Abstract
We examined how asparagine-linked glycans within and adjacent to the V3 loop (C2 and C3 regions) and within the immunologically silent face (V4, C4, and V5 regions) of the human immunodeficiency virus (HIV) SF612 envelope affect the viral phenotype. Five of seven potential glycosylation sites are utilized when the virus is grown in human peripheral blood mononuclear cells, with the nonutilized sites lying within the V4 loop. Elimination of glycans within and adjacent to the V3 loop renders SF162 more susceptible to neutralization by polyclonal HIV+-positive and simian/human immunodeficiency virus-positive sera and by monoclonal antibodies (MAbs) recognizing the V3 loop, the CD4- and CCR5-binding sites, and the extracellular region of gp41. Importantly, our studies also indicate that glycans located within the immunologically silent face of gp120, specifically the C4 and V5 regions, also conferred on SF162 resistance to neutralization by anti-V3 loop, anti-CD4 binding site, and anti-gp41 MAbs but not by antibodies targeting the coreceptor binding site. We also observed that the amino acid composition of the V4 region contributes to the neutralization phenotype of SF162 by anti-V3 loop and anti-CD4 binding site MAbs. Collectively, our data support the proposal that the glycosylation and structure of the immunologically silent face of the HIV envelope plays an important role in defining the neutralization phenotype of HIV type 1.
Entry of human immunodeficiency virus types 1 and 2 (HIV-1 and HIV-2, respectively) and simian immunodeficiency virus (SIV) into host cells is mediated by the viral envelope glycoprotein, which consists of two noncovalently associated subunits, gp120 and gp41, derived by the proteolytic cleavage of a gp160 precursor (2, 5, 42, 54, 62, 83, 100). The extracellular gp120 subunit mediates viral binding to the host cell surface CD4 antigen (25, 37, 60). Upon binding of gp120 to CD4, the gp120 molecule undergoes conformational changes enabling it to bind to the chemokine receptor (CCR5 or CXCR4) (1, 21, 26-29, 87). This results in the exposure of the fusion peptide in the membrane-anchored gp41 subunit and its insertion into the target cell membrane, mediating fusion of the viral and cell membranes (14, 30, 54, 106).
The gp120 subunit consists of five variable regions, V1 to V5, interspersed between five conserved regions (C1 to C5) (49, 96). Both N- and O-linked glycans are present on the HIV envelope glycoprotein. O-linked glycans are present on several unidentified serine or threonine residues in gp120, but little is known about their role in defining the phenotype of HIV or SIV (6, 12). In contrast, asparagine-linked (N-linked) glycosylation has been more extensively studied, and it is known that N-linked glycans comprise about 50% of the mass of gp120 (49). N-linked glycans are added cotranslationally as the viral envelope protein passes through the endoplasmic reticulum and are subsequently modified in the Golgi apparatus, giving rise to three types of glycans, termed high-mannose, hybrid, and complex glycans (41). In general, complex glycans are present within the variable regions of gp120, with high-mannose or hybrid glycans being present within the conserved regions (49, 110). The addition of N-linked glycans is essential for the correct folding and correct processing of the viral envelope protein (46, 49, 50). While the exact number of N-linked glycosylation sites varies among HIV-1 isolates, many sites are highly conserved (31, 53, 102). For the T-cell-line-adapted IIIB and SF2 HIV-1 isolates, the type of glycan present at each N-linked glycosylation site has been determined, and where a glycosylation site is conserved in position in both isolates, the same type of glycan is present (49, 110).
Glycosylation of the HIV and SIV envelope proteins limits their immunogenicity and in addition restricts the binding of certain antibodies to their epitopes on the virion surface (80, 104). In macaques experimentally infected with SIV or simian/human immunodeficiency virus (SHIV), the emergence of neutralization-resistant viruses is associated with changes in the envelope glycosylation pattern. Some of these changes comprise the removal, repositioning, and addition of potential glycosylation sites, resulting in the masking of certain neutralization epitopes (12, 13, 15, 55, 66, 69, 70, 79, 80, 84). A recent study in humans on the mechanism of HIV escape from antibody-mediated neutralization led to the proposal of an evolving “glycan shield” in which the repositioning of glycans on the HIV envelope limits envelope recognition by neutralizing antibodies while maintaining the ability of the envelope protein to interact with the CD4 and coreceptor molecules on the target cell membrane (101).
However, not every potential N-linked glycosylation site is utilized on SIV and HIV (48, 68), and the roles that individual glycans have in protecting the virus from neutralization most likely differ. Our group previously reported that on the background of the primary CCR5-tropic HIV-1 isolate SF162, the elimination of two N-linked glycosylation sites, one adjacent to the amino-terminal and the second adjacent to the carboxy-terminal side of the V2 loop, at positions 154 and 195, respectively, increases the susceptibility of this virus to neutralization by heterologous sera from HIV-infected patients and by certain monoclonal antibodies (MAbs) that bind to the V3 loop and the CD4 binding site (56). In contrast, removal of the only N-linked glycosylation site within the V2 loop of SF162 itself, at position 186, has a less pronounced effect on the susceptibility of the virus to neutralization. Therefore, in the case of SF162, the presence of glycans at the base of the V2 loop exerts a stronger phenotypic effect than the presence of the glycan within the V2 loop.
In the studies presented here, we extended this previous research on the SF162 isolate and examined the role that conserved N-linked glycosylation within and adjacent to the V3 loop and within the immunologically silent face of gp120 might play in controlling viral replication, coreceptor usage, and susceptibility to antibody-mediated neutralization. To this end, we disrupted potential N-linked glycosylation sites at the amino and carboxy termini of the V3 loop (in C2 and C3, respectively) and within the V3 loop, and in the V4, C4, and V5 regions, which comprise the immunologically silent face of gp120 (104).
The V3 loop itself contains epitopes for strain-restricted neutralizing antibodies, it is a major determinant for viral tropism and coreceptor usage, and its orientation is such that it partially masks the CD4 and chemokine receptor binding sites (11, 19-21, 36, 72, 82, 91, 92, 103, 104). We wished to determine if the glycans within the V3 loop or at the base of the V3 loop might play a role in any of the above processes. The V3 loop of most HIV-1 isolates contains a single N-linked glycosylation site (position 299 on the SF162 background) that is highly conserved among viral isolates from most subtypes, with the exception of subtype D (3, 31, 47, 53). Similarly, the glycan in C3 (position 329 on the SF162 background), at the carboxy-terminal side of the V3 loop, is highly conserved in viral isolates from most subtypes, the exception being subtype E, while the glycan in C2 (position 293 on the SF162 background), at the amino-terminal side of the V3 loop, is highly conserved among subtype B isolates and less so among subtype A and C isolates (3, 31, 47, 53, 85, 102). This degree of conservation across diverse viral subtypes suggests an important role for these three glycans in controlling the viral phenotype, and it is known that the glycans in C2 and C3, adjacent to the V3 loop, comprise part of the epitope for the broadly neutralizing MAb 2G12 (9, 85, 88).
The immunologically silent face of gp120 is heavily glycosylated in all HIV isolates and appears to be minimally immunogenic. Most HIV-1 isolates, irrespective of their subtype, contain potential N-linked glycosylation sites that are relatively conserved in position in the C4 (position 438 on the SF162 background) and V5 (position 454 on the SF162 background) regions of gp120. In contrast, most viral isolates in diverse subtypes contain four or five potential N-linked glycosylation sites within the V4 loop with three of the sites being conserved in position (31, 53, 88, 102). The C4 region contributes to the formation of the CD4 binding site, which consists of a deeply recessed pocket on the gp120 surface that is devoid of glycans but is flanked by regions of considerable glycosylation that are predicted to have a protective effect (43, 104, 106). Therefore, it is possible that glycans in the V4, C4, and V5 regions of gp120 might play a role in masking the CD4 binding site. In addition, they might also play a role in masking other conserved neutralization epitopes.
The present studies indicate that specific glycans within and adjacent to the V3 loop and within the immunologically silent face of gp120 protect SF162 from neutralization by antibodies recognizing epitopes within the V3 loop and the CD4 binding sites. We also observed that some of these glycans prevent the binding of neutralizing antibodies to the coreceptor binding site and to the gp41 envelope subunit. Overall, our data indicate an important role for the immunologically silent face of HIV gp120 in defining the neutralization phenotype of this virus.
MATERIALS AND METHODS
Cells.
Human peripheral blood mononuclear cells (PBMC) were prepared by Ficoll gradient centrifugation, from the peripheral blood of healthy HIV-negative donors (Puget Sound Blood Center). PBMC were cultured in RPMI 1640 medium (Cellgro) supplemented with 10% fetal bovine serum (FBS) (Cellgro), penicillin (100 U/ml; Cellgro), streptomycin (100 μg/ml; Cellgro), glutamine (2 mM; Cellgro) and interleukin-2 (20 U/ml; Hoffmann-La Roche, Inc.). Prior to use, PBMC were stimulated for 3 days with 2 μg of phytohemagglutinin (PHA; Sigma) per ml as previously described (16).
The U87 human astroglioma cell line (from N. R. Landau, Salk Institute, La Jolla, Calif.) expressing various combinations of human receptors—CD4 and CCR5, CD4 and CXCR4, CCR5 alone, and CXCR4 alone—were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS, penicillin (100 U/ml), streptomycin (100 μg/ml), glutamine (2 mM), and puromycin (1 μg/ml; Cellgro). The U87 cell line expressing CD4 (AIDS Research and Reference Reagent Program Catalog, National Institutes of Health [NIH]) was cultured in DMEM supplemented with 10% FBS, penicillin (100 U/ml), streptomycin (100 μg/ml), and glutamine (2 mM).
Antibodies.
The following reagents were obtained through the AIDS Research & Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, NIH: HIV-1 gp41 monoclonal antibody 2F5 from H. Katinger (7, 76, 77) and HIV-1 gp41 monoclonal antibody 50-69 from S. Zolla-Pazner (33, 73, 98, 99, 108). D. S. Dimitrov provided Fab X5 (64) and Fab M18 (109a). MAbs 17b and 48d were provided by J. Robinson (97, 107), MAbs 447D and 391-95D were provided by S. Zolla-Pazner (22, 32, 34, 89), MAb immunoglobulin G1 (IgG1) b12 was provided by D. R. Burton (8, 71, 86), IgG-CD4 was obtained from Genentech (San Francisco, Calif.) (10), and MAb G3.4 was provided by M. Fung (Tanox, Inc.) (35, 109).
Elimination by mutagenesis of potential N-linked glycosylation sites.
The motif for an N-linked glycosylation site is Asn-X-Thr/Ser, where X can be any amino acid except proline (59). Elimination of potential N-linked glycosylation sites was performed as previously described (56, 80), by changing an asparagine (N) to a glutamine (Q) with the QuikChange site-directed-mutagenesis kit (Stratagene). The template for mutagenesis was the 3′ half of the SF162 genome cloned into pUC19. The asparagine residues were changed to glutamine at the following positions: 293 in the C2 region, 299 in the V3 loop, 329 in the C3 region, 398 and 401 in the V4 loop, 438 in the C4 region, and 454 in the V5 loop of the SF162 envelope (Fig. 1). The positions on the SF162 envelope are numbered, with 1 as the initiator methionine residue. Potential N-linked glycosylation sites were also eliminated by changing a threonine (T) to an alanine (A) at positions 400 and 403. We designate the glycosylation mutant (GM) envelopes as follows: an envelope with N293 replaced by Q is GM293 (C2), one with N299 replaced by Q is GM299 (V3), etc. The introduction of mutations was verified by sequencing. Two clones of each mutant envelope were amplified and used for generation of infectious virions. The primers used for mutagenesis are listed in Table 1. The potential N-linked glycosylation sites listed above were also eliminated on the full-length envelope (gp160) of SF612 cloned into the expression vector pEMC* (74). Asparagine was changed to glutamine by site-directed mutagenesis, as described above, and the entire envelope was sequenced to confirm the presence of the mutation and to ensure that no additional mutations had been introduced.
