Abstract
Cells, even those having identical genotype, exhibit variability in their response to external stimuli. This variability arises from differences in the abundance, localization, and state of cellular components. Such nongenetic differences are likely heritable between successive generations and can also be influenced by processes such as cell cycle, age, or interplay between different pathways. To address the contribution of nongenetic heritability and cell cycle in cell-to-cell variability we developed a high-throughput and fully automated microfluidic platform that allows for concurrent measurement of gene expression, cell-cycle periods, age, and lineage information under a large number of temporally changing medium conditions and using multiple strains. We apply this technology to examine the role of nongenetic inheritance in cell heterogeneity of yeast pheromone signaling. Our data demonstrate that the capacity to respond to pheromone is passed across generations and that the strength of the response correlations between related cells is affected by perturbations in the signaling pathway. We observe that a ste50Δ mutant strain exhibits highly heterogeneous response to pheromone originating from a unique asymmetry between mother and daughter response. On the other hand, fus3Δ cells were found to exhibit an unusually high correlation between mother and daughter cells that arose from a combination of extended cell-cycle periods of fus3Δ mothers, and decreased cell-cycle modulation of the pheromone pathway. Our results contribute to the understanding of the origins of cell heterogeneity and demonstrate the importance of automated platforms that generate single-cell data on several parameters.
Keywords: single cell, microscopy, image analysis
Cells must detect biochemical cues and respond appropriately to changing environmental conditions. However, under identical chemical stimuli not all cells respond the same. Heterogeneity in “cellular decision making” exists across all levels of life, from bacteria (1, 2) to simple eukaryotes (3–5) to mammalian cells (6, 7), and may be important in cellular adaptation to quickly changing microenvironments (8), in differentiation programs during development (6), and in sensitivity to drugs (9, 10). In addition to the intrinsic stochastic nature of gene expression (1, 3), each cell has an inherent capacity to respond, which is determined by the state and abundance of cellular components. This extrinsic variability may be passed across multiple generations, producing a nongenetic but heritable source of variation. Additional sources that contribute to extrinsic variability include asynchrony and interplay between different signaling pathways (11, 12), cell-cycle effects (3, 13), asymmetry in cellular divisions (14), and cellular aging (13).
Investigating these effects requires tracking the genealogy of multiple generations of cells, followed by stimulation and quantitative single-cell measurement of response, including growth kinetics and gene expression. Although improved image analysis, microscopy, and fluorescent reporter strategies have enabled elegant cell lineaging experiments under static or in vivo conditions (15, 16); conventional microwell or agar pad formats provide only crude capabilities for the modulation of medium conditions and are thus not ideal for the study of dynamic processes such as signaling. Microfluidic systems provide an attractive platform for such experiments and offer advantages of precise temporal control over medium conditions, the physical confinement of cells to facilitate tracking (17–19) and assignment of lineage relationships (20, 21), and scalability to obtain statistically meaningful measures of population variability in multiple conditions and genotypes (18, 22). Despite the demonstrated utility of microfluidic formats for cell-lineage analysis (18, 20, 21), the realization of automated and high-throughput reconstruction of full lineage trees has not been previously achieved. This task is particularly difficult for asymmetrically dividing cells, such as budding yeast, where the small size of the budding daughter cells makes segmentation and assignment challenging. The reliance on manual or quasi-automated reconstruction of cell lineage relationships has presented a major bottleneck that has reduced analysis to only a few lineage trees and conditions or limited investigation to partial lineage trees. For this reason there is a paucity of data looking at the role of nongenetic inheritance in signaling heterogeneity.
To address this need we implemented a fully automated microfluidic live-cell imaging system capable of measuring single-cell responses of multiple yeast strains to a range of dynamically changing medium conditions, while at the same time tracking cell-lineage relationships at a throughput of 25,000 cells per run and for up to eight generations. We applied this system to examine how nongenetic heritability and cell-cycle effects contribute to cellular heterogeneity in signaling response, using the pheromone pathway of the yeast Saccharomyces cerevisiae as a model system.
The pheromone pathway is an archetypical model of mitogen-activated protein kinase (MAPK) signaling, a superfamily of extracellular signal-regulated kinases that is evolutionarily conserved across higher eukaryotes including humans (23), and governs numerous biological processes including differentiation, oncogenesis, proliferation, and growth. In haploid yeast the pheromone MAPK pathway is used for detection and conjugation with the opposite mating types, either a-type or α-type, producing a chemotropic- and concentration-dependent response that is initiated by the binding of pheromone ligand, either α-factor or a-factor, to a membrane-localized G protein-coupled receptor. Activation results in an MAPK phosphorylation cascade that culminates in growth arrest and activation of the transcription factor Ste12, which initiates a transcriptional mating program involving hundreds of genes (24).
The mating pathway of S. cerevisiae is a particularly interesting system for the study of nongenetic heritability in signaling due to the inherent asymmetry in cellular divisions. Cells divide through a budding process, which produces a daughter cell that is smaller than the mother cell. Due to the coupling of cell volume and cell cycle, daughter cells must grow to a critical size before division (25). This results in a mother–daughter asymmetry in cell-cycle kinetics, with daughter cells having a longer G1 phase than mothers. In addition, several genes, including those involved in the separation of cell walls and in the switching of the mating type, are known to be expressed only in daughters and are regulated by daughter-specific genetic programs (26, 27). Finally, although daughters retain a full replicative potential, mothers have a finite life span and progressively undergo replicative aging and accumulate damaged proteins and extrachromosomal DNA material (28, 29).
