Summary
Class I histone deacetylases (HDAC1, HDAC2, and HDAC3) are recruited by cognate corepressor proteins into specific transcriptional repression complexes that target HDAC activity to chromatin resulting in chromatin condensation and transcriptional silencing. We previously reported the structure of HDAC3 in complex with the SMRT corepressor. This structure revealed the presence of inositol-tetraphosphate [Ins(1,4,5,6)P4] at the interface of the two proteins. It was previously unclear whether the role of Ins(1,4,5,6)P4 is to act as a structural cofactor or a regulator of HDAC3 activity. Here we report the structure of HDAC1 in complex with MTA1 from the NuRD complex. The ELM2-SANT domains from MTA1 wrap completely around HDAC1 occupying both sides of the active site such that the adjacent BAH domain is ideally positioned to recruit nucleosomes to the active site of the enzyme. Functional assays of both the HDAC1 and HDAC3 complexes reveal that Ins(1,4,5,6)P4 is a bona fide conserved regulator of class I HDAC complexes.
Graphical Abstract
Highlights
-
•
Inositol phosphates are bona fide regulators of class I HDAC corepressor complexes
-
•
The ELM2-SANT motif is a conserved HDAC corepressor assembly module
-
•
MTA1 is a dimer that recruits two HDACs into the NuRD complex
-
•
The MTA1-BAH domain is positioned to recruit chromatin to the HDAC active site
Introduction
The acetylation of lysine residues is an important secondary modification that regulates protein function and is controlled by the action of acetylase and deacetylase enzymes. Class I histone deacetylases (HDACs) are recruited to, and activated by, cognate corepressor proteins that target HDAC activity to particular chromatin loci, resulting in the spatial and temporal control of gene expression. These large multiprotein complexes silence target genes through the removal of the acetyl groups from lysine residues in histone tails. The deacetylation of chromatin results in the formation of a higher-order, more condensed structure, leading to the repression of gene transcription (Grunstein, 1997; Struhl, 1998; Shogren-Knaak et al., 2006).
Class I HDACs have been reported to be associated with at least four major corepressor complexes. HDAC3 is recruited uniquely to the SMRT/NCoR repression complex (Guenther et al., 2000; Wen et al., 2000), whereas HDAC1 and HDAC2 are activated through recruitment into several corepressor complexes including the Sin3A (Laherty et al., 1997), CoREST (Humphrey et al., 2001), and NuRD (Xue et al., 1998; Zhang et al., 1999) complexes. Understanding the assembly of HDAC complexes is important since inhibitors of HDACs have an increasing number of therapeutic applications. Currently, inhibitors of class I HDACs are used in the clinic to treat cutaneous T cell lymphoma and may be useful in the treatment of other cancers and Alzheimer’s disease (Marks and Xu, 2009; Xu et al., 2011). The development of selective inhibitors that are specific for particular complexes may be important for effective clinical application.
We recently reported the structure of HDAC3 in complex with the extended SANT domain from the SMRT corepressor (Watson et al., 2012). This structure revealed that the interaction between HDAC3 and the SMRT-SANT domain requires the presence of a D-myo-inositol-1,4,5,6-tetrakisphosphate [Ins(1,4,5,6)P4] molecule sandwiched at the interface of the two proteins. We showed that Ins(1,4,5,6)P4 is essential for the interaction of HDAC3 with the SMRT-SANT domain, but it remained unclear whether Ins(1,4,5,6)P4 is simply a structural cofactor or a bona fide regulator of complex assembly (and hence HDAC3 activity). However, it is important to note that there is no evidence for a pool of free HDAC3 awaiting Ins(1,4,5,6)P4 to mediate assembly with corepressors.
