Abstract
The presence of acetylated histone H3K56 (H3K56ac) in human ES cells (ESCs) correlates positively with the binding of Nanog, Sox2, and Oct4 (NSO) transcription factors at their target gene promoters. However, the function of H3K56ac there has been unclear. We now report that Oct4 interacts with H3K56ac in mouse ESC nuclear extracts and that perturbing H3K56 acetylation decreases Oct4–H3 binding. This interaction is likely to be direct because it can be recapitulated in vitro in an H3K56ac-dependent manner and is functionally important because H3K56ac combines with NSO factors in chromatin immunoprecipitation sequencing to mark the regions associated with pluripotency better than NSO alone. Moreover, reducing H3K56ac by short hairpin Asf1a decreases expression of pluripotency-related markers and increases expression of differentiation-related ones. Therefore, our data suggest that H3K56ac plays a central role in binding to Oct4 to promote the pluripotency of ESCs.
The core transcriptional network, which contains transcription factors, chromatin regulators, and signaling proteins, helps determine self-renewal and pluripotency in ES cells (ESCs) and cell fate upon their differentiation (1, 2). This network is defined in part through the action of the key transcriptional regulators Nanog, Sox2, and Oct4 (NSO) that recognize defined DNA sequence motifs at target genes of the core transcriptional network. There, the key regulators are involved in the activation of pluripotency-related genes and repression of differentiation related ones (3–5). Consequently, changes in the expression levels of the key factors perturb this transcriptional circuitry, leading to the loss of pluripotency (6). Of the key factors, Oct4, a homeodomain transcription factor of the POU family, is unique (7). Although ectopic expression of four factors (Oct4, Sox2, Klf4, and c-Myc) together can generate induced pluripotent stem cells from somatic cells (8), only Oct4 is essential and sufficient in reprogramming adult neural stem cells to pluripotency (9).
Our previous work using Agilent microarrays found that histone H3K56ac is associated with upstream regulatory DNA sequences of the core transcriptional network in human ESCs (10). H3K56ac is conserved from yeast to mammals and is very abundant in yeast, marking ∼30% of total histone H3. Yeast H3K56ac is involved in gene transcription (11), replication-independent histone assembly (12), DNA repair (13), silencing at heterochromatin (14), transcription elongation (15), and life span regulation (16). However, its role in mammalian cells where H3K56ac marks less than 1% of total H3 (10, 17, 18) is less clear and is the focus of this article. Remarkably, we found that H3K56ac interacts with Oct4 both in vivo in mouse ESCs and directly in vitro. This interaction is in addition to that provided by the Oct4 recognition element in DNA. Our data indicate that H3K56ac has a central function in ESCs, where it binds the key transcription factor Oct4 to promote pluripotency.
Results
H3K56ac Presence Is Correlated with That of Key Transcriptional Regulators in Mouse ESCs.
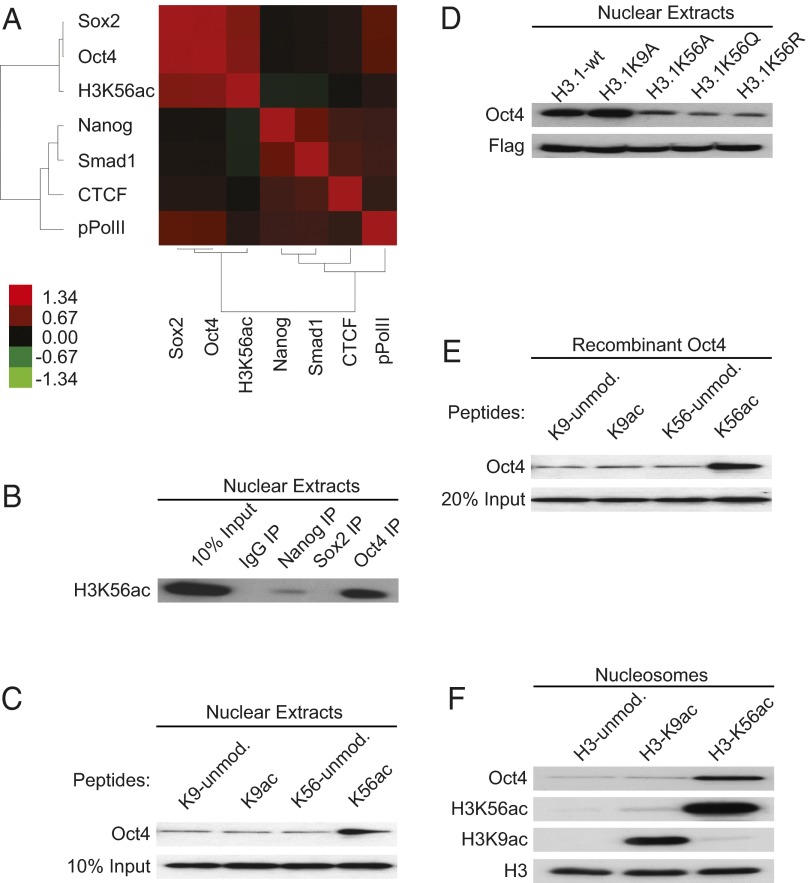
