Abstract
Paramyxoviruses are the leading cause of respiratory disease in children. Several paramyxoviruses possess a surface glycoprotein, the hemagglutinin-neuraminidase (HN), that is involved in attachment to sialic acid receptors, promotion of fusion, and removal of sialic acid from infected cells and progeny virions. Previously we showed that Newcastle disease virus (NDV) HN contained a pliable sialic acid recognition site that could take two states, a binding state and a catalytic state. Here we present evidence for a second sialic acid binding site at the dimer interface of HN and present a model for its involvement in cell fusion. Three different crystal forms of NDV HN now reveal identical tetrameric arrangements of HN monomers, perhaps indicative of the tetramer association found on the viral surface.
Viruses of the Paramyxoviridae family are the leading cause of respiratory disease in children. The human parainfluenza viruses are members of the Paramyxovirinae subfamily, which includes mumps virus, Newcastle disease virus (NDV), Sendai virus, and simian virus type 5 (27). The paramyxoviruses have two surface glycoproteins, a hemagglutinin-neuraminidase (HN) protein and a fusion (F) protein. HN has three functions: it recognizes sialic acid-containing receptors on cell surfaces, it promotes the fusion activity of the F protein, allowing the virus to penetrate the cell surface, and it acts as a neuraminidase (sialidase), removing sialic acids from progeny virus particles to prevent viral self-agglutination (27). Many studies show that only homotypic HN and F can induce fusion (19, 26, 27), suggesting that there is a specific interaction between HN and F, involving both the stalk and globular head regions of HN. The multifunctional HN molecule makes it an attractive target for structure-based drug design for diseases caused by paramyxoviruses.
We previously reported the crystal structure of NDV HN and its complexes with N-acetylneuraminic acid (Neu5Ac) and its unsaturated derivative, the inhibitor 2-deoxy-2,3-dehydro-N-acetylneuraminic acid (Neu5Ac2en) (14). The Neu5Ac2en complex revealed a tight, dimeric association of HN monomers and identified the active site sitting at the center of the β-propeller structure. This crystal form, obtained at pH 6.3, could only be obtained in the presence of ligand, suggesting that the ligand induced a structural change in HN leading to the stable dimer. A second crystal form could be obtained in the absence of ligand at pH 4.6, but this showed a dramatically different monomer association. Soaking these crystals in sialyllactose [Neu5Acα(2,3)Galβ(1,4)Glc] resulted in occupation of the active site by the β-anomer of N-acetylneuraminic acid: either the neuraminidase activity or the low pH led to acid hydrolysis of the substrate, and the site was flexible enough to accommodate the energetically more stable β-anomer formed by mutarotation of the released N-acetylneuraminic acid. Comparison of the two crystal forms revealed major changes in the active site, suggesting a pliable site that could switch between a sialic acid binding site and a catalytic site. Subsequent site-directed mutagenesis supported this hypothesis; mutation of most of the amino acids at the site severely reduced neuraminidase activity and also had a significant effect on hemagglutinin activity (an assay for sialic acid binding) and fusion activity (12). Further mutagenesis studies on NDV HN also suggested that the dimer interface is involved in the fusion process (13, 42).
The picture emerging from these experiments is that the pliable active site and the dimer association are somehow involved in the promotion of fusion. For paramyxoviruses, fusion of the viral and host cell membranes occurs at the cell surface, in contrast to the influenza virus, which undergoes endocytosis when the lower endosomal pH triggers a dramatic structural change in the influenza virus hemagglutinin (6). In the case of paramyxoviruses, binding of HN to sialic acid receptors must trigger the fusion protein into its fusogenic state through an association between HN and F (26).
In order to further define the switchable sialic acid recognition site, we reasoned that we should be able to trap N-acetylneuraminic acid in its α-anomeric form through the use of a nonhydrolyzable substrate. The thiosialoside Neu5Ac-2-S-α(2,6)Gal1OMe (Fig. 1) was cocrystallized with NDV HN and surprisingly revealed the inhibitor Neu5Ac2en bound in the active site. In addition, the complete disaccharide was found bound at two previously unidentified symmetrical sites at the HN dimer interface. A model that uses the release of sialic acid from sialic acid-containing receptors to drive a structural change in HN that triggers the fusion protein is developed. The role of this second sialic acid binding site is discussed in relation to this model. In addition, the results presented here suggest a tetrameric arrangement of HN that may represent a form found on the viral surface.
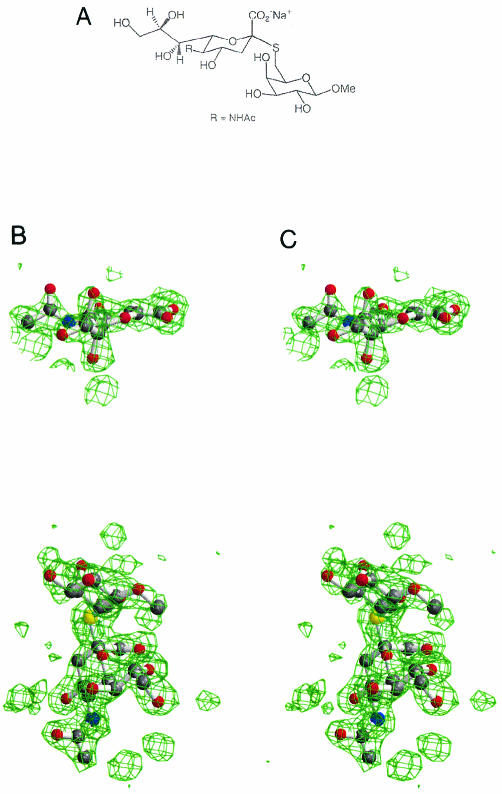
FIG. 1.
Thiosialoside ligand and its appearance in the electron density maps. (A) The thiosialoside Neu5Ac-2-S-α(2,6)Gal1OMe. (B) Stereo Fo-Fc electron density map for one of the active sites of the orthorhombic crystal form. (C) Stereo Fo-Fc electron density map at one of the symmetrical sialic acid binding sites at the dimer interface. Both electron density maps were contoured at 2.5 σ, and the refined ligand is superimposed.