Sequence conservation suggests that other class I HDAC complexes may also bind inositol phosphates. To explore this possibility, we investigated the reported interaction between HDAC1 and metastasis-associated protein 1 (MTA1) from the NuRD complex (Toh et al., 2000; Manavathi and Kumar, 2007) and determined the structure of HDAC1 in complex with the adjacent ELM2 and SANT domains from MTA1. The structure reveals that this complex also has what appears to be an inositol phosphate binding pocket at the interface between the MTA1-SANT domain and HDAC1, suggesting that this is a common feature in class I HDAC:corepressor complexes.
Importantly, we found that the ELM2 domain from MTA1 mediates assembly of the HDAC1 complex independently of inositol phosphate. By analogy, we found that a similarly extended region of SMRT is able to mediate interaction with HDAC3 in the absence of Ins(1,4,5,6)P4. Addition of Ins(1,4,5,6)P4 to both complexes results in a dramatic increase in HDAC activity. Together, these findings suggest that class I HDACs are constitutively assembled into corepressor complexes and that their activity is regulated by freely dissociating inositol phosphates. The role of the inositol phosphate is likely to involve “engaging” the SANT domain with the HDAC catalytic domain.
Comparison of the HDAC1 complex with that of HDAC3 reveals the stereochemical basis for the specificity of complex assembly. The ELM2 domain mediates dimerization of the complex and also has a conserved arm that wraps completely around the HDAC that is ideally positioned to allow the adjacent amino-terminal BAH domain to present nucleosomal substrates to the catalytic site of HDAC1, hence determining substrate specificity.
Results
Structure of the HDAC1:MTA1 Complex
To determine the structure of the HDAC1:MTA1 complex, HDAC1 was coexpressed with the ELM2-SANT domain from MTA1 (residues 162–335) in suspension grown HEK293 cells. Small crystals (15 μm) of the purified complex were obtained in 2 M ammonium sulfate. Diffraction data from three crystals were merged giving a complete data set to 3 Å (Table 1). The structure was solved by molecular replacement using the structure of HDAC3:SMRT (PDB code 4A69). The resulting maps were remarkably clear, and the complete ELM2-SANT domain from MTA1 could be built without ambiguity (Figure S1, available online, and Table 1).
Table 1.
Data Collection and Refinement Statistics
| Data Collection | |
|---|---|
| Space group | P 32 2 1 |
| Cell dimensions: a, b, c (Å) | 108.2, 108.2, 133.2 |
| Cell dimensions: α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 76.8–3.0 (3.18–3.0) |
| Rsym or Rmerge | 16.3 (65.7) |
| I/σI | 6.0 (1.8) |
| Completeness (%) | 96.1 (97.6) |
| Redundancy | 3.0 (3.1) |
| Refinement | |
| Resolution (Å) | 76.6–3.0 |
| No. of reflections | 16916 |
| Rwork/Rfree | 21.3/26.1 |
| No. of atoms: protein | 4,235 |
| No. of atoms: ligand/ion | 27 |
| No. of atoms: water | 0 |
| B factor: protein | 53.8 |
| B factor: Zn ions, K ions, and acetate | 56.8 |
| Rmsd: bond lengths (Å) | 0.008 |
| Rmsd: bond angles (°) | 1.023 |
See also Figure S1 for information about protein expression and crystallization.
∗The highest resolution shell is shown in parentheses.
