (Fig. 5 F and G; and Fig. S5B). To rule out the possibility that the lack of overlap is due to the polyploidy process in salivary glands, we costained insect S2 cells using anti-H3T3ph or anti-H3T6ph antibodies and antibodies against the amino or C terminus of UpSET. Again, we found that UpSET is excluded from regions enriched in H3T3ph and H3T6ph (Fig. 5G).
It has been shown that UpSET is required for gametogenesis (17). To explore whether the phospho-methyl switch can mediate levels of chromatin-bound UpSET during oogenesis, we examined the localization of UpSET and H3T6ph in developing Drosophila ovaries. We found that H3T6ph is highly abundant in the germarium and early stages (S1–S4) of oogenesis, but its level is gradually reduced after stage S4–S5 and is undetectable after stage S6–S7 (Fig. 5H). Concomitant with the reduction of H3T6ph, levels of UpSET increased from undetectable before stage S3 to high at stage S9. This anticorrelation and the fact that UpSET staining does not overlap with H3T6ph suggest that H3T6 phosphorylation can control the association and activity of UpSET at chromatin in a developmentally regulated manner.
Discussion
We have demonstrated that MLL5 is recruited to actively transcribed regions in myoblasts. MLL5 binding relies on a single PHD finger, which binds strongly to H3K4me3. Structural analyses suggest that MLL5 possesses a unique spatial organization, which allows the specific recognition of H3K4me3. The PHD domain of MLL5 and its orthologs share conserved amino acids that are key for the efficient interaction, that is impaired through phosphorylation of H3T3 and H3T6.
MLL5 knockout mice exhibit phenotypes similar to those found in patients with MDS, and have revealed that this protein is a key regulator of hematopoietic stem cell fitness (2–4). However, the genomic distribution of MLL5 had remained elusive due to the lack of ChIP-grade antibodies and the lethal effect of its overexpression. Our DamID chromatin profiling results show that MLL5 is preferentially recruited immediately downstream of active TSSs, despite its lack of catalytic activity (9). MLL5 orthologs UpSET and Set3 are similarly located at Drosophila and yeast TSS, respectively (17, 18). Our biochemical data indicate that the PHD domain is required to target active regions via recognition of H3K4me3 and suggest the transcriptional role of MLL5. We had previously shown that the Drosophila ortholog UpSET interacts with histone deacetylases to modulate open chromatin and fine-tune gene expression (17). Lack of UpSET leads to an up-regulation of off-target genes, which correlates with impaired gametogenesis. MLL5 KO mice also exhibit gametogenetic abnormalities, including male sterility (3). The murine MLL5 protein can partially rescue female sterility in flies, suggesting conserved functions. Additionally, RNAi-based genetic screenings suggest that MLL5 may be part of the histone deacetylase complex N-CoR (19). Thus, based on its binding and the available evidence, it is possible that MLL5 may be required to fine-tune gene expression in mammalian cells by targeting H3K4me3 of transcribed genes. It remains to be studied whether these functions could lead to, or be consequences of, certain MDS or leukemia with poor prognosis.
However, these are likely not the only functions encoded by the MLL5 gene, because H3K4me3 is also present in genomic locations other than promoter regions (Table S1). The small isoform MLL5s has been shown to be catalytically active and methylates H3K4 (9). Although we have not analyzed MLL5s binding in vivo, it is possible that this isoform binds chromatin in a similar fashion, because the PHD domain is present in both variants. The MLL5 chromatin profile suggests that only ∼60% of RNA polymerase II and H3K4me3-containing promoters are targeted by the full-length protein, and an attractive possibility is that the remaining promoters are targeted by MLL5s, which would suggest independent functions for each isoform.
MLL5 interaction with chromatin is quite dynamic during the mammalian cell cycle. We show that both MLL5 isoforms detach from mitotic chromosomes in dividing myoblasts. Upon detachment, MLL5 colocalizes at centrosomes, suggesting additional roles independent of its association with chromatin. Several PMTs can affect the recognition of H3K4me3 by the MLL5 PHD finger—in particular, H3T3ph and H3T6ph. In mammals, these modifications are introduced during mitosis, correlating with MLL5 dissociation from chromatin. We found that this PMT-based detachment is a conserved mechanism to block the binding of the PHD domain of this family of proteins. To date, only a few kinases, including Haspin and PKCβ(I), have been described to mediate the respective phosphorylation of H3T3 or H3T6 during mitosis (20, 21). Our analyses in Drosophila suggest that this phospho-methyl switch could also be used as a mechanism to exclude cofactors from specific nuclear compartments (i.e., nucleolus) or from heterochromatic regions, or for controlling the functions of PHD finger-containing proteins during developmental processes like oogenesis.
