Abstract
Medium-chain fatty acids (MCFAs, 4–12 carbons) are valuable as precursors to industrial chemicals and biofuels, but are not canonical products of microbial fatty acid synthesis. We engineered microbial production of the full range of even- and odd-chain–length MCFAs and found that MCFA production is limited by rapid, irreversible elongation of their acyl-ACP precursors. To address this limitation, we programmed an essential ketoacyl synthase to degrade in response to a chemical inducer, thereby slowing acyl-ACP elongation and redirecting flux from phospholipid synthesis to MCFA production. Our results show that induced protein degradation can be used to dynamically alter metabolic flux, and thereby increase the yield of a desired compound. The strategy reported herein should be widely useful in a range of metabolic engineering applications in which essential enzymes divert flux away from a desired product, as well as in the production of polyketides, bioplastics, and other recursively synthesized hydrocarbons for which chain-length control is desired.
Keywords: biochemicals, synthetic biology
Long-chain (16–18 carbon) fatty acids are required to synthesize the essential lipids that maintain bacterial membrane integrity (1). In-depth genetic and biochemical understanding of fatty acid synthesis in bacteria (2, 3) has made it possible to engineer microbes for increased fatty acid production, and catalyzed efforts to industrialize the process (4). As fatty acids can be transformed into a variety of biofuels and industrially useful oleo-chemicals, engineered microbes are a promising renewable alternative to petroleum-based chemical feedstocks (4–8).
Recent efforts have focused on engineering Escherichia coli fatty acid synthesis for production of free fatty acids (FFAs) (reviewed in ref. 4), alcohols (5), esters (6), and alkanes (7). Although most such studies have focused on long-chain (C14–C18) compounds because of their natural abundance, medium-chain fatty acids (MCFAs, 4–12 carbons), which are derived from low-abundance acyl-ACPs, are nevertheless valuable industrial chemicals (9–12), and their shorter chain lengths are associated with improved fuel quality (8, 13). Tailoring chain-length specificity in fatty-acid–producing microbes is therefore an important challenge in the production of renewable chemicals and biofuels.
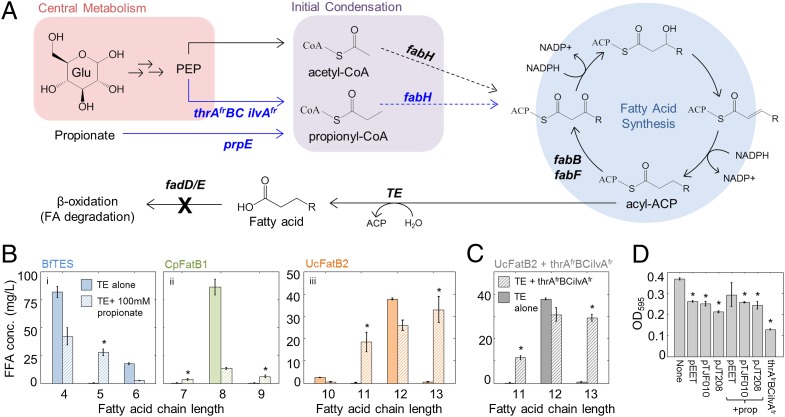
Chain-length control is exerted at many steps throughout fatty acid synthesis. Fatty acid synthesis begins with the condensation of acetyl-CoA and malonyl-ACP by the ketoacyl synthase (KAS) FabH to yield acetoacetyl-ACP, a four-carbon β-ketoacyl-ACP that is reduced to yield a four-carbon acyl-ACP (Fig. 1A). In subsequent rounds of fatty acid synthesis, this acyl-ACP is condensed with additional malonyl-ACP molecules by long-chain KAS enzymes FabB and FabF, then reduced to an acyl-ACP with two additional carbons. In this way, E. coli fatty acid synthesis builds acyl-ACPs two carbons at a time, in a recursive process yielding a range of even-chain–length acyl-ACPs (2). Odd-chain acyl-ACPs can also be produced when cellular propionyl-CoA levels are high, because of their incorporation in place of acetyl-CoA in the initial step of fatty acid synthesis (14–16) (Fig. 1A). In part because of KAS chain-length specificity, acyl-ACP elongation ends with the production of long-chain (16–18 carbon) acyl-ACPs, which are used by the cell to synthesize membrane lipids (reviewed in ref. 2).
Fig. 1.
Engineering production of all even- and odd-length MCFAs in E. coli. (A) The KAS FabH elongates acetyl-CoA to form a four-carbon β-ketoacyl ACP, which is reduced to C4 acyl-ACP. In subsequent rounds of fatty acid synthesis, the KAS’s FabB and FabF elongate acyl-ACPs two carbons at a time to yield a range of even-chain–length fatty acyl-ACPs. Incorporation of propionyl-CoA in place of acetyl-CoA causes production of odd-chain acyl-ACPs; propionyl-CoA can be produced from propionate (prpE) or from expression of a genetic cassette that increases flux through the isoleucine pathway (thrAfrBC ilvAfr) (15). Acyl-ACPs can be hydrolyzed to FFAs by an appropriate thioesterase (TE). Fatty acid degradation can be blocked by knocking out the β-oxidation enzymes fadD or fadE. (B) GC-MS analysis of FFA production by strain S001 [BL21*(DE3) ∆fadD] containing plasmid pEET (BfTES) (i), pTJF010 (CpFatB1) (ii), or pJT208 (UcFatB2) (iii) in M9 +0.5% glycerol alone (solid bars) or supplemented with 100 mM propionate (striped bars) 24 h after IPTG induction (n = 3, error bars = SEM). FFAs shown for the no-propionate experiments accounted for 67% (i), 85% (ii) and 72% (iii) of total FFAs (full chain-length profiles in Fig. S1). FFAs shown for the propionate experiments accounted for (i) 79%, (ii) 65%, and (iii) 84% of the total (Fig. S1). (C) Production of odd-chain FFAs by S001-pJT208-pCOLAthrAfrBCilvAfr (n = 3, error bars = SEM). FFAs shown accounted for 81% of total. Asterisks in B and C indicate significantly increased odd-chain production (P < 0.05, one-tailed Student t test). (D) Final OD595 of strains in this figure, with or without propionate supplementation (n = 3, error bars = SEM). Asterisks indicate decreased OD595 compared with strain S001 alone (P < 0.05, one-tailed Student t test).