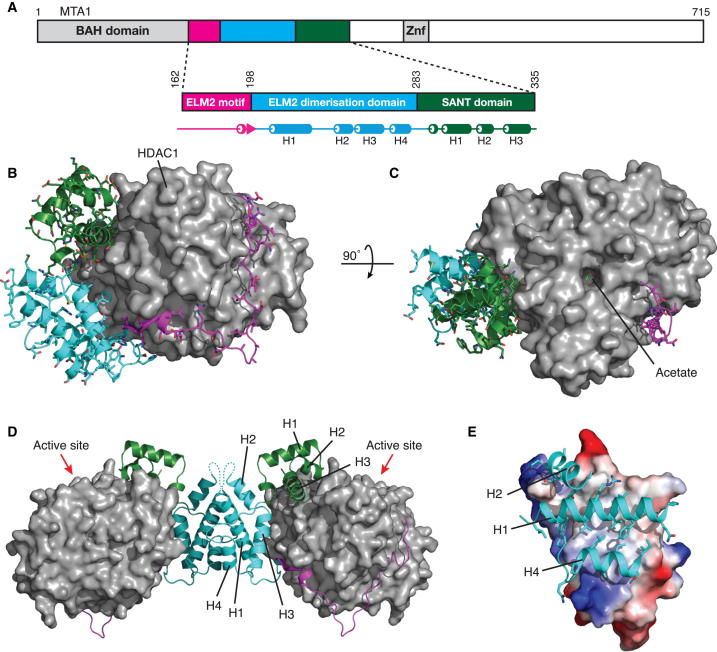
Remarkably, the MTA1 corepressor wraps completely around the structured catalytic core of HDAC1 and makes extensive contacts to the HDAC through both the ELM2 and SANT domains (Figures 1A–1C). The MTA1 binding interface with HDAC1 is large and characterized by a total solvent-excluded surface of 5,185 Å2. The interaction of MTA1 with HDAC1 can be rationalized by dividing MTA1 into three regions: the carboxy-terminal SANT domain (residues 283–335, green in Figure 1), a central helical domain (residues 199–282, cyan in Figure 1), and an amino-terminal ordered region lacking extensive secondary structure (residues 162–198, magenta in Figure 1). Unexpectedly, the central helical domain mediates dimerization of the HDAC complex and is termed henceforth the ELM2 dimerization domain (Figures 1D and 1E).
Figure 1.
Overall Structure of the HDAC1:MTA1 Complex
(A) MTA1 domain structure highlighting the ELM2-SANT and its secondary structure elements.
(B) A cartoon representation of MTA1 wrapped around HDAC1 (gray). MTA1 domains are shown in magenta (ELM2 specific motif), cyan (ELM2 dimerization), and green (SANT).
(C) View from (B), rotated by 90°, showing an acetate molecule (light green) bound in the active site.
(D) MTA1 homodimerization brings two HDAC1 molecules into the complex. Red arrows indicate the HDAC1 active sites.
(E) The MTA1 dimer interface with one molecule shown as an electrostatic surface and the other shown as a cartoon.
See also Figure S1 for information about expression and crystallization, Figure S3 for biochemical characterization of the ELM2 dimerization domain, and Figure S4 for an alignment of ELM2-SANT domains.
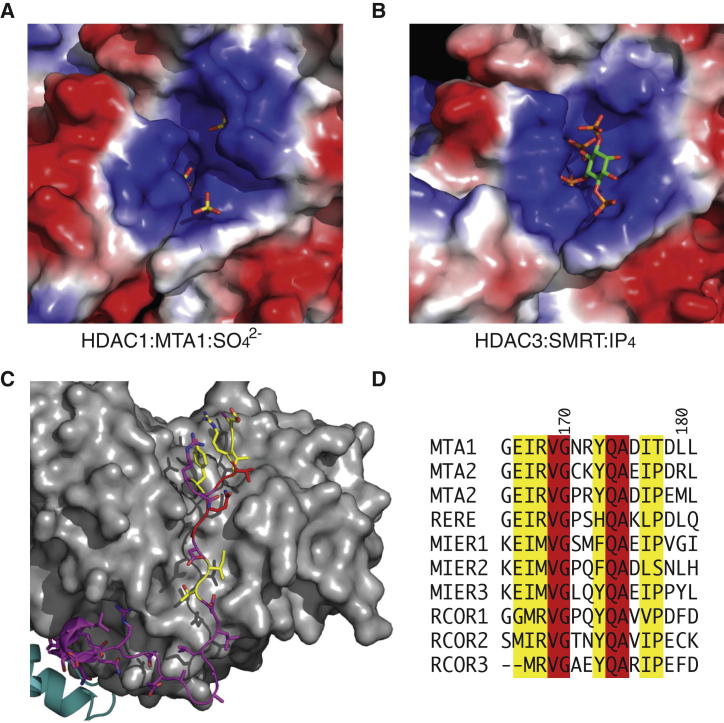
As was observed in the HDAC3:SMRT complex, there is a highly basic pocket at the interface between HDAC1 and the SANT domain of MTA1. This pocket accommodates three partly ordered sulfate groups rather than the inositol tetraphosphate molecule observed in the HDAC3 complex (Figures 2A and 2B). Importantly, the crystallization conditions contain 2 M sulfate ions that may have displaced any bound inositol phosphate. However, evidence presented below suggests that bound Ins(1,4,5,6)P4 is lost during purification of the complex, and we show that Ins(1,4,5,6)P4 can be added back to restore full HDAC1 activity.