To determine whether H3K56ac is associated with the pluripotency network in mouse ESCs, we used massively parallel ChIP-DNA sequencing using Illumina HiSeq-2000 to examine the genome-wide distribution of H3K56ac in E14Tg2a ESCs. Approximately 85.5 million reads uniquely mapped to the mouse genome and 92,549 H3K56ac significantly enriched regions were identified. We then wished to determine whether H3K56ac presence is associated with the key transcriptional factors genome-wide. By comparing the correlation of H3K56ac and known transcription factors using our H3K56ac chromatin immunoprecipitation sequencing (ChIP-Seq) results and published data (4), we found using Cistrome (19) that H3K56ac correlates most positively with the presence of Oct4 and Sox2 and much less so with Nanog (Fig. 1A; Pearson’s correlation values in SI Appendix, Table S1). This is consistent with the frequent association of Oct4 and Sox2 at their regulatory regions (4, 20) and the less abundant interaction of these factors with Nanog (21, 22). Therefore, mouse H3K56ac correlates best genome wide with the presence of Oct4 and Sox2.
Fig. 1.
H3K56ac interaction with Oct4 in mouse E14Tg2a cells and in vitro. (A) Correlation heat map: the degree of target cooccupancy of factors in the mouse genome is shown. Pearson’s correlation values were calculated by Cistrome (19). Red indicates more frequent colocalization of each pair of factors as shown using Cluster 3.0 (47). (B) Co-IP shows the interactions between IgG, Nanog, Sox2, and Oct4 with H3K56ac in nuclear extracts of E14Tg2a cells. A total of 2 μg of α-IgG, α-Nanog, α-Sox2, and α-Oct4 antibodies was used for each co-IP and immunoblotted with antibody against H3K56ac. Ten percent E14Tg2a cell nuclear extracts was loaded as input control. (C) In vivo peptide pull-down to probe the interaction between peptides containing H3K56ac and Oct4 in mouse E14Tg2a nuclear extracts. Biotinylated histone peptides that are either unmodified or modified at K9 or K56 were immobilized on Streptavidin beads. Experimental details are described in SI Appendix, Methods. Ten percent cell nuclear extracts was loaded as input control and probed using Western blots and antibody against Oct4. (D) Flag-IP to show the effect of mutations in K9 to A (Ala) or K56 to A (Ala), Q (Gln), and R (Arg) on the interactions between histone H3.1 and Oct4 in mouse ESCs. Histone H3.1 with C-terminal Flag tag was transfected into E14Tg2a cells for 48 h, α-Flag antibody was used for immunoprecipitation, and the precipitate was then immunoblotted with indicated antibodies. (E) In vitro peptide pull-down to probe the interaction between K56ac containing peptide and recombinant Oct4 protein. A total of 100 ng of recombinant Oct4 protein was used for each pull-down experiment. K9ac peptide and unmodified K9 and K56 containing peptides were used as controls. Twenty percent recombinant input Oct4 protein was loaded as input control and probed with α-Oct4 antibody. (F) Mononucleosome immunoprecipitation using recombinant Oct4, histone H3–unmodified nucleosomes, or H3K9ac- or H3K56ac-modified nucleosomes. Mononucleosomes were assembled by salt dilution using histone octamers and biotin-labeled DNA as described in SI Appendix, Methods. Proteins were pulled down with streptavidin beads and immunoblotted with indicated antibody.
H3K56ac Interaction with Oct4 in Mouse E14Tg2a Cells and in Vitro.