MATERIALS AND METHODS
Crystallization.
Prior to crystallization, 10 volumes of NDV HN (12 mg/ml) were incubated at room temperature for 2 h with 2 volumes of 100 mM thiosialoside, diluted in 0.1 M HEPES buffer (pH 6.3); 15 and 20% polyethylene glycol 3350 in 0.1 M HEPES buffer (pH 6.3)-2 mM CaCl2 (mother liquor) were used as a precipitant, mixed at 1:1 and 2:1 ratios with the protein solution in the drops. Two crystal forms were obtained under these conditions. Form 1 crystals appeared in a week in the form of thin plates and typically grew to dimensions of 0.1 by 0.2 by 0.5 mm. These crystals also grew if we used a 1.7 to 2.0 mM cocktail of lanthanide salts (Eu, Pr, and Ho) as an additive in the precipitant solution. Form 2 crystals grew in the same conditions in the shape of tetragonal bipyramids but over a longer time than form 1 (up to 1 month). The typical dimensions of the crystals were 0.15 by 0.15 by 0.3 mm.
Data collection and structure solution.
All data were collected from crystals flash frozen at 100 K. Crystals were mounted in cryoloops by a soaking protocol with a stepwise increasing concentration of a cryoprotectant (12 to 18% glycerol solution in mother liquor). Crystals of form 1 diffracted to 2.6 Å on an in-house rotating anode and to 2.0 Å on station 14.2 at the the synchotron radiation source (SRS), Daresbury Laboratory, Warrington, United Kingdom, at a wavelength of 0.979 Å. Form 1 crystals belong to the orthorhombic space group P21212, and a dimer per asymmetric unit corresponds to a Matthews coefficient of 2.9 Å3/Da and a solvent content of 57%. Crystals of form 2 diffracted to 3.4 Å in-house and to 2.7 Å on station ID-14.2, European Synchotron Radiation Facility (ESRF), Grenoble, France, at a wavelength of 0.933 Å. Form 2 crystals belong to the tetragonal space group P41212, and for a tetramer per asymmetric unit, we obtained a Matthews coefficient of 2.4 Å3/Da and a solvent content of 48.4%. Data were processed with MOSFLM (28) and the CCP4 package (10).
Initial phase information was obtained from the molecular replacement by using AMoRE in the CCP4 suite and an HN monomer as a search model. Clear solutions resulted in a dimer and a tetramer in the asymmetric unit for the orthorhombic and tetragonal crystals, respectively, as predicted. Structures were refined with CNS version 1.1 (5). The principal statistics of the synchrotron data and refinement are given in Table 1.
TABLE 1.
X-ray data and refinement statistics
| Characteristic | Orthorhombic form | Tetragonal form |
|---|---|---|
| Unit cell size (Å) | ||
| a | 175.48 | 115.07 |
| b | 99.26 | 115.07 |
| c | 64.33 | 283.96 |
| Space group | P21212 | P41212 |
| Resolution (Å) | 2.0 | 2.7 |
| No. of observations | 471,352 | 454,555 |
| No. of unique reflections | 89,733 | 39,099 |
| Completeness (%) | 98.5 (96.6) | 90.4 (88.4) |
| Rmerge | 0.083 (0.293) | 0.079 (0.254) |
| <1/σ(I)> | 7.6 (2.8) | 6.8 (2.2) |
| No. of atoms | 7,944 | 13,790 |
| R (%)/Rfree (%) | 0.191/0.227 | 0.225/0.289 |
| Bond length/anglea (Å/°) | 0.006/1.45 | 0.007/1.46 |
| <B> (Å2), protein/Neu5Ac2en/ thiosialoside | 23.9/22.4/23.7 | 26.3/31.1/52.9 |
Root mean square deviations from ideality.
Figures were drawn with Bobscript (17) and GRASP (35). Atomic coordinates have been deposited and given accession codes 1USR and 1USX for the P21212 and P41212 crystal forms complexed with the thiosialoside, respectively.
RESULTS
The thiosialoside (Fig. 1) was cocrystallized with NDV HN and produced two crystal forms at pH 6.3 (Table 1) which were different from the previously observed hexagonal crystal form grown under similar conditions in the presence of N-acetylneuraminic acid (43). In both new crystal forms, the inhibitor Neu5Ac2en was present in the active site (Fig. 1B). Mass spectroscopy analysis of the thiosialoside did not indicate the presence of any contaminant Neu5Ac2en, and therefore HN appears to be able to release the thioaglycon by a β-elimination process, leading to the formation of its own inhibitor, Neu5Ac2en. An identical β-elimination process with the same thiosialoside resulting in the formation of Neu5Ac2en has also been reported for the sialidase from Trypanosoma rangeli (1). Generation of Neu5Ac2en from N-acetylneuraminic acid was first reported for the influenza virus neuraminidase as a by-product of the catalytic reaction (7).
New sialic acid binding site.
Clear electron density revealed the thiosialoside bound at the dimer interface (Fig. 1C). In the 2.0-Å electron density map of the orthorhombic crystal form, which has a dimer in the asymmetric unit, the two symmetrical binding sites had clear density for the α-anomer of the thiosialoside, but only one site had clear density for the substituted galactose, as its position is stabilized by interactions with a symmetry-related HN monomer. Figure 2 shows the relationship between the catalytic sites and the new sialic acid binding site.
