Abstract
The serotonin transporter (SERT) is a major regulator of serotonergic neurotransmission and anxiety-related behaviors. SERT is expressed in two alternative polyadenylation forms that differ by an evolutionarily conserved element in the 3′ untranslated region of its mRNA. Expression of SERT mRNA containing the distal polyadenylation element is associated with decreased anxiety-related behaviors in mice and humans, suggesting that this element has behaviorally relevant modulatory effects on SERT expression. We have identified heterogeneous nuclear ribonucleoprotein K (hnRNPK), a protein known to integrate multiple signal transduction pathways with gene expression, as a SERT distal polyadenylation element binding protein. This interaction is functionally meaningful because genetic manipulation of hnRNPK alters expression of the SERT protein. Furthermore, the trophic factor S100β induces Src-family kinase-mediated tyrosine phosphorylation of hnRNPK and increased SERT expression. These results identify a previously unknown mechanism of regulated SERT expression and provide a putative mechanism by which the SERT distal polyadenylation element modulates anxiety-related behaviors.
Alternative polyadenylation creates structural and functional diversity in the expression of protein coding messenger RNA (mRNA) through the formation of mRNA species that differ in the sequence content of their 3′ untranslated regions (UTRs) (1–3). The 3′ UTRs of mRNAs have become recognized as major sources of regulated gene expression through sequence-specific microRNA (miR) and ribonucleoprotein (RNP) binding that can alter the stability, translational activity, and subcellular localization of mRNAs (4–6). As a result, alternative polyadenylation forms of mRNA species may display discrete biological properties (7, 8).
The serotonin transporter (SERT; SLC6A4) plays a central role in regulating serotonergic neurotransmission, and the mRNA coding for SERT occurs in two phylogenetically conserved alternative polyadenylation forms that differ by the presence or absence of an ∼125-bp element (9, 10). We have reported that expression of the longer, or distal, polyadenylation form of the SERT mRNA confers anxiolytic properties on behavior (11, 12). Specifically, males express higher levels of the distal polyadenylation form of SERT than females, who display a two- to threefold higher risk for anxiety disorders than males; a common polymorphism that is associated with expression of the SERT polyadenylation forms in human postmortem brain is also associated with risk for panic disorder, trait anxiety, and the ability of human subjects to retain fear extinction learning, a validated endophenotype of anxiety and its disorders (11, 12). Finally, treatment of mice with the SERT-selective antidepressant/anxiolytic drug fluoxetine, which has also been shown to enhance fear extinction retention, increases expression of the distal polyadenylation form of SERT (12, 13).
To advance understanding of how the SERT distal polyadenylation sequence modulates anxiety-related behavior, we screened for proteins that bind to RNA probes containing the SERT distal polyadenylation sequence and identified heterogeneous nuclear ribonucleoprotein K (hnRNPK) in lysates from several cell and tissue types. RNA immunoprecipitation (RIP) confirmed that hnRNPK interacts selectively with SERT mRNA species containing the distal polyadenylation element in cultured cells and mouse brain. Genetic manipulation of hnRNPK expression in cells led to altered expression of SERT protein, demonstrating a role for hnRNPK in the physiologic regulation of SERT expression. Finally, treatment of both serotonergic RN46A cells and C6 astroglioma cells with the neurotrophic protein S100β (S100B) increased tyrosine phosphorylation of hnRNPK and expression of SERT protein, both of which were blocked by the Src-family kinase inhibitor 3-(4-chlorophenyl) 1-(1,1-dimethylethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine (PP2). These results identify a previously unknown mechanism for the regulated expression of SERT protein and may provide an explanation for associations of the distal polyadenylation sequence with extinction retention and other anxiety-related behaviors.
Results
Identifying SERT Distal Polyadenylation Sequence-Binding Activity.
The sequence of the SERT distal polyadenylation element is highly conserved from mouse to human (Fig. 1), providing a priori evidence that it has important biological properties (14). To screen for the presence of proteins that bind to the SERT distal polyadenylation element, we synthesized two overlapping biotinylated oligoribonucleotide probes representing the human sequence and spanning its most highly conserved region (Fig. 1A and Fig. S1). These probes were combined with cytosolic extracts from mouse midbrain tissue containing the raphe nuclei, mouse frontal cortex, rat C6 astroglioma cells (15), primary mouse astrocytes, and human embryonic kidney (HEK) cells. Proteins binding the biotinylated probes were cross-linked by exposure to UV light at 254 nm. All four lysates contained similar proteins that were cross-linked to the biotlinylated probes (Fig. 2). Sequence specificity of the cross-linking reaction was assessed by inclusion of a 100-fold excess unbiotinylated oligoribonucleotide probe. Although the precise pattern of cross-linking varied by probe and lysate source, both probes were cross-linked to proteins running at ∼60 kDa and 76 kDa by SDS polyacrylamide gel electrophoresis (PAGE) that were competed away by a 100-fold excess unlabeled probe in cultured cell lysates (SERT1 probe) or all lysates (SERT2 probe). These results demonstrate that similar protein(s) specifically bind the SERT distal polyadenylation element in diverse cell and tissue types.
Fig. 1.
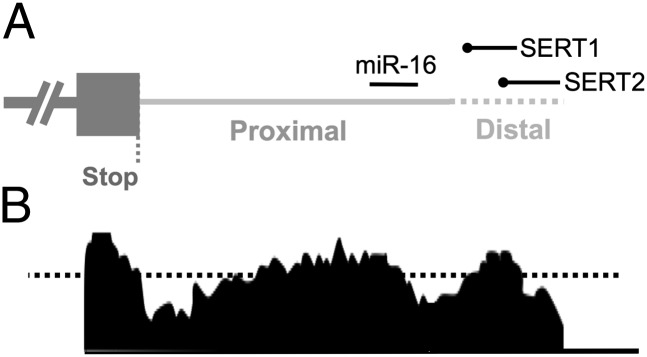
Schematic representation of the SERT 3′ UTR. (A) The 3′ end of the SERT transcript is presented 5′ to 3′ from left to right. The proximal (solid gray) and distal (dashed gray) polyadenylation elements are labeled. The binding site for miR-16 is in the proximal element present in all SERT mRNA species. The positions of the overlapping SERT1 probe and SERT2 probe are labeled with the 5′ biotin-TEG modification represented by black circles. (B) Conservation of the SERT 3′ UTR. Mouse/human conservation is presented based on alignment from the VISTA browser (http://pipeline.lbl.gov/cgi-bin/gateway2). The dotted black line represents the default threshold for functionally relevant conserved noncoding elements (70% identity across a sliding 100-bp window). See also Fig. S1.
