Abstract
Ca2+ efflux by Ca2+ cation antiporter (CaCA) proteins is important for maintenance of Ca2+ homeostasis across the cell membrane. Recently, the monomeric structure of the prokaryotic Na+/Ca2+ exchanger (NCX) antiporter NCX_Mj protein from Methanococcus jannaschii shows an outward-facing conformation suggesting a hypothesis of alternating substrate access for Ca2+ efflux. To demonstrate conformational changes essential for the CaCA mechanism, we present the crystal structure of the Ca2+/H+ antiporter protein YfkE from Bacillus subtilis at 3.1-Å resolution. YfkE forms a homotrimer, confirmed by disulfide crosslinking. The protonated state of YfkE exhibits an inward-facing conformation with a large hydrophilic cavity opening to the cytoplasm in each protomer and ending in the middle of the membrane at the Ca2+-binding site. A hydrophobic “seal” closes its periplasmic exit. Four conserved α-repeat helices assemble in an X-like conformation to form a Ca2+/H+ exchange pathway. In the Ca2+-binding site, two essential glutamate residues exhibit different conformations compared with their counterparts in NCX_Mj, whereas several amino acid substitutions occlude the Na+-binding sites. The structural differences between the inward-facing YfkE and the outward-facing NCX_Mj suggest that the conformational transition is triggered by the rotation of the kink angles of transmembrane helices 2 and 7 and is mediated by large conformational changes in their adjacent transmembrane helices 1 and 6. Our structural and mutational analyses not only establish structural bases for mechanisms of Ca2+/H+ exchange and its pH regulation but also shed light on the evolutionary adaptation to different energy modes in the CaCA protein family.
Keywords: Ca2+ transport, H+ coupling
The Ca2+ cation antiporter proteins (CaCAs) comprise a large transporter superfamily existing ubiquitously throughout all biological kingdoms, including animals, insects, plants, fungi, and bacteria (1). They promote Ca2+ extrusion by using an electrochemical gradient of Na+ or H+, regulating Ca2+-mediated signaling processes (2). In animals, all characterized CaCA proteins exclusively use an inward Na+ gradient for Ca2+ efflux. Na+/Ca2+ exchanger proteins (NCXs) are important for maintaining cardiac contractility in cardiac muscle and for neuronal transmission in brain (3). Dysfunction of Na+/Ca2+-K+ exchangers (NCKXs) impairs skin and retinal pigmentation, as well as synaptic plasticity (4).
Unlike those in animal cells, CaCA proteins in plants and yeast use a H+ gradient for coupled Ca2+ transport (1). In Arabidopsis, H+/Ca2+ exchangers (CAXs) on the tonoplast move cytoplasmic Ca2+ to acidic vacuoles, facilitating endoplasmic nutrient partitioning and storage (5). In contrast to eukaryotic members, prokaryotic CaCA proteins exhibit diverse modes of energy dependence. NCX_Mj from Methanococcus jannaschii catalyzes a Na+/Ca2+ exchange similar to eukaryotic NCXs (6), whereas YfkE from Bacillus subtilis has been characterized as a Ca2+/H+ antiporter (7). YfkE shares 60% protein sequence homology with Arabidopsis CAX1 and may contribute to cell differentiation processes during sporulation.
CaCAs are integral membrane proteins containing 10–11 predicted transmembrane helices (TMs) (1) (SI Appendix, Fig. S1). Despite the divergence in cation driving force, the superfamily is defined by the presence of two highly conserved α-repeat motifs in TMs 2–3 (α-1) and TMs 7–8 (α-2). The importance of these conserved motifs for Ca2+/cation exchange has been well documented in all characterized CaCA proteins (8–10), implying the conservation of the Ca2+/cation translocation mechanism in the CaCA superfamily. Recently, Liao et al. reported the crystal structure of the NCX_Mj protein (6). The structure shows a large cavity opening on the periplasmic surface (outward-facing conformation) with both Ca2+ and Na+ bound at the α-repeat regions, suggesting the following hypotheses: (i) a substrate alternating access mechanism in which Ca2+ and the counter transported cation access the ion-binding sites from each side of the membrane surface; and (ii) transition between outward-facing and inward-facing conformations is required to achieve Ca2+/cation exchange. Although the structure of the outward-facing conformation of NCX_Mj is available, structural evidence for the conformational change essential for alternating substrate access has not yet been obtained for any CaCA member. In addition, how the CaCA superfamily proteins modify their Ca2+-translocation pathways to use the different cation-gradient driving forces, H+ vs. Na+, remains a fundamental and interesting question.
To address these important questions, here, we report the crystal structure of an inward-facing conformation of the YfkE protein. This structure, together with protein kinetic analysis, not only provides an important structural characterization of the mechanism of Ca2+ efflux across the cell membrane but also sheds light on the different modes of energy coupling used by members of the CaCA protein superfamily.
Results
Ca2+-Transport Specificity and Structural Determination of YfkE.
YfkE catalyzes a H+-coupled Ca2+ influx in everted vesicles (SI Appendix, Fig. S2A). Na+, Li+, or Mg2+ do not inhibit its transport activity. Modest inhibition was only observed at higher concentrations of Mn2+, Cd2+, Co2+, or Sr2+, confirming the Ca2+ selectivity of YfkE (SI Appendix, Fig. S3). YfkE is strongly regulated by intracellular pH. Michaelis–Menten kinetic analysis revealed two distinct pH dependencies for its Ca2+-transport activity or substrate binding (SI Appendix, Fig. S2B). The Vmax values of YfkE vs. pH exhibit a Gaussian distribution, with an optimal pH of 7. In contrast, its substrate binding appears to be alkaline-dependent [i.e., the apparent Km value at pH 8.5 (51 µM) is dramatically increased to 320 µM at pH 7.5 and becomes undetectable below pH 6].