It is known that despite their functional association on chromatin, Oct4 and Sox2 do not necessarily coimmunoprecipitate (23–25). Therefore, we wished to determine by coimmunoprecipitation (co-IP) whether Oct4 or Sox2 separately interact with H3K56ac. We found that, unlike Sox2, Oct4 coimmunoprecipitates strongly with H3K56ac in E14Tg2a cell nuclear extracts (Fig. 1B). We also noticed the trace interaction between Nanog and H3K56ac, an interaction we have not pursued. We conclude that of the key transcription factors, Oct4 interacts most with H3K56ac in nuclear extracts.
We next asked whether the acetylation of H3K56 enhances the interaction between histone H3 peptides and Oct4 by peptide pull-down assays (26). Biotinylated H3 peptides containing acetylated K56 (or acetylated K9 used as a control) were used to pull down Oct4 in nuclear extracts of E14Tg2a cells. We found that acetylation of K56 in peptides improves the efficiency of Oct4 binding compared with unmodified peptides and control peptides containing acetylated K9 (Fig. 1C). We further confirmed the importance of H3K56 in Oct4 binding using substitutions of histone H3K56. Plasmids that express wild type (WT) histone H3.1 with a C-terminal Flag tag and similar constructs bearing substitutions at K56 (H3.1K56A, H3.1K56Q, and H3.1K56R) or at K9 (H3.1K9A) were transfected into E14Tg2a cells. We then immunoprecipitated H3.1-Flag and examined the immunoprecipitate for Oct4 presence using Western blots. Although H3.1-Flag and Oct4 co-IP are not decreased by substitution of H3.1K9A, substitution of H3.1K56 by A, Q, or R decreased Oct4 co-IP considerably (Fig. 1D). Collectively, our data indicate that H3K56ac is required for the interaction between histone H3 and Oct4 in vivo.
To determine whether there is a possible direct interaction between H3K56ac and Oct4, purified recombinant Oct4 protein was incubated with H3 peptides that were unacetylated (K9-unmod, K56-unmod) or acetylated (K9ac, K56ac) for peptide pull down. We found that recombinant Oct4 protein binds best in vitro to the H3 peptide in which K56 is acetylated (Fig. 1E). We then asked whether a recombinant nucleosome in which the only posttranslational modification is the acetylation of H3 at K56 promotes binding to recombinant Oct4 in vitro. Nucleosomes in which only H3K56 or H3K9 is acetylated were prepared as described (27, 28). Using the in vitro binding assay, we found that acetylation of H3K56 promotes the binding of Oct4 to nucleosomes much more strongly than to the unmodified nucleosomes or to nucleosomes containing H3K9ac (Fig. 1F). Thus, acetylation of H3K56 not only decreases nucleosome stability (29), increases unwrapping of nucleosomal DNA ends, and may affect histone-histone interactions (27, 30), but H3K56ac may also free H3K56 sufficiently from local nucleosomal constraints to enable H3K56ac recognition by Oct4. Taken together, our data indicate that H3K56ac interacts with Oct4 in vivo in mouse ESCs and directly in vitro.
H3K56ac Links NSO Factors to Pluripotency.
To investigate the spatial and functional relationships between NSO proteins and H3K56ac genome wide, we used ChIP-Seq to select and center all Oct4 peaks in the mouse genome including regions ±5 kb from the Oct4 peak centers. We then plotted the relative enrichment of Nanog, Sox2, and H3K56ac at the Oct4 peak regions and clustered them based on the combinatorial binding of NSO and H3K56ac. Four major clusters were identified that represent the enrichment of one or more of the NSO proteins and H3K56ac (Fig. 2A). Interestingly, gene ontology (GO) analysis using Genomic Regions Enrichment of Annotations Tool (GREAT) (31) showed that the genes of cluster 4 containing NSO at Oct4 peak centers but not H3K56ac were not enriched in any functional categories. In contrast, the copresence of NSO and H3K56ac (in cluster 1) caused strong functional enrichment for genes involved in early embryogenesis and pluripotency (Fig. 2B and SI Appendix, Table S2). For example, Oct4, Klf4, Nanog, and Nodal proteins are all associated with pluripotency (4) and are members of cluster 1. The copresence of H3K56ac and NSO proteins at these cluster 1 upstream regulatory sequences is shown in SI Appendix, Fig. S1. We find in a similar manner that the regions marked by Oct4 and Sox2 alone or Oct4 and Nanog alone (in the relevant portions of cluster 2) are enriched in pluripotency associated functions only when they are also marked by the copresence of H3K56ac (Fig. 2C and D and SI Appendix, Tables S3 and S4). We conclude that H3K56ac links genes marked by NSO proteins to pluripotency in mouse ESCs.