Fig. 2.
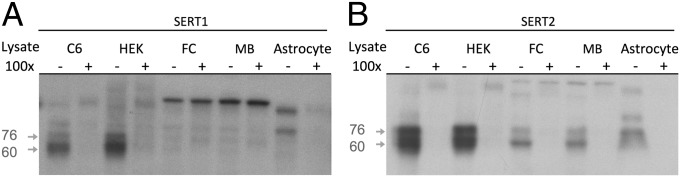
UV cross-linking identifies SERT distal polyadenylation element binding proteins in cytosolic extracts. (A and B) Biotinylated oligoribonucleotide probes (1 nM) were incubated with cytosolic extracts from: C6 rat astroglioma cells (C6), HEK, mouse frontal cortex (FC), mouse midbrain tissue (MB), and primary mouse astrocyte culture (astrocyte). Probes were covalently cross-linked to bound proteins by exposure to UV light (254 nm). Specific binding was assessed by comparing reactions with probe only (−) to reactions with 100-fold excess (100 nm) unlabeled probe (+). (A) SERT1 probe. Prominent specific bands of ∼60 kDa and 76 kDa (gray arrows) were present in C6 and HEK extracts. Less consistent cross-linking of similar bands was detected in FC, midbrain, and primary astrocyte extracts. (B) SERT2 probe. Consistent specific cross-linking to bands of ∼60 kDa and 76 kDa was observed in all extracts.
hnRNPK Is a SERT Distal Polyadenylation Sequence Binding Protein.
We performed avidin affinity chromatography on the UV cross-linked C6 lysates by using both biotinylated probes to purify the distal polyadenylation sequence binding proteins. Both probes substantially enriched proteins migrating at ∼60 kDa and ∼76 kDa (Fig. S2). These bands were excised from silver-stained polyacrylamide gels run with each probe followed by trypsin digest. Peptides from the two bands purified with each probe were separately analyzed by liquid chromatography/mass spectroscopy (LC/MS). Multiple peptides consistent with hnRNPK were present in samples from both the ∼60-kDa and ∼76-kDa bands purified with both probes. hnRNPK is a multifunctional docking protein that integrates several signal transduction pathways with a variety of gene expression regulatory mechanisms including transcription and translation by binding to chromatin and RNA, respectively (16). The presence of multiple peptides consistent with hnRNPK in all assayed samples combined with hnRNPK’s known RNA binding properties led us to prioritize it for subsequent analyses.
hnRNPK Interacts with SERT mRNA in Cells and Brain Tissue.
To confirm that hnRNPK binds to the endogenous SERT mRNA, we performed RIP on lysates from C6 (15) and SV40 T-antigen transformed serotonergic rat embryonic mid-brain cells (RN46A) (17), both of which express SERT mRNA, and whole brain from male mice. Using quantitative PCR (qPCR), we assessed the interaction of hnRNPK with specific mRNA species by comparing the abundance of target sequences in hnRNPK immunoprecipitates with their abundance in lysates before RIP (Fig. 3). Total SERT mRNA was quantified by using primers targeting the proximal polyadenylation sequence, which is present in all SERT mRNA species. Distally polyadenylated SERT mRNA was quantified by using primers targeting the distal polyadenylation sequence. SERT mRNA was significantly enriched in hnRNPK immunoprecipitates compared with the hnRNPK mRNA used as a negative control in C6, RN46A, and mouse brain lysates (Fig. 3). Moreover, the distal SERT polyadenylation sequence was enriched significantly more than total SERT mRNA in C6 and RN46A cells, suggesting that hnRNPK interacts selectively with the subset of SERT mRNA species that contain that element (Fig. 3 A and B). In male mouse brain, the degree of enrichment of the SERT distal polyadenylation sequence was similar to total SERT (Fig. 3C), which suggests that a greater proportion of total SERT mRNA is distally polyadenylated in brain tissue than in C6 or RN46A cells, which is consistent with our earlier report that male brain has a particularly high proportion of distal versus proximal polyadenylated SERT mRNA compared with females (11). As a positive control, we assessed the degree of enrichment of the cyclooxygenase 2 (COX2) mRNA, which is known to bind and be regulated by hnRNPK (18). We found that the relative enrichment of the SERT distal polyadenylation sequence in hnRNPK immunoprecipitates was similar to COX2 (Fig. 3A). To assess whether hnRNPK interacts with other serotonin-related genes, we performed qPCR by using primers targeting the vesicular monoamine transporter 2 (VMAT2), tryptophan hydroxylase (TPH) 1 and 2, and the serotonin 1A and 2B receptors (5HT1AR, 5HT2BR). All of the tested messages that were present in inputs from C6 and RN46A cells were immunoprecipitated by the hnRNPK antibody significantly more than the hnRNPK message as a negative control (Fig. S3), suggesting that hnRNPK may have coordinated effects on the serotonergic phenotype.
Fig. 3.