Figure 2.
Details of the HDAC:Corepressor Interface
(A) The positively charged inositol phosphate binding pocket of HDAC1:MTA1 is occupied by ordered sulfate ions. Figure S2 shows that mutations in this pocket perturb the interaction of the MTA1-SANT domain with HDAC1.
(B) The HDAC3:SMRT binding pocket contains Ins(1,4,5,6)P4.
(C) The ELM2 specific motif binds in a groove along the HDAC1 surface. Residues are colored according to their conservation (identical, red; conserved, yellow; nonconserved, magenta).
(D) Sequence alignment of ELM2 specific motifs.
See also Figures S2 and S4.
A particularly striking feature of the HDAC1:MTA1 structure is the way in which the conserved amino-terminal portion of the ELM2 domain (consensus: EIRVGxxYQAxI), further termed the ELM2-specific motif, occupies an extended groove on the side of HDAC1 (Figures 2C and 2D). This binding surface, which is also fully conserved in HDAC2, allows MTA1 to adopt an extended conformation wrapping around the HDAC1 such that the amino terminus is positioned on the opposite side of the active site to the carboxy-terminal SANT domain and the inositol phosphate (IP) binding pocket. There is rather little regular secondary structure within this region with the exception of a single α-helical turn followed by a short beta strand. This strand forms part of and extends the core beta sheet of HDAC1. The ELM2-specific motif is located immediately carboxy-terminal to the BAH domain. This has implications for substrate recruitment to the HDAC active site (see below).
The Role of Inositol Phosphate in HDAC1 and HDAC3 Corepressor Complexes
We previously demonstrated that Ins(1,4,5,6)P4, which copurified with the complex from mammalian cells, was essential for the interaction between HDAC3 and the extended SANT domain from the SMRT corepressor (Watson et al., 2012). In a similar fashion, we were only able to obtain a complex between the isolated MTA1-SANT domain and HDAC1 when the two proteins were coexpressed in mammalian cells (Figures 3A and 3B), raising the possibility that inositol phosphate may be required to mediate interaction of the HDAC1:MTA1-SANT complex. Indeed the mutation of residues in MTA1 that we expect to coordinate Ins(1,4,5,6)P4 leads to a reduced interaction with HDAC1 (Figure S2).