The YfkE protein was purified from cell membranes using the detergent n-dodecyl-β-maltoside. The purified protein exists in an oligomeric conformation estimated by its gel-filtration elution profile (SI Appendix, Fig. S10B). To facilitate protein expression and crystallization, the mutation K116A was introduced within the loop between TMs 3 and 4. This mutant exhibits a similar transport activity to that of wild type (see Fig. 4C). An additional mutation L77M was introduced for crystallization of the selenomethionine-substituted (Se-Met) protein. Crystals of both native and Se-Met proteins were obtained at pH 4 and diffracted to a resolution of 3.1 Å (SI Appendix, Table S1). The YfkE structure was determined using the multiple-wavelength anomalous-dispersion method combined with the phases of five additional methionine mutants (SI Appendix, Fig. S4 and Table S2). The refined models comprise amino acid residues 4–351, with the missing residues occurring in two cytoplasmic loops between TMs 1–2a (54–57) and between TMs 5–6 (179–201) (SI Appendix, Fig. S5).
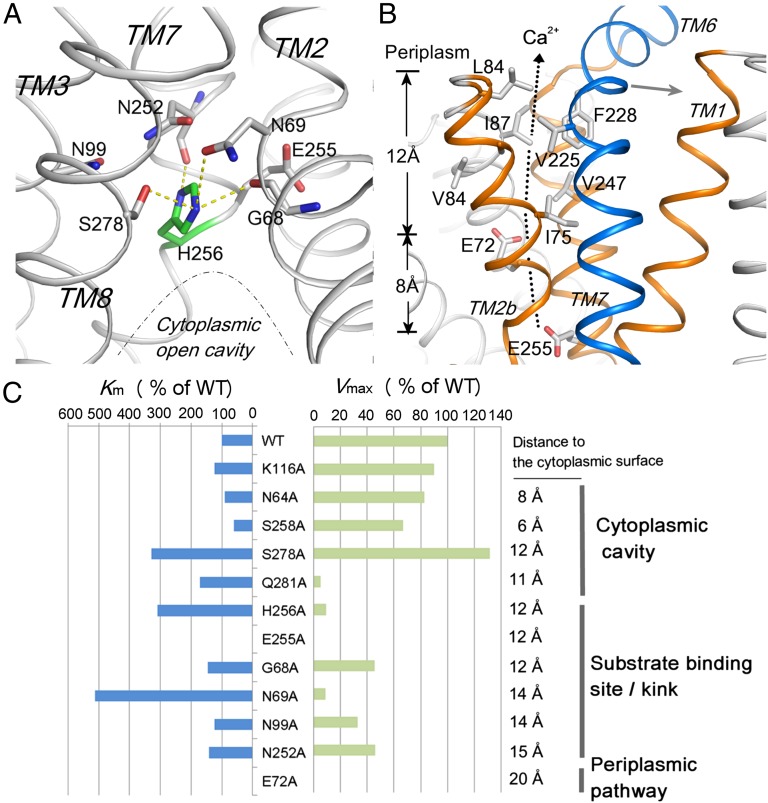
Fig. 4.
Ca2+-binding site and periplasmic closed tunnel conformation. (A) Oxygen umbrella conformation formed by the residues (gray sticks) from the α-repeat helices in the middle of the membrane. The residue H256 (green) is stabilized under the umbrella by hydrogen bond interactions (yellow dash lines). (B) Periplasmic funnel formed by TMs 1, 2b, and 7a (orange) is blocked by TM6 (blue) at the periplasmic exit. The hydrophobic residues along the closed periplasmic tunnel are depicted as sticks. (C) Km (blue bar) and Vmax (green bar) shifts of the YfkE mutants compared with that of wild type. Their position in the Ca2+-translocation pathway and their distance to the cytoplasmic surface are marked.
Overall Structure of YfkE Trimer.
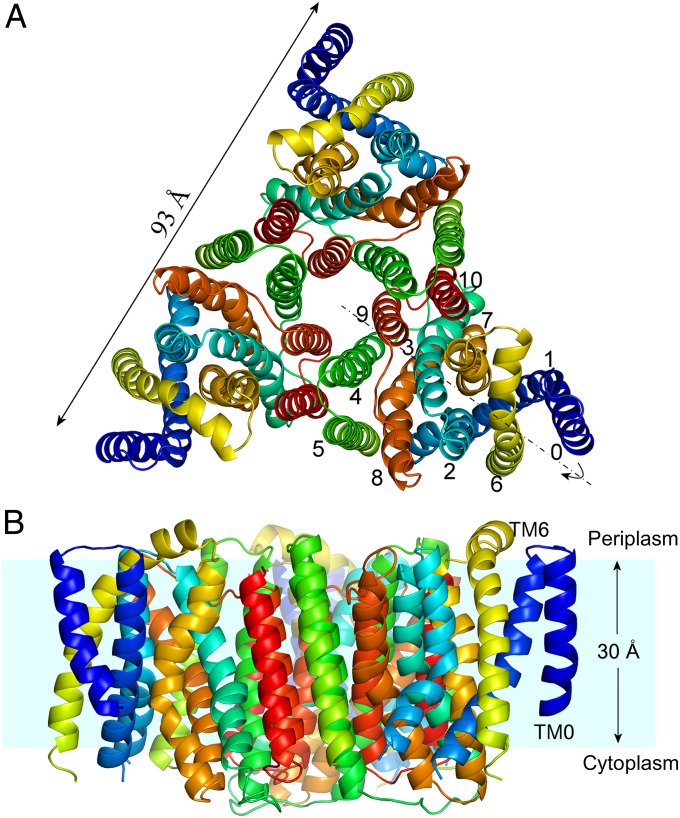
In sharp contrast to the monomeric NCX_Mj structure (6), YfkE crystallizes as a threefold symmetric trimer, reminiscent of an equilateral triangle viewed from the periplasmic side of the membrane (Fig. 1 A and B). Two anti-parallel–stacked trimers determine crystal packing in addition to limited contact between adjacent trimers (SI Appendix, Fig. S6). The “triangle” extends 50 Å from the central threefold axis to its triangular tips with each edge spanning 93 Å on the plane of the membrane. Each YfkE protomer contains 11 TMs (TMs 0–10) (SI Appendix, Fig. S5), in contrast to the 10 TMs of NCX_Mj. They can be characterized as two contiguous helical bundles, TMs 1–5 and TMs 6–10, and an N-terminal TM0 located at the distal tip of the triangle. The two helical bundles exhibit an antiparallel orientation with a pseudo twofold symmetry perpendicular to the membrane plane (Fig. 1A). Whereas its C terminus resides at the periplasmic surface, its N terminus is embedded in the membrane near the cytoplasm owing to the shortest helix TM0 (Fig. 1B). The sequence of TM0 is the least-conserved (SI Appendix, Fig. S1). In NCXs and NCKXs, the analogous fragment is thought to be a cleaved signal peptide, probably generating 10 TMs (3). In plant CAXs, a larger N-terminal segment serves an autoinhibitory function to regulate its Ca2+-transport activity (11). Despite the poor protein sequence homology (18% identity) between YfkE and NCX_Mj, a YfkE protomer can be superimposed on the NCX_Mj structure with a root mean square deviation (rmsd) value of 1.8 Å, demonstrating the structural conservation of the CaCA protein superfamily (Fig. 2 A–C).