Engineered fatty acid production in E. coli is typically achieved by expressing a cytosolic derivative of the E. coli thioesterase TesA (5), which hydrolyzes long-chain acyl-ACPs to yield long-chain free fatty acids (Fig. 1A) (17). High yields are a result of both the natural abundance of long-chain acyl-ACPs in the cell and thioesterase-mediated depletion of the long chain acyl-ACP pool, which feedback-inhibits upstream enzymes in fatty acid synthesis, such as FabH (16, 17). Although MCFAs can be produced by expressing thioesterases with substrate specificity for medium-chain acyl-ACPs (5, 18–21), yields are generally lower than for long-chain fatty acids, likely because medium-chain acyl-ACPs are not abundant (22, 23) and their depletion does not resolve an existing buildup of inhibitory long-chain acyl-ACPs.
In this work, we engineered E. coli to produce free fatty acids with all even and odd chain lengths from 4 to 13 carbons, and rationally modified fatty acid synthesis to favor the production of medium-chain acyl-ACPs. Treatment of MCFA-producing cells with the KAS inhibitor cerulenin (22) increased yields, demonstrating that MCFA production is limited by overly rapid acyl-ACP elongation. To test whether this strategy could be implemented genetically to increase the production of a specific MCFA, octanoic acid, we replaced the KAS FabF with a mutant incapable of elongating beyond eight carbons (24). We then engineered FabB to degrade in response to a chemical inducer (25), such that elongation beyond C8 acyl-ACP could be slowed on-demand, and flux redirected from lipid synthesis to octanoate production. These interventions increased octanoate yield and demonstrated the utility of inducible degradation to degrade essential genes and redirect metabolic flux (26). Our results demonstrate that altering the chain-length specificity of microbial fatty acid synthesis requires concerted changes in both the thioesterase and fatty acid synthesis machinery itself, and suggest strategies for the production of intermediate-length products in other recursive biosynthetic systems (27, 28).
Results
Our strategy for engineering MCFA synthesis was to first demonstrate the production of MCFAs with all even and odd chain lengths from 4 to 13 carbons, then identify factors that limit MCFA yield. After finding that elongation rates were too rapid for optimal MCFA production, we genetically engineered fatty acid synthesis to slow elongation in response to a chemical inducer, and thereby direct fatty acid synthesis toward the production of a specific MCFA, octanoic acid. Finally, we tested a set of knockout mutants for their ability to increase carbon flux into fatty acid synthesis and maximize yields in our engineered strain.
Comprehensive Production of all Even- and Odd-Chain MCFAs by Engineered E. coli.
We engineered production of MCFAs with 4–13 carbons in E. coli by varying the expressed thioesterase and adding propionate to the culture medium. Three thioesterases capable of hydrolyzing a range of medium-chain acyl-ACPs were chosen: BfTES [EET61113 from Bryantella formatexigens (21), CpFatB1 (20), and UcFatB2 (18)]. Strain S001 [BL21*(DE3) ∆fadD] (Table S1) was transformed with plasmids encoding codon-optimized versions of each thioesterase (pEET, pTJF010, and pJT208, respectively), grown in M9 + 0.5% glycerol, and induced with isopropyl-β-d-thiogalactopyranoside (IPTG) for 24 h (Materials and Methods). GC-MS analysis of free fatty acids showed that thioesterase expression increased the total moles of fatty acid produced (Fig. S1), and that each even-chain fatty acid from 4 to 12 carbons was produced by at least one thioesterase, with BfTES, CpFatB1, and UcFatB2 producing primarily butanoic, octanoic, and dodecanoic acids, respectively (Fig. 1B and Fig. S1). Propionate supplementation has been shown to cause odd-chain fatty acid production via incorporation of propionyl-CoA into the initial step of fatty acid synthesis (14, 19) (Fig. 1A). Addition of 100 mM propionate to the culture medium caused all three thioesterases to produce odd-chain fatty acids, with each odd-chain fatty acid in the 5–13 carbon range produced by at least one thioesterase (Fig. 1B and Fig. S1). These experiments demonstrated that the full range of 4–13 carbon MCFAs could be produced by engineered E. coli.
Odd-chain MCFAs could be produced without the need for propionate supplementation by genetically encoding a propionyl-CoA production pathway. Building on recent work in which endogenous propionyl-CoA production was engineered to synthesize a series of valuable small molecules (15, 29), we transformed S001-pJT208 with plasmid pCOLA-thrAfrBCilvAfr (15), which produces propionyl-CoA primarily by increasing flux through the isoleucine biosynthetic pathway (Fig. 1A). Strain S001-pJT208-pCOLA-thrAfrBCilvAfr produced the odd-chain fatty acids undecanoate and tridecanoate from glycerol as a sole carbon source (Fig. 1C), with similar selectivity to S001-pJT208 with added propionate (Fig. 1 B, iii). Thus, E. coli can be engineered to produce both even- and odd-chain fatty acids from a single carbon source, without the need for propionate supplementation.