In this paper, we analyze cell-to-cell variability of transcriptional response to pheromone and measure response correlations between related cells in wild type and seven deletion strains. We reveal two obvious phenotypes, fus3Δ with the highest heritability and ste50Δ with the highest heterogeneity, and we show that they are linked to the alterations of the cell cycle and age asymmetry.
Results and Discussion
Microfluidic Platform for High-Throughput Cell Lineaging.
To assess the interplay of cell cycle, age, and lineage history with signaling response we modified a previously described microfluidic device (18), and optimized this platform for long-term time-lapse imaging with a high numerical aperture objective (63×, NA 1.4), (see www.phas.ubc.ca/~chansen/autoCAD%20designs.rar for the device design). This microfluidic format implements parallel imaging experiments in 128 separate chambers (434 × 165 × 2 μm; w × l × h) arranged in 8 columns, each of which may be loaded with a different yeast strain, and 16 rows, through which independent sequences of medium conditions may be applied in a programmable fashion (Fig. 1A). Within each chamber, growing cells are immobilized within an agarose gel and constrained by the chamber height to grow in a monolayer. Growth rate and cell morphology in the microfluidic device was stable throughout the experiment and similar to the exponential growth in tubes when measured in the same media and temperature (doubling time on chip = 110 min, doubling time off chip = 125 min, Figs. S1 and S2).
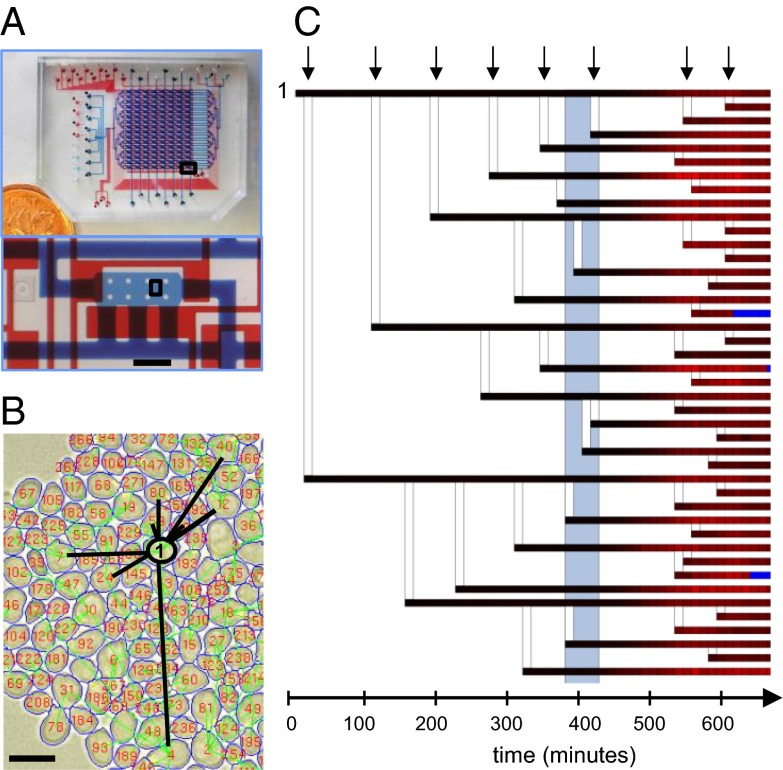
Fig. 1.
Microfluidic platform and automated lineage-tracking technology: (A) Picture of the microfluidic device used in this study. A Canadian penny is included for scale. Bottom shows an optical micrograph of one cell imaging chamber. The flow channels (blue) and control channels (red) have been filled with dye for visualization. The scale bar is 200 μm. (B) An example of image analysis of the indicated chamber area at the bottom of A. Image shows segmentation boundaries of cells (blue lines) that are labeled (red numbers) and tracked through all time frames. Lineage relationships, connecting each cell with its mother are depicted by green lines. For the purpose of visualization all of the progeny of a single cell (cell 1) are highlighted by black lines. The scale bar is 10 μm. (C) Lineage tree of cell 1 from B and transcriptional response of all cells within the tree during a 1-h pulse of 100-nM α-factor. Each horizontal line corresponds to one cell. Each square in the line represents one 12-min time point. Vertical lines connect each mother cell with its progeny. The intensity of red color correlates with transcriptional response. The gray rectangle represents a period of pheromone induction. Time points during which cells were not recognized by lineage tracking algorithm are indicated in blue.
Yeast strains were constructed with three fluorescent markers designed to facilitate automated cell lineaging and assess pheromone-signaling response: mCherry under the control of a promoter containing pheromone-responsive elements (PRE) (18, 22) was used as a transcriptional reporter of pheromone signaling pathway activation; YFP fused to the budneck protein Cdc10 was used to facilitate automated assignment of mother–daughter relationships (21); and CFP under the control of a constitutive promoter of actin was used for accurate segmentation of cells during image processing. To ensure repeatable image quality and robust analysis, we developed image acquisition and analysis software specifically optimized for our microfluidic imaging format and fluorescent marker strategy (available at www.phas.ubc.ca/~chansen/Lineage%20tracking%20software.rar). We have tested that cell growth is neither affected by the integration of fluorescent markers into the genome of the strains (Fig. S1A), nor by periodic imaging in fluorescent channels (Fig. S3) and that the response of cells does not depend on the position in the microfluidic array (Fig. S4).