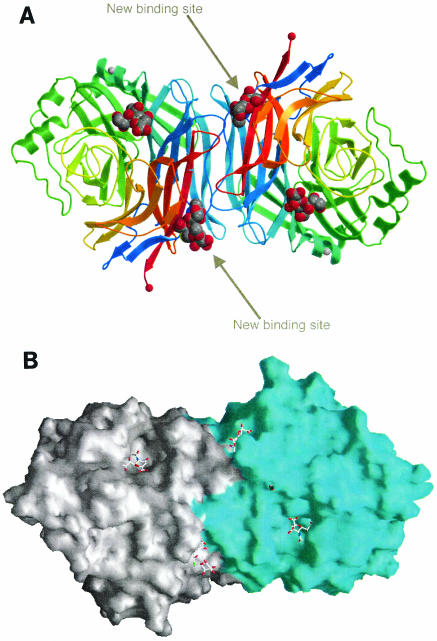
FIG. 2.
HN dimer and relationship between the ligand binding sites viewed down the twofold axis. (A) Schematic drawing of the HN dimer showing the location of the active sites with Neu5Ac2en bound and the sialic acid binding site with Neu5Ac or the complete thiosialoside bound. (B) GRASP image, with the monomers in different colors.
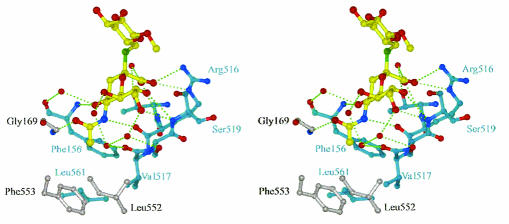
The new site was made up of residues from both monomers and involved interactions with the sialic acid and not the galactose, as shown in Fig. 3. Common to both sites were five direct hydrogen bonds to main-chain atoms and several water-mediated interactions involving six water molecules whose positions were conserved in both binding sites. At the site where the complete thiosialoside was visible, one of the carboxylate oxygen atoms of the sialic acid made two hydrogen bonds to the side chain of Arg516 (Fig. 3). At the other site, Arg516 had swung away, and an additional interaction was made between O-8 and the main-chain amide group of Ser519. At both sites, the acetamido methyl group sat in a hydrophobic pocket formed from Gly169, Leu552, and Phe553 from one monomer and Phe156, Val517, and Leu561 from the other monomer. In the tetragonal crystal form, containing four HN monomers in the asymmetric unit, the thiosialoside was present in all four binding sites, bound in an identical manner with the side chain of Arg516 away from the carboxyl group.
FIG. 3.
Stereo view of the sialic acid binding site. The thiosialoside is shown with yellow bonds. Residues belonging to monomer A are shown with cyan bonds, and residues belonging to monomer B are shown with grey bonds. Water molecules are shown as red spheres.
The fact that most of the hydrogen bond interactions occurred with main-chain atoms means that there is no conserved amino sequence defining this site across all paramyxoviruses. Arg516 is conserved across all NDV, Sendai virus, and human parainfluenza virus type 1 and 4 viruses, but as discussed above, the interaction with the side chain is not conserved. The hydrophobic pocket does appear to be conserved across the paramyxoviruses. We can conclude, therefore, that it is likely that this sialic acid binding site is conserved across the paramyxoviruses. A sialic acid binding site in addition to the active site was discovered for certain avian strains of influenza virus neuraminidase (NA), defined by a conserved sequence motif (49). There is, however, no similarity between the HN and NA sialic acid binding sites; the sites are at different locations on the surface relative to the active site, the NA site is not at an interface between influenza virus neuraminidase monomers, and the HN site does not have the conserved NA sequence motif.
HN tetramer.
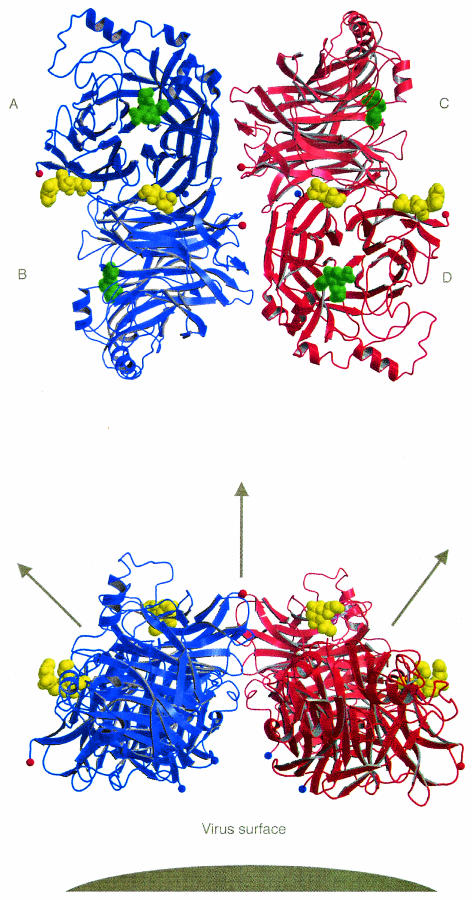
HN is known to form an oligomer consisting of homodimers, sometimes disulfide linked, that form a noncovalently linked tetramer (30, 33). The procedure used in this study to isolate the globular HN head involves chymotrypsin cleavage at Trp123 (43). Residue 123 in NDV HN strains is either a tryptophan or a cysteine, which can form a disulfide bridge with another monomer. Residues 124 are close together in the NDV HN dimer, suggesting that a disulfide bridge could easily form in strains having Cys123. The tetragonal crystal form reported here has a tetrameric arrangement of HN monomers in the asymmetric unit (Fig. 4). Both the orthorhombic crystal form presented here and the previously reported hexagonal crystal form (14) have a dimer in the asymmetric unit, but an identical tetrameric arrangement is formed by the application of crystal symmetry. We therefore have three quite different crystal forms all showing the same tetrameric arrangement, suggesting that this is a stable oligomer. As shown in Fig. 4, the N termini of all four monomers are on the same face of the tetramer, where the stalk regions (residues 1 to 123) would attach to the viral membrane. The sialic acid binding sites are on the opposite face, where they are able to interact with cell surfaces.
FIG. 4.