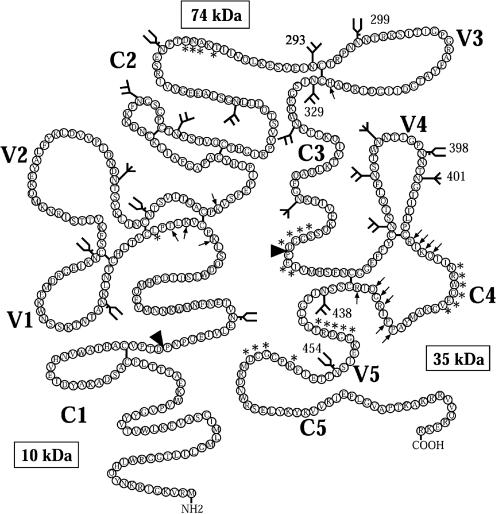
FIG. 1.
Schematic representation of SF162 gp120 showing the conserved regions (C1-C5) and the variable loops (V1-V5), based on the sequence of SF162 from Cheng-Mayer et al. (16) and figure adapted from Leonard et al. (49). The positions and types of potential N-linked glycosylation sites are indicated as follows:  , high-mannose or hybrid glycan;
, high-mannose or hybrid glycan;  , complex glycan; and
, complex glycan; and  , unknown type of glycan. Arrowheads indicate the positions where glacial acetic acid cleaves gp120 to produce three fragments of approximately 10, 35, and 74 kDa; arrows indicate amino acids outside the V3 loop that are involved in coreceptor binding (81, 82); asterisks indicate amino acids involved in CD4 binding (43).
, unknown type of glycan. Arrowheads indicate the positions where glacial acetic acid cleaves gp120 to produce three fragments of approximately 10, 35, and 74 kDa; arrows indicate amino acids outside the V3 loop that are involved in coreceptor binding (81, 82); asterisks indicate amino acids involved in CD4 binding (43).
TABLE 1.
List of primers used in site-directed mutagenesis to eliminate potential N-linked glycosylation sites in the SF162 envelope
| Primer | Sequencea |
|---|---|
| GM293 | F; 5′ GAATCTGTAGAAATTCAGTGTACAAGACC 3′ |
| R; 5′ GGTCTTGTACACTGAATTTCTACAGATTC 3′ | |
| GM299 | F; 5′ CAAGACCTAACCAGAATACAAGAAAAAG 3′ |
| R; 5′ CTTTTTCTTGTATTCTGGTTAGGTCTTG 3′ | |
| GM329 | F; 5′ GACAAGCACATTGTCAAATTAGTGGAG 3′ |
| R; 5′ CTCCACTAATTTGACAATGTGCTTGTC 3′ | |
| GM398 | F; 5′ CTATAGGGCCACAGAACACTAATGGAAC 3′ |
| R; 5′ GTTCCATTAGTGTTCTGTGGCCCTATAG 3′ | |
| GM400 | F; 5′ GGGCCAAATAACGCGAATGGAACTATCACACTCCC 3′ |
| R; 5′ GGGAGTGTGATAGTTCCATTCGCGTTATTTGGCCC 3′ | |
| GM401 | F; 5′ GGCCAAATAACACTCAGGGAACTATCAC 3′ |
| R; 5′ GTGATAGTTCCCTGAGTGTTATTTGGCC 3′ | |
| GM403 | F; 5′ GGGCCAAATAACACTAATGGAGCGATCACACTCCCATGC 3′ |
| R; 5′ GCATGGGAGTGTGATCGCTCCATTAGTGTTATTTGGCCC 3′ | |
| GM438 | F; 5′ GATGCTCATCACAGATTACAGGACTGC 3′ |
| R; 5′ GCAGTCCTGTAATCTGTGATGAGCATC 3′ | |
| GM454 | F; 5′ AAAGAGATCAGTCAAACCACCGAGATC 3′ |
| R; 5′ GATCTCGGTGGTTTGACTGATCTCTTT 3′ |
F, forward primer; R, reverse primer. Underlining indicates nucleotides that were changed to generate the glycosylation mutants.
Generation of infectious virions.
In PBMC, the generation of infectious, replication-competent virions was performed as previously described (56). Briefly, 4 × 105 human embryonic kidney 293T cells were added per well in a six-well dish, and 24 h later they were cotransfected with 5 μg of a pUC19 plasmid containing the 3′ half of the SF162 viral genome (either the parental or the mutagenized SF162 envelope gene) and 5 μg of a pUC19 plasmid containing the 5′ half of the SF162 viral genome, using DMRIE (Invitrogen), according to the manufacturer's instructions. At 72 h posttransfection, the cell supernatants were collected, subjected to centrifugation (5 min at 2,000 rpm at room temperature), and used to inoculate PHA-stimulated PBMC. Following 3 h of incubation at 37°C, the viral inoculum was removed, and the cells were cultured in RPMI medium, at a density of 3 × 106 cells per ml. Virus production was monitored by determining the concentration of p24 antigen in the cell supernatant every 3 to 4 days, using an in-house p24 detection enzyme-linked immunosorbent assay (ELISA). Supernatants with high p24 content were collected and stored as 0.5-ml aliquots at −80°C. These viral stocks were titrated in PBMC and used for all subsequent experiments.
Luciferase reporter viruses capable of only a single round of replication expressing either the parental SF162 or the deglycosylated SF162 envelope were generated by cotransfecting 293T cells with pNL-Luc-E−R− (provided by N. Landau) (24) and with the pEMC* vector expressing either the parental or deglycosylated SF162 gp160. 293T cells (1.8 × 106) were plated in a 10-cm2 plate, and 24 h later the cells were cotransfected with 12 μg of pNL-Luc-E−R− and 12 μg of the pEMC* vector, using DMRIE (Invitrogen), according to the manufacturer's instructions. After 6 to 7 h the medium was removed and 7.5 ml of fresh medium was added. At 72 h posttransfection, the cell supernatants were collected, subjected to centrifugation (5 min at 913 × g at room temperature), and assayed for p24 and gp120 content. The supernatants were stored as 0.5-ml aliquots at −80°C.
Electrophoretic mobility analysis of virion-associated gp120 proteins.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and subsequent Western blotting were performed on denatured viral proteins from both the parental SF162 and the glycosylation mutants, as previously described (56). Briefly, intact infectious viral particles produced in PBMC or pseudovirus produced in 293T cells was pelleted by centrifugation at 14,000 rpm for 2 h at 4°C. The viral pellet was resuspended in 30 μl of lysis buffer (50 mM Tris-HCl, 100 mM 2-mercaptoethanol, 2% SDS, 0.1% bromophenol blue, 20% glycerol), subjected to boiling for 5 min, and centrifuged (16,000 × g for 5 min at room temperature), and the supernatant was subjected to SDS-PAGE on 5% SDS gels. Proteins were then transferred to Immobilon P membranes (Millipore) and incubated overnight at 4°C with goat-anti-env 2-3 SF2 gp120 serum antibodies (Chiron; 1:1,000 dilution) and for 90 min at room temperature with protein G-horseradish peroxidase (Bio-Rad; 1:1,000 dilution). Visualization of the gp120 envelope molecules was performed with the use of enhanced chemiluminescence reagents (Amersham).
Acid hydrolysis of soluble monomeric and virion-associated gp120 molecules.
Acid hydrolysis was performed as previously described (79). Briefly, recombinant monomeric SF162 gp120 protein produced in CHO cells was hydrolyzed by incubation for 66 h at 40°C in a total volume of 100 μl of 10% glacial acetic acid. The hydrolyzed protein was then dried and dissolved in 30 μl of the above lysis buffer, boiled for 5 min, centrifuged for 5 min, and loaded onto 10% SDS gels. PAGE and subsequent Western blotting were performed as described above. Hydrolysis of virion-associated gp120 was performed as described above with the following modifications: intact infectious viral particles produced in PBMC were first pelleted by centrifugation, as described above, and the viral particles were resuspended in 100 μl of 10% glacial acetic acid. Following the 66-h incubation at 40°C, the hydrolyzed samples were incubated at 56°C for 1 h, prior to drying and resuspension in lysis buffer.
Sequencing of the viral envelope from infected PBMC.
Human PBMC were infected in vitro with either the parental or deglycosylated viruses. Four days postinfection, the PBMC were harvested, and the DNA was extracted with the QIAamp DNA blood kit (Qiagen). The viral envelope was then amplified in two rounds of PCR using the Expand high-fidelity PCR system (Roche). The primers used were E0 (5′ TAGAGCCCTGGAAGCATCCAGGAAGTCAGCCTA 3′) and env8R (5′ CACAATCCTCGCTGCAATCAAG 3′) for the first round of PCR and gp160F (5′ GGACCATAGTGTACATAGAATACAG 3′) and env6R (5′ CTTGCCCACTTATCCAATTC 3′) for the second round, amplifying a region of 2,070 bp containing 96 bp 5′ to gp120, all of gp120, and the first 470 bp of gp41. The PCR products were cloned into the pCR3.1 eukaryotic TA bidirectional expression vector (Invitrogen) or the pCR2.1-Topo vector (Invitrogen) and the gp120 region of the clones obtained was sequenced. At least five clones were sequenced for each virus.
Viral replication assays.
The viral replication potential of the mutant viruses was assessed by using PHA-activated PBMC. A total of 30 × 106 PBMC in 3 ml of RPMI were treated with 6 μg of Polybrene (Sigma) for 30 min at 37°C, divided into three tubes, and subjected to centrifugation (913 × g for 5 min at room temperature), and the supernatant was removed. The cells were inoculated for 3 h at 37°C with 100 50% tissue culture infectious doses of each virus in a total volume of 1 ml (multiplicity of infection [MOI] of 10−5). Viral replication kinetics were also performed at an MOI of 3 × 10−3. Following the 3-h incubation, the viral inoculum was removed by aspiration, and the cells were cultured in RPMI medium at a density of 3 ×106 cells per ml. Viral replication was monitored by determining the concentration of p24 antigen in the supernatant every 3 to 4 days. All experiments were repeated at least three times with pooled cells from multiple different donors to minimize potential donor-specific effects on viral infectivity.
Fusogenic potential of SF162 and mutant envelopes.
U87 CD4+ CCR5+ cells were added to flat-bottom 96-well plates at a density of 105 cells per ml of medium (104 cells per well). Twenty-four hours later the cells were treated with Polybrene (2 μg/ml) and were inoculated in triplicate with each virus (1 ng or 4 ng of p24) in a total volume of 100 μl of complete DMEM, and after a 3-h incubation at 37°C the inoculum was removed and 100 μl of complete DMEM was added. Seventy-two hours after inoculation, the medium was aspirated and the cells were lysed in 100 μl of 1× lysis buffer (Promega) at room temperature for 2 h. A 40-μl portion of this lysate and 100 μl of luciferase assay substrate (Promega) were used to determine the luciferase levels (relative light units) by using the Fluroskan Ascent FL (Thermo Labsystems).
Assay of IgG-CD4 binding to virions.
This assay was performed as previously described with a few modifications (94). Briefly, virions produced in PBMC were incubated with increasing concentrations of IgG-CD4 (3.2 ng/ml to 2 μg/ml diluted in RPMI medium) for 2 h at room temperature, centrifuged at 20,000 × g for 2 h at 4°C, the supernatant was aspirated, and the pellet was resuspended in lysis buffer (Tris-buffered saline containing 4% milk and 1% NP-40) for 2 h at room temperature. A 200-μl portion of each lysate was added to ELISA wells precoated with the anti-gp120 sheep antibody 6205 (5 μg/ml) (International Enzymes, Inc.). The relative amount of IgG-CD4 bound to captured gp120 was determined by ELISA as previously described in detail (94). The optical density signals at 490 nm were corrected appropriately by taking into consideration differences in the concentration of envelope molecules present in each virion sample with the use of pooled human sera from patients infected with clade B isolates.
Determination of coreceptor usage of mutant envelopes.