Fig. 1.
Overall view of trimeric YfkE structure. (A) Cartoon representation of an YfkE trimer viewed from the periplasmic side of the membrane with TMs 0–10 colored in rainbow (from blue of TM0 to red of TM10). The pseudo twofold symmetry is highlighted as the dashed line. (B) View from the membrane plane. Hydrophobic membrane area (cyan) is estimated based on multiple tryptophan residue positions near TM termini.
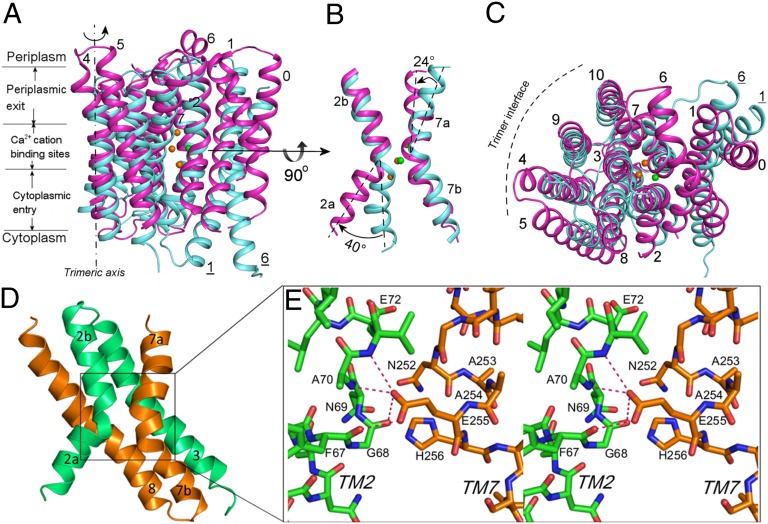
Fig. 2.
Conserved conformation of a YfkE protomer. (A–C) Structural superimposition of a YfkE protomer (magenta) on NCX_Mj (cyan) viewed from the membrane plane (A) and from the periplasm (C). Spheres represent Ca2+ (green) and Na+ (orange) from the NCX_Mj structure, which are also used in other figures. TMs from NCX_Mj are labeled by underlining. (B) Conformational difference of the “X” conformation formed by TMs 2 and 7. (D) The X-like conformation formed by α-1 (green) and α-2 (orange) motifs. (E) Stereoview of the kink conformations of TMs 2 (green) and 7 (orange). The hydrogen bond interactions are depicted as red dashed lines.
The YfkE trimeric conformation is stabilized by extensive hydrophobic interactions and shape complementation between TMs 4–5 of one protomer and TMs 9–10 of the adjacent protomer with a large interface area of more than 1,200 Å2 (Fig. 1B). Along the trimeric axis, three copies of TMs 4 and 9 assemble a vase-like cavity of 4 Å in diameter, which is opened on the periplasmic surface and sealed near the intracellular surface (SI Appendix, Fig. S7). To further examine the trimeric conformation of YfkE, we generated two double-cysteine mutants, I133C/A322C and I162C/I343C (SI Appendix, Fig. S8A). In each mutant, the two adjacent residues from TM4/9 or TM5/10 face each other on the trimeric interface separated by a ∼7-Å distance between their Cα atoms. Disulfide crosslinking under oxidizing condition leads to complete trimer formation by the two mutant proteins both in the cell membrane and in the detergent-solubilized form (SI Appendix, Fig. S8 B and C). No crosslinking was observed with another double-cysteine mutant I133C/I343C, consistent with their greater separation (14 Å) on the interface. These specific crosslinking results strongly suggest that the trimeric conformation demonstrated in the crystal structure represents the physiological condition of YfkE in the cell membrane.
Inward-Facing Conformation.