Wild-Type Fatty Acid Elongation Rates Are Nonoptimal for MCFA Production.
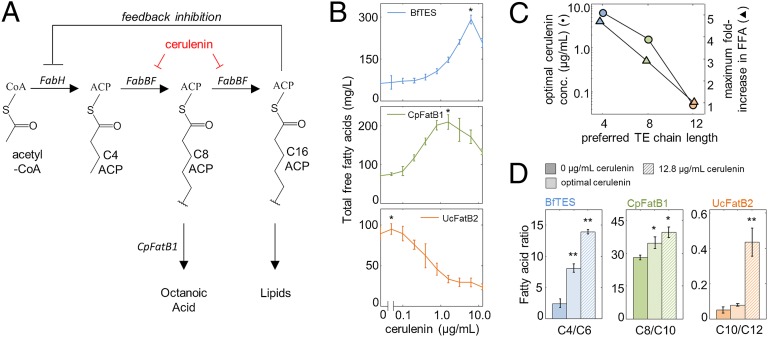
One challenge in MCFA production is the low concentration of medium-chain acyl-ACPs in growing cells (22, 23). Treatment of E. coli with cerulenin, an antibiotic that targets the KAS enzymes FabB and FabF, inhibits acyl-ACP elongation without inhibiting the initial condensing enzyme FabH (Fig. 2A) and causes accumulation of medium-chain acyl-ACPs in vivo (22). This accumulation is likely a result of two factors: (i) decreased elongation of medium-chain acyl-ACPs to long-chain acyl-ACPs; and (ii) the resulting decrease in long-chain acyl-ACPs, which would relieve feedback inhibition of FabH (16, 17) and increase flux into fatty acid synthesis (Fig. 2A). We hypothesized that there should be an optimal level of cerulenin at which elongation is slow enough to increase the concentration of medium-chain acyl-ACPs and rate of MCFA production, but not so slow that elongation itself becomes rate-limiting.
Fig. 2.
Chemical inhibition of fatty acid elongation can increase MCFA production. (A) Schematic diagram of fatty acid elongation. FabH performs the slow initial elongation step in fatty acid biosynthesis to generate C4-ACP; FabB and FabF more rapidly elongate the acyl-ACP 2 carbons at a time to produce 16–18 carbon acyl-ACPs, which are precursors to lipid synthesis and inhibit FabH more strongly than other acyl-ACPs (16, 17). Cerulenin inhibits FabB and FabF, but not FabH. (B) Total FFA produced by strain S001 expressing the indicated thioesterases over a range of cerulenin concentrations from 0 to 12.8 µg/mL, 24 h after IPTG induction, as measured by GC-MS (n = 3, error bars = SEM). Long-chain FFAs (C14–C18) accounted for ≤ 12 mg/L in any given measurement. Asterisks indicate the point at which mean fatty acid production is highest; the increase in production is significant for BfTES and CpFatB1 (P < 0.05, one-tailed Student t test), but not for UcFatB2. OD595 data are provided in Fig. S2. (C) Filled circles: cerulenin concentration at which fatty acid production is maximal as a function of each thioesterase’s preferred chain length. C4 for BfTES (blue); C8 for CpFatB1 (green); C12 for UcFatB2 (orange). Filled triangles indicate the maximum fold-increase in FFA production over the no-cerulenin control (calculated as the ratio of maximal FFA production to that of the no-cerulenin control). (D) Ratio of the two most abundant FFAs produced by each thioesterase (shorter/longer), as a function of cerulenin concentration (n = 3, error bars = SEM). Single asterisks indicate that the given bar is significantly different from the no-cerulenin control with P < 0.05 (two-tailed Student t test). Double asterisks indicate that the given bar is significantly different from both other bars in the panel.
Adding cerulenin to MCFA-producing cultures demonstrated that FFA yields produced by the shorter-chain thioesterases BfTES and CpFatB1 could be increased by inhibiting elongation (Fig. 2B). Yield increases came primarily from MCFAs rather than other fatty acids, because long-chain fatty acids were detected at < 12 mg/L in all samples, and biomass generally did not change with cerulenin concentration (Fig. S2). Cerulenin also increased the MCFA production rate (Fig. S3A). Given that cerulenin has been previously shown to increase the concentration of medium-chain acyl-ACPs (22), and assuming the MCFA production rate increases with the concentration of its substrate acyl-ACPs, one possible explanation of this result is that the increased MCFA production rate is because of an increased cellular concentration of medium-chain acyl-ACPs. These data suggested that wild-type fatty acid elongation rates are not optimal for the production of MCFAs in our strains and demonstrated, counterintuitively, that inhibiting fatty acid synthesis can increase the production of fatty acids.
The maximal increase in FFA yield was greater for shorter chain-length–specific thioesterases, as was the amount of cerulenin needed to achieve it (Fig. 2C); this is consistent with a model in which the distribution of acyl-ACPs is a function of elongation rate, with slower elongation rates yielding shorter acyl-ACP pools and enhancing production of shorter fatty acids. GC-MS analysis offered further evidence for this model. First, cerulenin treatment caused all three thioesterases to shift their production toward shorter products (Fig. 2D), which is most easily explained by an increase in the fraction of shorter acyl-ACPs in the cell. Second, although cerulenin decreased total and C12-specific FFA yields in the UcFatB2 strain, it increased the yield of C10 FFAs (Fig. S4), suggesting differential effects of cerulenin on shorter and longer acyl-ACP pools.