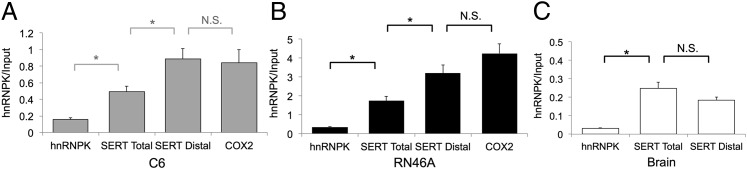
RNA immunoprecipitation confirms hnRNPK as a SERT distal polyadenylation element binding protein. Cell or tissue lysates were immunoprecipitated (IP) with an anti-hnRNPK antibody. Relative abundance of target sequences in hnRNPK immunoprecipitates (hnRNPK) was assessed by qPCR and compared with their abundance in total lysate (input) to determine the degree of hnRNPK interaction with the various target mRNA species. The hnRNPK and COX2 mRNA’s were quantified as negative and positive controls, respectively. (A) C6 cells. The proximal polyadenylation sequence of SERT present in all SERT mRNAs (SERT Total) is significantly enriched in hnRNPK IPs relative to the hnRNPK mRNA. SERT mRNAs containing the distal polyadenylation element (SERT Distal) are significantly more enriched in hnRNPK IPs than total SERT mRNA. COX2, a known hnRNPK-binding mRNA, is enriched to a similar degree as SERT distal. *P < 0.05, N.S., nonsignificant. Error bars, SEM. (B) RN46A cells. Total SERT mRNA is enriched in hnRNPK immunoprecipitates relative to hnRNPK and distal polyadenylation element-containing messages are significantly enriched relative to total SERT mRNA. *P < 0.05. Error bars, SEM. (C) Mouse brain homogenates. Both total and distal-containing SERT mRNAs are enriched relative to hnRNPK but are similar to one another. *P < 0.05, N.S., nonsignificant. Error bars, SEM.
Genetic Manipulation of hnRNPK Alters Expression of SERT Protein.
Having confirmed that hnRNPK binds to the physiologic SERT mRNA, we assessed the functional implications of this interaction. We manipulated hnRNPK expression by using mammalian expression plasmids containing the wild-type hnRNPK ORF or an hnRNPK-targeting shRNA construct. Transfection of C6 and RN46A cells with wild-type hnRNPK significantly increased its own expression and that of SERT protein (Fig. 4). Conversely, the shRNA construct significantly knocked down hnRNPK protein levels and also reduced expression of SERT protein. These results demonstrate that hnRNPK regulates expression of SERT. Binding of hnRNPK to the 3′ UTRs of different genes can either increase or decrease their expression (18–21). In the case of the SERT 3′ UTR, it appears that interaction with hnRNPK increases expression of the SERT protein.
Fig. 4.
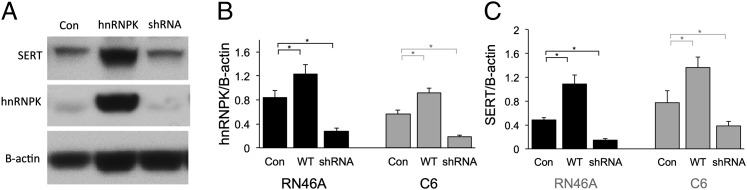
Genetic manipulation of hnRNPK alters SERT expression. (A) Representative Western blot of lysates from untreated cells (Con), cells transfected with wild-type hnRNPK (hnRNPK), and cells transfected with an anti-hnRNPK shRNA (shRNA) construct. (B) Quantification of hnRNPK protein after genetic manipulation confirms that transfection of wild-type hnRNPK increased its expression, and shRNA knocked down its expression in both C6 and RN46A cells. Optical density (OD) of hnRPK immunoblots were normalized to the OD of B-actin for each lane. All treatments were run three times in triplicate. *P < 0.05. Error bars, SEM. (C) Quantification of SERT protein after genetic manipulation of hnRNPK. SERT protein is significantly increased by hnRNPK overexpression and reduced by hnRNPK knockdown in C6 and RN46A cells. OD of SERT immunoblots were normalized to the OD of B-actin for each lane. All treatments were run three times in triplicate. *P < 0.05. Error bars, SEM.
S100B Induces SERT Expression in C6 Cells Through Src-Family Kinase Phosphorylation of hnRNPK.
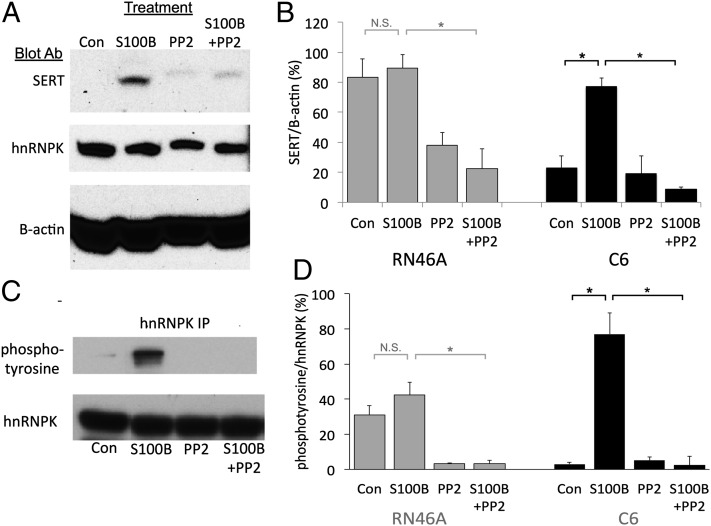
Having established that hnRNPK binds to the SERT distal polyadenylation element and facilitates expression of SERT protein, we wished to understand the physiologic regulation of this process. As an initial step, we focused on the effects of S100B in C6 cells. S100B is a dimeric calcium binding protein that has both intracellular and extracellular signaling properties (22, 23). We focused on S100B as a possible regulator of SERT expression because it is enriched in astrocytes in the brain and in culture where it acts as a paracrine and autocrine trophic factor (24, 25), S100B can increase SERT expression in hippocampal and locus coeruleus neurons, S100B has been linked to the antidepressant effects of fluoxetine (26–30), and has trophic effects on serotonergic neurons (31). Moreover, S100B regulates COX2 expression through activation of hnRNPK by binding to the 3′ UTR of its message (18). Treating C6 cells with S100B (1 nM) for 48 h caused increased expression of SERT protein without altering the expression of hnRNPK protein (Fig. 5 A and B). S100B increased SERT expression through a translational mechanism, as S100B did not increase expression of total SERT mRNA (Fig. S4).