Fig. 2.
H3K56ac links NSO factors to mark pluripotency. (A) The distribution of Oct4, Nanog, Sox2, and H3K56ac at the Oct4-binding regions are shown as heat maps. Purple indicates that the factor is enriched at those regions. All regions were grouped into four clusters based on combinatorial presence of Oct4, Nanog, Sox2, and H3K56ac. (B–D) Functional annotation of H3K56ac/Oct4/Sox2/Nanog–enriched elements in cluster 1 (B), H3K56ac/Oct4/Sox2 (C), and H3K56ac/Oct4/Nanog–enriched (D) elements in cluster 2 in mouse ESCs were performed using GREAT (31). Mouse Genome Informatics (expression detected) that indicates the information on tissue and developmental stage–specific expression in mouse is used as GO category. The x axes values (in logarithmic scale) correspond to the binomial raw P values.
Relationship Among H3K56ac, Oct4, and Oct4-Binding Motifs.
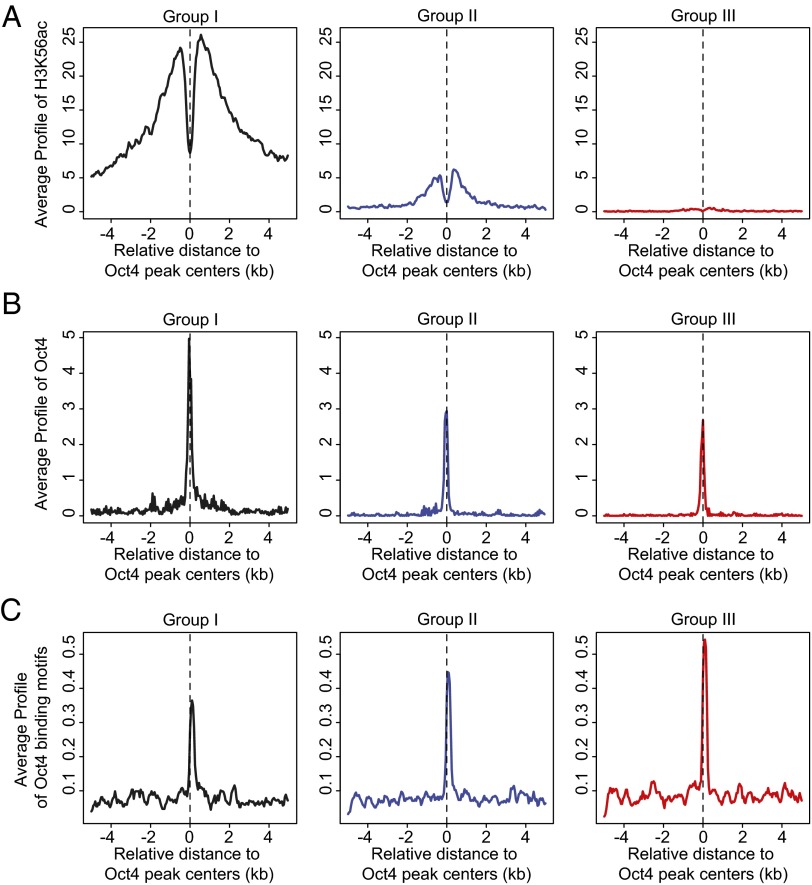
We then probed the extent to which Oct4 presence in cluster 1 correlates with H3K56ac in addition to its association with Oct4-binding motifs. To do so, we divided cluster 1 regions into three groups based on the enrichment of H3K56ac (Fig. 3A). We found that the enrichment of Oct4 at Oct4 peak centers (Fig. 3B) is highest in group I that also contains the highest level of H3K56ac. In contrast, group III contains lower enrichment of both H3K56ac and Oct4. Interestingly, the low presence of H3K56ac in group III is not accompanied by lower enrichment of Oct4-binding motifs [calculated by Cistrome (19)]. Instead, group III contains a high level of Oct4-binding motifs and a lower but significant level of Oct4 at the Oct4 peak centers. This argues for the use of Oct4 motifs in binding a lower level of Oct4 when H3K56ac is absent. A similar relationship between H3K56ac and Oct4 presence is seen when comparing the four clusters of Fig. 2A. As shown in SI Appendix, Fig. S2, the greatest enrichment of H3K56ac (in clusters 1 and 2) is accompanied by the highest levels of Oct4. In contrast, the lowest levels of H3K56ac in clusters 3 and 4 are accompanied by the lowest levels of Oct4. Oct4-binding motif presence is similar in all four clusters. Furthermore, when we analyzed the functions associated with each of the three groups within cluster 1, we found that regions highly enriched for H3K56ac in group I are most associated with pluripotency (SI Appendix, Fig. S3A and Table S5), whereas the lower enrichment of H3K56ac in groups II and III is more associated with differentiation (SI Appendix, Fig. S3B and Tables S6 and S7).