These results suggest that BfTES and CpFatB1’s FFA yields increase in response to cerulenin treatment because of a shift of the acyl-ACP distribution toward shorter (4–10 carbon) acyl-ACPs, which are more effectively hydrolyzed by these thioesterases; FFA production by UcFatB2, however (which prefers C12 acyl-ACPs), may fail to increase for the same reason. These apparent shifts in the acyl-ACP distribution are consistent with the expectation that cerulenin would simultaneously increase the rate at which acyl-ACPs are initiated by FabH and decrease the rate at which they elongate, and with previous results demonstrating that medium-chain acyl-ACPs accumulate in response to cerulenin treatment (22). In all, these results suggest that inhibiting fatty acid elongation increases the concentration of medium-chain acyl-ACPs with ≤ 10 carbons, a useful strategy for increasing MCFA yield.
Targeted Genetic Production of MCFAs via Inducible Degradation.
Having shown that inhibiting elongation can increase MCFA yield, we attempted to engineer production of a specific MCFA, octanoic acid, by implementing two genetic interventions: (i) replacing FabF with a mutant, FabF*, incapable of elongating beyond C8 because of the insertion of a bulky phenylalanine into its fatty acid binding pocket (24); and (ii) engineering FabB with a C-terminal SsrA DAS+4 tag, enabling inducible degradation by the E. coli ClpXP system via overexpression of the adaptor protein SspB (25, 30) (Fig. 3A). The inducible degradation system was necessary because fabB is required for unsaturated fatty acid synthesis, and is essential for growth on minimal media (31). We hypothesized that FabF* would allow high rates of octanoyl-ACP production, and that inducing degradation of FabB would slow elongation beyond C8 acyl-ACP, similar to the effect of cerulenin. This process should cause octanoyl-ACP to accumulate and increase fatty acid yields in strains expressing CpFatB1, which primarily produces octanoic acid (Fig. 1B and Fig. S1).
Fig. 3.
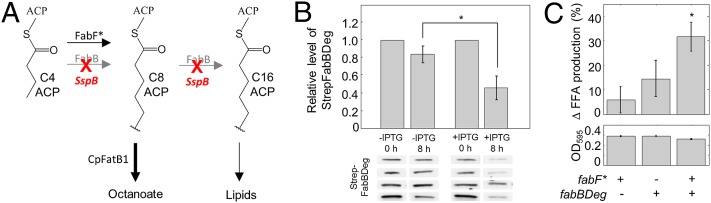
Engineering KAS for increased octanoic acid production. (A) Schematic indicating the elongation reactions carried out by ketoacyl synthases FabB and FabF. Wild-type FabB and FabF elongate acyl-ACPs with ≥ 4 carbons up to a length of 16–18 carbons. In our engineered system, FabBDeg can elongate up to 16–18 carbons but is degraded by the E. coli ClpXP system upon SspB expression. FabF* can only elongate up to 8 carbons. (B) StrepFabBDeg degradation in strain S007-SspBpET21b. Strain S007-SspBpET21b was induced (+IPTG, 1 mM) or not (−IPTG) at time 0 and StrepFabBDeg detected via Western blotting and densitometry at 0 and 8 h (n = 4, error bars = SEM). Loading was normalized to OD600, and reported values are normalized to StrepFabBDeg levels at t = 0 h. StrepFabBDeg bands are shown below the bar graph; original blots are shown in Fig. S5. An asterisk indicates a significant decrease in StrepFabBDeg between the ± IPTG samples at 8 h (P < 0.05, one-tailed Student t test). (C) FFA production [as determined by the Free Fatty Acids, Half Micro Test (Roche)] and final culture OD595 44 h after induction, for strains expressing CpFatB1 and SspB in S002 [BL21*(DE3) ΔfadE] with the indicated modifications (n = 24; error bars = SEM). FFA production is represented as the percent increase over FFA production in the parent strain (S002). An asterisk indicates a significant increase in FFA production compared with S002 (P < 0.05 by one-tailed Student t test).
Expressing CpFatB1 (pTJF038) and SspB (SspBpET21b) (25) in strain S003 [BL21*(DE3)ΔfadE fabF*] did not result in significantly increased FFA yields over those of S002 [BL21*(DE3)ΔfadE] (Fig. 3C), suggesting that the octanoyl-ACP generated by FabF* was not accumulating sufficiently to increase octanoate production, likely because of elongation by FabB. We therefore appended an SsrA DAS+4 degradation tag (25) to the endogenous fabB in both S002 and S003, generating strains S004 [BL21*(DE3) ΔfadE fabBDeg] and S005 [BL21*(DE3) ΔfadE fabF* fabBDeg], respectively. SspB-induced degradation of FabBDeg was demonstrated via Western blot of strain S007, a derivative of S004 in which FabBDeg is appended with an N-terminal strep tag to facilitate detection (Fig. 3B and Fig. S5).