Using this microfluidic platform every 13-h-long experiment is capable of generating 128 time-lapse imaging series, each represented by a sequence of images taken in two fields of view, each having three fluorescent images and three bright-field images. Our microscopy system allowed for imaging at a maximum frequency of ∼0.85 Hz, corresponding to ∼60,000 images per run and a temporal resolution of 20 min when imaging the entire array. Higher temporal resolution, with images taken every 12 min, was found to be optimal for lineage-tracking experiments; and this was achieved by collecting data on only nine rows of the device, leaving the remaining seven rows for controls or end-point analysis. When starting from low initial seeding densities we were able to routinely perform lineage-tracking experiments for up to eight generations, which is comparable to previously published work (20, 21) but at much higher experimental throughput. Using 72 chambers, a typical run generates data for ∼25,000 cells, and identifies over 9,000 mother–daughter pairs. The performance of the image-analysis software was manually verified on randomly selected cells in every time point of a 13-h-long experiment. The accuracy of segmentation and tracking algorithm exceeded 98%, whereas lineage assignment was correct in 83% of all assessed cases, deteriorating toward the end of the experiment (Fig. S5, SI Materials and Methods). An example of the lineage tracking analysis of a time-lapse series collected from one field of view is demonstrated in Movie S1. A representative image of segmentation and tracking together with a corresponding lineage tree of a fraction of tracked cells are shown in Figs. 1 B and C. The lineage tree diagram displays both the level of PRE-dependent mCherry expression, indicated by the color of each branch, and the genealogical relationship of each cell in the population. In addition, this analysis also provides precise measurements of age and cell-cycle periods for each cell in the population.
Nongenetic Heritability and Heterogeneity of MAPK-Signaling Capacity.
Because signaling response is governed by protein network structure (24), we hypothesized that perturbed networks should exhibit variable effects of nongenetic heritability. To test this we assessed the impact of nongenetic variability in pheromone response of S. cerevisiae both in wild-type cells and across a collection of seven strains having deletions in nonessential components of pheromone response pathway: msg5Δ, kss1Δ, fus3Δ, ste50Δ, far1Δ, ptp2Δ, and mpt5Δ. Strains were first loaded into the microfluidic device and grown for several generations in rich media to record lineage relationships. Cells were then stimulated with 100 nM α-factor for 1 h, followed by time-course analysis of response. Using reconstructed lineage relationships of all cells, we then quantified the correlation of response between mother–daughter pairs, as well as the mean response and total population heterogeneity (Fig. 2 A and B). On average 1,100 mother–daughter pairs were measured per strain.
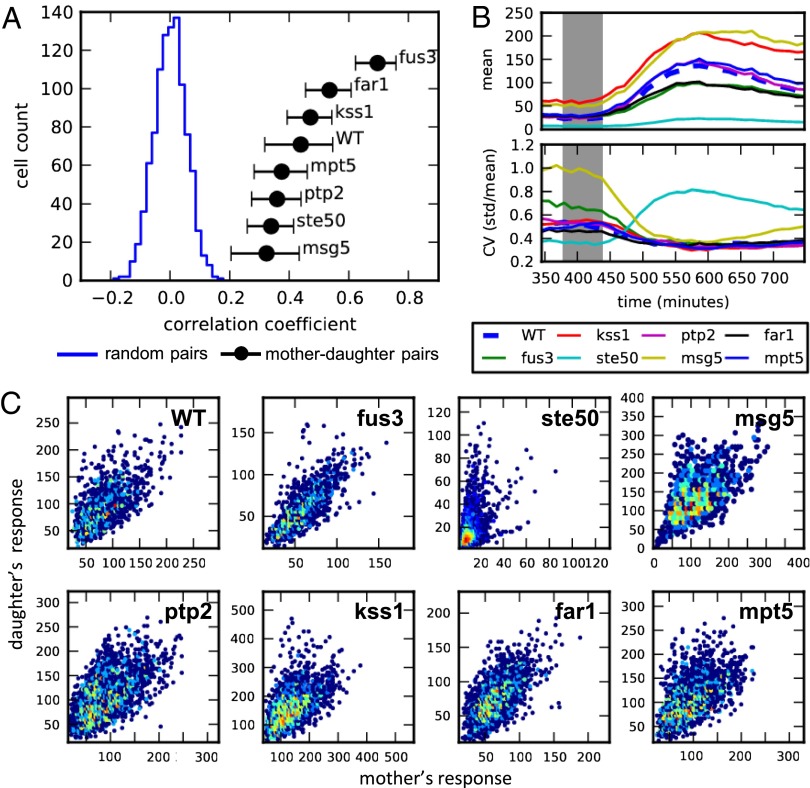
Fig. 2.