Fig. 3.
Relationship between H3K56ac, Oct4, and Oct4-binding motifs. Cluster 1 regions (from Fig. 2A) are divided into five groups based on the enrichment of H3K56ac surrounding regions ±5 kb from the Oct4 peak centers. Group I contains 20% of Oct4 regions with highest enrichment of H3K56ac; group II contains 20% of Oct4 regions in the middle (40–60%) with moderate enrichment of H3K56ac; and group III contains 20% of Oct4 regions with lowest enrichment of H3K56ac. (A) H3K56ac, (B) Oct4 and (C) Oct4-binding motifs (MA0142 in JASPAR) (48). Signal distribution relative to the center of group I, II, and III regions was determined by Cis-regulatory Element Annotation System (CEAS)-sitepro (49). The x axis represents the distance to the center of Oct4 peaks in mouse ESCs. The y axis represents the signals normalized by ChIP-Seq tag count numbers.
These data are consistent with a role for H3K56ac in recruiting or stabilizing Oct4 in vivo in addition to the interaction between Oct4 DNA–binding motifs and Oct4 protein at pluripotency-related regions. To further test this possibility, we cloned a single Oct4 DNA–binding site (ATTTG-N5-CAAAT) (32), 16 bp from the 3′ end of the 601 nucleosome positioning sequence (Fig. 4A) (33). This construct was assembled with histone octamers using salt dilution as described (34). Only histone H3K56 was acetylated in the assembled nucleosome. We then performed gel shift experiments with recombinant Oct4 protein (Origene TP311998) as described (28, 35) to probe the interaction of Oct4 protein to its DNA recognition element with and without an adjacent nucleosome acetylated at H3K56. We found that recombinant Oct4 protein slows the mobility in the native gel of the construct containing the H3K56 unmodified nucleosome (II) and shifts it to a new position (III). In contrast, mobility of the construct containing both the Oct4 DNA–binding site and the H3K56ac nucleosome is slowed further (to position IV). We conclude that acetylation of H3K56 promotes increased Oct4 binding to the construct containing a nucleosome adjacent to the Oct4-binding site (Fig. 4B).
Fig. 4.
Native gel mobility analysis for Oct4 binding to H3K56ac-modified nucleosomes adjacent to Oct4-binding motif. (A) DNA template for nucleosome assembly containing 601 nucleosome positioning sequence adjacent to a 16-bp spacer and a single Oct4-binding site (prepared by PCR from pGEM3Z601R plasmids) (33, 34). (B) Approximately 25 ng of nucleosomes were incubated with indicated recombinant Oct4 protein for 2 h at room temperature in each reaction. Position I, unassembled nucleosomal DNA template; II, assembled nucleosomal DNA template; III, assembled nucleosomal DNA template with Oct4 molecules recruited to the adjacent Oct4-binding site; IV, assembled nucleosomal DNA template containing H3K56ac with Oct4-recruited to H3K56ac histones and to Oct4-binding site.
Decreased H3K56ac Promotes the Loss of Pluripotency of Mouse ESCs.