Inducing SspB and CpFatB1 expression in strain S005, which contains both fabF* and fabBDeg, resulted in a significant, 32% increase in FFA yield to 118 mg/L and a 12% decrease in final OD595 compared with parent strain S002 (Fig. 3C). This finding suggests that FabB degradation shunts fatty acid synthesis to FabF* and increases the pool of octanoyl-ACP for hydrolysis by CpFatB1. Consistent with this, strain S005 (fabF* fabBDeg) had a greater rate of FFA production than strain S003 (fabF* alone) (Fig. S3B). Importantly, because fabBDeg alone (S004) does not significantly alter yield or OD595, it is unlikely that yield increases in the fabF* fabBDeg strain (S005) are because of general effects on growth or fatty acid synthesis caused by SspB synthesis or FabB degradation.
Optimization of the Engineered Strain.
We further optimized octanoic acid production in our engineered strain by: (i) screening a series of knockout mutations for their ability to increase yield, and (ii) titrating the level of SspB induction to optimize FabB degradation and FFA production by CpFatB1.
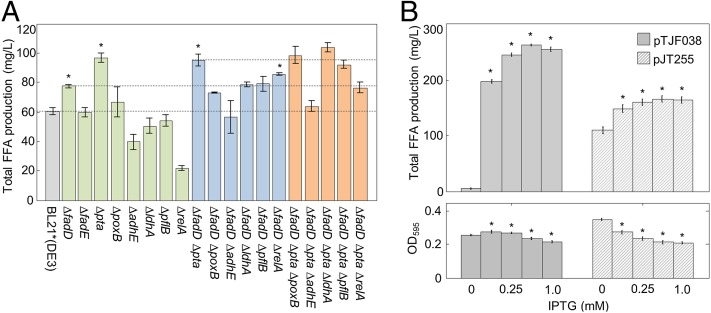
We expressed CpFatB1 in BL21*(DE3) derivatives with knockouts in fatty acid degradation (fadD, fadE), fermentation of undesirable products (pta, poxB, adhE, ldhA, pflB) (3), and the stringent response (relA), which inhibits lipid synthesis during starvation (32), and measured the total FFA production. Because strains with fadD and pta showed the greatest improvement in FFA among the single-knockout strains (Fig. 4A, green), we constructed all possible double mutants in a ΔfadD background (Fig. 4A, blue), as well as all possible triple mutants in a ΔfadD Δpta background (Fig. 4A, orange). The strain with the highest yield (ΔfadD Δpta ΔldhA) showed little increase in FFA production beyond that of the ΔfadD Δpta double knockout. ΔfadD and Δpta were therefore used to further improve FFA yields in our strains engineered with fabF* and fabBDeg.
Fig. 4.
Optimizing octanoic acid yields via metabolic knockouts and SspB titration. (A) Total FFA production in BL21*(DE3) knockout strains. Each strain was grown in M9 + 0.5% glucose, CpFatB1 was induced from pTJF010 with 1 mM IPTG for 44 h, and total FFA measured with the Free Fatty Acids, Half Micro Test (Roche) (n = 3, error bars = SEM). Green, blue, and orange bars indicate single knockouts, double knockouts, and triple knockouts, respectively. Dashed lines indicate fatty acid production in the parent strains BL21*(DE3), BL21*(DE3) ∆fadD, and BL21*(DE3) ∆fadD ∆pta. An asterisk indicates FFA production significantly greater than the parent strain (P < 0.05 by one-tailed Student t test). OD595 for these strains is shown in Fig. S6. (B) FFA production and OD595 as a function of IPTG concentration (0, 0.125, 0.25, 0.5 or 1.0 mM). CpFatB1 and SspB expressed from pTJF038 and SspBpET21b respectively were cotitrated in S006 [BL21*(DE3)ΔfadD Δpta ΔlacY fabF* fabBDeg] (solid gray bars) with IPTG for 24 h in M9 + 0.5% glucose (n = 18, error bars = SEM). SspB alone was titrated with IPTG in S006 expressing SspB from SspBpET21b and CpFatB1 from aTc-inducible pJT255 induced with 400 ng/mL aTc (striped bars, n = 18, error bars = SEM). An asterisk indicates significantly different FFA production, or altered OD595, versus the 0 mM IPTG control (P < 0.05 by two-tailed Student t test).
Our improved strain S006 [BL21*(DE3) ΔfadD Δpta ΔlacY fabf* fabBDeg], which included a ΔlacY mutation to allow titratable IPTG induction (33), increased FFA production ∼twofold over S005 to 263mg/L (Fig. 4B, solid bars) at the optimal level of IPTG induction of SspBpET21b and pTJF038 (CpFatB1). GC-MS analysis showed that FFA production by S006 was highly selective for octanoate (92.1 ± 0.2%) (Fig. S7), with a > 130 mg/L increase in octanoate production over the nonoptimized parent strain (S002). We estimated from OD595 data that S006 produced only 15 mg/L less lipid-bound long-chain fatty acid than S002 (SI Materials and Methods); our genetic interventions therefore increased the total moles of fatty acid produced primarily by increasing MCFA production.
Because IPTG induction controlled both CpFatB1 and SspB expression, it was unclear whether the increase in yield was a result of altering CpFatB1 expression, altering degradation, or a combination of the two; we therefore generated a strain in which we could induce CpFatB1 and SspB separately. Strain S006 was transformed with an anhydrotetracycline (aTc)-inducible, CpFatB1-expressing plasmid (pJT255) and an IPTG-inducible, SspB-expressing plasmid (SspBpET21b). This strain was induced with 400 ng/mL aTc, and IPTG titrated from 0 to 1 mM (Fig. 4B, striped bars). FFA yields increased significantly at lower doses of IPTG (110–166 mg/L) and OD595 decreased, but plateaued beyond this point. Our highest yield in this strain was 166 mg/L. Although this is lower than the yields achieved with pTJF038 and SspBpET21b, these results nevertheless demonstrate that optimizing inducible degradation of FabB can increase yields over those achieved through more traditional knockout strategies, and that it is important to tune degradation of FabB to achieve optimal production while limiting effects on cell growth.