Fig. 5.
S100B induces tyrosine phosphorylation of hnRNPK and SERT translation. (A) Representative immunoblot of lysates from C6 cells. Untreated cells (Con), cells treated with 1 nM S100b (S100B), 10 µM 4-Amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimidine (PP2), or both (S100B+PP2) were cultured for 48 h. (B) Quantification of SERT protein in C6 and RN46A cell lysates. OD of immunblots for SERT was normalized to B-actin for each sample. S100B induced expression of SERT protein (S100B), which was blocked by PP2 (S100B+PP2). PP2 alone had no effect on expression of SERT (PP2). *P < 0.05. Error bars, SEM. (C) Representative immunoblot of anti-hnRNPK immunoprecipitated C6 proteins. Blot was probed with an antiphosphotyrosine antibody then stripped and reprobed with the anti-hnRNPK antibody. (D) Quantification of tyrosine phosphorylation in hnRNPK immunoprecipitates. The ODs of antiphosphotyrosine bands were divided by the OD of anti-hnRNPK bands. S100B induced tyrosine phosphorylation of hnRNPK (S100B), which was blocked by coincubation with PP2 (S100B+PP2) in C6 cells. PP2 alone did not affect tyrosine phosphorylation of hnRNPK (PP2). *P < 0.05. Error bars, SEM.
In astrocytes and pulmonary artery smooth muscle cells, S100B effects on morphology and migration are mediated by the Src kinase, which can alter hnRNPK activity through phosphorylation of several of its tyrosine residues (32–35). Stimulation of SERT translation by S100B in C6 cells also requires tyrosine phosphorylation of hnRNPK by a Src-family kinase because S100B had no effects on SERT expression in the presence of PP2, an antagonist of Src-family kinases. We confirmed that S100B induces tyrosine phosphorylation of hnRNPK in C6 cells by probing a Western blot of anti-hnRNPK immunoprecipitates from S100B-treated C6 cells with an antiphosphotyrosine antibody (Fig. 5 C and D). S100B-induced tyrosine phosphorylation of hnRNPK was also blocked by PP2.
hnRNPK Interaction with SERT mRNA Reduces miR-16 Binding.
miR-16 binds to and represses translation of the SERT mRNA (26, 36). The target sequence for miR-16 binding to the SERT mRNA occurs in the proximal portion of the 3′ UTR present in both forms of the SERT mRNA (Fig. 1A and Fig. S1) adjacent to the distal sequence element that binds hnRNPK. The gene for COX2 displays regulation of mRNA expression through miR-16, which is antagonized by hnRNPK binding to an adjacent site in its 3′ UTR (18). We therefore tested the effects of hnRNPK expression on binding of miR-16 to the SERT 3′ UTR to determine whether SERT expression displayed similar reciprocal regulation by miR-16 and hnRNPK (Fig. 6A). A biotinylated SERT 3′ UTR containing the distal polyadenylation element was produced by in vitro transcription with the inclusion of biotin-UTP and mixed with radiolabeled miR-16 and lysates from C6 cells. After pulldown of the 3′ UTR using avidin beads, miR-16 binding was assessed by scintillation counting. Binding of miR-16 to the biotinylated SERT 3′ UTR was significantly greater when mixed with C6 lysates after hnRNPK knockdown than when mixed with mock-transfected C6 lysates (Fig. 6B). This effect was similar to what we observed with the COX2 3′ UTR as a positive control (Fig. 6C). miR-16 pulldown was sequence specific because there was significantly less miR-16 present in pulldowns run without 3′ UTRs added, and the reverse complement sequence of miR-16 also displayed significantly lower binding than miR-16 itself. Thus, it appears that hnRNPK regulates SERT expression by antagonizing miR-16 binding and derepressing translation. However, S100B has also been reported to enhance expression of SERT protein in neurons by reducing expression of miR-16, raising the possibility that similar reductions of miR-16 by S100B in C6 cells contribute independently to the increases we see in SERT protein (26, 27). To determine whether S100B decreased expression of miR-16 in C6 cells, we quantified miR-16 by qPCR in C6 and RN46A cells with and without S100B treatment (Fig. 6D and Fig. S5). In C6 and RN46A cells, S100B increased expression of miR-16, suggesting that decreased miR-16 expression does not explain S100B’s effects on SERT expression in those cell types; rather, the data indicate that SERT has COX2-like reciprocal regulation of translation by hnRNPK and miR-16.
Fig. 6.
hnRNPK inhibits miR-16 binding to the SERT 3′ UTR. (A) Schematic diagram of miR-16 binding assay. Biotinylated 3′ UTRs were produced by in vitro transcription including biotinylated UTP and mixed with [32P]–end-labeled miR-16 or anti–miR-16 in the presence of C6 cell lysates. The 3′ UTRs were pulled down with avidin beads, and miR-16 binding to 3′ UTR’s was assessed by scintillation counting of bead pellets after washing. (B) miR-16 binding to the SERT 3′ UTR. Knockdown of hnRNPK in C6 cell lysates (hnRNPK shRNA) increases binding of miR-16 to the SERT 3′ UTR compared with control C6 lysates (Control). Binding of miR-16 is specific because there is significantly less miR-16 pulldown in the absence of biotinylated 3′ UTR (no 3′ UTR) or when the reverse complement of miR-16 is radiolabeled (Anti-miR-16). ***P < 0.005, #P < 0.001 vs. Control. Error bars, SEM. (C) miR-16 binding to the COX2 3′ UTR. The effects of hnRNPK knockdown on miR-16 binding to the COX2 3′ UTR as a positive control are shown. Reciprocal binding of miR-16 and hnRNPK has been reported for COX2. ***P < 0.005, #P < 0.001 vs. Control. Error bars, SEM. (D) S100B induces expression of miR-16 in C6 cells. miR-16 and GAPDH were quantified in C6 cell lysates. Normalized miR-16 expression (miR-16/GAPDH) is increased by the same S100B treatment that induces SERT translation. ***P < 0.005. Error bars, SEM.