Finally, we wished to determine whether H3K56ac promotes pluripotency in mouse ESCs by examining the effect of reduced H3K56ac level on the expression of markers for pluripotency and differentiation of ESCs. The histone chaperone Asf1 is required for the acetylation of H3K56, whereas the effect of Asf1 deletion on other histone acetylation sites is relatively minor as determined by Western blots and MS (18, 36, 37). Using Western blots, we found that knockdown of Asf1a with shAsf1a in mouse E14Tg2a cells decreases H3K56ac significantly but has very little effect on other acetylation sites in histone H3 (Fig. 5A). We also used quantitative PCR to examine the expression of pluripotency related markers after shAsf1a transfection. We observed the down-regulation of NSO pluripotency related markers and the up-regulation of the marker genes for endoderm, ectoderm, and mesoderm 10 d after transfection using the protocol described (38, 39) (Fig. 5B). Thus, our results argue that decreasing H3K56ac through loss of Asf1a results in the dissolution of the pluripotency network to initiate the expression of differentiation-related markers of mouse ESCs, indicating that H3K56ac helps maintain the pluripotency of ESCs.
Fig. 5.
Decreased H3K56ac promotes the loss of pluripotency of mouse ESCs. (A) Down-regulation of Asf1a by shAsf1a results in decreased H3K56ac with little apparent effect on other sites of acetylation in histone H3. shAsf1a treatment of mouse E14Tg2a ESCs for 48 h was followed by Western blot with indicated antibodies. Short hairpin GFP (shGFP) was used as a control. Histone H3 and α-Tubulin were used as loading controls. (B) Mouse E14Tg2a cells were transfected with plasmids that express shAsf1a or shGFP and were selected with puromycin from day 4 after transfection. Prolonged decrease of H3K56ac by shAsf1a plasmids for 10 d caused the reduction of pluripotency markers and up-regulation of differentiated makers for endoderm, ectoderm, and mesoderm.
Discussion
We have shown previously that H3K56ac is enriched at NSO target gene promoters in human ESCs (10). However, the data presented here support a special role for H3K56ac in recognizing Oct4 in mouse ESCs. Oct4 and Sox2 are associated with each other at their targets in the core transcriptional network (4), and we find that H3K56ac presence correlates with that of Oct4 and Sox2. However, Oct4 and Sox2 recognize different DNA motifs and do not coimmunoprecipitate (23–25), and we find that H3K56ac coimmunoprecipitates only with Oct4 in nuclear extracts. We cannot exclude the possibility that other histone acetylation sites also interact with Oct4. For example, H3K27 and H3K56 are both acetylated by the histone acetyltransferase p300 and H3K27ac is also associated with NSO cooccupancy (40). However, although the interaction between Oct4 and H3K56ac is specific in that H3K9ac, which marks enhancers, is not required for Oct4 binding (Fig. 1), we do not know whether H3K27ac binds to Oct4 or to other regulators such as Sox2 and Nanog. Nevertheless, the interaction between Oct4 and H3K56ac is unique. Previous studies have demonstrated that H3K56ac displaces nucleosomal DNA in vitro to allow DNA “breathing” (27) and access of a LexA DNA-binding protein to nucleosomal DNA (41, 42). Surprisingly, our data argue not for an interaction between Oct4 and nucleosomal DNA, but for a direct interaction between Oct4 and nucleosomal H3K56ac.
How H3K56 becomes acetylated in a localized manner to bind Oct4 is unclear. Localized H3K56ac may result from the presence of the histone acetyltransferase p300 that colocalizes often at enhancers with NSO proteins (4, 40), resulting in acetylation of H3K56 in a bimodal manner surrounding p300 (SI Appendix, Fig. S4). Thus, histone chaperone Asf1a and p300 may collaborate to acetylate H3K56 on histone H3 (17), which might subsequently replace unacetylated H3K56 in chromatin (12). This may target H3K56ac to sites surrounding Oct4-binding sites to enable H3K56ac–Oct4 interactions.
It is known that NSO proteins target not only genes of the core transcriptional network but also many genes related to differentiation (1, 2). This helps explain why the clustering of NSO protein presence at Oct4 peak regions is not associated by GO analysis preferentially with pluripotency. In contrast, when H3K56ac is clustered with NSO, those regions are strongly enriched functionally for pluripotency (Fig. 2; cluster 1) and regions in this cluster with the highest enrichment of H3K56ac were shown to be most enriched for Oct4 (Fig. 3) and most associated with pluripotency (SI Appendix, Fig. S3). In support, shAsf1a, which is expected to decrease H3K56ac, specifically (Fig. 5) decreased the levels of NSO mRNAs pluripotency related markers while promoting the presence of differentiation related ones. Collectively, these data argue for the biological importance of the interaction between Oct4 and H3K56ac in regulating pluripotency.