Coreceptor usage of viruses expressing mutant envelopes was determined using the following cell lines: U87 CD4+, U87 CCR5+, U87 CXCR4+, U87 CD4+ CCR5+, and U87 CD4+ CXCR4+. Single-round replication-competent viruses (1 ng of p24) expressing luciferase and either the parental SF162 or the deglycosylated viral envelopes were used to inoculate the above cells (104 cells per well of a 96-well plate). The extent of entry was determined by measuring the level of luciferase in the various U87 cell lines after infection, as described above. Background luciferase levels were determined from cells that were inoculated with luciferase-expressing virions lacking viral envelopes.
Neutralization assays.
Neutralization assays were performed as previously described (56, 93, 95). Briefly, each virus produced and titrated in activated human PBMC (50 50% tissue culture infective doses in 25 μl of RPMI medium) was mixed with an equal volume of serially diluted MAbs or heat-inactivated sera (56°C for 45 min; from patients infected with clade B isolates or from a rhesus macaque infected with SHIVSF162P4) (17) in triplicate wells of a U-bottom 96-well plate and incubated for 1 h at 37°C. Virus was also incubated in the absence of HIV-positive serum or MAb or in the presence of serum from the rhesus macaque collected prior to infection with SHIVSF162P4. These wells served as positive controls. PBMC (2 × 105 in 50 μl of RPMI medium) were then added to each well. Approximately 24 h later, half the cell supernatant of each well was replaced with fresh medium, and this procedure was performed twice. The percent neutralization in the presence of each serum or MAb dilution was determined as previously described (95). Neutralization experiments were performed three times with pooled PBMC from multiple donors in order to minimize potential donor-specific effects on viral infectivity.
RESULTS
Glycosylation of specific asparagine residues on the SF162 envelope.
The positions of all the potential N-linked glycosylation sites on the SF162 envelope gp120 subunit are shown in Fig. 1, and they are numbered with 1 as the initiator methionine residue. Given that the type of glycan at each position in the envelope of the HIV-1 IIIB and SF2 isolates has been determined and that where a glycan is conserved in position in both isolates, the same type of glycan is found (49, 110), we assumed that where a glycan is conserved in position between IIIB and SF162, the same type of glycan will be found at that position in the SF162 envelope. Where a glycan is not conserved between the SF162 and IIIB envelopes, the glycan is marked as being of unknown composition. By changing asparagine to glutamine, we eliminated the potential N-linked glycosylation sites on the SF162 gp120 subunit at positions 293 (C2), 299 (V3), 329 (C3), 398 (V4), 401 (V4), 438 (C4), and 454 (V5). Based on the above assumption, we expect that high-mannose or hybrid glycans are found at positions 293 (C2), 329 (C3), and 438 (C4) and that complex glycans are found at positions 299 (V3), 398 (V4), and 454 (V5) of the SF162 gp120 envelope (Fig. 1).
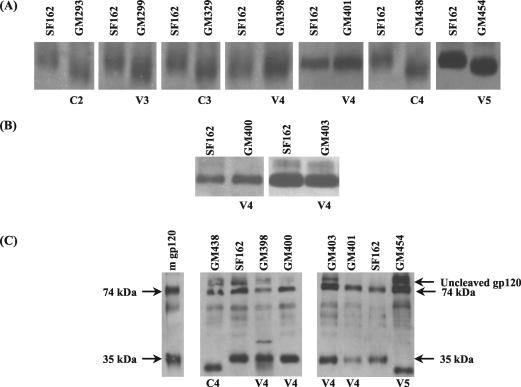
To examine if the N-linked glycosylation sites that we eliminated are in fact utilized during the in vitro replication of SF162 in PBMC, we compared the electrophoretic mobilities of virion-associated gp120 from the parental SF162 and the glycosylation mutants (Fig. 2A). It is expected that the elimination of an individual utilized N-linked glycosylation site will reduce the molecular mass of the gp120 protein by approximately 2.5 kDa (41). This change in molecular mass will result in an increase in the electrophoretic mobility of the mutated gp120 compared to the fully glycosylated parental gp120. The gp120 molecules from GM293 (C2), GM299 (V3), GM329 (C3), GM438 (C4), and GM454 (V5) all migrated faster than those from the parental SF162 virus, while the gp120 molecules from GM398 (V4) and GM401 (V4) migrated at the same rate as the parental SF162 gp120 molecules. Similar results were obtained when we compared the electrophoretic mobility of virion-associated gp120 from pseudovirions, produced in 293T cells, containing either the SF162 or a glycosylation mutant envelope (data not shown). These data suggest that of the seven potential N-linked glycosylation sites that we disrupted, five are utilized when SF162 is propagated in human PBMC. These are the asparagines at positions 293 (C2), 299 (V3), 329 (C3), 438 (C4), and 454 (V5).
FIG. 2.
Electrophoretic mobility of virion-associated gp120 molecules during SDS-PAGE, as described in Materials and Methods. No attempt was made to correct for differences in the gp120 content in any of the samples. (A) Comparison of virion-associated gp120 molecules from SF162 and the indicated glycosylation mutants generated by changing asparagine to glutamine; (B) comparison of virion-associated gp120 molecules from SF162 and the indicated glycosylation mutants generated by changing threonine to alanine; (C) acid hydrolysis of recombinant monomeric SF162 gp120 (m gp120) and comparison of acid-hydrolyzed virion-associated gp120 from SF162 and some glycosylation mutants. The 74-kDa band contains regions C1 to C3 of gp120, and the 35-kDa band contains regions V4 to C5 of gp120 (Fig. 1). The 10-kDa band has been intentionally run off the gel.
The fact that the gp120 molecules from GM398 (V4) and GM401 (V4) migrated similarly to the parental SF162 gp120 suggests that the asparagines at these positions are not glycosylated when SF162 propagates in human PBMC in vitro. To eliminate the possibility that the substitution of asparagine by glutamine at these particular positions alters the structure of the gp120 proteins in a way that restores their electrophoretic mobility, we mutated the threonine at the third position of these two potential N-linked glycosylation sites to alanine, generating mutants GM400 and GM403. The electrophoretic mobility of virion-associated gp120 from these mutants was compared to that of the parental SF162 gp120. The gp120 molecules from GM400 and GM403 migrated at the same rate as those from the parental SF162 virus (Fig. 2B). These data support the observation that the asparagines at positions 398 and 401 in the V4 loop are not glycosylated when SF162 is propagated in human PBMC in vitro. However, an alternative explanation for the observed results is that although the asparagines at positions 398 and 401 are glycosylated when SF162 is propagated in human PBMC in vitro (and are eliminated by mutagenesis), during the replication of GM398 and GM401 in PBMC a new glycosylation site is introduced elsewhere on gp120, to compensate for the loss of the glycosylation sites at positions 398 and 401. The presence of such a glycosylation site would therefore restore the electrophoretic mobility of the mutant gp120 envelope to that of the parental gp120 envelope.
To address this possibility, we performed acid hydrolysis on virion-associated gp120 from both the parental and mutant viruses and compared the electrophoretic mobilities of smaller envelope fragments. Glacial acetic acid cleaves proteins between aspartic acid and proline residues (79), cleaving SF162 gp120 at positions 77 and 364, thus producing fragments of approximately 10 kDa (containing the amino-terminal region of C1), 74 kDa (containing the remainder of C1 to the end of C3), and 35 kDa (containing the V4 loop and the remaining C-terminal part of gp120) (Fig. 1). Because the fragments produced by acid hydrolysis are shorter than gp120, small changes in electrophoretic mobility due to modifications in glycosylation would be more easily detectable. Firstly, acid hydrolysis was performed on recombinant SF162 gp120 protein, produced in CHO cells, and the predicted 74-kDa, 35-kDa (Fig. 2C) and 10-kDa (not shown, because it was intentionally run off the gel) fragments were obtained. We then performed acid hydrolysis on GM438 (C4) and GM454 (V5) virion-associated envelope molecules, since we knew that these glycosylation sites are utilized (Fig. 2A) and these sites lie within the 35-kDa fragment. As expected, the 35-kDa fragment from these mutants migrates faster than that from the parental SF162 envelope (Fig. 2C). In contrast, the 35-kDa fragment from the V4 loop mutants GM398, GM400, GM401, and GM403 migrated at the same rate as the 35-kDa fragment from parental SF162. Similarly, the 74-kDa fragment migrated at the same rate as the corresponding SF162 gp120 fragment (Fig. 2C). This again is an indication that the potential N-linked glycosylation sites at positions 398 and 401 are not utilized when SF162 is propagated in human PBMC in vitro.
In order to further disprove the possibility that new glycosylation sites were not introduced elsewhere within the 74- and 35-kDa fragments, as well as the 10-kDa fragment, during replication of SF162 in human PBMC, the extracellular part of the envelope was amplified by PCR from human PBMC infected with SF162, GM398, GM400, GM401, or GM403 and was cloned and sequenced. Analysis of five clones of the glycosylation mutants confirmed that the introduced glycosylation mutation was indeed present and that no additional glycosylation sites had been introduced within gp120 (data not shown). All of the above data indicate that the asparagines at positions 398 (V4) and 401 (V4) are not glycosylated when SF162 is propagated in human PBMC in vitro. We refer to the asparagine-to-glutamine mutants with mutations at positions 398 and 401 as M398 and M401, respectively, and to the alanine-to-threonine mutants with mutations at positions 400 and 403 as M400 and M403, respectively.
Viral replication in PBMC.
The replication kinetics of all the mutant viruses described above were compared to those of the parental SF162 virus to determine whether the disruption of individual N-linked glycosylation sites at positions 293, 299, 329, 438, and 454, the mutation of the asparagine residues at positions 398 and 401, or the mutation of the threonine residues at positions 400 and 403 alters the replication potential of SF162 in human PBMC. With the exception of GM299 (V3), all the viruses expressing the mutant envelopes were capable of efficiently replicating in human PBMC, with kinetics similar to those of the parental SF162 virus (Fig. 3A).
FIG. 3.
Replication kinetics in activated human PBMC. (A) The replication of the mutant viruses (open boxes) was compared to that of the parental SF162 virus (solid boxes), as described in Materials and Methods, using a MOI of 10−5. Values are mean p24 concentrations from triplicate wells, and error bars indicate the standard deviations from the means. The data are from one of four independent experiments, with different pools of PBMC used in each experiment. (B) The replication of glycosylation mutant virus GM299 (V3) (open boxes) was compared to that of the parental SF162 virus (solid boxes) as described for panel A, except that the MOI of each virus was increased to 3 × 10−3.
Although increasing the MOI from 1 × 10−5 to 3 × 10−3 (similar to what is used during the neutralization experiments; see below) resulted in relatively higher GM299 titers in the cell supernatant, these titers were not as high as those recorded with the parental SF162 (Fig. 3B). These results indicate that while some glycans seem to be dispensable for SF162 replication in human PBMC, the presence of the glycan within the V3 loop (GM299) is important for viral infectivity.
Binding of virion-associated mutant envelopes to CD4.
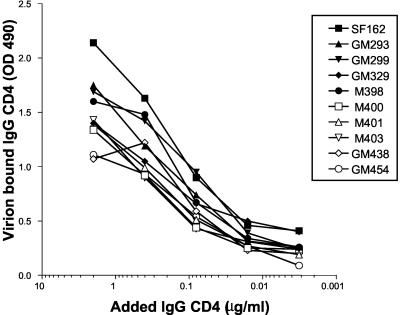
The failure of glycosylation mutant GM299 (V3) to efficiently support viral replication is not due to fewer envelope molecules being incorporated into GM299 virions relative to the parental SF162 virions. The numbers of envelope protein spikes on the parental SF162 and mutant viral particles were similar (7 to 15) and in agreement with previous observations (18). The inefficient replication of GM299 virions could be explained by a decreased ability of the mutant envelope glycoprotein to bind to CD4. Since it has been previously shown that removal of a single N-linked glycosylation site in HIV-2 gp120 can reduce CD4 binding affinity (63), we examined whether CD4 binding was impaired in GM299 (V3) and in the other mutants. As the results in Fig. 4 show, all virion-associated mutant envelopes bound to IgG-CD4 with a similar relative binding affinity (within onefold) to that of the virion-associated parental SF162 envelope.