Discussion
In this report, we have characterized the regulatory properties of the alternative polyadenylation element of the SERT mRNA. In an unbiased screen for proteins that selectively bind the distal polyadenylation sequence, we identified hnRNPK in extracts from physiologically diverse cells and tissues. Furthermore, we have demonstrated that hnRNPK binds to SERT mRNA, expression of SERT protein increases with tyrosine phosphorylation of hnRNPK, and a major astrocyte-derived trophic factor, S100B, can regulate this process. These results have identified an important form of regulation of SERT expression and provide an initial step to understanding the apparent anxiolytic properties of the SERT distal polyadenylation element.
hnRNPK is a ubiquitously expressed protein containing discreet domains involved in DNA and RNA binding, protein–protein interactions, and regulation by a number of signal transduction pathways (16, 37). hnRNPK contains three K protein homology (KH) domains that are named after hnRNPK in which they were first identified. These domains facilitate interaction with both regulatory regions in the genomic structure of genes and with transcribed mRNA species, although the specific sequences that hnRNPK targets for binding can vary greatly (38, 39), preventing bioinformatic target prediction. A number of other proteins contain KH binding domains and could therefore also target the SERT distal polyadenylation element; however, we did not identify any other known KH proteins in our LC/MS data. Our results suggest that hnRNPK can interact with other messages involved in serotonin signaling, suggesting that it may coordinate a functional program of serotonergic gene expression, but this possibility must be investigated further.
hnRNPK contains proline-rich domains that interact with Src homology 3 (SH3) domains in a number of other proteins. In addition to localizing hnRNPK to specific subcellular compartments, these protein–protein interactions also facilitate regulation of hnRNPK by multiple kinases, including the Src-family tyrosine kinases and serine/threonine kinases such as protein kinase C, extracellular signal-regulated kinase 1/2, and c-Jun N-terminal kinase. The diversity of binding partners and regulatory mechanisms for hnRNPK suggest that the translational control of SERT may be open to an array of physiologic signals potentially including serotonin itself and additional trophic factors other than S100B (20, 40–43).
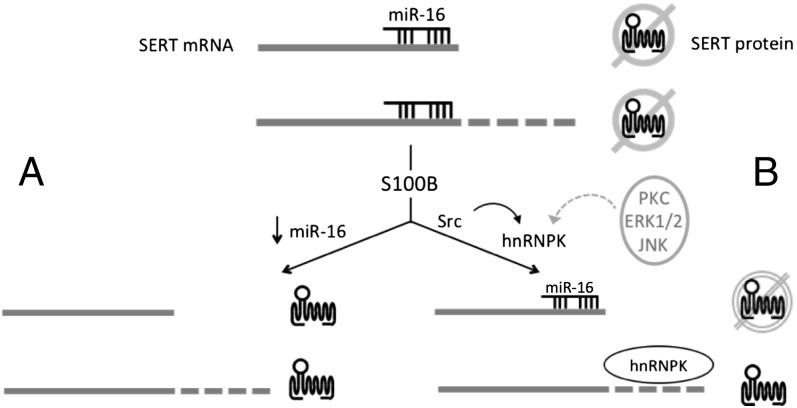
The 3′ UTR of the SERT mRNA contains a target sequence for miR-16 in its proximal portion present in all SERT messages; binding of miR-16 to this target represses translation of the SERT mRNA (26, 36) (Fig. 7). Nonserotonergic neurons in the locus coeruleus and hippocampus can express SERT in response to treatments that reduce miR-16 expression in those cells including S100B (26, 27) (Fig. 7A). This exogenous expression of SERT has been linked to the antidepressant effects of fluoxetine (26, 44). Our results demonstrate that distally polyadenylated SERT mRNAs possess an additional regulatory mechanism through binding of hnRNPK that displaces miR-16 and derepresses their translation (Fig. 7B). The fact that we identified hnRNPK as a SERT mRNA-binding protein using distal element-specific probes and the fact that the distal polyadenylation element is more highly enriched by hnRNPK-specific RIP than the coding region suggest that this mechanism preferentially targets distal polyadenylated SERT mRNA and, therefore, contributes to their apparent anxiolytic properties; however, our experiments do not conclusively rule out a role for hnRNPK regulation of proximally polyadenylated messages. Reciprocal regulation of SERT expression by miR-16 and hnRNPK is similar to regulation of COX2 expression (18). Remarkably, this coordinate regulation of COX2 expression is also mediated by S100B, which activates hnRNPK binding and decreases miR-16 binding to its 3′ UTR (18), suggesting that this mechanism of translational regulation may apply to additional genes. This possibility is supported by our findings that hnRNPK interacts with the messages of other serotonin-related genes.
Fig. 7.
Diversity of translational regulation of SERT expression through alternative polyadenylation. Under basal conditions in cells expressing miR-16 translation of both proximal and distal polyadenylation forms of the SERT mRNA is blocked as indicated by slashed out topological model of SERT protein. (A) In some cell types (26, 27), S100B decreases expression of miR-16 derepressing translation of SERT protein from both proximal and distal polyadenylated mRNAs. (B) hnRNPK confers additional regulation of translation that our data suggests is specific to distal polyadenylated SERT mRNAs; however, we cannot rule out that hnRNPK may also regulate translation of proximal polyadenylated SERT mRNA as reflected in B by the dashed circle. S100B activates a Src-family kinase, which tyrosine phosphorylates hnRNPK, displacing miR-16 from distal polyadenylated messages and activating their translation. Additional signal transduction mechanisms can also modify hnRNPK function and may provide additional diversity of SERT translational regulation (dotted gray arrow).