One of the most remarkable features of the YfkE structure is the symmetric conformation of its two α-repeat motifs. TMs 2–3 and TMs 7–8 form two anti-parallel “hairpins” that ride each other from each side of the membrane (Fig. 2D). Whereas TMs 3 and 8 are tilted in parallel, kinked TMs 2 and 7 form an X-shaped conformation. Such conformation was also observed in the NCX_Mj structure (6) but not in other transporter proteins, indicating the unique scaffold of the α-repeat regions within the CaCA proteins. Whereas TMs 3 and 8 are well superimposable between NCX_Mj and YfkE structures, TMs 2 and 7 exhibit different conformations at their kink angles (Fig. 2B). Compared with the NCX_Mj structure, TM2a of YfkE rotates clockwise around its kink angle by 40°, generating a larger split toward the cytoplasmic side of the membrane. Meanwhile, its TM7a undergoes an anticlockwise rotation by 24° to approach toward TM2b on the periplasmic side of the molecule. The residue E255 located at the TM7 kink apparently stabilizes this inside-opening conformation. The detailed interaction of this highly conserved residue cannot be resolved at this resolution. Based on the electron density map, the carboxylate side chain of E255 extends toward the kink angle of TM2 to interact with four residues (F67, G68, N69 and A70), stabilizing the TM2 kink conformation (Fig. 2E).
Adjacent to the X-conformation of TMs 2 and 7, TMs 1 and 6 exhibit the major conformational difference between YfkE and NCX_Mj. The entire TM1 helix of YfkE is moved ∼15 Å from the TM6 position of NCX_Mj. TM6, the longest helix (45 Å), traverses the V-shaped groove formed by TMs 1 and 2b to bend toward TM7a (Fig. 1 A and B). One-third of TM6 extends above the projected membrane boundary. It may lie on the periplasmic membrane surface, given the opposing polarity of its two helical surfaces. The conformational differences of TMs 1 and 6 appear to be associated with changes in the kink angle of their adjacent TMs 2a and 7a occurring on each side of the membrane. In YfkE, TMs 6–7a lie toward TM3, resulting in closure of the large periplasmic cavity that is observed in the NCX_Mj structure (6). Meanwhile, its TMs 1–2a swing away from the X-like conformation by widening the TM2 kink angle, generating a large funnel-like cavity opened at the cytoplasmic side of the YfkE molecule (Figs. 2C and 3A; see below).
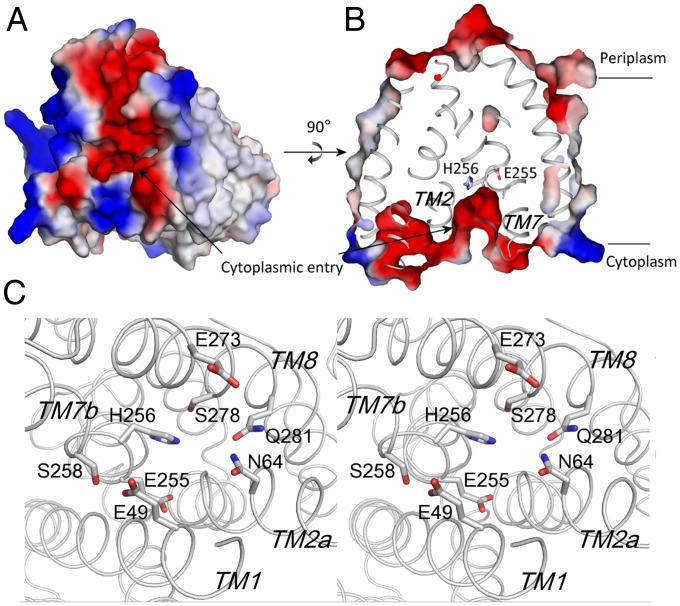
Fig. 3.
Inward-facing conformation of YfkE. (A) Surface representation of electrostatic potential (positive in blue and negative in red) of a YfkE protomer viewed from the cytoplasmic side of the membrane. (B) Cross-section view through the membrane plane showing the inward open conformation. (C) Stereoview of the open cytoplasmic cavity formed by polar residues depicted as sticks.
Intracellular Substrate Entry.
Of major interest regarding the structure of YfkE is the possibility of identifying the Ca2+-translocation pathway and the Ca2+-binding site. The native YfkE crystal was obtained in the presence of 2 mM Ca2+. There was no significant change in the overall protein conformation of the structures with or without substrate. Efforts to obtain anomalous signals by collecting the native crystal data at a wavelength of 1.6 Å did not localize any metal ion in the structure. Therefore, the YfkE structure obtained at pH 4 may represent a protonated inactive conformation.
To transport Ca2+, a hydrophilic ion-translocation pathway is predicted. In YfkE, the trimetric central cavity would readily exclude any ion because of its hydrophobicity (SI Appendix, Fig. S7). A Ca2+/Na+-translocation pathway is formed by the four α-repeat helices in the NCX_Mj structure (6). We hypothesize that Ca2+ efflux occurs through a similar pathway in each protomer of YfkE based on the following. (i) The residues along the pathway are highly conserved among Ca2+/H+ antiporter proteins and the CaCA superfamily. Many of them contribute their side-chain oxygen atoms to form the sole hydrophilic tunnel across the membrane in YfkE (Fig. 3 and SI Appendix, Fig. S1). (ii) The alkaline dependence of Ca2+ binding also suggests involvement of carboxylate residues in the binding site (SI Appendix, Fig. S2B). Each YfkE protomer has 16 Asp/Glu residues, among which 14 locate on the periplasmic or cytoplasmic surface (SI Appendix, Fig. S9). E72 and E255, two CaCA fingerprint residues each from the α-1 or α-2 motif, reside in the middle of the pathway. (iii) Single alanine substitution of these two residues completely abolishes Ca2+-transport activity in everted vesicles (Fig. 4C). To exclude any effect of protein expression and folding, all YfkE mutant proteins were confirmed using Western blot analysis of the vesicles (SI Appendix, Fig. S10A) and further purified by size-exclusion chromatography (SEC). The SEC profiles and CD spectroscopic analyses suggest that all mutants exist in an overall protein conformation similar to that of wild type (SI Appendix, Fig. S10 B and C).