Two views of the HN tetramer. (Top) View looking down towards the viral surface. (Bottom) Side view of the tetramer, with the viral surface indicated in grey. The active sites contain Neu5Ac2en (green). The sialic acid binding sites contain Neu5Ac (yellow) or complete thiosialoside. The blue spheres show the locations of residue 124 at the N terminus of each monomer; the red spheres show the locations of the C terminus of each monomer. The arrows indicate the direction of the twofold axes of symmetry relating the monomers within a dimer and relating two dimers.
The tetramer has dimensions of approximately 100 by 100 by 50 Å, similar to that seen in electron microscopy for Sendai virus HN that had been proteolytically removed from viruses at residue 131 (46). The tetramer is unusual, as it does not have the fourfold symmetry seen for influenza virus neuraminidase or tetrahedral symmetry; rather, the two folds of the AB and CD dimers are located 33° on either side of the two fold relating the two dimers to each other (Fig. 4). The AB (or CD) dimer buries 1,750 Å2 of surface area for each monomer, whereas the association of the dimers in the tetramer is much less extensive. The contact between the B and C monomers buries 312 Å2, the contact between A and C (or B and D) buries only 73 Å2, and there is no association between A and D.
Despite this small interaction between the dimers, we did observe exactly the same tetramer formation in three different crystal forms with different packing arrangements. The dimensions determined for the tetramer agree with electron microscopy observations, and the tetramer has all the N termini on one face and all the sialic acid recognition sites on the opposite face. Interestingly, after extensive crystal trials, we have been unable to grow crystals of the tetrameric form in the absence of ligand. In addition, all three crystal forms containing the tetramer have Neu5Ac2en in the HN active sites, having been cocrystallized with sialic acid (in the case of the hexagonal crystal form) or the thiosialoside. This suggests that in isolation at physiological pH and unable to associate with F, HN may exist in a metastable state that becomes stabilized upon cleavage of sialic acid-containing receptors. We have also crystallized ligand-free HN at pH 7.5 with 20% polyethylene glycol 3350 and 0.1 M HEPES buffer as a precipitant (data not shown). The crystals are in the same space group and show the same association of monomers as obtained at pH 4.6. We can conclude that the presence of ligand in the crystallization buffer is essential for the formation of the HN dimer association that leads to the tetrameric arrangements reported here.
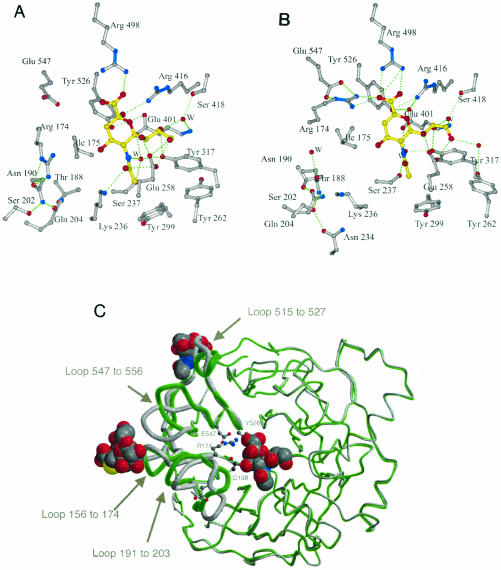
Changes in the active site.
Our previous studies on NDV HN revealed a pliable active site, in contrast to other viral and bacterial neuraminidases, which show a rigid active site (45). There is a large cavity in the active site around the O-4 position of Neu5Ac or Neu5Ac2en, which appears to allow large movements of Arg174, Lys236, Tyr526, and Glu547, residues conserved across all paramyxoviruses. Arg174 is one of three arginines that interact with the carboxylate group of the ligand, Glu547 interacts with Arg174, Lys236 is part of the conserved hexapeptide Asn-Arg-Lys-Ser-Cys-Ser, and Tyr526 is implicated in the catalytic mechanism through its homology with other neuraminidases. In the pH 4.6 crystal form, the active site appears to be switched off, with Tyr526 shifted away from the C-1-C-2 bond of the ligand and Arg174 swung 90° away from the ligand carboxylate group (Fig. 5A and B). Another residue whose position changes dramatically is Asp198, the homologue of Asp151 in the influenza virus neuraminidase that has been implicated in hydrolysis (9, 50). In the pH 4.6 form, Asp198 is poised in the same position seen in other neuraminidases, above the sugar ring of the substrate. In all pH 6.3 crystal forms, the loop containing Asp198 has moved, and Asp198 points away from the active site. The dimer association seen in the pH 4.6 form is totally different from that seen in the three pH 6.3 crystal forms, although, as stated above, we have grown ligand-free crystals at pH 7.5 and obtained the same dimer as was found at pH 4.6.
FIG. 5.
Changes in the two states of the HN monomer. (A) The pH 4.6 active site with the β-anomer of sialic acid bound. (B) The pH 6.3 active site with Neu5Ac2en bound. (C) Superposition of the pH 4.6 (grey) and pH 6.3 (green) monomers. Residues Asp198, Arg174, Glu547, and Tyr526 are shown, as are Neu5Ac2en at the active site and Neu5Ac or the complete thiosialoside at the dimer interface. The loops whose positions changed significantly are drawn with thicker lines and labeled.
Model for triggering of fusion.
As seen in Fig. 5A and B, there are multiple interactions between HN and the right-hand side of the ligand; in particular, all three hydroxyls of the glycerol group interacted with residues invariant among paramyxovirus HNs. Two of these residues, Glu258 and Tyr262, are on a small helix stabilized by a calcium ion. Such a complex interaction with the glycerol group is unique among the neuraminidases studied to date. Thus, one can imagine that sialic acid binds initially to the off state (Fig. 5A) through multiple interactions on the right-hand side of the active site. The presence of the sialic acid carboxylate group at the site would then trigger a series of side chain movements: Arg174 would move to interact with one of the carboxylate oxygen atoms and Glu547, which might also lead to movement of Ile175 and Tyr526, and Lys236 would move to interact with O-10 of the ligand (Fig. 5B). Asp198 is initially in the correct position to participate in catalysis, but postcatalysis, the loop containing Asp198 moves (Fig. 5C).