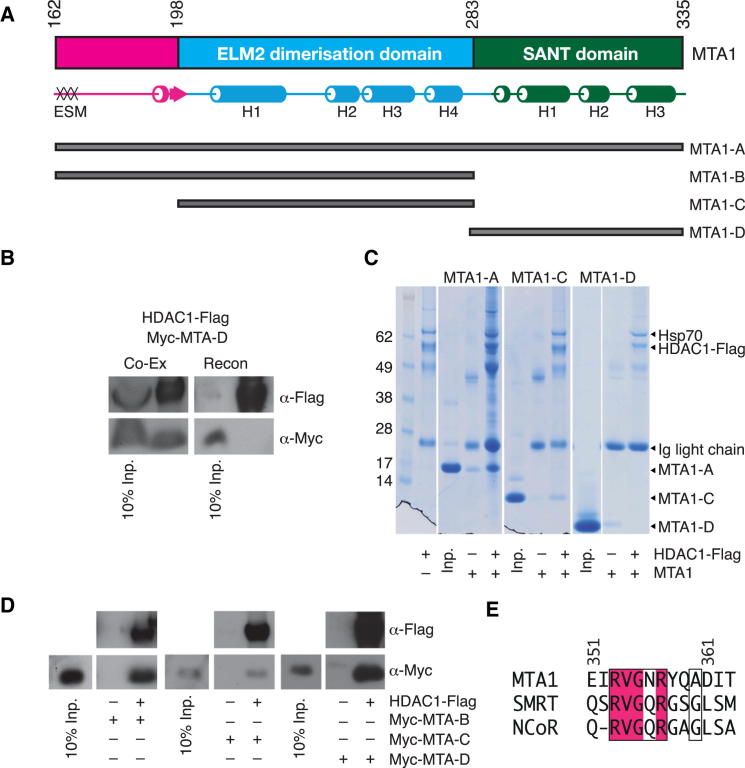
Figure 3.
Assembly of the HDAC1:MTA1 Complex
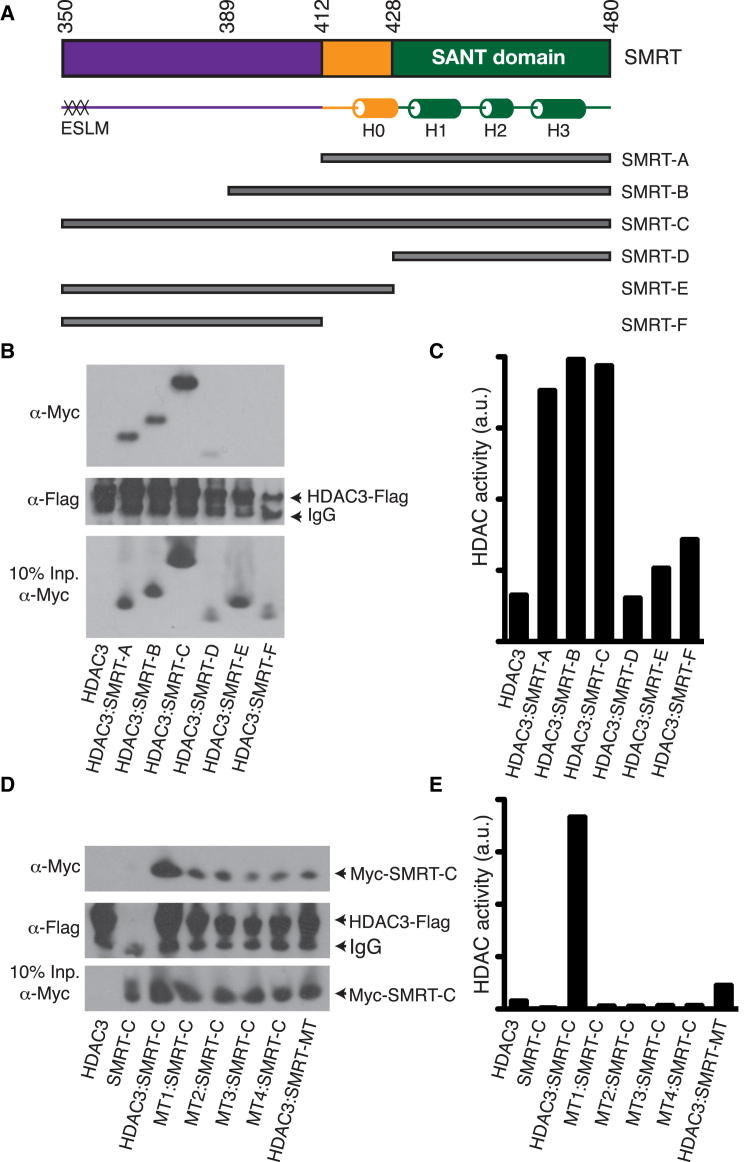
(A) MTA1 constructs (A to D) used in the interaction studies. The position of the ELM2-specific motif (ESM) is indicated.