Nongenetic inheritance and variability in pheromone response of eight deletion strains. (A) Plot showing the distribution of correlation coefficients for WT random pairs (blue) and the mean correlation coefficients of mother–daughter pairs of all tested strains (black). Cells were first grown over 6 h in media to allow for several divisions and then exposed to 1 h of 100-nM α-factor. Average mCherry fluorescence of each cell was measured 3 h after the beginning of the pheromone induction and used to calculate the correlation of mother–daughter pairs and random pairs born prior induction. Error bars represent SEs. (B) Mean response (Top) and coefficient of variation (Bottom) of each strain over time. Gray area shows period of pheromone stimulation. (C) Heatmaps of the mother–daughter correlations composed of data measured at five consecutive time points between 2 and 3 h after stimulation.
For all strains tested, the response of mother–daughter pairs, measured 3 h after the beginning of stimulation, was found to be significantly correlated compared with that of random cell pairings (Spearman correlation; P < 0.01) (Fig. 2A). Mother–daughter correlations in fus3Δ strains were consistently the highest of all strains tested and had significantly enhanced correlations over all other strains (P < 10−4). Strains deleted for FAR1 and KSS1 were found to have the second highest correlation, although the measured values were significantly higher than WT in only two of three experiments for both strains. On the other hand, rank order analysis consistently placed msg5Δ as the least correlated of all strains. The observed differences in mother–daughter correlations across all eight strains are not explained by the mean response or the overall population variability in response [coefficient of variation (CV) = standard deviation/mean] (Fig. 2B). Both fus3Δ and msg5Δ strains were found to have significantly higher variability in basal expression levels before stimulation and converged to similar values following induction. In terms of mean population expression, msg5Δ and kss1Δ were both found to be hypersensitive to pheromone whereas fus3Δ, ste50Δ, and far1Δ showed diminished sensitivity relative to WT (18, 22, 24). Note that the differences in mother–daughter correlations did not track with either the mean or the variability in cell areas (Fig. S2) and that the mean growth rate before pheromone induction was similar in all strains (Fig. S6).
Inspection of plots of expression variability also revealed a distinct phenotype in ste50Δ mutant. Although the CV of all other strains decreased relative to basal levels following stimulation and converged to a common value, ste50Δ cells exhibited a pronounced increase that resulted in CV approximately twice that of the other strains tested. Time-course measurements of response for individual cells showed that this large variability, which was not apparent in mother–daughter correlation analysis, is due to a generally attenuated pheromone response, but the persistence of a minority population of cells that retain responsiveness similar to nonsterile mutants such as fus3Δ (Fig. S7). This heterogeneity, which is only apparent in single-cell studies, may account for ambiguity in the sterile phenotype of ste50Δ cells (24).
To further explore the origin of ste50Δ heterogeneity we generated scatterplots comparing mother and daughter PRE-dependent expression and compared this to similar plots generated for WT cells and the other deletion strains (Fig. 2C and Fig. S8). This analysis revealed an asymmetric response, unique to ste50Δ mutants, in which daughters showed higher response than their mothers. This asymmetry was found to be correlated with the replicative age of cells; the mean response of “young” daughter cells, defined as those that have not divided before induction, was significantly higher than that of mother cells (mean daughter = 27.26, mean mother = 15.61, P value = 9.71e-08, two-sample Kolmogorov–Smirnov test). Again, this observation highlights the use of lineage tracking in identifying nonrandom sources of heterogeneity that arise through asymmetric cell division.
fus3Δ Preserves High Response Heritability Across Several Generations.
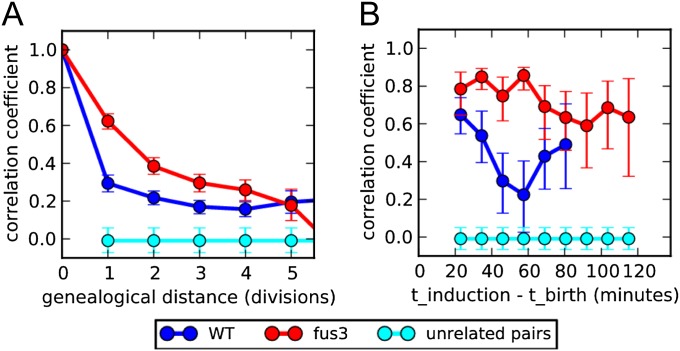
The large enhancement of mother–daughter correlation in fus3Δ strains relative to WT prompted us to further investigate this phenotype. Lineage analysis was used to plot the correlation of transcriptional response between cells separated by increasing genealogical distance for both WT and fus3Δ cells (Fig. 3A). Correlation of signaling response in both WT and fus3Δ cells was found to persist over multiple divisions, and the enhanced correlation of fus3Δ relative to WT remained significant up to four generations. Fig. 3A suggests that the divergence of correlation measures between the two strains is most pronounced during the first cell cycle, with the correlation of WT cells decaying more rapidly. We asked whether this decay arises from noise introduced during the cellular division process or from divergence of cell-signaling capacity between divisions. To discriminate between these two sources we examined the correlation decay during the first cell cycle by plotting the correlation of mother–daughter response as a function of daughter age at the time of induction (Fig. 3B). No difference in mother–daughter correlations between WT and fus3Δ cells were observed when induction occurred immediately after birth. However, as the delay between division and stimulation increased, the correlation of fus3Δ cells remained high, while that of WT cells progressively decayed. Thus, the enhanced correlation of fus3Δ is due to preservation of mother–daughter correlations through the first division.