Figure 5C shows that Arg174, Tyr526, and Glu547 are located, in each case, a few residues from the loops that form the new sialic acid binding site: a loop involving residue 169; a loop involving residues 516, 517, and 519; and a loop involving residues 552 and 553. The positions of these loops are quite different in the pH 4.6 and pH 6.3 structures. As we have argued previously, it is possible that the changes in the active site upon binding and cleavage of the sialic acid-containing receptor or synthetic substrate could propagate changes to the dimer interface, in particular, changes in the three loops (42). The loop containing Asp198 also shifts position, as shown in Fig. 5C, and is adjacent to the residue 169-containing loop. Such changes might alter the HN dimer interface and create the new sialic acid binding site and in the process allow the fusion protein to undergo a structural transition. The importance of the HN dimer interface in fusion has been shown by recent mutagenesis studies on NDV HN (13, 42).
Based on our structural studies to date, we propose a model for triggering fusion and a role for the second sialic acid binding site (Fig. 6). (i) HN and F associate on the viral surface in a way that holds both proteins in their off states. That is, the active site of HN is switched off, as we observed in the pH 4.6 crystal form (Fig. 5A), and F is in a state with its fusion peptide masked from exposure to solvent. HN binds to sialic acid-containing receptors via its active site. HN is triggered into its on state by movement of Arg174, Tyr526, and Glu547 to form a catalytic site that can facilitate release of the sialic acid from the sialic acid-containing receptor or a synthetic substrate such as thiosialoside. In the case of the thiosialoside, the enzyme releases N-acetylneuraminic acid by a β-elimination process, leading to the formation of its own inhibitor, Neu5Ac2en. The movement of these three residues and others, including Lys236 and Asp198, propagates conformational changes to the HN dimer interface. (ii) The changes at the HN dimer interface drive the F protein into its fusogenic on state and lead to exposure of the fusion peptide, presumably resulting in a six-helix bundle structure for F, as reported for several viruses (11), including paramyxoviruses (2). The altered HN dimer also results in the creation of a new sialic acid binding site. (iii) The new sialic acid binding site allows the virus to remain attached to sialic acid-containing receptors, while the F protein's fusion peptide is embedded in the cell membrane, and fusion proceeds.
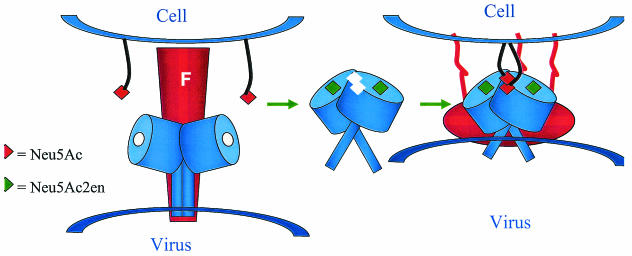
FIG. 6.
Model for fusion. (Left) F and HN hold each other in the switched-off state, with the region from 124 to 151 binding to the membrane-proximal HR2a region of F (18). HN binds to sialic acid receptors. (Middle) The sialic acid is released from the sialic acid-containing receptor, Neu5Ac2en is bound at the active site, the dimer association changes, and a new sialic acid binding site is created (F was removed for clarity). (Right) The changes in HN promote F into its fusogenic state, releasing its fusion peptide into the cell membrane while the virus is held proximal to the membrane by the sialic acid binding site. The structure(s) adopted by the HN stalk region is unknown.
DISCUSSION
There is confusion in the literature over whether there are separate NA and HA sites on HN. Several studies on monoclonal antibody binding and the sequencing of escape mutants supported two separate sites for HA and NA activities (34). Early studies suggested that a single site was responsible for both functions (31, 39). Other studies have suggested that there are two sites in proximity (47), and a kinetic analysis of isolated NDV HN suggested that binding of substrates to an HA site affects the activity of the NA site (38). We previously suggested a single, switchable site that was consistent with the monoclonal antibody binding data (14). Mutagenesis of the HN active-site residues showed that, in most cases, a loss of neuraminidase activity was accompanied by a loss in hemagglutination activity and in some cases a loss in fusion activity (12, 15, 21, 22, 24, 32, 41).
Our finding of the second binding site on HN may help to clarify the confusion. Mutation of conserved amino acids around the NA active site resulted in elimination of NA activity but showed variable effects on hemadsorption activities (12, 21). The variable effects on HA seem to be dependent on the location of the mutated residues. Mutations at residues Tyr526 and Glu547 resulted in the loss of both NA and HA activities. These residues are located between the NA active site and the second binding site, suggesting that these mutations affect the structure of both sites and result in the elimination of both functions. In contrast, mutations at Glu258 and Tyr299, located far from the second binding site, eliminated only the NA activity (12, 21). Furthermore, a recent report showed that some of the mutations at the dimer interface (Phe220, Ser222, and Leu224) severely impaired the ability of HN to adsorb to red blood cells at 37°C but not at 4°C (13). The loss of the receptor-binding activity at 37°C could be explained by the loss of the second binding site caused by these mutations. At 4°C, both the NA active site and the second binding site may function to bind receptors on cells. At 37°C, however, any receptor-binding ability at the NA active site will be reduced because of the enhanced NA activity at the site at that temperature. If the HN retains the second binding site, HN may still adsorb to the red blood cells even at 37°C. Although these residues are not located close to the second binding site, mutations at the dimer interface may affect the interaction between the two HN molecules that is required for the formation of the second binding site.
The results of NA and HA inhibition by monoclonal antibodies also caused some confusion about the presence of two separate sites on HN. Most anti-NDV HN monoclonal antibodies that inhibit NA also inhibit attachment (20, 24, 36, 52). However, monoclonal antibodies that inhibit NA but not attachment have been reported for Sendai virus (37). Considering the relative proximity of the two binding sites (Fig. 2) and the molecular footprint of an antibody, it is not difficult to conceive that most monoclonal antibodies that bind close to the NA active site also cover the second sialic acid binding site on HN. In fact, monoclonal antibodies that recognize antigenic site 23 block both NA and HA activities. This antigenic site includes residues 174, 193, 194, 200, 201, 203, and 547, which covers an area between the NA active site and dimer interface that is close to the second binding site (21, 23-25, 29).