In summary, MLL5 and its orthologs establish preferential association with H3K4me3, which can be modulated by PTMs flanking the trimethylated lysine. Our findings highlight MLL5 as a key transcriptional regulator, however, future functional studies are necessary to establish the role of this unusual SET domain-containing protein in homeostasis and pathogenesis.
Materials and Methods
See SI Materials and Methods for details on cell culture and fly stocks; DamID chromatin profiling and analysis; immunofluorescence; X-ray crystallography; NMR; peptide microarrays; and proximity ligation assays. Atomic coordinates for the structure have been deposited in Protein Data Bank (4L58). The raw data have been deposited into the Gene Expression Omnibus (GEO) database (GSE47100).
Supplementary Material
Acknowledgments
We thank Agnes Telling for help with experiments, and Jay Nix at beamline 4.2.2 of the Advanced Light Source in Berkeley for help with data collection. This research is supported by National Institutes of Health Grants GM096863 and GM101664 (to T.G.K.), HL65440 (to M.G.), GM097083 (to S.M.P.), and GM068088 (to B.D.S.); and Singapore National Medical Research Council Grant R183000293213 (to L.-W.D.). S.B.R. is supported by University of North Carolina Lineberger Comprehensive Cancer Center Basic Sciences Training Program Grant T32CA09156 and Postdoctoral Fellowship PF-13-085-01-DMC from the American Cancer Society.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4L58). The raw data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE47100).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310156110/-/DCSupplemental.
References
- 1.Emerling BM, et al. MLL5, a homolog of Drosophila trithorax located within a segment of chromosome band 7q22 implicated in myeloid leukemia. Oncogene. 2002;21(31):4849–4854. doi: 10.1038/sj.onc.1205615. [DOI] [PubMed] [Google Scholar]
- 2.Heuser M, et al. Loss of MLL5 results in pleiotropic hematopoietic defects, reduced neutrophil immune function, and extreme sensitivity to DNA demethylation. Blood. 2009;113(7):1432–1443. doi: 10.1182/blood-2008-06-162263. [DOI] [PubMed] [Google Scholar]
- 3.Madan V, et al. Impaired function of primitive hematopoietic cells in mice lacking the mixed-lineage-leukemia homolog MLL5. Blood. 2009;113(7):1444–1454. doi: 10.1182/blood-2008-02-142638. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, et al. MLL5 contributes to hematopoietic stem cell fitness and homeostasis. Blood. 2009;113(7):1455–1463. doi: 10.1182/blood-2008-05-159905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng LW, Chiu I, Strominger JL. MLL 5 protein forms intranuclear foci, and overexpression inhibits cell cycle progression. Proc Natl Acad Sci USA. 2004;101(3):757–762. doi: 10.1073/pnas.2036345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Cheng F, Deng LW. MLL5 maintains genomic integrity by regulating the stability of the chromosomal passenger complex through a functional interaction with Borealin. J Cell Sci. 2012;125(Pt 19):4676–4685. doi: 10.1242/jcs.110411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sebastian S, et al. MLL5, a trithorax homolog, indirectly regulates H3K4 methylation, represses cyclin A2 expression, and promotes myogenic differentiation. Proc Natl Acad Sci USA. 2009;106(12):4719–4724. doi: 10.1073/pnas.0807136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang P, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29(22):6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujiki R, et al. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009;459(7245):455–459. doi: 10.1038/nature07954. [DOI] [PubMed] [Google Scholar]
- 10.Pijnappel WW, et al. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 2001;15(22):2991–3004. doi: 10.1101/gad.207401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogel MJ, Peric-Hupkes D, van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc. 2007;2(6):1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- 12.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25(10):1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asp P, et al. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc Natl Acad Sci USA. 2011;108(22):E149–E158. doi: 10.1073/pnas.1102223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Musselman CA, Kutateladze TG. Handpicking epigenetic marks with PHD fingers. Nucleic Acids Res. 2011;39(21):9061–9071. doi: 10.1093/nar/gkr613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawicka A, Seiser C. Histone H3 phosphorylation—a versatile chromatin modification for different occasions. Biochimie. 2012;94(11):2193–2201. doi: 10.1016/j.biochi.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi X, et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem. 2007;282(4):2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rincon-Arano H, Halow J, Delrow JJ, Parkhurst SM, Groudine M. UpSET recruits HDAC complexes and restricts chromatin accessibility and acetylation at promoter regions. Cell. 2012;151(6):1214–1228. doi: 10.1016/j.cell.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009;137(2):259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kittler R, et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol. 2007;9(12):1401–1412. doi: 10.1038/ncb1659. [DOI] [PubMed] [Google Scholar]
- 20.Dai J, Sullivan BA, Higgins JM. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev Cell. 2006;11(5):741–750. doi: 10.1016/j.devcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 21.Metzger E, et al. Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4. Nature. 2010;464(7289):792–796. doi: 10.1038/nature08839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.