Discussion
MCFAs and their derivatives are important precursors to high-quality fuel and industrial molecules (3, 8–12). In this work, we engineered the production of a range of MCFAs, and showed that their yields increase in response to chemical inhibition of fatty acid elongation. By engineering ketoacyl synthases to selectively block long-chain acyl-ACP elongation, we demonstrated that we can substantially increase the yield of a particular MCFA, octanoic acid. In principle, this strategy can be adapted for production of any MCFA with 4–13 carbons.
In addition to even-chain MCFA production, we were able to produce odd-chain MCFAs, which are industrially useful as plasticizers, as herbicides, and in the fragrance industry (11, 12, 34). Odd-chain fatty acid production has previously been achieved, but required propionate supplementation (14, 19). Endogenous propionyl-CoA production has also been achieved (15, 29), but not in conjunction with fatty acid production. We synthesized these strategies to engineer production of odd-chain MCFAs from a single carbon source in the absence of propionate. Importantly, this result was achieved using thioesterases known only to produce even-chain fatty acids, and some thioesterases produced odd chains better than others. Among the wide range of known even-chain thioesterases (21), some may therefore be capable of effectively or even selectively producing odd-chain fatty acids.
An important finding of our work was that application of the KAS inhibitor cerulenin increased the yield of some MCFAs, suggesting that the optimal rate of acyl-ACP elongation for MCFA production is slower than the wild-type elongation rate. Our results suggest that this yield increase is because of accumulation of medium-chain acyl-ACPs, which may be a consequence of KAS inhibition both (i) slowing loss of medium-chain acyl-ACPs to elongation, and (ii) indirectly increasing the rate of new acyl-ACP synthesis by FabH. Although the goal of metabolic engineering is often to increase flux through a naturally occurring pathway, our results suggest that in recursive systems like fatty acid synthesis, intermediate-length products may be produced more efficiently by slowing the native pathway.
We demonstrated that we could genetically engineer KASs to inhibit unwanted elongation of medium-chain acyl-ACPs, and thereby increase the yield of a specific MCFA, octanoic acid. Although a large body of work has been amassed on attempts to increase flux to fatty acid precursors (8, 35, 36), to prevent fatty acid degradation (5, 35), and to optimize the thioesterase (5, 8) (reviewed in refs. 3 and 4), there have been few attempts to engineer the fatty acid synthesis machinery itself. Reasons for this may include the toxicity of FabF overexpression (37) and the essentiality of FabB on minimal medium (31). Nevertheless, we were able to mimic the effect of cerulenin supplementation by coupling expression of the octanoyl-ACP–specific FabF* (24) with inducible degradation of FabB. These interventions presumably slowed the elongation of octanoyl-ACP produced by FabF*, thereby increasing octanoyl-ACP levels and octanoate production by CpFatB1. Through these means we achieved 12% theoretical yield of octanoate (Fig. 4B). This strategy should be generally applicable to other MCFAs, although the optimal degradation rate and specificity of FabF is likely to be chain-length–dependent.
Degradation tags have previously been used to decrease the basal level of metabolic enzymes (38) and to dynamically degrade regulatory proteins (39). We engineered our strains to inducibly degrade an essential enzyme and thereby redirect metabolic flux on demand. Inducible degradation is an attractive metabolic engineering strategy because it allows an essential gene’s activity to be decreased or knocked out entirely, without blocking the strain’s ability to grow. Inducible degradation of FabB allowed us to achieve our highest yields. This approach should be generally applicable to all essential metabolic genes in E. coli, opening up additional opportunities for flux modulation.
This study presents a strategy to increase fatty acid production that combines KAS manipulation and more traditional metabolic engineering. It demonstrates the utility of engineering the fatty acid synthesis machinery itself, and not just the thioesterase or other enzymes that feed into the pathway, to enhance the yield of specific fatty acids. Given the interest in producing diverse fatty acid-derived compounds for use as biofuels and industrial chemicals, it is likely that the engineering of KASs and the other enzymes of fatty acid synthesis will become increasingly important in microbial fatty acid engineering. We expect the strategy of dynamically inhibiting essential enzymes to be broadly useful as a tool for metabolic engineering and synthetic biology. In particular, dynamically inhibiting polymer elongation may be a useful approach for increasing production or tuning the chain length of other valuable molecules synthesized by recursive biosynthetic pathways, such as iteratively synthesized polyketides, polysaccharides, and bioplastics.
Materials and Methods
Plasmid and Strain Construction.
pEET and SspBpET21b were obtained from refs. 21 and 25, respectively. All other plasmids and strains used in this work, and the primers used to generate them, are listed in Tables S1 and S2. Details on plasmid and strain construction are provided in SI Materials and Methods.
Growth and Induction.
Briefly, fatty acid production experiments were performed at 30 °C either in shake flasks or 96-well plates inoculated from overnight LB cultures. Shake flasks (Fig. 1) (M9 + 0.5% glycerol) were grown to OD 0.4–0.6 and induced with 1 mM IPTG. The 96-well plates (M9 + 0.5% glucose) were inoculated, grown for 3.5 h, and induced with 1 mM IPTG (unless otherwise noted) and aTc (where indicated). Full experimental details are provided in SI Materials and Methods.