Fig. 3.
The fus3Δ exhibits high heritability due to the preserved correlation within the first division: (A) Response correlation of WT (blue) and fus3Δ (red) related cells as a function of their genealogical distance measured as the number of divisions separating two cells from the common ancestor. The correlation of unrelated cells is shown in cyan. (B) WT and fus3Δ correlation of mothers and daughters as a function of daughter’s age at the time of stimulation (time interval between daughter’s birth and pheromone induction). Cell birth was defined at the time point when the image analysis algorithm recognizes a new bud, before bud separation.
Fus3 Plays a Role in Cell-Cycle Modulation of Pheromone Pathway.
In WT cells the interaction between cell cycle and mating response is two-way: On the one hand, pheromone pathway activation inhibits progression into S phase mainly through the cell cycle inhibitor Far1 that inactivates complexes of cyclin-dependent kinases (CDKs) with G1 cyclins (Clns) (30). On the other hand the Cln/CDK activity in cells in S phase targets Far1 for degradation and inhibits recruitment of MAPK scaffold protein Ste5 to the membrane resulting in inhibition of pheromone signaling (31, 32).
We speculated that the decay of MD correlation in WT cells may be attributed to asynchrony in the cell cycle, which in turn modulates pheromone response, and that the deletion of FUS3 somehow alleviates this source of noise. To test this we used cell tracking to directly compare the coupling of cell cycle and mating response in fus3Δ cells. Growth arrest of WT and fus3Δ strain was compared by measuring the extension of cell-cycle periods of cells entering a transient 1-h pheromone stimulation (Fig. 4A, blue line). WT cells exhibited a characteristic bimodal distribution of cell-cycle periods, arising from growth arrest of all cells excluding those that were in S phase at the time of induction (Fig. 4C). This bimodal response was not resolved in fus3Δ cells, which instead displayed a broadened distribution, albeit with pronounced increase in the mean cell-cycle period. The absence of a definitive nonresponsive subpopulation in the fus3Δ cells motivated us to further examine cell-cycle kinetics in the absence of pheromone. This revealed another striking difference: unlike WT or any other tested strain, where mothers have a period that is a quarter to a third shorter than newborn daughters, mother fus3Δ cells have an extended cell-cycle period and divide almost as slowly as newborn daughters (Fig. 4A, gray and black dotted line; Fig. S9).
Fig. 4.
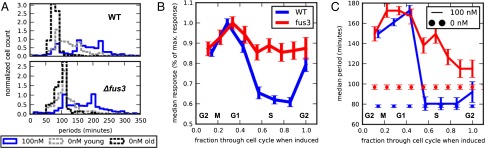
FUS3 deletion alters normal cell cycle progression and affects cell-cycle modulation of the pheromone signaling: (A) Distributions of WT (Top) and fus3Δ (Bottom) cell-cycle periods of young daughters (gray) and mothers (black) under normal conditions and of all cells (combined mother and young daughter cells) under 1 h stimulation of 100-nM αF (blue). Only cell-cycle periods that started before stimulation were measured. (B) Median response, plotted as percentage of maximum response, and median cell-cycle period (C) of WT and fus3Δ cells as a function of their position through the cell cycle at the time of stimulation (measured as percent of the progression through the cell-cycle period). The beginning of the cell cycle was defined as the time point when the image algorithm recognizes a new bud, corresponding approximately to G2 phase. Estimated cell-cycle phases are depicted in the plot.
We then measured the transcriptional response capacity of cells as a function of the cell-cycle phase, estimated using the elapsed time between the latest division and induction (Fig. 4B). WT cells showed a pronounced modulation of pheromone response through the cell cycle, with maximum sensitivity occurring in the heart of G1, where pheromone signaling is least inhibited by Cln/CDKs, and minimum sensitivity observed in S phase where cells are committed to DNA synthesis and therefore inhibit the mating response (30, 31). By comparison, fus3Δ response remained considerably stable throughout the cell cycle and did not drop below 85% of its maximum response, indicating reduced cell-cycle regulation of the pheromone pathway. In correspondence with measured transcriptional response, fus3Δ cells were also less resistant to pheromone-induced growth arrest during S phase compared with WT (Fig. 4C).
To obtain further evidence that the coupling of cell cycle and mating is compromised in fus3Δ mutants we measured transcriptional response in populations of cells synchronized for different positions in the cell cycle. Cells, first grown under normal medium conditions to track lineages, were synchronized in S phase using a 2.5 h exposure to hydroxyurea (HU), followed by a transient stimulation with pheromone at varying delays designed to capture S, G2/M, and G1 phases of the cell cycle (Fig. 5A, scheme). Although hydroxyurea treatment reduced overall response compared with the nonsynchronized condition, synchronization experiment confirmed our data from cell-cycle tracking analysis (Fig. 5B). Time-course measurements of average fluorescence in synchronized cells show that the cell-cycle modulation of pheromone response observed in WT cells is completely absent in fus3Δ cells. In addition, consistent with it being the second most correlated mutant, HU-synchronized far1Δ cells also showed no modulation of mating response by the cell cycle (Fig. S10). This is in agreement with a well-established role of Far1 in cell-cycle arrest mediated through Fus3 (33). We note that this phenotype was not observed in any other strain including the deletion of Kss1, a kinase that is redundant with Fus3 in the final step of signaling transmission through the phosphorylation of Ste12, despite its relatively high mother–daughter correlation.