Along the pathway, the intracellular cavity is formed by TMs 1, 2a, 7b, 8, and 10 (Fig. 3C). The cavity has a water-accessible entry 5 Å in diameter calculated by MOLE (12) and gradually narrows to the intersection of TMs 2a and 7b in the middle of the membrane (Fig. 3B and SI Appendix, Fig. S11). Several polar residues including E49, N64, S258, E273, S278, and Q281 contribute to the net electronegativity of the cavity (Fig. 3A). Mutation of Q281 to alanine nearly abrogated Ca2+-transport activity, whereas the N64A or S258A mutants remained largely unaffected, suggesting that the residues deeply embedded in the membrane are more important for Ca2+-transport activity than those near the cytoplasmic entry (Fig. 4C). Therefore, one would expect Ca2+ diffusion in the cavity to reach a binding position near residue Q281.
Ca2+-Binding Site and Closed Periplasmic Exit.
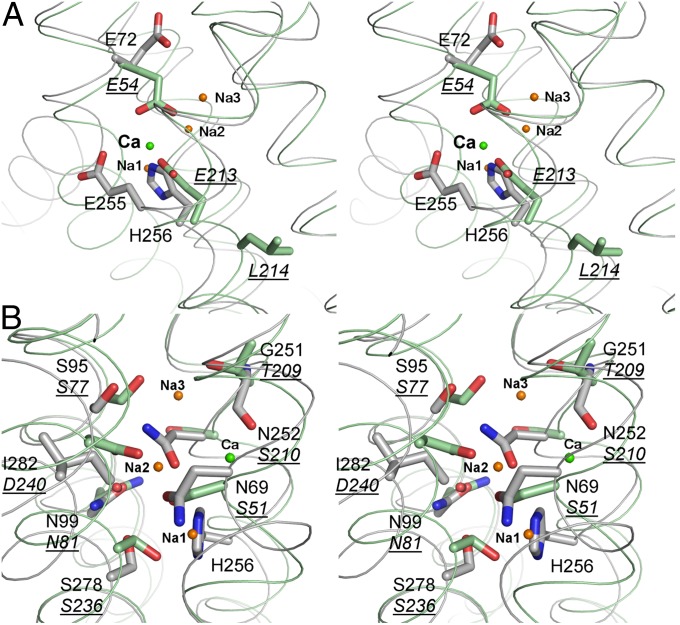
At the end of the cytoplasmic cavity, six conserved residues from the four α-repeat helices, G68 and N69 from TM2, N99 from TM3, N252 and E255 from TM7, and S278 from TM8, assemble an oxygen “umbrella” conformation in the middle of the membrane (Fig. 4A). Alanine scanning of the umbrella suggests that each asparagine residue, particularly N69, is important in maintaining Ca2+-transport activity (Fig. 4C), supporting the hypothesis that Ca2+ binding takes place at a position near the oxygen umbrella. In the NCX_Mj structure, these conserved residues or their counterparts form an ion binding region to coordinate 3 Na+ and 1 Ca2+ (6). However, in contrast to NCX_Mj, these ion-binding sites undergo significant conformational changes in the YfkE structure: (i) two glutamate residues (E72 and E255) coordinating Ca2+ in NCX_Mj exhibit different conformations in the YfkE structure (Fig. 5A); and (ii) the Na+-binding sites are replaced by amino acid substitutions to remove available ligands and occlude the binding sites (Fig. 5B). E72 rotates its carboxylate side chain away from the Ca2+-binding site toward the periplasmic side, whereas H256 moves along the helix to occupy the position of E213 (NCX_Mj numbering), the counterpart residue of E255 in YfkE, and places its imidazole ring on the Na1 position. This residual translocation leads to a helical distortion at the TM7 kink region (Fig. 5A). S95, N99, and S278 are well superimposed with their counterpart residues involved in the Na+-binding sites of NCX_Mj, whereas N69 replaces S51 that coordinates Na-1. I282 substitutes D240 in the Na-2–binding site; N252 (S210 in NCX_Mj) inserts its side chain onto the Na-2 position. T209 involved in the Na-3 binding is also replaced by G251 in the YfkE structure (Fig. 5B).
Fig. 5.
Stereoviews of the Ca2+-binding site of YfkE (gray) superimposed on NCX_Mj (green). Conformational changes of the residues in the Ca2+-binding site (A) and in the abolished Na+-binding sites (B). The residues from both structures are labeled and the counterpart residues from NCX_Mj are underlined.
Right above the umbrella, there is no obvious path opening to the periplasmic surface, because of the tightly packed helices (Fig. 4B). Direct Ca2+ passage along TMs 3 and 8 is prevented because of the tilting of TMs 2b–3 on TM8 and their hydrophobic interface (SI Appendix, Fig. S12). TM5 and the membrane-buried loop between TMs 8 and 9 also restrict any large movement of TM8. Instead, the tilted TMs 2b–3 generate another funnel-like cavity together with TMs 1 and 7a that is connected to the Ca2+-binding site (Fig. 4B). The location of the essential residue E72 suggests Ca2+ passage through this cavity. Compared with the open cytoplasmic entry, the periplasmic tunnel is completely blocked by TM6 lying at the exit. Furthermore, unlike the overall polarity of the cytoplasmic entry, the periplasmic tunnel is more hydrophobic. Eight residues lined along the closed tunnel or located at the periplasmic exit (including I75 and A79 from TM2b; L84, I87, and V88 from TM3; V225 and F228 from TM6; and V247 from TM7a) form a hydrophobic “seal,” eliminating water accessibility to the Ca2+-translocation pathway from the periplasmic side of the membrane.