Although H3K56ac is generally important for Oct4 occupancy, there are also DNA regions containing Oct4-binding motifs, at which Oct4 binds in the absence of H3K56ac. In either case, the binding of Oct4 uses Oct4 recognition motifs, and we have found that Oct4 can bind its recognition motifs to a lower, albeit significant, extent in the absence of H3K56ac [Figs. 3 (group III) and 4]. These different binding mechanisms are both important functionally. When Oct4 enrichment occurs through DNA-binding motifs and H3K56ac, those Oct4-bound regions are preferentially associated with pluripotency (SI Appendix, Fig. S3). When Oct4 enrichment occurs through its DNA-binding motifs only, those Oct4-bound regions are preferentially associated with differentiation. Therefore, the presence of H3K56ac delineates Oct4 function biologically.
Interestingly, although our data (Figs. 1 and 4) support a direct interaction between Oct4 and H3K56ac, the H3K56ac modification is generally absent at the actual Oct4-binding peaks (Figs. 2A and 3) that contain Oct4 recognition motifs and may exclude histones. Instead, H3K56ac surrounds the Oct4 peak centers and DNA recognition motifs in a bimodal manner at pluripotency-associated regions (SI Appendix, Fig. S4). Therefore, we propose that the DNA recognition motifs may determine the sites of Oct4 binding, whereas adjacent nucleosomes acetylated at H3K56 stabilize Oct4 at those sites. In support, our gel shift experiment indicates that a nucleosomal construct containing H3K56ac placed adjacent to an Oct4-binding site is likely to interact with more Oct4 than the construct that is not acetylated at H3K56 (Fig. 4). How this occurs in vivo in the presence of Oct4 homodimers, Oct4-Sox2 heterodimers (32, 43–45), and multiple adjacent DNA recognition motifs (46) remains to be determined. Nevertheless, the unique role of H3K56ac in Oct4 binding is likely one factor that contributes to Oct4 stability and to the function of Oct4 as a master regulator of pluripotency.
Materials and Methods
All experiments performed with mouse ESCs were cultured under standard feeder-free conditions, and were approved by the University of California Los Angeles (UCLA) Embryonic Stem Cell Research Oversight Committee. ChIP DNA was sequenced with Illumina Genome Analyzer HiSeq2000 at the UCLA Broad Stem Cell Research Center high-throughput sequencing facility. For details, please refer to SI Appendix.
Supplementary Material
Acknowledgments
We thank members of the M.G. laboratory for comments and discussion throughout this work; Yongxin Yu, Arnie Berk, Kathrin Plath, Siavash Kurdistani, and Roberto Ferrari for critique of the manuscript; Gerald Crabtree for the gift of short hairpin GFP plasmids; Jason W. Chin for the plasmids used to express acetyl-histones; Michael Carey for plasmids used to express recombinant histones; the UCLA Broad Stem Cell Research Center High Throughput Sequencing core facility for sequencing services; and Roberto Ferrari for help with the analysis of ChIP-Seq data. This work was supported by National Institutes of Health Grant GM23674 and by a Broad Center of Regenerative Medicine and Stem Cell Research at UCLA grant (to M.G.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The primary sequencing data reported in this paper have been deposited in the Gene Expression Omnibus database (GEO), www.ncbi.nlm.nih.gov/geo (accession no. GSE47387).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309914110/-/DCSupplemental.