In light of the discovery of the new sialic acid binding site, the structural model needs to be modified. We propose that binding and catalysis of sialic acid receptors at the active site provide the energy to drive the fusion protein into its fusogenic state. How this is accomplished is not clear, as residues in the head and stalk regions of HN have been implicated in fusion (3, 4, 16, 40, 44, 48, 51). We propose that binding and catalysis at the active site involving conserved residues Lys236, Asp198, Arg174, Glu547, and Ty526 causes changes in the positions of the loops forming the dimer interface. The dimer interface changes may alter the association of the HN dimer or tetramer, which may also propagate a change in the stalk region and in doing so trigger the fusion protein. A connection between structural changes in the head and stalk regions of NDV HN is suggested by the observation that mutations in a conserved region of the stalk impair neuraminidase activity in the head (51). Another consequence of the changes at the dimer interface is the creation of a new sialic acid binding site, allowing the virus to remain proximal to the cell surface as fusion proceeds.
Mutation of Phe553, which forms part of the sialic acid binding site at the dimer interface, to an alanine abolished fusion promotion and a caused a 70% drop in hemoadsorption and neuraminidase activities compared to those of the wild type (42). It is difficult to predict the structural changes that such a mutation would produce; it may lead to reduced hydrophobic interactions with the methyl group of Neu5Ac; it may indirectly affect the active site, as Phe553 is only a few residues from Glu547; and it may disrupt the dimer association. Mutation of other dimer interface residues led to a weakening of the interaction between HN and its receptor (13). It may well be that mutations at the interface altered the interface sialic acid binding site, leading to reduced binding. This study also showed that the conformation-dependent monoclonal antibody mapping to antigenic site 23 (24), which lies close to the dimer interface, was still able to recognize the mutants. This suggested that, at least for the mutants made in this study, there was little disruption of structure in this region (residues 193 to 203), but this does not rule out changes that might have occurred at the interface sialic acid binding site. It is important to note, however, that the conformation-dependent antibodies probably recognize the intimate HN dimer of Fig. 2 and not an alternative dimeric form that may exist when HN is complexed with F. Both of these mutational studies do, however, emphasize the importance of the dimer interface in the fusion process.
Does the dimer form of NDV HN that we reported for the pH 4.6 crystal form have any physiological significance? As discussed above, we observed the same dimer formation in a crystal form grown at pH 7.5, suggesting that this form of the dimer is not a function of acid pH. One problem with this dimer is that the distance between residue 124 in each monomer is ≈60 Å. For strains of NDV that have a cysteine at residue 123, forming a disulfide bridge between the monomers, this is clearly impossible unless the structure adopted by residue 124 onwards takes a different structure. A recent report suggests that this might be the case (18). In this study on the interaction between NDV HN and the HR2a heptad repeat peptide of NDV F (residues 477 to 496), it was found that the HR2a peptide bound to residues 124 to 151 of HN. In all the crystal structures of NDV HN determined so far, this region forms a random coil beneath the β-propeller domain on the opposite face from the active site. It is possible that, when interacting with the HR2a region of F, this region adopts a different conformation that may lead to bringing residue 124 in the adjacent monomers closer together. The fact that the key residue, Asp198, is in the correct position for participating in the catalytic mechanism in the ligand-free structure of HN lends weight to this form's having some physiological significance.
In summary, viruses of the Paramyxovirinae subfamily of paramyxovirues infect host cells by the interaction of two surface glycoproteins, hemagglutinin-neuraminidase and the fusion protein. Unlike influenza viruses, which undergo endocytosis with fusion triggered by the low pH of the endosome, paramyxovirus fusion occurs at the cell surface. Recognition of sialic acid-containing receptors by HN somehow leads to a conformational change in F into its fusogenic state. HN both recognizes sialic acid, as measured by a hemagglutination assay, and hydrolyzes sialosides, as measured by a neuraminidase assay. In this study, we report the discovery of a second sialic acid binding site in addition to the active site. Interactions with sialic acid come mainly from backbone atoms and a conserved hydrophobic pocket that are probably present in all paramyxoviruses. The location of this new site at the HN dimer interface, together with observed changes in the HN structure that appear to be generated by catalysis, suggests a model for how sialic acid binding by HN might trigger conformational changes in F. In addition, the observation of a tetrameric form of HN in three different crystal forms suggests a model for how HN exists on the viral surface. How the fusion protein and HN associate and how changes in the head region of HN can propagate to the stalk region remain to be discovered. The structure of the prefusogenic NDV F protein is known (8), but only determination of the structure of a complex of HN and F will reveal the precise details of their association and how changes in HN precipitate changes in F. Nevertheless, the key role of the HN active site in sialic acid recognition, promotion of fusion, and spread of the virus makes it an attractive target for inhibitor design and the development of drugs against important childhood respiratory viruses.
Acknowledgments
We thank Ibrahim Moustafa for useful discussions. We thank the support staff at the Grenoble and Daresbury synchrotrons for their assistance in data collection.
This work was supported by the United Kingdom Medical Research Council, the Australian Research Council, an EU TMR/LSF grant for access to the ESRF synchrotron, and by U.S. NIAID grants AI-11949 and AI-38956. M.V.I. gratefully acknowledges the award of a Federation Fellowship.