FIG. 4.
IgG-CD4 binding by SF162 and the mutants. The ability of the glycosylation mutants to bind IgG-CD4 was compared to that of the parental SF162 virus by ELISA, as described in Materials and Methods. The results are from one of two independent experiments.
Effect of envelope-mutations on SF162 entry into target cells.
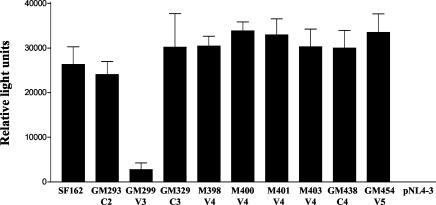
The inability of glycosylation mutant GM299 (V3) to efficiently replicate in human PBMC could be due therefore to post-CD4 binding steps involved in entry. To determine how efficiently the SF162 mutant envelopes support viral entry into cells, we generated single-round replication-competent luciferase reporter viruses expressing either the parental or the mutant SF162 envelope proteins. We compared their ability to infect U87 cells expressing CD4 and CCR5. With the exception of GM299 (V3), the remaining mutant envelopes supported virus entry into cells as efficiently as the parental SF162 envelope protein (Fig. 5). Therefore, the failure of GM299 (V3) to efficiently enter target cells is due not to a failure of its envelope glycoprotein to efficiently bind to CD4 but to a deficiency at some later step in the viral entry process. The inefficiency of the GM299 (V3) envelope to support virus entry into cells explains the poor replication kinetics observed for this mutant virus in PBMC (Fig. 3). These results suggest an important role for this V3 loop glycan in ensuring efficient viral entry into target cells.
FIG. 5.
Envelope-mediated entry into U87 CD4+ CCR5+ cells. The ability of the parental SF162 and the mutant envelopes to support viral entry into U87 CD4+ CCR5+ cells was determined by assessing the levels of luciferase expressed in the cell cytoplasm following viral entry, as described in Materials and Methods. Values are means from triplicate wells, and the error bars indicate the standard deviations from the means. One of three independent experiments is shown. pNL4-3 is a pseudovirus lacking any envelope and was used as a negative control.
Coreceptor usage.
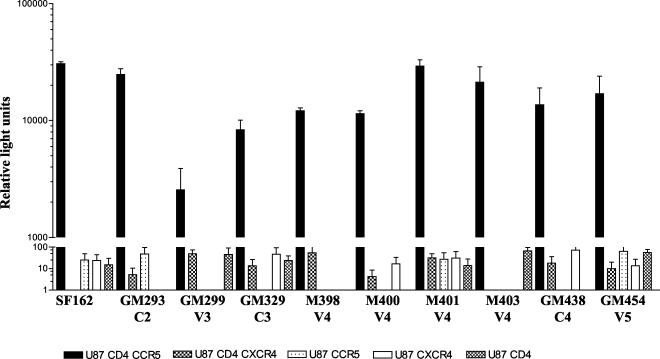
Its has been reported that changes in the glycosylation pattern of the HIV-1 envelope can result in altered coreceptor usage and in CD4 independence for some HIV and SIV isolates (39, 40, 44, 51, 75). We examined if any of the mutations we introduced altered the coreceptor usage of the SF162 envelope by evaluating the ability of single-round replication-competent luciferase reporter viruses expressing either the parental or mutant envelopes to enter into U87 target cells expressing either CD4 and CCR5, CD4 and CXCR4, CCR5, CXCR4, or CD4 alone. As for the parental SF162 virus, all the mutants, including GM299, could enter only cells expressing both CD4 and CCR5 indicating that no change in coreceptor usage had occurred (Fig. 6). Similar observations were made when N-linked glycans were eliminated from the V1V2 region of SF162 (56).
FIG. 6.
Coreceptor usage of the glycosylation mutants. The ability of the glycosylation mutant pseudoviruses to enter cells expressing either CCR5 or CXCR4 and/or CD4 was determined by assessing the luciferase activity of infected cells, as described in Materials and Methods. Values are the means from triplicate wells, and the error bars indicate the standard deviations from the means. The data are from one of two independent experiments. The background obtained with pNL4-3 pseudovirus lacking any envelope has been subtracted.
Effect of envelope deglycosylation on the neutralization phenotype of SF162.
Previous studies have demonstrated the important role of envelope glycosylation on the susceptibility of HIV and SIV to antibody-mediated neutralization (3, 12, 15, 57, 58, 69, 78, 80). Our group previously reported that on the background of SF162, elimination of two N-linked glycosylation sites, one at the amino-terminal and the second at the carboxy-terminal side of the V2 loop (154 and 195, respectively) (Fig. 1), increases the susceptibility of this virus to neutralization by HIV+ sera and MAbs that bind to the V3 loop and the CD4 binding site (56). To investigate whether other N-linked glycosylation sites on the SF162 envelope have the same effect on the susceptibility of this virus to neutralization, we evaluated the susceptibility of the glycosylation mutant viruses to neutralization by HIV+ sera and several MAbs that recognize well-defined epitopes on the HIV envelope.
(i) Neutralization by serum antibodies.
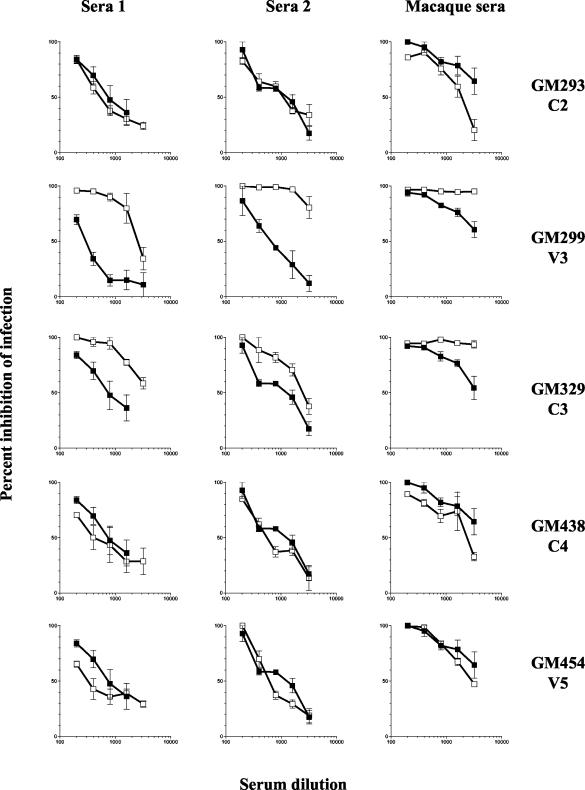
First, we used sera collected from two HIV patients infected with heterologous clade B isolates (serum 1 and serum 2). Of the five glycosylation mutants tested, GM299 (V3) and GM329 (C3) were more susceptible to neutralization than SF162 with both sera tested, whereas there was no change in the susceptibility of GM293 (C2), GM438 (C4), and GM454 (V5) to neutralization compared to SF162 (Fig. 7). We next used sera collected from a macaque infected with SHIVSF162P4 (17), a virus that expresses an envelope that is closely related to that of HIV-1 SF162. As expected, SF162 was more susceptible to neutralization by the macaque sera than by the heterologous human sera. Again, both GM299 (V3) and GM329 (C3) were more susceptible to neutralization than SF162, whereas there was no change in the susceptibility of GM293 (C2), GM438 (C4), and GM454 (V5) to neutralization compared to SF162 (Fig. 7). These studies suggest that while the glycan within the V3 loop (GM299) and in the C3 region (GM329), immediately adjacent to the carboxy-terminal side of the V3 loop, protect SF162 from antibody-mediated neutralization, glycans located in the C2, C4, and V5 regions are most likely not involved in defining the neutralization phenotype of this virus. Alternatively, the sera tested do not contain high titers of antibodies recognizing epitopes occluded by the glycans located within the C2, C4, and V5 regions.
FIG. 7.
Neutralization susceptibility of the N-linked glycosylation mutants. The neutralization susceptibility of the mutant viruses (open boxes) was compared to that of the parental virus (closed boxes), as described in Materials and Methods. Sera 1 and 2 were collected from two HIV-infected patients, and the macaque serum was collected from a rhesus macaque infected with SHIVSF162P4 (17). Values are the mean neutralization from triplicate wells, and the error bars indicate the standard deviations from the means. The data are from one of three independent experiments.
(ii) Neutralization by MAbs.
In an effort to determine the regions that are protected or exposed by the presence or absence of glycans within and adjacent to the V3 loop of SF162 gp120, and to determine whether the glycans present on the C2, C4, and V5 regions occlude specific neutralization epitopes, we used a panel of MAbs with known epitope specificities (Table 2). In contrast to what we observed with sera, all five glycosylation mutants had increased susceptibility to neutralization, relative to SF162, by both IgG-CD4 and IgG1 b12. The latter MAb recognizes a complex conformational epitope that overlaps the CD4 binding site (8, 10, 71, 86, 112). In addition, all five mutants were susceptible to neutralization by the anti-V3 loop MAb 391-95D, which recognizes a conformational epitope flanking the tip of the V3 loop (89, 90), whereas SF162 is resistant to neutralization by this MAb, at the highest concentration tested here (10 μg/ml) (95). The glycosylation mutants with mutations within and adjacent to the V3 loop, GM299 (V3), GM293 (C2), and GM329 (C3), demonstrated the greatest increase in susceptibility to neutralization by this MAb. Removal of these three glycans also resulted in an increase in susceptibility to neutralization by MAb 447D, whose core epitope is the GPXR motif at the tip of the V3 loop with some contribution from sequences outside this region (22, 32, 34). However, although removal of glycans in C4 (GM438) and in V5 (GM454) did increase the susceptibility of SF162 to neutralization by 391-95D, these mutant viruses were as resistant as the parental SF162 to 447D. All five glycosylation mutants, like SF162, were resistant to neutralization by the anti-V2 loop MAb G3.4 (35, 109). In contrast, we previously reported that removal of glycans from the base of the V2 loop of SF162 renders the virus highly susceptible to neutralization by anti-V3 loop MAbs (56).
TABLE 2.
Neutralization of SF162 glycosylation mutants by antibodies
| MAb or Fab | Epitope specificity | IC90, IC80a
|
|||||
|---|---|---|---|---|---|---|---|
| SF162 | GM293 (C2) | GM299 (V3) | GM329 (C3) | GM438 (C4) | GM454 (V5) | ||
| IgG-CD4 | CD4 binding site | 5.2, 4.1 | 0.3, 0.2 | 0.2, 0.1 | 1.0, 0.5 | 1.6, 1.1 | 1.7, 1.2 |
| IgG1 b12 | CD4 binding site | 6.1, 4.8 | 0.3, 0.2 | 0.9, 0.2 | 3.3, 2.1 | 2.0, 1.5 | 1.7, 1.1 |
| 391-95D | V3 loop | —, — | 0.3, 0.2 | <0.08, <0.08 | 0.6, 0.3 | 5.6, 2.7 | 6.0, 3.0 |
| 447D | V3 loop | —, — | 3.3, 1.6 | 0.3, 0.3 | 1.6, 1.0 | —, — | —, — |
| G3.4 | V2 loop | —, — | —, — | —, — | —, — | —, — | —, — |
| 48d | CD4i | —, — | —, — | —, — | —, — | —, — | —, — |
| 17b | CD4i | —, — | —, — | —, — | —, — | —, — | —, — |
| Fab X5 | CD4i | —, — | 4.2, 3.1 | 4.5, 3.3 | —, 4.0 | —, — | —, — |
| Fab M18 | Conserved, unknown | —, — | —, 6.4 | 3.7, 2.5 | —, — | —, — | —, — |
| 50-69 | gp41 | —, — | —, — | —, — | —, — | —, — | —, — |
| 2F5 | gp41 | —, — | 7.7, 5.3 | —, — | —, — | —, 5.0 | —, 6.7 |
IC90 and IC80, concentration of MAb or Fab (in micrograms per milliliter) that results in 90% (IC90) or 80% (IC80) inhibition of infection. Values are average from three independent experiments. —, less than 50% neutralization was achieved with the highest concentration of MAb used (10 μg/ml). CD4i, CD4-induced epitope. Bold indicates an increase in susceptibility to neutralization relative to the parental SF162.