References
- 1.Young RA. Control of the embryonic stem cell state. Cell. 2011;144(6):940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng HH, Surani MA. The transcriptional and signalling networks of pluripotency. Nat Cell Biol. 2011;13(5):490–496. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- 3.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133(6):1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 5.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132(6):1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24(4):372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 9.Kim JB, et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136(3):411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 10.Xie W, et al. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell. 2009;33(4):417–427. doi: 10.1016/j.molcel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu F, Zhang K, Grunstein M. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell. 2005;121(3):375–385. doi: 10.1016/j.cell.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 12.Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell. 2007;27(3):393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 13.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436(7048):294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 14.Xu F, Zhang Q, Zhang K, Xie W, Grunstein M. Sir2 deacetylates histone H3 lysine 56 to regulate telomeric heterochromatin structure in yeast. Mol Cell. 2007;27(6):890–900. doi: 10.1016/j.molcel.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Värv S, et al. Acetylation of H3 K56 is required for RNA polymerase II transcript elongation through heterochromatin in yeast. Mol Cell Biol. 2010;30(6):1467–1477. doi: 10.1128/MCB.01151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feser J, et al. Elevated histone expression promotes life span extension. Mol Cell. 2010;39(5):724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459(7243):113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan J, Pu M, Zhang Z, Lou Z. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle. 2009;8(11):1747–1753. doi: 10.4161/cc.8.11.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu T, et al. Cistrome: An integrative platform for transcriptional regulation studies. Genome Biol. 2011;12(8):R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailey TL. DREME: Motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27(12):1653–1659. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunarso G, et al. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42(7):631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 22.Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38(4):431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 23.Liang J, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat Cell Biol. 2008;10(6):731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- 24.Pardo M, et al. An expanded Oct4 interaction network: Implications for stem cell biology, development, and disease. Cell Stem Cell. 2010;6(4):382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van den Berg DL, et al. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6(4):369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wysocka J. Identifying novel proteins recognizing histone modifications using peptide pull-down assay. Methods. 2006;40(4):339–343. doi: 10.1016/j.ymeth.2006.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neumann H, et al. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36(1):153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuryan BG, et al. Histone density is maintained during transcription mediated by the chromatin remodeler RSC and histone chaperone NAP1 in vitro. Proc Natl Acad Sci USA. 2012;109(6):1931–1936. doi: 10.1073/pnas.1109994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreira H, Flaus A, Owen-Hughes T. Histone modifications influence the action of Snf2 family remodelling enzymes by different mechanisms. J Mol Biol. 2007;374(3):563–579. doi: 10.1016/j.jmb.2007.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon M, et al. Histone fold modifications control nucleosome unwrapping and disassembly. Proc Natl Acad Sci USA. 2011;108(31):12711–12716. doi: 10.1073/pnas.1106264108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Botquin V, et al. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev. 1998;12(13):2073–2090. doi: 10.1101/gad.12.13.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276(1):19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 34.Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell. 2006;24(3):481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li G, Widom J. Nucleosomes facilitate their own invasion. Nat Struct Mol Biol. 2004;11(8):763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 36.Adkins MW, Carson JJ, English CM, Ramey CJ, Tyler JK. The histone chaperone anti-silencing function 1 stimulates the acetylation of newly synthesized histone H3 in S-phase. J Biol Chem. 2007;282(2):1334–1340. doi: 10.1074/jbc.M608025200. [DOI] [PubMed] [Google Scholar]
- 37.Recht J, et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci USA. 2006;103(18):6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho L, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci USA. 2009;106(13):5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernardo AS, et al. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell. 2011;9(2):144–155. doi: 10.1016/j.stem.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.North JA, et al. Regulation of the nucleosome unwrapping rate controls DNA accessibility. Nucleic Acids Res. 2012;40(20):10215–10227. doi: 10.1093/nar/gks747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimko JC, North JA, Bruns AN, Poirier MG, Ottesen JJ. Preparation of fully synthetic histone H3 reveals that acetyl-lysine 56 facilitates protein binding within nucleosomes. J Mol Biol. 2011;408(2):187–204. doi: 10.1016/j.jmb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chew JL, et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol. 2005;25(14):6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saxe JP, Tomilin A, Schöler HR, Plath K, Huang J. Post-translational regulation of Oct4 transcriptional activity. PLoS ONE. 2009;4(2):e4467. doi: 10.1371/journal.pone.0004467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reményi A, et al. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003;17(16):2048–2059. doi: 10.1101/gad.269303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reményi A, Schöler HR, Wilmanns M. Combinatorial control of gene expression. Nat Struct Mol Biol. 2004;11(9):812–815. doi: 10.1038/nsmb820. [DOI] [PubMed] [Google Scholar]
- 47.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20(9):1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 48.Portales-Casamar E, et al. JASPAR 2010: The greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38(Database issue):D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin H, Liu T, Manrai AK, Liu XS. CEAS: Cis-regulatory element annotation system. Bioinformatics. 2009;25(19):2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.