REFERENCES
- 1.Amaya, M. F., A. Buschiazzo, T. Nguyen, and P. M. Alzari. 2003. The high resolution structures of free and inhibitor-bound Trypanosoma rangeli sialidase and its comparison with T. cruzi trans-sialidase. J. Mol. Biol. 325:773-784. [DOI] [PubMed] [Google Scholar]
- 2.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 3.Bousse, T., T. Takimoto, W. L. Gorman, T. Takahashi, and A. Portner. 1994. Regions on the hemagglutinin-neuraminidase proteins of human parainfluenza virus type-1 and Sendai virus important for membrane-fusion. Virology 204:506-514. [DOI] [PubMed] [Google Scholar]
- 4.Bousse, T., T. Takimoto, and A. Portner. 1995. A single amino-acid change enhances the fusion promotion activity of human parainfluenza virus type-1 hemagglutinin-neuraminidase glycoprotein. Virology 209:654-657. [DOI] [PubMed] [Google Scholar]
- 5.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 6.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza hemagglutinin at the pH of membrane-fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 7.Burmeister, W. P., B. Henrissat, C. Bosso, S. Cusack, and R. W. H. Ruigrok. 1993. Influenza-B virus neuraminidase can synthesize its own inhibitor. Structure 1:19-26. [DOI] [PubMed] [Google Scholar]
- 8.Chen, L., J. J. Gorman, J. McKimm-Breschkin, L. J. Lawrence, P. A. Tulloch, B. J. Smith, P. M. Colman, and M. C. Lawrence. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure (Camb.) 9:255-266. [DOI] [PubMed] [Google Scholar]
- 9.Chong, A. K. J., M. S. Pegg, N. R. Taylor, and M. Vonitzstein. 1992. Evidence for a sialosyl cation transition-state complex in the reaction of sialidase from influenza-virus. Eur. J. Biochem. 207:335-343. [DOI] [PubMed] [Google Scholar]
- 10.Collaborative Computing Project No. 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 11.Colman, P. M., and M. C. Lawrence. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 4:309-319. [DOI] [PubMed] [Google Scholar]
- 12.Connaris, H., T. Takimoto, R. Russell, S. Crennell, I. Moustafa, A. Portner, and G. Taylor. 2002. Probing the sialic acid binding site of the hemagglutinin-neuraminidase of Newcastle disease virus: identification of key amino acids involved in cell binding, catalysis, and fusion. J. Virol. 76:1816-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corey, E. A., A. M. Mirza, E. Levandowsky, and R. M. Iorio. 2003. Fusion deficiency induced by mutations at the dimer interface in the Newcastle disease virus hemagglutinin-neuraminidase is due to a temperature-dependent defect in receptor binding. J. Virol. 77:6913-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crennell, S., T. Takimoto, A. Portner, and G. Taylor. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 7:1068-1074. [DOI] [PubMed] [Google Scholar]
- 15.Deng, R. T., Z. Y. Wang, P. J. Mahon, M. Marinello, A. Mirza, and R. M. Iorio. 1999. Mutations in the Newcastle disease virus hemagglutinin-neuraminidase protein that interfere with its ability to interact with the homologous F protein in the promotion of fusion. Virology 253:43-54. [DOI] [PubMed] [Google Scholar]
- 16.Deng, R. T., Z. Y. Wang, A. M. Mirza, and R. M. Iorio. 1995. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology 209:457-469. [DOI] [PubMed] [Google Scholar]
- 17.Esnouf, R. M. 1997. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model. 15:112-113, 132-134. [DOI] [PubMed] [Google Scholar]
- 18.Gravel, K. A., and T. G. Morrison. 2003. Interacting domains of the HN and F proteins of Newcastle disease virus. J. Virol. 77:11040-11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, X. L., R. Ray, and R. W. Compans. 1992. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J. Virol. 66:1528-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iorio, R. M., and M. A. Bratt. 1984. Monoclonal-antibodies as functional probes of the HN glycoprotein of Newcastle disease virus—antigenic separation of the hemagglutinating and neuraminidase sites. J. Immunol. 133:2215-2219. [PubMed] [Google Scholar]
- 21.Iorio, R. M., G. M. Field, J. M. Sauvron, A. M. Mirza, R. Deng, P. J. Mahon, and J. P. Langedijk. 2001. Structural and functional relationship between the receptor recognition and neuraminidase activities of the Newcastle disease virus hemagglutinin-neuraminidase protein: receptor recognition is dependent on neuraminidase activity. J. Virol. 75:1918-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iorio, R. M., and R. L. Glickman. 1992. Fusion mutants of Newcastle disease virus selected with monoclonal antibodies to the hemagglutinin-neuraminidase. J. Virol. 66:6626-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iorio, R. M., R. L. Glickman, A. M. Riel, J. P. Sheehan, and M. A. Bratt. 1989. Functional and neutralization profile of 7 overlapping antigenic sites on the HN glycoprotein of Newcastle disease virus monoclonal-antibodies to some sites prevent viral attachment. Virus Res. 13:245-261. [DOI] [PubMed] [Google Scholar]
- 24.Iorio, R. M., R. J. Syddall, R. L. Glickman, A. M. Riel, J. P. Sheehan, and M. A. Bratt. 1989. Identification of amino-acid residues important to the neuraminidase activity of the HN glycoprotein of Newcastle disease virus. Virology 173:196-204. [DOI] [PubMed] [Google Scholar]
- 25.Iorio, R. M., R. J. Syddall, J. P. Sheehan, M. A. Bratt, R. L. Glickman, and A. M. Riel. 1991. Neutralization map of the hemagglutinin-neuraminidase glycoprotein of Newcastle disease virus: domains recognized by monoclonal antibodies that prevent receptor recognition. J. Virol. 65:4999-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamb, R. A. 1993. Paramyxovirus fusion—a hypothesis for changes. Virology 197:1-11. [DOI] [PubMed] [Google Scholar]
- 27.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 28.Leslie, A. G. W. 1992. Recent changes to the MOSFLM package for processing film and image plate data. In CCP4 newsletter on protein crystallography, no. 26. Daresbury Laboratory, Warrington, United Kingdom.