Discussion
In this study, we have determined the crystal structure of the H+-coupled Ca2+ transporter YfkE. The crystal structure supplemented by crosslinking analysis demonstrates that the CaCA protein exists as a trimer. A trimeric structure has been observed in other transporter structures, including AmtB and Gltph (13, 14), but is unexpected in the CaCA family because both mammalian NCXs and NCKXs are predicted to form a homodimer and NCX_Mj was crystallized as a monomer (15, 16). The YfkE structure provides structural evidence that varied oligomeric conformations may exist in the CaCA protein family. It is noteworthy that several aliphatic residues on the trimer interface, including I142, L158, L319, and I343, are conserved among the Ca2+/H+ exchanger proteins (SI Appendix, Figs. S1 and S8A). Whether such a trimeric conformation also exists in other CAX proteins may require further investigation. Additionally, whether the trimeric conformation plays a specific role in the Ca2+/H+ exchange mechanism remains an interesting question. As shown in Fig. 2C, the trimer-forming helices in the inward-facing YfkE structure are well superimposable with their counterparts in the outward-facing NCX_Mj structure, arguing that the trimer conformation is not directly involved in Ca2+ transport or responsible for their difference in energy coupling. Instead, the trimer may support Ca2+ transport by stabilizing the protein in cell membranes given that large conformational changes are expected to occur in other parts of the protein. Studying this aspect in the future could provide insights into the working mechanism of CaCA proteins in general.
The Mechanism of pH Regulation.
Ca2+ efflux via YfkE is tightly regulated by intracellular pH. Our structure obtained at pH 4 demonstrates an acid-locked conformation. Interestingly, the structure of the Na+/H+ antiporter NhaA from Escherichia coli crystallized at the same pH also exhibited an acid-inactive conformation (17). Despite the lack of sequence homology and structural similarity between these two H+-coupled ion-exporter proteins, their inactive states were both trapped at their inward-facing conformations, raising the hypothesis that the inward-facing conformation may be energetically favorable for acidic inactivation of H+-coupled transporter proteins. However, YfkE apparently uses a different mechanism for pH regulation than that of NhaA. In contrast to the acid-inactive conformation of NhaA, which is triggered by a structural element outside its ion-translocation pathway (17), the inactivation of YfkE likely takes place within its Ca2+-binding site. At acidic pH, the protonated E255 stabilizes the highly kinked TM2a to seize the protein in a widely open inward-facing conformation (Fig. 2 D and E). This stable interaction stabilizes TM7a and TM6 to attach to TM3, consequently blocking the periplasmic pathway (Fig. 4B). At alkaline pH, the deprotonated E255 may move away from the TM2 kink, liberating the X structure of TMs 2 and 7 for any conformational movement. Owing to their high sequence homology and similar pH-regulatory properties (18), this unique pH-regulatory mechanism may be shared by all members of the CAX protein family.
Ca2+/H+ Exchange Mechanism.
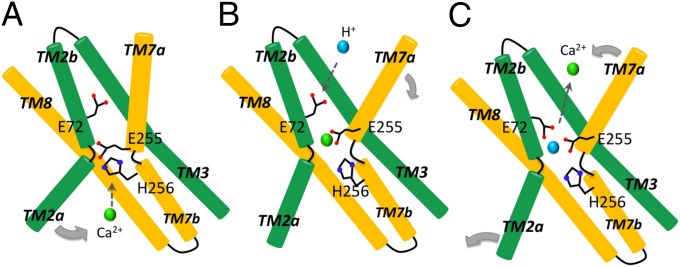
The inward-facing conformation of the YfkE structure and the outward-facing conformation of the NCX_Mj structure provide important structural information about the Ca2+/cation exchanging mechanism. The fact that protonated YfkE exists in an inward-facing conformation suggests that H+ influx resets the protein to a state ready for cytoplasmic Ca2+ access. Based on the Ca2+-bound NCX_Mj structure, Ca2+ binding in the cytoplasmic cavity would induce a closure rotation of TM2a around the TM2 kink to narrow the cytoplasmic cavity (Fig. 6A). Constraint of the kink rotation by mutating G68 to alanine causes a 50% reduction in the Vmax value, supporting this view (Fig. 4C). The deprotonated E255 liberated from the TM2 kink would then move toward the oxygen umbrella to join the Ca2+-binding site, as shown in the NCX_Mj structure (6) (Fig. 5A).
Fig. 6.
Proposed mechanism of Ca2+/H+ exchange by YfkE. (A) Inward open conformation stabilized by the protonated residue E255 for Ca2+ access. Conformational changes of the residues E255 and H256 may generate a path for Ca2+ binding and also shift TM2a for closure of the cytoplasmic entry. (B) Ca2+ binding at the oxygen umbrella leads to a rearrangement of the binding site, inducing conformational changes in TMs 7a and 6 (not shown for clarity) to transit the protein into an outward-facing conformation. (C) The residue E72 transfers H+ (blue sphere) from the periplasmic surface to the Ca2+-binding site. Ca2+ release is then triggered by protonation of E255 and/or H256, which may also reset the protein back to the inward-facing conformation.
In the YfkE inward-facing conformation, the residue H256 blocks the Ca2+-binding site under the oxygen umbrella (Fig. 4A). Attempts to remove this blocking residue resulted in 90% loss of the transport activity (Vmax) and threefold reduction in the Km value, suggesting its critical role for transport activity rather than substrate binding (Fig. 4C). Histidine is not a favored residue for Ca2+ coordination. This essential residue is highly conserved among all H+-driven CAX proteins but not within Na+-coupled NCXs/NCKXs (SI Appendix, Fig. S1), further supporting its specific role in H+ coupling rather than Ca2+ binding. The observation that the transport Vmax exhibits an optimal pH of 7 further implies that a histidine residue is involved in H+ transfer (SI Appendix, Fig. S2B). H256 is the only histidyl residue located on the Ca2+-translocation pathway, making it an ideal position for the H+ transfer (SI Appendix, Fig. S13). Assuming that the imidazole ring of H256 is in the protonated state at crystallization pH 4, it is stabilized as a plug under the oxygen umbrella (Fig. 4A). Upon its deprotonation, H256 would unlock its imidazole ring from the plug position, providing a specific position for Ca2+ binding.