Although all five glycosylation mutants and SF162 were resistant to neutralization (at the highest concentration tested [10 μg/ml]) by two MAbs, 48d and 17b, that recognize overlapping CD4 induced (CD4i) epitopes (97, 103, 105, 107), three glycosylation mutants, GM293 (C2), GM299 (V3), and GM329 (C3), all had increased susceptibility to neutralization by Fab X5, which also recognizes a CD4i epitope (64). This suggests that glycans within and adjacent to the V3 loop occlude regions of the envelope involved in post-CD4 binding, and possibly post-CCR5-binding, steps that lead to virus-cell fusion. In contrast, GM438 (C4) and GM454 (V5) were resistant to neutralization by Fab X5.
Fab M18 (109a) recognizes a currently unknown epitope which is conserved among HIV isolates, and both GM293 (C2) and GM299 (V3) are susceptible to neutralization by this Fab, whereas SF162, GM329 (C3), GM438 (C4), and GM454 (V5) are resistant to neutralization.
To investigate whether the removal of the above glycans from gp120 affects the susceptibility of SF162 to neutralization by antibodies recognizing epitopes in the gp41 extracellular domain, the susceptibility of the glycosylation mutant viruses to neutralization by two anti-gp41 MAbs, 50-69 and 2F5, was examined. 50-69 recognizes a conformational epitope in the extracellular part of gp41 (33, 73, 98, 99, 108), and similar to SF162, all the glycosylation mutants were resistant to neutralization by this MAb. 2F5 recognizes the conserved ELDKWA epitope in the extracellular part of gp41, adjacent to the transmembrane domain (7, 23, 65, 76, 77, 111). Although GM299 (V3) and GM329 (C3) were as resistant to 2F5 neutralization as SF162, GM293 (C2) and to a lesser extent GM438 (C4) and GM454 (V5) were more susceptible to 2F5 neutralization.
Susceptibility of the V4 loop mutant viruses to neutralization by MAbs.
Although the asparagines at positions 398 and 401, in the V4 loop, are not glycosylated during the replication of SF162 in human PBMC, their substitution by glutamine altered the susceptibility of SF162 to neutralization by certain MAbs (Table 3). The susceptibility of SF162 to neutralization by MAbs that target the CD4 binding site, IgG-CD4 and IgG1 b12, and by the anti V3-loop MAb 391-95D increased when asparagines were replaced by glutamines at positions 398 and 401 in the V4 loop. Interestingly, M401 had also increased susceptibility to neutralization by the anti-gp41 MAb 2F5. It appears that the observed changes in the neutralization phenotype of SF162 are due to the change of asparagine to glutamine and not to the elimination of a utilized N-linked glycosylation site, since replacement of threonine by alanine at positions 400 and 403 did not result in a similar increase in susceptibility to neutralization by these MAbs. This supports our observation (see above) that these positions are not glycosylated during replication of SF162 in PBMC. M398 and M401 did not become more susceptible to neutralization by the remaining MAbs and Fabs tested (Table 3).
TABLE 3.
Neutralization of V4 loop mutants by antibodies
| MAb or Fab | Epitope specificity | IC90, IC80a
|
||||
|---|---|---|---|---|---|---|
| SF162 | M398 (V4) | M400 (V4) | M401 (V4) | M403 (V4) | ||
| IgG-CD4 | CD4 binding site | 5.2, 4.1 | 2.1, 1.4 | 4.2, 2.6 | 2.1, 0.8 | 4.9, 3.5 |
| IgG1 b12 | CD4 binding site | 6.1, 4.8 | 0.9, 0.7 | 4.4, 2.8 | 1.9, 1.8 | 4.9, 3.7 |
| 391-95D | V3 loop | —, — | 3.8, 2.0 | —, — | 1.3, 0.5 | —, — |
| 447D | V3 loop | —, — | —, — | —, — | —, — | —, — |
| G3.4 | V2 loop | —, — | —, — | —, — | —, — | —, — |
| 48d | CD4i | —, — | —, — | —, — | —, — | —, — |
| 17b | CD4i | —, — | —, — | —, — | —, — | —, — |
| Fab X5 | CD4i | —, — | —, — | —, — | —, — | —, — |
| Fab M18 | Unknown | —, — | —, — | —, — | —, — | —, — |
| 50-69 | gp41 | —, — | —, — | —, — | —, — | —, — |
| 2F5 | gp41 | —, — | —, — | —, — | —, 7.8 | —, — |
IC90 and IC80, concentration of MAb or Fab (in micrograms per milliliter) that results in 90% (IC90) or 80% (IC80) inhibition of infection. Values are average from three independent experiments. —, less than 50% neutralization was achieved with the highest concentration of MAb used (10 μg/ml). CD4i, CD4-induced epitope. Bold indicates an increase in susceptibility to neutralization relative to the parental SF162.
DISCUSSION
The results presented in this study address the role that asparagine-linked glycosylation within and adjacent to the V3 loop and within the immunologically silent face of gp120 plays in controlling the replication, coreceptor usage, and susceptibility to antibody-mediated neutralization of HIV-1 SF162. Although there are 21 potential N-linked glycosylation sites within the gp120 subunit of the HIV-1 SF162 envelope, our results demonstrate that certain potential N-linked glycosylation sites are not utilized when SF162 is propagated in human PBMC (Fig. 2 and 8).
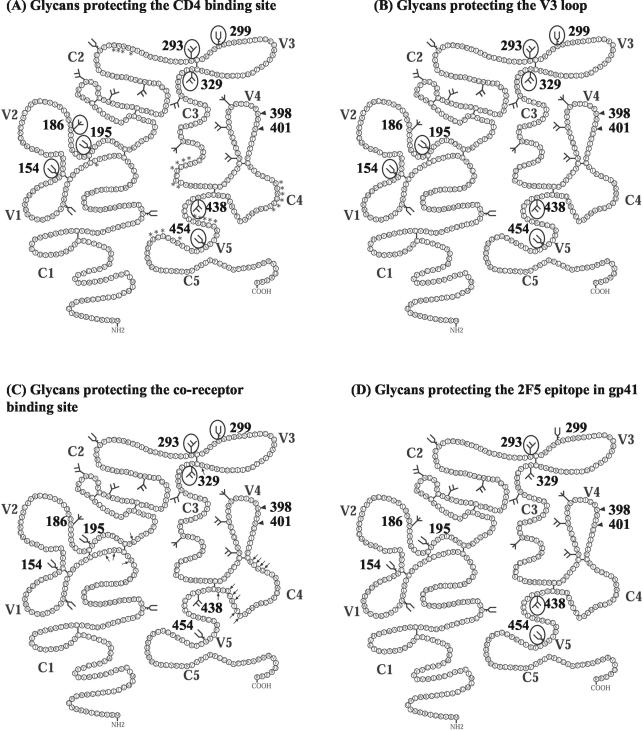
FIG. 8.
Summary of the glycans involved in protecting conserved neutralization epitopes in SF162. The numbered glycans are ones utilized when SF162 is grown in human PBMC (56; also this work), the arrowheads indicate positions of N-linked glycosylation sites that are not utilized when SF162 is grown in human PBMC, and the glycans that are not numbered indicate positions of potential N-linked glycosylation sites whose utilization is not yet known. Glycans that are circled prevent the binding of antibodies to CD4 binding site epitopes (the amino acids that contribute to the formation of the CD4 binding site are indicated by asterisks) (43) (A), V3 loop epitopes (B), CD4i epitopes (the amino acids outside the V3 loop that contribute to the coreceptor binding site are indicated by arrows) (81, 82) (C), and the gp41 epitope recognized by MAb 2F5 (D).
Interestingly, the two potential N-linked glycosylation sites that we found not to be utilized when SF162 replicates in human PBMC in vitro (i.e., in the absence of selective pressure exerted by neutralizing antibodies) are located within the V4 loop (amino acids 398 and 401) in close proximity to each other. These two sites are less conserved in position among diverse HIV-1 isolates than the other potential N-linked glycosylation sites within the V4 loop (31, 53). The nonutilization of potential N-linked glycosylation sites on the envelope glycoproteins of SIV and HIV-1 has been previously documented. When SIVmac239 replicates in MT4 cells, of the 23 potential N-linked glycosylation sites in the gp120 subunit, one site located in C2 is not utilized (68). Similarly, when HXB2 is produced in COS-1 cells, 2 of the 24 potential N-linked glycosylation sites in the gp120 subunit, one located in C2 and one located in V4, are not utilized (48). The exact reasons for the nonutilization of certain potential N-linked glycosylation sites are unknown but probably relate to the folding of the gp120 subunit, which may not allow the enzymatic access necessary for the addition of glycans during protein synthesis. Potential N-linked glycosylation sites need to be properly oriented and accessible and not occluded by adjacent amino acids for the addition of glycans to occur (41, 61). A second possibility has to do with the actual amino acid sequence of the potential glycosylation site and the adjacent regions. Glycosylation motifs containing threonine at the third position are reported to be glycosylated more frequently than those ending in serine, and the amino acid adjacent to the serine or threonine in the glycosylation motif can also be an important determinant of glycosylation efficiency (61). For asparagines at positions 398 and 401 of SF162, both glycosylation motifs end in threonine and are followed by favorable amino acids (asparagine and isoleucine, respectively), suggesting that steric constraints rather than unfavorable amino acids prevent these asparagines from being glycosylated on the SF162 envelope.
The present studies, in combination with those reported previously (56), indicate that while elimination of single N-linked glycosylation sites from the V1V2, C2, C3, V4, C4, and V5 regions does not reduce the ability of SF162 to replicate in human PBMC, the presence of the glycan within the V3 loop (GM299) is required for efficient SF162 replication. Despite the high level of conservation of this V3 loop glycan across diverse HIV and SIV isolates (3, 31, 47, 53), its role in infectivity is not conserved. Disruption of this conserved glycosylation site within the V3 loop of SIVmac239 renders this virus noninfectious (68). In contrast, presence of this glycan is not required for viral infectivity of HIV-1 HXB2 or BRU isolates (3, 51, 52), and the HIV-1 isolate SF33, which lacks this V3 loop glycan, is fully infectious (15, 57). Therefore, the role that this particular V3 loop glycan plays in viral infectivity appears to be isolate dependent. In the case of SF162, the failure of glycosylation mutant GM299 (V3) to efficiently enter target cells is not caused by an inability of the mutant envelope to bind to CD4 (Fig. 4) but is most likely due to a deficiency at some step that follows gp120-CD4 binding. Given that the base of the V3 loop lies adjacent to the bridging sheet in gp120 and that the bridging sheet is involved in coreceptor binding (43, 82, 104, 106), it is possible that removal of the V3 loop glycan alters the structure of SF162 gp120 such that coreceptor binding becomes inefficient. This is supported by data from Rizzuto and Sodroski (81), who demonstrated that an envelope glycoprotein derived from the YU2 primary HIV-1 isolate, which lacks this V3 loop glycan, no longer efficiently binds to CCR5, though it binds to CD4 with an efficiency similar to that of the parental protein. In addition, mutation of this V3 loop glycan in the dually tropic HIV-1 DH12 and SHIV89.6 isolates abolishes their ability to utilize CCR5 as a coreceptor and decreases their ability to utilize CXCR4 (51, 67). For the DH12 V3 loop mutant, it was also demonstrated that there was no loss in CD4 binding. However, our results regarding the role of this V3 loop glycan on SF162 infectivity contradict the findings of others (51, 57). It is possible that these differences are due to the different cell types used to examine viral entry in our study and those previously published. Different cell types express different numbers of CCR5 receptors on their surface. Since GM299 is defective in post-CD4 binding steps in entry, differences in CCR5 surface expression among the various target cells could affect the extent of GM299 entry. Further investigation is required to reconcile these differing results.