- 29.Mahon, P. J., R. T. Deng, A. M. Mirza, and R. M. Iorio. 1995. Cooperative neuraminidase activity in a paramyxovirus. Virology 213:241-244. [DOI] [PubMed] [Google Scholar]
- 30.Markwell, M. K., and C. F. Fox. 1980. Protein-protein interactions within paramyxoviruses identified by native disulfide binding or reversible chemical cross-linking. J. Virol. 33:152-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merz, D. C., P. Prehm, A. Scheid, and P. W. Choppin. 1981. Inhibition of the neuraminidase of paramyxoviruses by halide ions: a possible means of modulating the 2 activities of the HN-protein. Virology 112:296-305. [DOI] [PubMed] [Google Scholar]
- 32.Mirza, A. M., R. T. Deng, and R. M. Iorio. 1994. Site-directed mutagenesis of a conserved hexapeptide in the paramyxovirus hemagglutinin-neuraminidase glycoprotein: effects on antigenic structure and function. J. Virol. 68:5093-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrison, T. G. 1988. Structure, function, and intracellular processing of paramyxovirus membrane-proteins. Virus Res. 10:113-135. [DOI] [PubMed] [Google Scholar]
- 34.Morrison, T. G., and A. Portner. 1991. Structure, function, and intracellular processing of the glycoproteins of Paramyxoviridae, p. 347-382. In D. W. Kingsbury (ed.), The paramyxoviruses. Plenum Press, New York City, N.Y.
- 35.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 36.Nishikawa, K., T. Morishima, T. Toyoda, T. Miyadai, T. Yokochi, T. Yoshida, and Y. Nagai. 1986. Topological and operational delineation of antigenic sites on the HN glycoprotein of Newcastle disease virus and their structural requirements. J. Virol. 60:987-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Portner, A., R. A. Scroggs, and D. W. Metzger. 1987. Distinct functions of antigenic sites of the HN glycoprotein of Sendai virus. Virology 158:61-68. [DOI] [PubMed] [Google Scholar]
- 38.Sagrera, A., C. Cobaleda, I. Munoz Barroso, V. Shnyrov, and E. Villar. 1995. Modulation of the neuraminidase activity of HN protein from Newcastle disease virus by substrate binding and conformational change: kinetic and thermal denaturation studies. Biochem. Mol. Biol. Int. 37:717-727. [PubMed] [Google Scholar]
- 39.Scheid, A., and P. W. Choppin. 1974. Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis, and infectivity by proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology 57:470-490. [DOI] [PubMed] [Google Scholar]
- 40.Sergel, T., L. W. McGinnes, M. E. Peeples, and T. G. Morrison. 1993. The attachment function of the Newcastle disease virus hemagglutinin-neuraminidase protein can be separated from fusion promotion by mutation. Virology 193:717-726. [DOI] [PubMed] [Google Scholar]
- 41.Sheehan, J. P., and R. M. Iorio. 1992. A single amino-acid substitution in the hemagglutinin-neuraminidase of Newcastle disease virus results in a protein deficient in both functions. Virology 189:778-781. [DOI] [PubMed] [Google Scholar]
- 42.Takimoto, T., G. L. Taylor, H. C. Connaris, S. J. Crennell, and A. Portner. 2002. Role of the hemagglutinin-neuraminidase protein in the mechanism of paramyxovirus-cell membrane fusion. J. Virol. 76:13028-13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takimoto, T., G. L. Taylor, S. J. Crennell, R. A. Scroggs, and A. Portner. 2000. Crystallization of Newcastle disease virus hemagglutinin-neuraminidase glycoprotein. Virology 270:208-214. [DOI] [PubMed] [Google Scholar]
- 44.Tanabayashi, K., and R. W. Compans. 1996. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 70:6112-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor, G. 1996. Sialidases: structures, biological significance and therapeutic potential. Curr. Opin. Struct. Biol. 6:830-837. [DOI] [PubMed] [Google Scholar]
- 46.Thompson, S. D., W. G. Laver, K. G. Murti, and A. Portner. 1988. Isolation of a biologically active soluble form of the hemagglutinin-neuraminidase protein of Sendai virus. J. Virol. 62:4653-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson, S. D., and A. Portner. 1987. Localization of functional sites on the hemagglutinin neuraminidase glycoprotein of Sendai virus by sequence-analysis of antigenic and temperature-sensitive mutants. Virology 160:1-8. [DOI] [PubMed] [Google Scholar]
- 48.Tsurudome, M., M. Kawano, T. Yuasa, N. Tabata, M. Nishio, H. Komada, and Y. Ito. 1995. Identification of regions on the hemagglutinin-neuraminidase protein of human parainfluenza virus type-2 important for promoting cell-fusion. Virology 213:190-203. [DOI] [PubMed] [Google Scholar]
- 49.Varghese, J. N., P. M. Colman, A. vanDonkelaar, T. J. Blick, A. Sahasrabudhe, and J. L. McKimm Breschkin. 1997. Structural evidence for a second sialic acid binding site in avian influenza virus neuraminidases. Proc. Natl. Acad. Sci. USA 94:11808-11812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Varghese, J. N., J. L. McKimm Breschkin, J. B. Caldwell, A. A. Kortt, and P. M. Colman. 1992. The structure of the complex between influenza-virus neuraminidase and sialic-acid, the viral receptor. Proteins Struct. Funct. Genet. 14:327-332. [DOI] [PubMed] [Google Scholar]
- 51.Wang, Z. Y., and R. M. Iorio. 1999. Amino acid substitutions in a conserved region in the stalk of the Newcastle disease virus HN glycoprotein spike impair its neuraminidase activity in the globular domain. J. Gen. Virol. 80:749-753. [DOI] [PubMed] [Google Scholar]
- 52.Yusoff, K., M. Nesbit, H. McCartney, P. T. Emmerson, and A. C. R. Samson. 1988. Mapping of 3 antigenic sites on the hemagglutinin-neuraminidase protein of Newcastle disease virus. Virus Res. 11:319-333. [DOI] [PMC free article] [PubMed] [Google Scholar]