Although SERT is generally described as a transporter involved in reuptake of serotonin into cells that produce and release serotonin, expression of SERT has been described in cells that do not produce serotonin. Uptake of exogenous serotonin by thalamocortical neurons, which transiently express SERT during early postnatal development, is required for normal development of somatosensory cortex (45, 46). More recently, locus coeruleus and hippocampal neurons have been shown to display induced SERT translation through reduced expression of miR-16 in response to fluoxetine treatment (26, 27). In this report, we have shown that hnRNPK can induce expression of SERT in embryonic midbrain-derived serotonergic RN46A cells as well as nonserotonergic cell types, including astrocytic C6 cells, through modulation of miR-16 effects on SERT translation. Further studies in additional cell types will be required to determine the extent of this regulatory mechanism for SERT expression.
Expression of the distal polyadenylation sequence is associated with enhanced fear extinction retention, reduced risk for panic disorder, and chronic treatment with fluoxetine. These results are all consistent with the distal polyadenylation sequence regulating anxiety-related emotional responses. The results of this study provide a putative molecular mechanism by which these anxiolytic effects may occur; however, definitive demonstration of the anxiolytic properties of the distal polyadenylation sequence and hnRNPK-based regulation of SERT expression will require in vivo manipulation of the SERT polyadenylation forms.
Experimental Procedures
Detailed methods including antibodies, suppliers, and oligonucleotide sequences are available in SI Experimental Procedures. All experiments using animals were appoved by the Weill Cornell Institutional Animal Care and Use Committee.
Cell Culture.
Rat C6 astroglioma cells and HEK cells were obtained from American Type Culture Collection. Primary cortical astrocyte cultures were prepared from postnatal day (P)1–P3 mouse pups (47). RN46A cells were a generous gift from Scott Whittemore (Department of Neurological Surgery, University of Louisville School of Medicine, Louisville, KY) (17).
Transfections.
Cells were plated 24 h before transfection using Lipofectamine 2000 (Life Technologies) for 48 h before analyses (details of constructs and transfection protocol are available in SI Experimental Procedures).
Ribonucleotide Probe Cross-Linking Assays.
Probes.
Overlapping ribonucleotide (RNA) probes comprising the most highly conserved portion of the human SERT distal polyadenylation element were synthesized with a 5′ biotin modification and tetra-ethyleneglycol (TEG; 15 atom) spacer (IDT).
Preparation of cytosolic protein extracts.
Protein extracts were prepared from adult mouse brain and cultured cells. Mouse brains were dissected to obtain midbrain tissue including the serotonergic raphe nuclei or nonraphe containing frontal cortex. Cultured cells were trypsinized, harvested, and briefly centrifuged (500 × g for 5 min) to collect cells. Tissue or cells were homogenized in 2 volumes of ice-cold lysis buffer by using a polytron and then centrifuged at 14,000 × g for 30 min at 4 °C to remove debris. Supernatants were then divided into aliquots at a final concentration of 10 mg/mL in lysis buffer after determining the protein concentration with BCA protein assay kit (Pierce), and stored at −80 °C.
UV cross-linking.
Cytosolic extracts (10 µg) were combined with biotinlyated RNA probe (1 nM final concentration) in the presence or absence of 100× (100 nM) unlabeled probe in 1.5-cc microfuge tubes. Samples were UV cross-linked (254 nm, 1,650 mJ of total energy; Stratalinker, Agilent Technologies) on ice. Samples were then run on SDS/PAGE, cross-linked complexes were transferred to polyvinylidene fluoride membrane, and biotinlyated probes were detected using Brightstar biodetect (Ambion) by following the manufacturer's instructions.
Avidin affinity purification of cross-linked proteins.
Cytosolic extracts from C6 cells (15 mg of protein per reaction) were cross-linked to probes as described above, diluted into binding buffer, mixed with 50 μL of monomeric avidin magnetic beads, and incubated overnight at 4 °C with mixing. Proteins were eluted by incubating with 2 mM d-biotin in PBS for 10 min at room temperature. Eluates were pooled and run on SDS/PAGE. Designated bands were excised and subjected to in-gel tryptic digestion.
LC/MS.
Tryptic digest products were injected onto an LC/MS system consisting of an 1100 series HPLC, HPLC-Chip Cube MS interface, and 6520 Series Q-TOF mass spectrometer (Agilent Technologies). Swiss-Prot database (November 2011 release) was searched for tryptic peptides.
RNA immunoprecipitation.
Homogenates from one 10-cm culture dish (∼107 cells) or 100 mg of mouse brain tissue were used for immunoprecipitations. Magna RIP reagents (Millipore) were used according to the manufacturer’s standard protocol for all samples.
Quantitative PCR.
Species-specific assays were developed to target the distal polyadenylation element, the protein coding region of the SERT mRNA, additional assays targeted other genes involved in serotonin signaling, and hnRNPK and COX2 were negative and positive controls, respectively. All assays displayed specific and highly efficient (>95% per cycle) amplification of target sequences. The relative abundance of target sequences in immunoprecipitates was compared with their abundance in diluted input samples (IPs were volume adjusted to allow direct comparison of input and immunoprecipitated samples) for quantitative evaluation of target sequence immunoprecipitation.
Western blotting and immunoprecipitation.
For Western blotting and immunoprecipitation, protein lysates were prepared in RIPA lysis buffer and run on SDS/PAGE. Immunoreactive bands were detected by using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific). For immunoprecipitation, protein lysates (100 µg) were incubated with antibody and 30 µL of agarose beads at 4 °C overnight. Resulting immunocomplexes were washed five times with RIPA buffer and run on SDS/PAGE.
miR-16 Binding Assay.
The 3′ UTRs of the human serotonin transporter and COX2 were in vitro transcribed in the presence of biotin UTP. Synthetic oligoribonucleotides representing miR-16 or its reverse complement anti–miR-16 were end labeled with [32P]phosphate. miRs, 3′ UTRs, and C6 cellular extracts were mixed, and binding of oligoribonucleotides to the 3′ UTRs was assessed by pulldown using magnetic streptavidin-coated beads and scintillation counting.
Data Analysis.
All experiments were run in duplicate with triplicate measures within each experiment. Averaged data values are presented as mean ± SEM. All statistical analyses used unpaired Student’s t tests between conditions labeled in figures and/or as specified in figure legends.