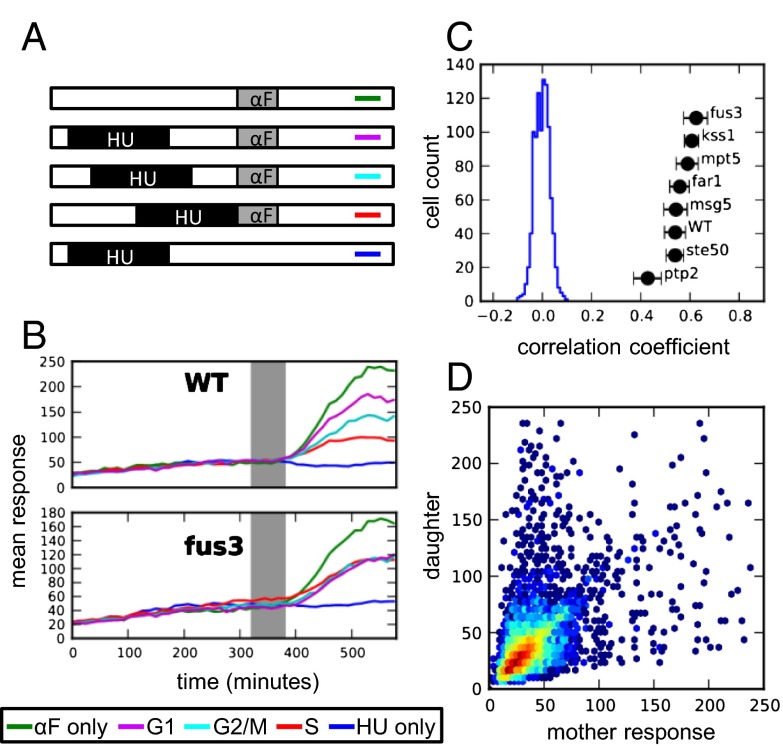
Fig. 5.
Cell-cycle modulation of pheromone signaling and response heritability in HU synchronized cells. (A) Schematic showing synchronization experiment using HU. Cells were synchronized by HU and released to media at different intervals before stimulating them with pheromone. Each line represents one condition that corresponds to the condition in B. HU = 2.5 h exposure of 250-mM hydroxyurea, αF = 1 h of 100-nM αF. (B) Mean response of WT and fus3Δ cells captured in different cell-cycle positions. (C) Response correlations of mother–daughter pairs (black) and a distribution of correlation coefficients of WT random pairs (blue) that were synchronized in G1 phase at the time of pheromone induction. (D) A heatmap of ste50Δ mother–daughter correlations upon HU synchronization in G1 phase.
The enhanced correlation of mother–daughter responses in fus3Δ, and also in far1Δ, is therefore a result of attenuating two major sources of noise: cell-cycle inhibition of MAPK signaling and asymmetry in the cell-cycle kinetics of mother–daughter pairs. Consistent with this, synchronization in G1 before pheromone induction resulted in dramatically improved correlation between mother–daughter pairs in all strains except fus3Δ and far1Δ (compare Figs. 5C and 2A). Moreover, HU synchronization was also found to resolve the mother–daughter asymmetry in ste50Δ response (compare Figs. 5D and 2C). We attribute this effect to the more symmetric division under HU synchronization; under HU arrest, newly formed buds grow to a size comparable to mother cells before division and the asymmetry of the response and of the cell-cycle periods between mothers and newborn daughters during pheromone induction is abolished (Figs. S11 and S12).
Conclusions
We have combined high-resolution microfluidic imaging, fluorescent reporter strategies, and cooptimized image analysis to implement an automated system for tracking cell-lineage relationships and responses under programmable medium conditions. This platform allows the simultaneous collection of single-cell data on gene expression, lineage relationships, cell cycle, and age, and at the same time provides the ability to achieve the experimental throughput needed for generating statistical measures of population variability across multiple conditions and genotypes. Using the yeast pheromone response as a model, we show how such experimental approaches can address biological questions that are otherwise intractable (18, 20–22).
Our data on fus3Δ provides an excellent example of how network structure impacts nongenetic heritability, and how quantitative cell-cycle and lineage analysis can reveal sources of heterogeneity that would otherwise be missed. In specific, we show that fus3Δ exhibits increased correlation of response between related cells that is attributable to reduced coupling of the cell cycle with mating response, and to altered cell-cycle kinetics that exist even in the absence of pheromone stimulation. Previous studies have implicated Fus3 in the suppression of the G1 to S transition under pheromone induction, either directly or through phosphorylation of Far1 (33, 34). This is consistent with fus3Δ having reduced sensitivity to pheromone and incomplete growth arrest upon pheromone stimulation measured on the whole population over extended period (Fig. S13) (33, 34). However, by synchronizing cells “in silico” we found that Fus3 also modulates cell cycle in the opposite direction. fus3Δ cells exhibited higher growth arrest in S phase at the beginning of pheromone stimulation compared with WT cells and cell-cycle periods of fus3Δ mothers were extended even under normal growth conditions. This points to a previously unrecognized role for Fus3 in the promotion and maintenance of the cell cycle. Although surprising, Fus3's implication in cell-cycle progression is consistent with previous observations in that it promotes budding and recovery from pheromone arrest (35) and that FUS3 deletion reduces long-term survival (36).