OD and Enzymatic FFA Measurements.
The OD595 of 100 μL of each culture was measured in 200-μL 96-well flat-bottom, nontreated, sterile polystyrene plates (Corning) on a Victor3V 1420 Multilabel Counter (Perkin-Elmer) using a 595/60-nm filter. FFAs in 5 μL of culture were measured using the enzymatic Free Fatty Acid Half Micro Test (Roche) according to the manufacturer’s instructions, scaled down to 1/10 volume in a 96-well plate. OD600 was measured using an Ultrospec 10 spectrophotometer (Amersham).
Fatty Acid Identification and Quantification Using GC-MS.
Fatty acids were extracted from 400 µL of acidified culture with ethyl acetate and esterified with ethanol. Fatty ethyl esters were extracted with hexane and run on an Agilent GC-MS 5975/7890 on an HP-5MS column. Compounds were identified via GC retention times and mass spectra, and quantified by normalizing to an internal standard (ethyl pentadecanoate) and comparing against a standard curve. See SI Materials and Methods for details.
Supplementary Material
Acknowledgments
We thank R. T. Sauer, K. L. Prather, P. J. Reilly, B. J. Nikolau, D. F. Savage, and D. C. MacKellar for plasmids and reagents; G. Webster, D. C. MacKellar, and D. C. Ducat for reading the manuscript; D. C. Ducat for helpful conversations throughout this work; and C P. Johnson for advice and assistance with mass spectrometry. This work was conducted with support from the Advanced Research Projects Agency–Energy ‘Electrofuels’ Collaborative Agreement DE-AR0000079 (to P.A.S.); National Science Foundation Graduate Research fellowships (to J.P.T. and T.J.F.); a Herchel Smith Graduate Research Fellowship (to J.P.T.); and the Ruth L. Kirschtein Nationalesearch Service Award program of Harvard Catalyst | The Harvard Clinical and Translational Science Center Award UL1 RR 025758 and financial contributions from Harvard University and its affiliated academic health care centers (to T.J.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.F.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307129110/-/DCSupplemental.
References
- 1.Cronan JE. Bacterial membrane lipids: Where do we stand? Annu Rev Microbiol. 2003;57:203–224. doi: 10.1146/annurev.micro.57.030502.090851. [DOI] [PubMed] [Google Scholar]
- 2.Magnuson K, Jackowski S, Rock CO, Cronan JE., Jr Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol Rev. 1993;57(3):522–542. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Handke P, Lynch SA, Gill RT. Application and engineering of fatty acid biosynthesis in Escherichia coli for advanced fuels and chemicals. Metab Eng. 2011;13(1):28–37. doi: 10.1016/j.ymben.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Lennen RM, Pfleger BF. Engineering Escherichia coli to synthesize free fatty acids. Trends Biotechnol. 2012;30(12):659–667. doi: 10.1016/j.tibtech.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steen EJ, et al. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463(7280):559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 6.Zhang F, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat Biotechnol. 2012;30(4):354–359. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- 7.Schirmer A, Rude MA, Li X, Popova E, del Cardayre SB. Microbial biosynthesis of alkanes. Science. 2010;329(5991):559–562. doi: 10.1126/science.1187936. [DOI] [PubMed] [Google Scholar]
- 8.Liu T, Vora H, Khosla C. Quantitative analysis and engineering of fatty acid biosynthesis in E. coli. Metab Eng. 2010;12(4):378–386. doi: 10.1016/j.ymben.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Ohlrogge JB. Design of new plant products: Engineering of fatty acid metabolism. Plant Physiol. 1994;104(3):821–826. doi: 10.1104/pp.104.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang ST. Bioprocessing for Value-Added Products from Renewable Resources: New Technologies and Applications. Amsterdam: Elsevier Science; 2006. [Google Scholar]
- 11.Braude GL. Preparation of Polymeric Plasticizers from Tall Oil Fatty Acids. 1967. US Patent 3337594. [Google Scholar]
- 12.Bauer K, Garbe D, Surburg H. Common Fragrance and Flavor Materials. Weinheim, Germany: Wiley-VCH; 2008. pp. 8–20. [Google Scholar]
- 13.Knothe G. Improving biodiesel fuel properties by modifying fatty ester composition. Energy Environ Sci. 2009;2(7):759–766. [Google Scholar]
- 14.Ingram LO, Chevalier LS, Gabba EJ, Ley KD, Winters K. Propionate-induced synthesis of odd-chain-length fatty acids by Escherichia coli. J Bacteriol. 1977;131(3):1023–1025. doi: 10.1128/jb.131.3.1023-1025.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tseng HC, Prather KL. Controlled biosynthesis of odd-chain fuels and chemicals via engineered modular metabolic pathways. Proc Natl Acad Sci USA. 2012;109(44):17925–17930. doi: 10.1073/pnas.1209002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heath RJ, Rock CO. Inhibition of beta-ketoacyl-acyl carrier protein synthase III (FabH) by acyl-acyl carrier protein in Escherichia coli. J Biol Chem. 1996;271(18):10996–11000. doi: 10.1074/jbc.271.18.10996. [DOI] [PubMed] [Google Scholar]