The removal of glycans from within and adjacent to the V3 loop and from the C4 and V5 regions of gp120, does not alter the coreceptor usage of the SF162 envelope and does not render it CD4-independent (Fig. 6). The fact that similar mutations on the background of other HIV or SIV isolates alter the coreceptor utilization of the viral envelope and in certain cases allow the envelope to mediate virus-cell fusion in a CD4-independent manner (39, 40, 44, 51, 75) suggests that the effect of envelope glycosylation on receptor usage is isolate specific. This observation is also indicative of subtle structural differences among envelope protein molecules derived from diverse HIV-1 isolates that have significant effects on envelope function.
Although the elimination of glycosylation sites from the C2, V3, C3, C4, and V5 regions of the SF162 gp120 envelope glycoprotein did not alter the CD4 binding potential of this protein (Fig. 4) and, with the exception of GM299 (V3 loop), did not reduce its fusogenic potential (Fig. 5), it differentially affected the susceptibility of SF162 to antibody-mediated neutralization (Table 2).
Neutralization studies performed with MAbs revealed that removal of the glycosylation sites within and adjacent to the V3 loop increased the viral susceptibility not only to neutralization by anti-V3 loop antibodies but also to neutralization by antibodies recognizing epitopes located within the CD4 binding site and epitopes participating in the post-CD4 binding steps of the HIV infection cycle (Table 2 and Fig. 8). The fact that Fab X5, but not MAbs 17b and 48d, was capable of neutralizing (at the concentrations tested here) GM293, GM299, and GM329 (Table 2), even though these three antibodies recognize overlapping CD4-inducible epitopes (64, 107), is most likely due to the smaller size of FabX5, which allows more efficient binding to CD4-induced epitopes (45). Alternatively, Fab X5 binds with a higher affinity to the SF162 envelope than MAbs 17b and 48d. Our results support those recently reported by Koch et al., where elimination of the glycan within the V3 loop on the background of the JR-FL or YU2 envelopes also resulted in an increase in susceptibility to neutralization by anti-V3 loop and anti-CD4 binding site antibodies (38). Thus, the sugar molecules on this V3 loop glycosylation site are positioned in a similar way on the trimeric envelopes derived from diverse primary HIV-1 isolates and thus mask similar neutralization epitopes.
Interestingly, the elimination of the glycosylation site adjacent to the amino terminus of the V3 loop (GM293, C2 region) rendered the virus more susceptible to the anti-gp41 MAb 2F5 (Table 2 and Fig. 8D). This raises the possibility that this glycan occludes part of the extracellular domain of gp41. Alternatively, this modification alters the orientation of the V3 loop in relation to the epitope recognized by MAb 2F5. In support of this finding, it was reported that in a virus clone derived from a chimpanzee-passaged HIV-1 IIIB isolate, mutations in and near the epitope for MAb 2F5 affect the recognition of the V3 loop by human sera and anti-V3 loop MAbs (4). Finally, it is also possible that elimination of this glycan results in global alterations of the gp120 subunit, affecting its interaction with gp41 and resulting in exposure of the epitope recognized by 2F5.
Although the glycans within and adjacent to the V3 loop protect SF162 from neutralization by anti-V3 loop, anti-CD4 binding site, and anti-CD4i antibodies, the present studies indicate that they do not mask the same epitopes (Table 2 and Fig. 7 and 8). For example, only GM299 and GM329 were more susceptible than SF162 to neutralization by human and macaque sera tested. In addition, both GM293 (C2) and GM299 (V3) were more susceptible to neutralization by Fab M18 than GM329 (C3) (Table 2). Finally, only GM293 (C2) was susceptible to neutralization by MAb 2F5.
Even though the envelope of GM299 (V3) binds efficiently to CD4, it is less effective in mediating virus cell entry than the envelope of the parental SF162 virus. It could be argued therefore that the increased susceptibility of GM299 to neutralization by certain MAbs and sera arises not because of increased exposure of neutralizing epitopes per se, but because of a delay in the kinetics of the envelope conformation cascade that lead to virus-cell fusion. Such a delay may result in prolonged exposure of certain neutralization epitopes and render such epitopes more susceptible to antibody-binding, leading to a more efficient neutralization. Although this possibility remains to be properly examined, we note that GM299 did not become susceptible to neutralization by all the MAbs tested (for example, by the anti-gp41 neutralizing MAb 2F5, Table 2) and while removal of this V3 loop glycan from the envelope of HXB2 does not affect viral infectivity it does result in increased susceptibility to neutralization by MAbs that target the CD4 binding site and the V3 loop (3). Similarly, elimination of this glycosylation site from the background of the JR-FL and YU2, does not hinder the fusogenic potential of these envelopes, but renders these viruses more susceptible to neutralization by anti-CD4 binding site antibodies (38).
Our neutralization studies revealed that the presence of the glycans in the C4 (GM438) and the V5 (GM454) regions of gp120 contribute to the neutralization phenotype of SF162. Removal of these glycans renders the virus more susceptible to anti-CD4 binding site MAbs and to certain anti-V3 loop antibodies (Table 2 and Fig. 8). Interestingly, elimination of glycans from the C4 and V5 regions of the SF162 envelope renders the virus more susceptible to the anti-gp41 MAb 2F5 (Table 2 and Fig. 8D). It is possible that the glycans at these two positions are oriented in such a way as to “mask” certain gp41 epitopes (including that of MAb 2F5). An alternative explanation for our results is that within the context of the oligomeric envelope configuration the deglycosylation of the C4 and V5 gp120 regions alters the orientation of regions of gp120 in relation to certain gp41 neutralization epitopes. The above results are, to our knowledge, the first direct demonstration in a primary HIV-1 isolate of a role for glycosylation of the C4 and V5 regions, which comprise part of the immunologically silent face of gp120, in protecting the virus from neutralization by antibodies targeting diverse epitopes within gp120 as well as gp41. Clearly, the effect that glycosylation of the immunologically silent face of the gp120 envelope has on the viral neutralization phenotype merits more extensive investigation.
In contrast to what we observed in the case of the C4 and V5 regions, the two potential glycosylation sites we examined in the V4 region are not utilized when SF162 replicates in human PBMC. Thus, although the replacement of the asparagines at positions 398 and 401 by glutamines was not expected to alter the susceptibility of the virus to neutralization, it has previously been shown that amino acid changes within the V4 loop affect the binding of CD4 and MAb IgG1 b12 to gp120 of JR-CSF (71). This suggests that amino acid changes in the V4 loop might alter the neutralization phenotype of HIV. Indeed, we observed an increase in the susceptibility of M398 and M401 to neutralization by anti-CD4 binding site MAbs and certain anti-V3 loop MAbs (Table 3). This increase was less significant than what we recorded in the case of the C2, V3, C3, C4, and V5 glycosylation mutants (Table 2). Therefore, mutations in V4 can indirectly affect the accessibility of the CD4 binding site and the V3 loop by antibodies and alter the neutralization phenotype of HIV. Interestingly, M401 also became more susceptible to the anti-gp41 MAb 2F5 (Table 3). In contrast, replacement of the threonine at positions 400 and 403 by alanine did not result in changes in the susceptibility of the virus to neutralization (Table 3). Therefore, it is likely that the observed differences in the neutralization susceptibility between M398 and M400 and between M401 and M403 are due to the greater structural changes of the V4 loop caused when asparagine is replaced by glutamine, compared with changes caused when threonine is replaced by alanine at these positions. The fact that replacement of asparagine by glutamine (in the Asn-X-Thr/Ser motif) affects the neutralization phenotype of the virus, while replacement of threonine by alanine does not, further supports our conclusions that the glycosylation sites at positions 398 and 401 are not utilized in SF162.
The above results, however, raise the possibility that the increase in neutralization susceptibility recorded upon elimination of a utilized glycosylation site is not solely due to the loss of carbohydrate molecules but is also due to the specific change of the asparagine to glutamine in the Asn-X-Thr/Ser motif. This possibility merits further investigation. Our results also indicate that although a change in neutralization susceptibility of HIV is recorded when an attempt is made to eliminate a potential N-linked glycosylation site, that does not necessarily mean that the site is actually utilized.
In summary, the present studies and those previously published (56) suggest that glycans present within and adjacent to the V1V2 and V3 loops protect SF162 from antibodies recognizing epitopes participating in CD4 binding steps of the HIV infection cycle. In addition, the present studies indicate that the glycosylation and amino acid composition of the immunologically silent face of gp120 (V4, C4, and V5 regions) protect the virus from neutralization by antibodies directed against the V3 loop and the CD4 binding site without affecting the ability of the envelope to bind to receptor molecules on the target surface. In this respect, our data are in agreement with the recently proposed model of an “evolving glycan shield” to explain HIV escape from antibody-mediated neutralization (101). Our studies suggest that HIV could selectively protect specific neutralization epitopes over the course of infection by altering the utilization of diverse N-linked glycosylation sites. In addition, our studies indicate that glycans in the immunologically silent face of gp120 hinder the binding of neutralizing antibodies to the gp41 transmembrane subunit. Finally, our studies demonstrate that some glycosylation sites will protect HIV from neutralization by antibodies that recognize diverse regions on the viral envelope glycoprotein, while others will have a more limited protective effect.
The knowledge from these studies could contribute toward the design of more effective envelope-based immunogens that will elicit broadly neutralizing antibodies. Given that N-linked glycans located in different regions of gp120 prevent antibodies from binding to the CD4 binding site, multiple glycosylation sites might have to be eliminated from envelope-based immunogens in order to elicit neutralizing antibodies against such conserved epitopes that are shared among diverse primary HIV isolates.
Acknowledgments
These studies were supported by NIH grants AI47708 and AI51217 (L.S.). L.S. also acknowledges SBRI's private donors for their financial support.
We thank all those who provided Fabs and MAbs for these studies.