Supplementary Material
Acknowledgments
This work was supported by The Hartwell Foundation (C.E.G.), National Institutes of Health Grant MH091401 (to C.E.G.), and National Natural Science Foundation of China Grant 31270874 (to G.X.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301485110/-/DCSupplemental.
References
- 1.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43(6):853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lutz CS, Moreira A. Alternative mRNA polyadenylation in eukaryotes: An effective regulator of gene expression. Wiley Interdiscip Rev RNA. 2011;2(1):22–31. doi: 10.1002/wrna.47. [DOI] [PubMed] [Google Scholar]
- 3.MacDonald CC, McMahon KW. Tissue-specific mechanisms of alternative polyadenylation: Testis, brain, and beyond. Wiley Interdiscip Rev RNA. 2010;1(3):494–501. doi: 10.1002/wrna.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Standart N, Jackson RJ. Regulation of translation by specific protein/mRNA interactions. Biochimie. 1994;76(9):867–879. doi: 10.1016/0300-9084(94)90189-9. [DOI] [PubMed] [Google Scholar]
- 5.Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci. 2003;28(4):182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 6.Goldie BJ, Cairns MJ. Post-transcriptional trafficking and regulation of neuronal gene expression. Mol Neurobiol. 2012;45(1):99–108. doi: 10.1007/s12035-011-8222-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oe S, Yoneda Y. Cytoplasmic polyadenylation element-like sequences are involved in dendritic targeting of BDNF mRNA in hippocampal neurons. FEBS Lett. 2010;584(15):3424–3430. doi: 10.1016/j.febslet.2010.06.040. [DOI] [PubMed] [Google Scholar]
- 8.An JJ, et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134(1):175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Battersby S, et al. Presence of multiple functional polyadenylation signals and a single nucleotide polymorphism in the 3′ untranslated region of the human serotonin transporter gene. J Neurochem. 1999;72(4):1384–1388. doi: 10.1046/j.1471-4159.1999.721384.x. [DOI] [PubMed] [Google Scholar]
- 10.Heils A, et al. Functional promoter and polyadenylation site mapping of the human serotonin (5-HT) transporter gene. J Neural Transm. 1995;102(3):247–254. doi: 10.1007/BF01281159. [DOI] [PubMed] [Google Scholar]
- 11.Gyawali S, et al. Association of a polyadenylation polymorphism in the serotonin transporter and panic disorder. Biol Psychiatry. 2010;67(4):331–338. doi: 10.1016/j.biopsych.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartley CA, et al. Serotonin transporter polyadenylation polymorphism modulates the retention of fear extinction memory. Proc Natl Acad Sci USA. 2012;109(14):5493–5498. doi: 10.1073/pnas.1202044109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karpova NN, et al. Fear erasure in mice requires synergy between antidepressant drugs and extinction training. Science. 2011;334(6063):1731–1734. doi: 10.1126/science.1214592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pennacchio LA, et al. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science. 2001;294(5540):169–173. doi: 10.1126/science.1064852. [DOI] [PubMed] [Google Scholar]
- 15.Benda P, Lightbody J, Sato G, Levine L, Sweet W. Differentiated rat glial cell strain in tissue culture. Science. 1968;161(3839):370–371. doi: 10.1126/science.161.3839.370. [DOI] [PubMed] [Google Scholar]
- 16.Bomsztyk K, Denisenko O, Ostrowski J. hnRNP K: One protein multiple processes. Bioessays. 2004;26(6):629–638. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- 17.White LA, et al. Distinct regulatory pathways control neurofilament expression and neurotransmitter synthesis in immortalized serotonergic neurons. J Neurosci. 1994;14(11 Pt 1):6744–6753. doi: 10.1523/JNEUROSCI.14-11-06744.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shanmugam N, Reddy MA, Natarajan R. Distinct roles of heterogeneous nuclear ribonuclear protein K and microRNA-16 in cyclooxygenase-2 RNA stability induced by S100b, a ligand of the receptor for advanced glycation end products. J Biol Chem. 2008;283(52):36221–36233. doi: 10.1074/jbc.M806322200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sataranatarajan K, Lee MJ, Mariappan MM, Feliers D. PKCdelta regulates the stimulation of vascular endothelial factor mRNA translation by angiotensin II through hnRNP K. Cell Signal. 2008;20(5):969–977. doi: 10.1016/j.cellsig.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Habelhah H, et al. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat Cell Biol. 2001;3(3):325–330. doi: 10.1038/35060131. [DOI] [PubMed] [Google Scholar]
- 21.Mukhopadhyay NK, et al. Heterogeneous nuclear ribonucleoprotein K is a novel regulator of androgen receptor translation. Cancer Res. 2009;69(6):2210–2218. doi: 10.1158/0008-5472.CAN-08-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donato R. S100: A multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int J Biochem Cell Biol. 2001;33(7):637–668. doi: 10.1016/s1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- 23.Donato R, et al. S100B’s double life: Intracellular regulator and extracellular signal. Biochim Biophys Acta. 2009;1793(6):1008–1022. doi: 10.1016/j.bbamcr.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 24.Van Eldik LJ, Wainwright MS. The Janus face of glial-derived S100B: Beneficial and detrimental functions in the brain. Restor Neurol Neurosci. 2003;21(3-4):97–108. [PubMed] [Google Scholar]