A second example of how new observations can be obtained by single cell-lineage analysis is the case of ste50Δ where we detected high heterogeneity within a minority cell population that is masked in bulk analysis. Comparison of responses between mother–daughter pairs shows that this heterogeneity is linked to the inherently asymmetric division process in yeast. Although the molecular mechanism of this asymmetry is not clear, the recovery of mother–daughter symmetry in HU-synchronized cells suggests that, as is the case with fus3Δ, this may also be linked to cell cycle (SI Results and Discussion).
The source of heterogeneity in monoclonal populations has been subject to heavy investigation. Stochastic gene expression (4) and more recently stochastic segregation of molecules at division (14) have been used to provide quantitative models of this phenomenon. However, there has been considerably less emphasis on the role of cell cycle and interplay between signaling pathways as a primary source of variability in cellular responses (3). Here we have shown that deletions in the pathway can impact cell-to-cell variability indirectly by modifying the coupling between pathways and either increasing or decreasing pathway sensitivity to preexisting variability between the cellular states. At one level this study complements existing frameworks for the modeling of signaling-network topology and response. For example, removal of the phosphatase Msg5, which acts in a negative feedback loop (37), was found to increase population variability and reduce mother–daughter correlations. However, our data also highlights how pathway interplay can dominate cell-to-cell variability and brings into question the utility of models that capture signaling networks in isolation. As proposed by others, the development of more comprehensive system-level analyses may ultimately be inescapable. This is a formidable challenge that may need to be met if we are to achieve a truly predictive and quantitative understanding of biological systems. Such understanding is likely to be of high practical importance for applications including the targeting of signaling pathways for cancer therapy (38), directing stem cell differentiation for regenerative medicine, and developing strategies to mitigate drug resistance (9, 39).
Materials and Methods
The list of strains used in this study is in the Table S1. Protocols describing fabrication and operation of the microfluidic device, strain construction, image acquisition, and image analysis are provided in SI Materials and Methods and Figs. S14 and S15. The design of the microfluidic device and the custom-made lineage-tracking software together with sample images and a manual that explains how to use the code is available at www.phas.ubc.ca/~chansen/autoCAD%20designs.rar and www.phas.ubc.ca/~chansen/Lineage%20tracking%20software.rar, respectively.
Supplementary Material
Acknowledgments
We thank H. Zahn for the construction of the microscope encapsulation; P. Hieter, I. Barret, and M. Filiatrault for use of equipment and supplies; J. Stoepel for helpful suggestions and training in micromanipulation technique; and A. Carter for PRE-mCherry vector construction and K. Lee for providing pKL973 plasmid. This work was supported by Canadian Institutes of Health Research Grant MOP-93571, Genome BC and Western Diversification. Salary support was provided by the Canadian Institutes of Health Research (to M.R., A.Q. and C.L.H.), the Michael Smith Foundation for Health Research (to C.L.H.), and Natural Sciences and Engineering Research Council (to M.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215850110/-/DCSupplemental.
References
- 1.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 2.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31(1):69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 3.Colman-Lerner A, et al. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437(7059):699–706. doi: 10.1038/nature03998. [DOI] [PubMed] [Google Scholar]
- 4.Raser JM, O’Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304(5678):1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaufmann BB, Yang Q, Mettetal JT, van Oudenaarden A. Heritable stochastic switching revealed by single-cell genealogy. PLoS Biol. 2007;5(9):e239. doi: 10.1371/journal.pbio.0050239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453(7194):544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tay S, et al. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466(7303):267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acar M, Mettetal JT, van Oudenaarden A. Stochastic switching as a survival strategy in fluctuating environments. Nat Genet. 2008;40(4):471–475. doi: 10.1038/ng.110. [DOI] [PubMed] [Google Scholar]
- 9.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459(7245):428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gefen O, Gabay C, Mumcuoglu M, Engel G, Balaban NQ. Single-cell protein induction dynamics reveals a period of vulnerability to antibiotics in persister bacteria. Proc Natl Acad Sci USA. 2008;105(16):6145–6149. doi: 10.1073/pnas.0711712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McClean MN, Mody A, Broach JR, Ramanathan S. Cross-talk and decision making in MAP kinase pathways. Nat Genet. 2007;39(3):409–414. doi: 10.1038/ng1957. [DOI] [PubMed] [Google Scholar]
- 12.Main H, et al. Interactions between Notch- and hypoxia-induced transcriptomes in embryonic stem cells. Exp Cell Res. 2010;316(9):1610–1624. doi: 10.1016/j.yexcr.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 13.Sumner ER, Avery AM, Houghton JE, Robins RA, Avery SV. Cell cycle- and age-dependent activation of Sod1p drives the formation of stress resistant cell subpopulations within clonal yeast cultures. Mol Microbiol. 2003;50(3):857–870. doi: 10.1046/j.1365-2958.2003.03715.x. [DOI] [PubMed] [Google Scholar]