REFERENCES
- 1.Alkhatib, G., C. Combadiere, C. Broder, Y. Feng, P. Kennedy, P. Murphy, and E. Berger. 1996. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 2.Allan, J., J. Coligan, F. Barin, M. McLane, J. Sodroski, C. Rosen, W. Haseltine, T. Lee, and M. Essex. 1985. Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science 228:1091-1094. [DOI] [PubMed] [Google Scholar]
- 3.Back, N., L. Smit, J. De Jong, W. Keulen, M. Schutten, J. Goudsmit, and M. Tersmette. 1994. An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology 199:431-438. [DOI] [PubMed] [Google Scholar]
- 4.Back, N., L. Smit, M. Schutten, P. Nara, M. Tersmette, and J. Goudsmit. 1993. Mutations in human immunodeficiency virus type 1 gp41 affect sensitivity to neutralization by gp120 antibodies. J. Virol. 67:6897-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barin, F., M. McLane, J. Allan, T. Lee, J. Groopman, and M. Essex. 1985. Virus envelope protein of HTLV-III represents major target antigen for antibodies in AIDS patients. Science 228:1094-1096. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein, H., S. Tucker, E. Hunter, J. Schutzback, and R. Compans. 1994. Human immunodeficiency virus type 1 envelope glycoprotein is modified by O-linked oligosaccharides. J. Virol. 68:463-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchacher, A., R. Predl, K. Strutzenberger, W. Steinfellner, A. Trkola, M. Purtscher, G. Gruber, C. Tauer, F. Steindl, A. Jungbauer, and H. Katinger. 1994. Generation of human monoclonal antibodies against HIV-1 proteins: electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retrovir. 10:359-369. [DOI] [PubMed] [Google Scholar]
- 8.Burton, D., J. Pyati, R. Koduri, S. Sharp, G. Thornton, P. Parren, L. Sawyer, R. Hendry, N. Dunlop, P. Nara, M. Lamacchia, G. E., E. Stiehm, Y. Bryson, Y. Cao, J. Moore, D. Ho, and C. Barbas. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 9.Calarese, D., C. Scanlan, M. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. Wormald, R. Stanfield, K. Roux, J. Kelly, P. Rudd, R. Dwek, H. Katinger, D. Burton, and I. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065-2071. [DOI] [PubMed] [Google Scholar]
- 10.Capon, D., S. Chamow, J. Mordenti, S. Marsters, T. Gregory, H. Mitsuya, R. Bryn, C. Lucas, F. Wurm, J. Groopman, S., and D. Smith. 1989. Designing CD4 immunoadhesins for AIDS therapy. Nature 337:525-531. [DOI] [PubMed] [Google Scholar]
- 11.Carrillo, A., and L. Ratner. 1996. Human lymphoid virus type 1 tropism for T-lymphoid cell lines: role of the V3 loop and C4 envelope determinants. J. Virol. 70:1301-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chackerian, B., L. Rudensey, and J. Overbaugh. 1997. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 71:7719-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakrabarti, L., T. Ivanovic, and C. Cheng-Mayer. 2002. Properties of the surface envelope glycoprotein associated with virulence of simian-human immunodeficiency virus SHIVSF33A molecular clones. J. Virol. 76:1588-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan, D., and P. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 15.Cheng-Mayer, C., A. Brown, J. Harouse, P. Luciw, and A. Mayer. 1999. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J. Virol. 73:5294-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng-Mayer, C., M. Quiroga, J. Tung, D. Dina, and J. Levy. 1990. Viral determinants of human immunodeficiency virus type 1 T-cell or macrophage tropism, cytopathogenicity, and CD4 antigen modulation. J. Virol. 64:4390-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cherpelis, S., I. Shrivastava, A. Gettie, X. Jin, D. Ho, S. Barnett, and L. Stamatatos. 2001. DNA vaccination with the human immunodeficiency virus type 1 SF162ΔV2 envelope elicits immune responses that offer partial protection from simian/human immunodeficiency virus infection to CD8+ T-cell-depleted rhesus macaques. J. Virol. 75:1547-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chertova, E., J. Bess, B. Crise, R. Sowder, T. Schaden, J. Hilburn, J. Hoxie, R. Benveniste, J. Lifson, L. Henderson, and L. Arthur. 2002. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 76:5315-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chesebro, B., J. Nishio, S. Perryman, A. Cann, W. O'Brien, I. Chen, and K. Wehrly. 1991. Identification of human immunodeficiency virus envelope gene sequences influencing viral entry into CD4-positive HeLa cells, T-leukemia cells, and macrophages. J. Virol. 65:5782-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chesebro, B., K. Wehrly, J. Nishio, and S. Perryman. 1996. Mapping of independent V3 envelope determinants of human immunodeficiency virus type 1 macrophage tropism and syncytium formation in lymphocytes. J. Virol. 72:9055-9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. Ponath, L., C. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 22.Conley, A. J., M. K. Gorny, J. A. Kessler II, L. J. Boots, M. Ossorio-Castro, S. Koenig, D. W. Lineberger, E. A. Emini, C. Williams, and S. Zolla-Pazner. 1994. Neutralization of primary human immunodeficiency virus type 1 isolates by the broadly reactive anti-V3 monoclonal antibody, 447-52D. J. Virol. 68:6994-7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conley, A., J. Kessler, L. Boots, J. Tung, B. Arnold, P. Keller, A. Shaw, and E. Emini. 1994. Neutralization of divergent human immunodeficiency virus type 1 variants and primary isolates by IAM-41-2F5, an anti gp41 human monoclonal antibody. Proc. Natl. Acad. Sci. USA 91:3348-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Connor, R., B. Chen, S. Choe, and N. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type 1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 25.Dalgleish, A., P. Beverley, P. Clapham, D. Crawford, M. Greaves, and R. Weiss. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763-767. [DOI] [PubMed] [Google Scholar]
- 26.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. diMarzio, S. Marmon, R. Sutton, C. Hill, C. Davis, S. Peiper, T. Schall, D. Littman, and N. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 27.Doranz, B., J. Rucker, Y. Yi, R. Smyth, M. Samson, S. Peiper, M. Parmentier, R. Collman, and R. Doms. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149-1158. [DOI] [PubMed] [Google Scholar]
- 28.Dragic, T., V. Litwin, G. Allaway, S. Martin, Y. Huang, K. Nagashima, C. Cayanan, P. Maddon, R. Koup, J. Moore, and W. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 29.Feng, F., C. Broder, P. Kennedy, and E. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 276:51-52. [DOI] [PubMed] [Google Scholar]
- 30.Freed, E., D. Myers, and R. Risser. 1990. Characterization of the fusion domain of the human immunodeficiency virus type 1 envelope glycoprotein gp41. Proc. Natl. Acad. Sci. USA 87:4650-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao, F., S. G. Morrison, D. L. Robertson, C. L. Thornton, S. Craig, G. Karlsson, J. Sodroski, M. Morgado, B. Galvao-Castro, H. von Briesen, S. Beddows, J. Weber, P. J. Sharp, G. M. Shaw, B. H. Hahn, and the WHO and NIAID Networks for HIV Isolation and Characterization. 1996. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. J. Virol. 70:1651-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorny, M., A. Conley, S. Karwowska, A. Buchbinder, J. Xu, E. Emini, S. Koenig, and S. Zolla-Pazner. 1992. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 66:7538-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorny, M., V. Gianakakos, S. Sharpe, and S. Zolla-Pazner. 1989. Generation of human monoclonal antibodies to human immunodeficiency virus. Proc. Natl. Acad. Sci. USA 86:1624-1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorny, M., J. Xu, S. Karwowska, A. Buchbinder, and S. Zolla-Pazner. 1993. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J. Immunol. 150:635-643. [PubMed] [Google Scholar]
- 35.Ho, D., M. Fung, Y. Cao, X. Li, C. Sun, T. Chang, and N. Sun. 1991. Another discontinuous epitope on glycoprotein gp120 that is important in human immunodeficiency virus type 1 neutralization is identified by a monoclonal antibody. Proc. Natl. Acad. Sci. USA 88:8949-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang, S. S., T. J. Boyle, H. K. Lyerly, and B. R. Cullen. 1991. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science 253:71-74. [DOI] [PubMed] [Google Scholar]
- 37.Klatzman, D., E. Champagne, S. Chamaret, J. Gruest, D. Guetard, T. Hercend, J. Gluckman, and L. Montagnier. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767-768. [DOI] [PubMed] [Google Scholar]
- 38.Koch, M., M. Pancera, P. Kwong, P. Kolchinsky, C. Grundner, L. Wang, W. Hendrickson, J. Sodroski, and R. Wyatt. 2003. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology 313:387-400. [DOI] [PubMed] [Google Scholar]
- 39.Kolchinsky, P., E. Kiprilov, P. Bartley, R. Rubinstein, and J. Sodroski. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J. Virol. 75:3435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolchinsky, P., T. Mirzabekov, M. Farzan, E. Kiprilov, M. Cayabyab, L. Mooney, H. Choe, and J. Sodroski. 1999. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J. Virol. 73:8120-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kornfeld, R., and S. Kornfeld. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54:631-634. [DOI] [PubMed] [Google Scholar]
- 42.Kowalski, M., J. Potz, L. Basiripour, T. Dorfman, W. Goh, E. Terwilliger, A. Dayton, C. Rosen, W. Haseltine, and J. Sodroski. 1987. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science 237:1351-1355. [DOI] [PubMed] [Google Scholar]
- 43.Kwong, P., R. Wyatt, J. Robinson, R. Sweet, J. Sodroski, and W. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LaBranche, C., T. Hoffman, J. Romano, B. Haggarty, T. Edwards, T. Matthews, R. Doms, and J. Hoxie. 1999. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity. J. Virol. 73:10310-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Labrijn, A., P. Poignard, A. Raja, M. Zwick, K. Delgado, M. Franti, J. Binley, V. Vivona, C. Grundner, C. Huang, M. Venturi, C. Petropoulos, T. Wrin, D. Dimitrov, J. Robinson, P. Kwong, R. Wyatt, J. Sodroski, and D. Burton. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Land, A., and I. Braakman. 2001. Folding of the human immunodeficiency virus type 1 envelope glycoprotein in the endoplasmic reticulum. Biochimie 83:783-790. [DOI] [PubMed] [Google Scholar]
- 47.LaRosa, G., J. Davide, K. Weinhold, J. Waterbury, A. Profy, J. Lewis, A. Langlois, G. Dreesman, R. Boswell, P. Shadduck, H. Holley, M. Karplus, D. Bolognesi, T. Matthews, E. Emini, and S. Putney. 1990. Conserved sequence and structural elements in the HIV-1 principal neutralizing determinant. Science 249:932-935. [DOI] [PubMed] [Google Scholar]
- 48.Lee, W., W. Syu, B. Du, M. Matsuda, S. Tan, A. Wolf, M. Essex, and T. Lee. 1992. Nonrandom distribution of gp120 N-linked glycosylation sites important for infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 89:2213-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leonard, C., M. Spellman, L. Riddle, R. Harris, J. Thomas, and T. Gregory. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265:10373-10382. [PubMed] [Google Scholar]
- 50.Li, Y., L. Luo, N. Rasool, and C. Kang. 1993. Glycosylation is necessary for the correct folding of human immunodeficiency virus gp120 in CD4 binding. J. Virol. 67:584-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li, Y., M. Rey-Cuille, and S. Hu. 2001. N-linked glycosylation in the V3 region of HIV type 1 surface antigen modulates coreceptor usage in viral infection. AIDS Res. Hum. Retrovir. 17:1473-1479. [DOI] [PubMed] [Google Scholar]
- 52.Losman, B., M. Biller, S. Olofsson, K. Schonning, O. Sogaard Lund, B. Svennerholm, J. Hansen, and A. Bolmstedt. 1999. The N-linked glycan of the V3 region of HIV-1 gp120 and CXCR4 dependent multiplication of a human immunodeficiency virus type 1 lymphocyte-tropic variant. FEBS Lett. 454:47-52. [DOI] [PubMed] [Google Scholar]
- 53.Louwagie, J., W. Janssens, J. Mascola, L. Heyndrickx, P. Hegerich, G. van der Groen, F. McCutchan, and D. Burke. 1995. Genetic diversity of the envelope glycoprotein from human immunodeficiency virus type 1 isolates of African origin. J. Virol. 69:263-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu, M., S. Blackow, and P. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 55.Lue, J., M. Hsu, D. Yang, P. Marx, Z. Chen, and C. Cheng-Mayer. 2002. Addition of a single gp120 glycan confers increased binding to dendritic cell-specific ICAM-3-grabbing nonintegrin and neutralization escape to human immunodeficiency virus type 1. J. Virol. 76:10299-10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ly, A., and L. Stamatatos. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malenbaum, S., D. Yang, L. Cavacini, M. Posner, J. Robinson, and C. Cheng-Meyer. 2000. The N-terminal V3 loop glycan modulates the interaction of clade A and B human immunodeficiency virus type 1 envelopes with CD4 and chemokine receptors. J. Virol. 74:11008-11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malenbaum, S., D. Yang, and C. Cheng-Meyer. 2001. Evidence for similar recognition of the conserved neutralization epitopes of human immunodeficiency virus type 1 envelope gp120 in humans and macaques. J. Virol. 75:9287-9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marshall, R. 1972. Glycoproteins. Annu. Rev. Biochem. 41:673-702. [DOI] [PubMed] [Google Scholar]