The movement of H256 and E255 may take place simultaneously with the helical distortion, because of their adjacent position (Fig. 5A), generating an active Ca2+-binding site in the middle of the α-repeat helices. This concurrent action occurring at the TM7 kink hinge may induce an outward rotation of the tilted TM7a together with the adjacent TM6 toward the exterior TM1 (Fig. 6B). These motions would subsequently open the Ca2+-translocation pathway by sliding the hydrophobic seal on the periplasmic surface, resulting in an outward-opening conformation providing access to the Ca2+-binding site.
Opening of the periplasmic exit may allow water access to the periplasmic tunnel to facilitate inward H+ flux. To accomplish H+/Ca2+ exchange at the Ca2+-binding site, the H+ has to travel a long distance of 20 Å from the periplasmic surface to residues E255 and H256 deeply buried in the middle of the membrane (Fig. 4B). Passive diffusion may not afford a rapid H+ uptake given the hydrophobic nature of the periplasmic tunnel. Instead, residue E72 residing along the tunnel may more efficiently facilitate H+ influx given its remarkable mobility illustrated in the two structures. Shifting upward of the carboxylate group of E72 shortens its distance to the periplasmic surface to 12 Å, making it an ideal candidate to import periplasmic H+. The protonated carboxylate group of E72 may flip toward the Ca2+-binding site to transfer the H+ to residues E255 and/or H256 (Fig. 6C). In fact, the structural conformation of residue E54 interacting with both Ca2+ and Na+ in the NCX_Mj structure is in agreement with our hypothesis (6) (Fig. 5A).
In the NCX_Mj structure, both Na+ and Ca2+ are bound to central binding sites (6). In YfkE, we predict that E72 and E255 are involved in binding an alternate substrate because a protonated glutamate residue may not be able to coordinate with Ca2+. Although we cannot exclude partial substrate occupancy of the binding site, Ca2+ release can be immediately triggered by protonating the Ca2+-binding site. E72 may also assist Ca2+ escape toward the periplasmic surface by flipping its side chain back to the uplift conformation. The protonated H256 residue may return to the plug position under the umbrella. As a consequence, TMs 7a and 6 move back to close the periplasmic tunnel and reset the protein to the inward conformation. It is still unknown how the H+ releases from the Ca2+-binding site to the cytoplasm. Any water molecule network stabilized in the cytoplasmic cavity may form a H+-relaying pathway. Alternatively, two carboxylate residues (E49 and E273) located at the cytoplasmic entry may facilitate H+ delivery in the cavity (Fig. 3C). However, conformational changes in these residues would be necessary for direct H+ relay from the Ca2+-binding site given their distances of more than 10 Å from E255 and H256.
This substrate-alternating access model is consistent with the outward-facing conformation seen in the NCX_Mj structure (6), suggesting that the CaCA proteins share a conserved mechanism for Ca2+/cation exchange. The structures of YfkE and NCX_Mj provide two examples of the inward-facing and outward-facing conformations. Whether any other conformational state(s), such as the Ca2+-bound conformation or the partially protonated state that Liao et al. proposed for NCX_Mj (6), is/are available during one Ca2+/H+-exchanging cycle is unknown. Based on the two structures, transition from the inward-facing to the outward-facing conformation is triggered by alternative substrate binding at the kinks of the α-repeat helices to alter the kink angles of TMs 2 and 7 (Fig. 2B). These helical rotations occurring on each side of the membrane may be coupled via their adjacent TMs 1 and 6 (Fig. 2 A and C). These two interactive helices move together on the exterior of the trimer, resulting in alternative opening and closing of the Ca2+/H+-exchanging pathway at each side of the molecule.
One of the most intriguing questions regarding the YfkE structure is how the different CaCA proteins adapt different driving forces while maintaining their Ca2+ specificity. Despite the possibility that Ca2+ binding may change the conformation of the binding site, comparison of the apo form of YfkE with the Ca2+/Na+-binding site of NCX_Mj provides insight into this fundamental question (Fig. 5). In addition to the two conserved glutamate residues, other residues coordinating the three Na+ ions in NCX_Mj undergo significant modifications, which apparently abolish the Na+-binding sites. Strikingly, these residues reassemble to form a cluster of side-chain oxygen atoms to generate a more compact binding site that would only accommodate Ca2+. These sequence differences conserved in either the Na+-coupled NCXs and NCKXs or the H+-driven CAX family (SI Appendix, Fig. S1) may reflect evolutionary adaptation to different energy driving forces while maintaining substrate specificity.
Methods and Materials
Both native and SeMet-YfkE were expressed in E. coli with a polyhistidine tag inserted within the cytoplasmic loop between TMs 5 and 6, extracted by dodecylmaltoside, and purified by a metal-affinity column. The proteins were further purified by SEC and concentrated to 8 mg⋅mL−1 for crystallization. Crystals were obtained using the sitting-drop vapor-diffusion method. Both native and Se-Met crystals were diffracted to 3.1-Å resolution after dehydration treatment. The crystal diffraction data were processed using XDS (19). The structures were determined using the multiple-wavelength anomalous dispersion method using AutoSHARP (20). The structures were built using Coot (21) and refined to the final R/Rfree values (%) of 22/26 for the native structure and 24/27 for the Se-Met structure using Refmac (22). Transport analyses were carried out by measuring 45Ca2+ influx into everted vesicles prepared as described previously (23). Disulfide crosslinking was performed using an iodine-catalyzed approach (24). The details of methods and materials are provided in SI Appendix.