- 17.Jiang P, Cronan JE., Jr Inhibition of fatty acid synthesis in Escherichia coli in the absence of phospholipid synthesis and release of inhibition by thioesterase action. J Bacteriol. 1994;176(10):2814–2821. doi: 10.1128/jb.176.10.2814-2821.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voelker TA, Davies HM. Alteration of the specificity and regulation of fatty acid synthesis of Escherichia coli by expression of a plant medium-chain acyl-acyl carrier protein thioesterase. J Bacteriol. 1994;176(23):7320–7327. doi: 10.1128/jb.176.23.7320-7327.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dellomonaco C, Clomburg JM, Miller EN, Gonzalez R. Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals. Nature. 2011;476(7360):355–359. doi: 10.1038/nature10333. [DOI] [PubMed] [Google Scholar]
- 20.Dehesh K, Edwards P, Hayes T, Cranmer AM, Fillatti J. Two novel thioesterases are key determinants of the bimodal distribution of acyl chain length of Cuphea palustris seed oil. Plant Physiol. 1996;110(1):203–210. doi: 10.1104/pp.110.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jing F, et al. Phylogenetic and experimental characterization of an acyl-ACP thioesterase family reveals significant diversity in enzymatic specificity and activity. BMC Biochem. 2011;12:44. doi: 10.1186/1471-2091-12-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackowski S, Rock CO. Acetoacetyl-acyl carrier protein synthase, a potential regulator of fatty acid biosynthesis in bacteria. J Biol Chem. 1987;262(16):7927–7931. [PubMed] [Google Scholar]
- 23.Rock CO, Jackowski S. Regulation of phospholipid synthesis in Escherichia coli. Composition of the acyl-acyl carrier protein pool in vivo. J Biol Chem. 1982;257(18):10759–10765. [PubMed] [Google Scholar]
- 24.Val D, Banu G, Seshadri K, Lindqvist Y, Dehesh K. Re-engineering ketoacyl synthase specificity. Structure. 2000;8(6):565–566. doi: 10.1016/s0969-2126(00)00146-5. [DOI] [PubMed] [Google Scholar]
- 25.McGinness KE, Baker TA, Sauer RT. Engineering controllable protein degradation. Mol Cell. 2006;22(5):701–707. doi: 10.1016/j.molcel.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 26.Way JC, Davis JH. Methods and Molecules for Yield Improvement Involving Metabolic Engineering. 2010. US Patent Appl PCT/US2010/036902. [Google Scholar]
- 27.Staunton J, Weissman KJ. Polyketide biosynthesis: A millennium review. Nat Prod Rep. 2001;18(4):380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 28.Suriyamongkol P, Weselake R, Narine S, Moloney M, Shah S. Biotechnological approaches for the production of polyhydroxyalkanoates in microorganisms and plants—A review. Biotechnol Adv. 2007;25(2):148–175. doi: 10.1016/j.biotechadv.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Tseng HC, Harwell CL, Martin CH, Prather KL. Biosynthesis of chiral 3-hydroxyvalerate from single propionate-unrelated carbon sources in metabolically engineered E. coli. Microb Cell Fact. 2010;9:96. doi: 10.1186/1475-2859-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farrell CM, Grossman AD, Sauer RT. Cytoplasmic degradation of ssrA-tagged proteins. Mol Microbiol. 2005;57(6):1750–1761. doi: 10.1111/j.1365-2958.2005.04798.x. [DOI] [PubMed] [Google Scholar]
- 31.Cronan JE, Jr, Birge CH, Vagelos PR. Evidence for two genes specifically involved in unsaturated fatty acid biosynthesis in Escherichia coli. J Bacteriol. 1969;100(2):601–604. doi: 10.1128/jb.100.2.601-604.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heath RJ, Jackowski S, Rock CO. Guanosine tetraphosphate inhibition of fatty acid and phospholipid synthesis in Escherichia coli is relieved by overexpression of glycerol-3-phosphate acyltransferase (plsB) J Biol Chem. 1994;269(42):26584–26590. [PubMed] [Google Scholar]
- 33.Jensen PRWH, Westerhoff HV, Michelsen O. The use of lac-type promoters in control analysis. Eur J Biochem. 1993;211(1–2):181–191. doi: 10.1111/j.1432-1033.1993.tb19885.x. [DOI] [PubMed] [Google Scholar]
- 34.Miller TW. Natural herbicides and amendments for organic weed control. In: Felsot AS, Racke KD, editors. Crop Protection Products for Organic Agriculture. Washington, DC: ACS Publications; 2006. pp. 174–175. [Google Scholar]
- 35.Zhang F, et al. Enhancing fatty acid production by the expression of the regulatory transcription factor FadR. Metab Eng. 2012;14(6):653–660. doi: 10.1016/j.ymben.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 36.Davis MS, Solbiati J, Cronan JE., Jr Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherichia coli. J Biol Chem. 2000;275(37):28593–28598. doi: 10.1074/jbc.M004756200. [DOI] [PubMed] [Google Scholar]
- 37.Subrahmanyam S, Cronan JE., Jr Overproduction of a functional fatty acid biosynthetic enzyme blocks fatty acid synthesis in Escherichia coli. J Bacteriol. 1998;180(17):4596–4602. doi: 10.1128/jb.180.17.4596-4602.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doroshenko VG, et al. Construction of an L-phenylalanine-producing tyrosine-prototrophic Escherichia coli strain using tyrA ssrA-like tagged alleles. Biotechnol Lett. 2010;32(8):1117–1121. doi: 10.1007/s10529-010-0265-1. [DOI] [PubMed] [Google Scholar]
- 39.Huang D, Holtz WJ, Maharbiz MM. A genetic bistable switch utilizing nonlinear protein degradation. J Biol Eng. 2012;6(1):9. doi: 10.1186/1754-1611-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.