- 60.McDougal, J., M. Kennedy, J. Sligh, S. Cort, A. Mawle, and J. Nicholson. 1986. Binding of HTLV-III/LAV to T4+ T-cells by a complex of the 110K viral protein and the T4 molecule. Science 231:382-385. [DOI] [PubMed] [Google Scholar]
- 61.Mellquist, J. L., L. Kasturi, S. L. Spitalnik, and S. H. Shakin-Eshleman. 1998. The amino acid following an Asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry 37:6833-6837. [DOI] [PubMed] [Google Scholar]
- 62.Montagnier, L., F. Clavel, B. Krust, S. Chamaret, F. Rey, F. Barre-Sinoussi, and J. Chermann. 1985. Identification and antigenicity of the major envelope glycoprotein of lymphadenopathy-associated virus. Virology 144:283-289. [DOI] [PubMed] [Google Scholar]
- 63.Morikawa, Y., J. Moore, A. Wilkinson, and I. Jones. 1991. Reduction in CD4 binding affinity associated with removal of a single glycosylation site in the external glycoprotein of HIV-2. Virology 180:853-856. [DOI] [PubMed] [Google Scholar]
- 64.Moulard, M., S. Phogat, Y. Shu, A. Labrijn, X. Xiao, J. Binley, M. Zhang, I. Sidorov, C. Broder, J. Robinson, P. Parren, D. Burton, and D. Dimitrov. 2002. Broadly cross-reactive HIV-1 neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc. Natl. Acad. Sci. USA 99:6913-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Narayan, S., S. Mukherjee, F. Jia, Z. Li, C. Wang, L. Foresman, C. McCormack-Davis, E. Stephens, S. Joag, and O. Narayan. 1999. Characterization of a neutralization-escape variant of SHIVku-1, a virus that causes acquired immune deficiency syndrome in pig-tailed macaques. Virology 156:54-63. [DOI] [PubMed] [Google Scholar]
- 67.Ogert, R., M. Lee, W. Ross, A. Buckler-White, M. Martin, and M. Cho. 2001. N-linked glycosylation sites adjacent to and within the V1/V2 and the V3 loops of dualtropic human immunodeficiency virus type 1 isolate DH12 affect coreceptor usage and cellular tropism. J. Virol. 75:5998-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ohgimoto, S., T. Shioda, K. Mori, E. Nakayama, H. Hu, and Y. Nagai. 1998. Location-specific, unequal contribution of the N glycans in simian immunodeficiency virus gp120 to viral infectivity and removal of multiple glycans without disturbing infectivity. J. Virol. 72:8365-8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Overbaugh, J., and L. Rudensey. 1992. Alterations in potential sites for glycosylation predominate during evolution of the simian immunodeficiency virus envelope gene in macaques. J. Virol. 66:5937-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Overbaugh, J., L. Rudensey, M. Papenhausen, R. Benveniste, and W. Morton. 1991. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J. Virol. 65:7025-7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pantophlet, R., E. Ollmann Saphire, P. Poignard, P. Parren, I. Wilson, and D. Burton. 2003. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 77:642-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Parren, P., J. Moore, D. Burton, and Q. Sattentau. 1999. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13:S137-S162. [PubMed] [Google Scholar]
- 73.Pinter, A., W. Honnen, S. Tilley, C. Bona, H. Zaghouani, M. Gorny, and S. Zolla-Pazner. 1989. Oligomeric structure of gp41, the transmembrane protein of human immunodeficiency virus type 1. J. Virol. 63:2674-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Planelles, V., A. Haislip, E. Withers-Ward, Y. Xie, and I. Chen. 1995. A new reporter system for detection of viral infection. Gene Ther. 2:369-376. [PubMed] [Google Scholar]
- 75.Puffer, B. A., S. Pohlamnn, A. L. Edinger, D. Carlin, M. D. Sanchez, J. Reitter, D. D. Watry, H. S. Fox, R. C. Desrosiers, and R. W. Doms. 2002. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J. Virol. 76:2595-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Purtscher, M., A. Trkola, A. Grassauer, P. Schilz, A. Klima, S. Dopper, G. Gruber, A. Buchacher, T. Muster, and H. Katinger. 1996. Restricted variability of the epitope recognized by the neutralizing antibody 2F5. AIDS 10:587-593. [DOI] [PubMed] [Google Scholar]
- 77.Purtscher, M., A. Trkola, G. Gruber, A. Buchacher, R. Predl, F. Steindl, C. Tauer, R. Berger, N. Barrett, A. Jungbauer, and H. Katinger. 1994. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 10:1651-1658. [DOI] [PubMed] [Google Scholar]
- 78.Quinones-Kochs, M., L. Bunonocore, and J. Rose. 2002. Role of N-linked glycans in a human immunodeficiency virus envelope glycoprotein: effects on protein function and the neutralizing antibody response. J. Virol. 76:4199-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reitter, J., and R. Desrosiers. 1998. Identification of replication-competent strains of simian immunodeficiency virus lacking multiple attachment sites for N-linked carbohydrates in variable regions 1 and 2 of the surface envelope protein. J. Virol. 72:5399-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reitter, J., R. Means, and R. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 81.Rizzuto, C., and J. Sodroski. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum. Retrovir. 16:741-749. [DOI] [PubMed] [Google Scholar]
- 82.Rizzuto, C., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. Kwong, W. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 83.Robey, W., B. Safi, S. Oroszlan, L. Arthur, M. Gonda, R. Gallo, and P. Fischinger. 1985. Characterization of envelope and core structural gene products of HTLV-III with sera from AIDS patients. Science 228:593-595. [DOI] [PubMed] [Google Scholar]
- 84.Rudensey, L., J. Kimata, A. Long, B. Chackerian, and J. Overbaugh. 1998. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of simian immunodeficiency virus SIVMne infection affect neutralizing antibody recognition, syncytium formation, and macrophage tropism but not replication, cytopathicity, or CCR-5 coreceptor recognition. J. Virol. 72:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sanders, R., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. Lloyd, P. Kwong, and J. Moore. 2002. The mannose-dependent epitope for the neutralization antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saphire, E., P. Parren, R. Pantophlet, M. Zwick, G. Morris, P. Rudd, R. Dwek, R. Stanfield, D. Burton, and I. Wilson. 2001. Crystal structure of a neutralizing human IgG against HIV-1: a template design for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 87.Sattentau, Q., and J. Moore. 1993. Conformational changes induced in the envelope glycoproteins of human and simian immunodeficiency viruses by soluble receptor binding. J. Virol. 67:7383-7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Scanlan, C., R. Pantophlet, M. Wormald, E. Saphire, R. Stanfield, I. Wilson, H. Katinger, R. Dwek, P. Rudd, and D. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1-2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Seligman, S., J. Binley, M. Gorny, D. Burton, S. Zolla-Pazner, and K. Sokolowski. 1996. Characterization by serial deletion competition ELISAs of HIV-1 V3 loop epitopes recognized by monoclonal antibodies. Mol. Immunol. 33:737-745. [DOI] [PubMed] [Google Scholar]
- 90.Sharon, M., N. Kessler, R. Levy, S. Zolla-Pazner, M. Gorlach, and J. Anglister. 2003. Alternative conformations of HIV-1 V3 loops mimic β hairpins in chemokines, suggesting a mechanism for coreceptor selectivity. Structure 11:225-236. [DOI] [PubMed] [Google Scholar]
- 91.Shioda, T., J. Levy, and C. Cheng-Mayer. 1991. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature 349:167-169. [DOI] [PubMed] [Google Scholar]
- 92.Shioda, T., J. Levy, and C. Cheng-Mayer. 1992. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 89:9434-9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stamatatos, L., and C. Cheng-Mayer. 1998. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J. Virol. 72:7840-7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stamatatos, L., and C. Cheng-Mayer. 1995. Structural modulations of the envelope gp120 glycoprotein of human immunodeficiency virus type 1 upon oligomerization and differential V3 loop epitope exposure of isolates displaying distinct tropism upon virion-soluble receptor binding. J. Virol. 69:6191-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stamatatos, L., S. Zolla-Pazner, M. Gorny, and C. Cheng-Mayer. 1997. Binding of antibodies to virion associated gp120 molecules of primary-like human immunodeficiency virus type 1 (HIV-1) isolates: effect on HIV-1 infection of macrophages and peripheral blood mononuclear cells. Virology 229:360-369. [DOI] [PubMed] [Google Scholar]
- 96.Starich, B., B. Hahn, G. Shaw, P. McNeely, S. Modrow, H. Wolf, E. Parks, W. Parks, S. Josephs, R. Gallo, and F. Wong-Staal. 1986. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell 45:637-648. [DOI] [PubMed] [Google Scholar]
- 97.Thali, M., J. Moore, C. Furman, M. Charles, D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Till, M., V. Ghetie, R. May, P. Auerback, S. Zolla-Pazner, M. Gorny, T. Gregory, J. Uhr, and E. Vitetta. 1990. Immunoconjugates containing ricin A chain and either human anti-gp41 of C4 kill H9 cells infected with different isolates of HIV, but do not inhibit normal T or B cell function. J. Acquir. Immune Defic. Syndr. 3:609-614. [PubMed] [Google Scholar]
- 99.Tyler, D., S. Stanley, S. Zolla-Pazner, M. Gorny, P. Shadduck, A. Langlois, T. Matthews, D. Bolognesi, T. Palker, and K. Weinhold. 1990. Identification of sites within gp41 that serve as targets for antibody-dependent cellular cytotoxicity by using human monoclonal antibodies. J. Immunol. 145:3276-3282. [PubMed] [Google Scholar]
- 100.Veronese, F., A. DeVico, T. Copeland, S. Oroszlan, R. Gallo, and M. Sarngadharan. 1985. Characterization of gp41 as the transmembrane protein coded by the HTLV-III/LAV envelope gene. Science 229:1402-1405. [DOI] [PubMed] [Google Scholar]
- 101.Wei, X., J. Decker, S. Wang, H. Hui, J. Kappes, X. Wu, J. Salazar-Gonzalez, M. Salazar, J. Kilby, M. Saag, N. Komarova, M. Nowak, B. Hahn, P. Kwong, and G. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 102.Willey, R., R. Rutledge, D. Dias, T. Folks, T. Theodore, C. Buckler, and M. Martin. 1986. Identification of conserved and divergent domains within the envelope gene of the acquired immunodeficiency syndrome retrovirus. Proc. Natl. Acad. Sci. USA 83:5038-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu, L., N. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 104.Wyatt, R., P. Kwong, E. Desjardin, R. Sweet, J. Robinson, W. Hendrickson, and J. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 105.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 107.Xiang, S., N. Doka, R. Choudhary, J. Sodroski, and J. Robinson. 2002. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res. Hum. Retrovir. 18:1207-1217. [DOI] [PubMed] [Google Scholar]
- 108.Xu, J.-Y., M. K. Gorny, T. Palker, S. Karwowska, and S. Zolla-Pazner. 1991. Epitope mapping of two immunodominant domains of gp41, the transmembrane protein of human immunodeficiency virus type 1, using ten human monoclonal antibodies. J. Virol. 65:4832-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ysohiyama, H., H. Mo, J. Moore, and D. Ho. 1994. Characterization of mutants of human immunodeficiency virus type 1 that have escaped neutralization by a monoclonal antibody to the gp120 V2 loop. J. Virol. 68:974-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109a.Zhang, M., Y. Shu, S. Phogat, X. Xiao, F. Cham, P. Bouma, A. Choudhary, Y. Feng, I. Sanz, S. Rybak, C. Broder, G. Quinnan, L. Evans, and D. Dimitrov. 2003. Broadly cross-reactive HIV neutralizing human monoclonal antibody Fab selected by sequential antigen panning of a phage display library. J. Immunol. Methods 283:17-25. [DOI] [PubMed] [Google Scholar]
- 110.Zhu, X., C. Borchers, R. Bienstock, and K. Tomer. 2000. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry 39:11194-11204. [DOI] [PubMed] [Google Scholar]
- 111.Zwick, M., A. Labrijn, M. Wang, C. Spenlehauer, E. Saphire, J. Binley, J. Moore, G. Stiegler, H. Katinger, D. Burton, and P. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zwick, M., P. Parren, E. Saphire, S. Church, M. Wang, J. Scott, P. Dawson, I. Wilson, and D. Burton. 2003. Molecular features of the broadly neutralizing immunoglobulin G1 b12 required for recognition of human immunodeficiency virus type 1 gp120. J. Virol. 77:5863-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]