- 25.Zimmer DB, Cornwall EH, Landar A, Song W. The S100 protein family: History, function, and expression. Brain Res Bull. 1995;37(4):417–429. doi: 10.1016/0361-9230(95)00040-2. [DOI] [PubMed] [Google Scholar]
- 26.Baudry A, Mouillet-Richard S, Schneider B, Launay JM, Kellermann O. miR-16 targets the serotonin transporter: A new facet for adaptive responses to antidepressants. Science. 2010;329(5998):1537–1541. doi: 10.1126/science.1193692. [DOI] [PubMed] [Google Scholar]
- 27.Launay JM, Mouillet-Richard S, Baudry A, Pietri M, Kellermann O. Raphe-mediated signals control the hippocampal response to SRI antidepressants via miR-16. Transcult Psychiatry. 2011;1:e56. doi: 10.1038/tp.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tramontina AC, et al. Secretion of S100B, an astrocyte-derived neurotrophic protein, is stimulated by fluoxetine via a mechanism independent of serotonin. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32(6):1580–1583. doi: 10.1016/j.pnpbp.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Manev R, Uz T, Manev H. Fluoxetine increases the content of neurotrophic protein S100beta in the rat hippocampus. Eur J Pharmacol. 2001;420(2-3):R1–R2. doi: 10.1016/s0014-2999(01)00989-x. [DOI] [PubMed] [Google Scholar]
- 30.Akhisaroglu M, Manev R, Akhisaroglu E, Uz T, Manev H. Both aging and chronic fluoxetine increase S100B content in the mouse hippocampus. Neuroreport. 2003;14(11):1471–1473. doi: 10.1097/00001756-200308060-00013. [DOI] [PubMed] [Google Scholar]
- 31.Azmitia EC, Dolan K, Whitaker-Azmitia PM. S-100B but not NGF, EGF, insulin or calmodulin is a CNS serotonergic growth factor. Brain Res. 1990;516(2):354–356. doi: 10.1016/0006-8993(90)90942-5. [DOI] [PubMed] [Google Scholar]
- 32.Brozzi F, Arcuri C, Giambanco I, Donato R. S100B Protein Regulates Astrocyte Shape and Migration via Interaction with Src Kinase: IMPLICATIONS FOR ASTROCYTE DEVELOPMENT, ACTIVATION, AND TUMOR GROWTH. J Biol Chem. 2009;284(13):8797–8811. doi: 10.1074/jbc.M805897200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddy MA, et al. Key role of Src kinase in S100B-induced activation of the receptor for advanced glycation end products in vascular smooth muscle cells. J Biol Chem. 2006;281(19):13685–13693. doi: 10.1074/jbc.M511425200. [DOI] [PubMed] [Google Scholar]
- 34.Ostareck-Lederer A, et al. c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol Cell Biol. 2002;22(13):4535–4543. doi: 10.1128/MCB.22.13.4535-4543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ostrowski J, et al. Role of tyrosine phosphorylation in the regulation of the interaction of heterogenous nuclear ribonucleoprotein K protein with its protein and RNA partners. J Biol Chem. 2000;275(5):3619–3628. doi: 10.1074/jbc.275.5.3619. [DOI] [PubMed] [Google Scholar]
- 36.Moya PR, Wendland JR, Salemme J, Fried RL, Murphy DL. miR-15a and miR-16 regulate serotonin transporter expression in human placental and rat brain raphe cells. Int J Neuropsychopharmacol. 2013;16(3):621–629. doi: 10.1017/S1461145712000454. [DOI] [PubMed] [Google Scholar]
- 37.Bomsztyk K, Van Seuningen I, Suzuki H, Denisenko O, Ostrowski J. Diverse molecular interactions of the hnRNP K protein. FEBS Lett. 1997;403(2):113–115. doi: 10.1016/s0014-5793(97)00041-0. [DOI] [PubMed] [Google Scholar]
- 38.Paziewska A, Wyrwicz LS, Bujnicki JM, Bomsztyk K, Ostrowski J. Cooperative binding of the hnRNP K three KH domains to mRNA targets. FEBS Lett. 2004;577(1-2):134–140. doi: 10.1016/j.febslet.2004.08.086. [DOI] [PubMed] [Google Scholar]
- 39.Thisted T, Lyakhov DL, Liebhaber SA. Optimized RNA targets of two closely related triple KH domain proteins, heterogeneous nuclear ribonucleoprotein K and alphaCP-2KL, suggest Distinct modes of RNA recognition. J Biol Chem. 2001;276(20):17484–17496. doi: 10.1074/jbc.M010594200. [DOI] [PubMed] [Google Scholar]
- 40.Assender JW, Irenius E, Fredholm BB. 5-Hydroxytryptamine, angiotensin and bradykinin transiently increase intracellular calcium concentrations and PKC-alpha activity, but do not induce mitogenesis in human vascular smooth muscle cells. Acta Physiol Scand. 1997;160(3):207–217. doi: 10.1046/j.1365-201X.1997.00157.x. [DOI] [PubMed] [Google Scholar]
- 41.Gil C, Najib A, Aguilera J. Serotonin transport is modulated differently by tetanus toxin and growth factors. Neurochem Int. 2003;42(7):535–542. doi: 10.1016/s0197-0186(02)00187-0. [DOI] [PubMed] [Google Scholar]
- 42.Schullery DS, et al. Regulated interaction of protein kinase Cdelta with the heterogeneous nuclear ribonucleoprotein K protein. J Biol Chem. 1999;274(21):15101–15109. doi: 10.1074/jbc.274.21.15101. [DOI] [PubMed] [Google Scholar]
- 43.Habelhah H, et al. Identification of new JNK substrate using ATP pocket mutant JNK and a corresponding ATP analogue. J Biol Chem. 2001;276(21):18090–18095. doi: 10.1074/jbc.M011396200. [DOI] [PubMed] [Google Scholar]
- 44.Baudry A, Mouillet-Richard S, Launay JM, Kellermann O. New views on antidepressant action. Curr Opin Neurobiol. 2011;21(6):858–865. doi: 10.1016/j.conb.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 45.Lebrand C, et al. Transient uptake and storage of serotonin in developing thalamic neurons. Neuron. 1996;17(5):823–835. doi: 10.1016/s0896-6273(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 46.Lebrand C, et al. Transient developmental expression of monoamine transporters in the rodent forebrain. J Comp Neurol. 1998;401(4):506–524. [PubMed] [Google Scholar]
- 47.Yamasaki K, et al. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum Mol Genet. 2003;12(8):837–847. doi: 10.1093/hmg/ddg106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.