- 14.Huh D, Paulsson J. Non-genetic heterogeneity from stochastic partitioning at cell division. Nat Genet. 2011;43(2):95–100. doi: 10.1038/ng.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eilken HM, Nishikawa S-I, Schroeder T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature. 2009;457(7231):896–900. doi: 10.1038/nature07760. [DOI] [PubMed] [Google Scholar]
- 16.Young JW, et al. Measuring single-cell gene expression dynamics in bacteria using fluorescence time-lapse microscopy. Nat Protoc. 2012;7(1):80–88. doi: 10.1038/nprot.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferry MS, Razinkov IA, Hasty J. Microfluidics for synthetic biology: from design to execution. Methods Enzymol. 2011;497:295–372. doi: 10.1016/B978-0-12-385075-1.00014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falconnet D, et al. High-throughput tracking of single yeast cells in a microfluidic imaging matrix. Lab Chip. 2011;11(3):466–473. doi: 10.1039/c0lc00228c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paliwal S, et al. MAPK-mediated bimodal gene expression and adaptive gradient sensing in yeast. Nature. 2007;446(7131):46–51. doi: 10.1038/nature05561. [DOI] [PubMed] [Google Scholar]
- 20.Rowat AC, Bird JC, Agresti JJ, Rando OJ, Weitz DA. Tracking lineages of single cells in lines using a microfluidic device. Proc Natl Acad Sci USA. 2009;106(43):18149–18154. doi: 10.1073/pnas.0903163106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charvin G, Cross FR, Siggia ED. A microfluidic device for temporally controlled gene expression and long-term fluorescent imaging in unperturbed dividing yeast cells. PLoS ONE. 2008;3(1):e1468. doi: 10.1371/journal.pone.0001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor RJ, et al. Dynamic analysis of MAPK signaling using a high-throughput microfluidic single-cell imaging platform. Proc Natl Acad Sci USA. 2009;106(10):3758–3763. doi: 10.1073/pnas.0813416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Widmann C, Gibson S, Jarpe MB, Johnson GL. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79(1):143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 24.Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides. 2005;26(2):339–350. doi: 10.1016/j.peptides.2004.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartwell LH, Unger MW. Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. J Cell Biol. 1977;75(2 Pt 1):422–435. doi: 10.1083/jcb.75.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bobola N, Jansen RP, Shin TH, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84(5):699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- 27.Colman-Lerner A, Chin TE, Brent R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell. 2001;107(6):739–750. doi: 10.1016/s0092-8674(01)00596-7. [DOI] [PubMed] [Google Scholar]
- 28.Sinclair DA, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91(7):1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 29.Aguilaniu H, Gustafsson L, Rigoulet M, Nyström T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299(5613):1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 30.Chang F, Herskowitz I. Identification of a gene necessary for cell cycle arrest by a negative growth factor of yeast: FAR1 is an inhibitor of a G1 cyclin, CLN2. Cell. 1990;63(5):999–1011. doi: 10.1016/0092-8674(90)90503-7. [DOI] [PubMed] [Google Scholar]
- 31.Strickfaden SC, et al. A mechanism for cell-cycle regulation of MAP kinase signaling in a yeast differentiation pathway. Cell. 2007;128(3):519–531. doi: 10.1016/j.cell.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henchoz S, et al. Phosphorylation- and ubiquitin-dependent degradation of the cyclin-dependent kinase inhibitor Far1p in budding yeast. Genes Dev. 1997;11(22):3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyers M, Futcher B. Far1 and Fus3 link the mating pheromone signal transduction pathway to three G1-phase Cdc28 kinase complexes. Mol Cell Biol. 1993;13(9):5659–5669. doi: 10.1128/mcb.13.9.5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elion EA, Brill JA, Fink GR. FUS3 represses CLN1 and CLN2 and in concert with KSS1 promotes signal transduction. Proc Natl Acad Sci USA. 1991;88(21):9392–9396. doi: 10.1073/pnas.88.21.9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cherkasova V, Lyons DM, Elion EA. Fus3p and Kss1p control G1 arrest in Saccharomyces cerevisiae through a balance of distinct arrest and proliferative functions that operate in parallel with Far1p. Genetics. 1999;151(3):989–1004. doi: 10.1093/genetics/151.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cherkasova VA, McCully R, Wang Y, Hinnebusch A, Elion EA. A novel functional link between MAP kinase cascades and the Ras/cAMP pathway that regulates survival. Curr Biol. 2003;13(14):1220–1226. doi: 10.1016/s0960-9822(03)00490-1. [DOI] [PubMed] [Google Scholar]
- 37.Andersson J, Simpson DM, Qi M, Wang Y, Elion EA. Differential input by Ste5 scaffold and Msg5 phosphatase route a MAPK cascade to multiple outcomes. EMBO J. 2004;23(13):2564–2576. doi: 10.1038/sj.emboj.7600250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bessard A, et al. RNAi-mediated ERK2 knockdown inhibits growth of tumor cells in vitro and in vivo. Oncogene. 2008;27(40):5315–5325. doi: 10.1038/onc.2008.163. [DOI] [PubMed] [Google Scholar]
- 39.Niepel M, Spencer SL, Sorger PK. Non-genetic cell-to-cell variability and the consequences for pharmacology. Curr Opin Chem Biol. 2009;13(5-6):556–561. doi: 10.1016/j.cbpa.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.