Supplementary Material
Acknowledgments
We acknowledge the assistance of the staff at Swiss Light Source beamline X06SA and X06DA and Jay Nix from Advanced Light Source beamline 4.2.2 for data collection. We thank Julia Lever and John Spudich for comments on the manuscript. This study was supported by National Institute of Health Grants R01GM097290 and R01GM098572 (to L.Z.) and American Heart Association postdoctoral fellowships (to M. Wu and S.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4KJR and 4KJS).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302515110/-/DCSupplemental.
References
- 1.Cai X, Lytton J. The cation/Ca(2+) exchanger superfamily: Phylogenetic analysis and structural implications. Mol Biol Evol. 2004;21(9):1692–1703. doi: 10.1093/molbev/msh177. [DOI] [PubMed] [Google Scholar]
- 2.Clapham DE. Calcium signaling. Cell. 2007;131(6):1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 3.Philipson KD, Nicoll DA. Sodium-calcium exchange: A molecular perspective. Annu Rev Physiol. 2000;62:111–133. doi: 10.1146/annurev.physiol.62.1.111. [DOI] [PubMed] [Google Scholar]
- 4.Altimimi HF, Schnetkamp PP. Na+/Ca2+-K+ exchangers (NCKX): Functional properties and physiological roles. Channels (Austin) 2007;1(2):62–69. doi: 10.4161/chan.4366. [DOI] [PubMed] [Google Scholar]
- 5.Hirschi K. Vacuolar H+/Ca2+ transport: Who’s directing the traffic? Trends Plant Sci. 2001;6(3):100–104. doi: 10.1016/s1360-1385(00)01863-x. [DOI] [PubMed] [Google Scholar]
- 6.Liao J, et al. Structural insight into the ion-exchange mechanism of the sodium/calcium exchanger. Science. 2012;335(6069):686–690. doi: 10.1126/science.1215759. [DOI] [PubMed] [Google Scholar]
- 7.Fujisawa M, Wada Y, Tsuchiya T, Ito M. Characterization of Bacillus subtilis YfkE (ChaA): A calcium-specific Ca2+/H+ antiporter of the CaCA family. Arch Microbiol. 2009;191(8):649–657. doi: 10.1007/s00203-009-0494-7. [DOI] [PubMed] [Google Scholar]
- 8.Nicoll DA, Hryshko LV, Matsuoka S, Frank JS, Philipson KD. Mutation of amino acid residues in the putative transmembrane segments of the cardiac sarcolemmal Na+-Ca2+ exchanger. J Biol Chem. 1996;271(23):13385–13391. doi: 10.1074/jbc.271.23.13385. [DOI] [PubMed] [Google Scholar]
- 9.Winkfein RJ, et al. Scanning mutagenesis of the alpha repeats and of the transmembrane acidic residues of the human retinal cone Na/Ca-K exchanger. Biochemistry. 2003;42(2):543–552. doi: 10.1021/bi026982x. [DOI] [PubMed] [Google Scholar]
- 10.Kamiya T, Maeshima M. Residues in internal repeats of the rice cation/H+ exchanger are involved in the transport and selection of cations. J Biol Chem. 2004;279(1):812–819. doi: 10.1074/jbc.M309726200. [DOI] [PubMed] [Google Scholar]
- 11.Pittman JK, Hirschi KD. Regulation of CAX1, an Arabidopsis Ca(2+)/H+ antiporter. Identification of an N-terminal autoinhibitory domain. Plant Physiol. 2001;127(3):1020–1029. [PMC free article] [PubMed] [Google Scholar]
- 12.Petrek M, Kosinová P, Koca J, Otyepka M. MOLE: A Voronoi diagram-based explorer of molecular channels, pores, and tunnels. Structure. 2007;15(11):1357–1363. doi: 10.1016/j.str.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 13.Zheng L, Kostrewa D, Bernèche S, Winkler FK, Li XD. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc Natl Acad Sci USA. 2004;101(49):17090–17095. doi: 10.1073/pnas.0406475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yernool D, Boudker O, Jin Y, Gouaux E. Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature. 2004;431(7010):811–818. doi: 10.1038/nature03018. [DOI] [PubMed] [Google Scholar]
- 15.Ren X, Nicoll DA, Galang G, Philipson KD. Intermolecular cross-linking of Na+-Ca2+ exchanger proteins: Evidence for dimer formation. Biochemistry. 2008;47(22):6081–6087. doi: 10.1021/bi800177t. [DOI] [PubMed] [Google Scholar]
- 16.Schwarzer A, Kim TS, Hagen V, Molday RS, Bauer PJ. The Na/Ca-K exchanger of rod photoreceptor exists as dimer in the plasma membrane. Biochemistry. 1997;36(44):13667–13676. doi: 10.1021/bi9710232. [DOI] [PubMed] [Google Scholar]
- 17.Hunte C, et al. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature. 2005;435(7046):1197–1202. doi: 10.1038/nature03692. [DOI] [PubMed] [Google Scholar]
- 18.Pittman JK, Shigaki T, Hirschi KD. Evidence of differential pH regulation of the Arabidopsis vacuolar Ca2+/H+ antiporters CAX1 and CAX2. FEBS Lett. 2005;579(12):2648–2656. doi: 10.1016/j.febslet.2005.03.085. [DOI] [PubMed] [Google Scholar]
- 19.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 21.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 23.Rosen BP, Tsuchiya T. Preparation of everted membrane vesicles from Escherichia coli for the measurement of calcium transport. Methods Enzymol. 1979;56:233–241. doi: 10.1016/0076-6879(79)56026-1. [DOI] [PubMed] [Google Scholar]
- 24.Wu J, Kaback HR. A general method for determining helix packing in membrane proteins in situ: Helices I and II are close to helix VII in the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 1996;93(25):14498–14502. doi: 10.1073/pnas.93.25.14498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.