(B) HDAC1-Flag binds to Myc-MTA1-D when cotransfected in HEK293F cells but not when both components are expressed separately and then reconstituted.
(C) HDAC1-Flag (expressed in HEK293F cells) recruits bacterially expressed MTA1-A and MTA1-C, but not MTA1-D.
(D) Myc-MTA1-B, Myc-MTA1-C, and Myc-MTA1-D bind to HDAC1-Flag when coexpressed in HEK293F cells.
(E) Alignment of the ELM2-specific motif (ESM) from MTA1 with the ELM2-specific-like motif (ESLM) of SMRT and NCoR.
While the isolated MTA1-SANT domain only forms a complex with HDAC1 when coexpressed in mammalian cells, the extended interaction between the ELM2 domain of MTA1 and HDAC1 would suggest that this region of the interface should mediate interaction independently of bound Ins(1,4,5,6)P4. Indeed, pull-down assays show that the ELM2 domain is sufficient to mediate interaction with HDAC1 when the complex is reconstituted in the absence of Ins(1,4,5,6)P4 (Figures 3C and 3D). This means that MTA1 will be bound to HDAC1 whether or not inositol phosphate is present at the interface with the SANT domain. Thus, the SANT domain is “tethered” to but not “engaged” with HDAC1 in the absence of Ins(1,4,5,6)P4.
As mentioned above, the conserved ELM2 specific motif of MTA1 interacts in an extended nonpolar groove on the surface of HDAC1. Significantly, this groove is essentially identical to a corresponding groove in the surface of HDAC3. This prompted us to ask whether there might be a similar motif in the SMRT corepressor that could interact in an analogous fashion. Figure 3E shows an alignment of the ELM2 specific motif with a motif in the amino-terminal region of SMRT (residues 351–363) and NCoR that we proposed might contribute to the interaction with HDAC3. To test this possibility, we made a number of extended constructs of the SMRT corepressor and explored their ability to interact with, and activate, wild-type HDAC3 (Figures 4A–4C). We also tested whether the longest of these SMRT constructs (residues 350–480) could interact with various mutant HDAC3 proteins that we have previously shown no longer interact with the extended SANT domain (residues 389–480). These HDAC3 mutants also lack HDAC activity, presumably as a consequence of impaired Ins(1,4,5,6)P4 binding. Significantly, we found that this longer SMRT construct is able to interact with all the HDAC3 mutants tested (Figures 4D and 4E). This supports the hypothesis that in vivo, SMRT will be tethered to HDAC3 even in the absence of Ins(1,4,5,6)P4.
Figure 4.
Assembly of the HDAC3:SMRT Complex
(A) SMRT constructs (A to F) used in the interaction studies. The position of the ELM2-specific-like motif (ESLM) is indicated.
(B) Interaction of SMRT constructs coexpressed with HDAC3.
(C) SMRT-A, SMRT-B, and SMRT-C bind and activate HDAC3.
(D) SMRT-C interacts with wild-type and mutant HDAC3 (MT1, R265P and L266M; MT2, R265P; MT3, H17C, G21A, and K25I; and MT4, H17C, G21A, K25I, R265P, L266M, and R301A), and HDAC3 interacts with mutant SMRT-C (SMRT-MT, K474A and K475A).
(E) The wild-type HDAC3:SMRT complex shows HDAC activity, whereas the complexes with a mutated Ins(1,4,5,6)P4 binding site do not have HDAC activity.
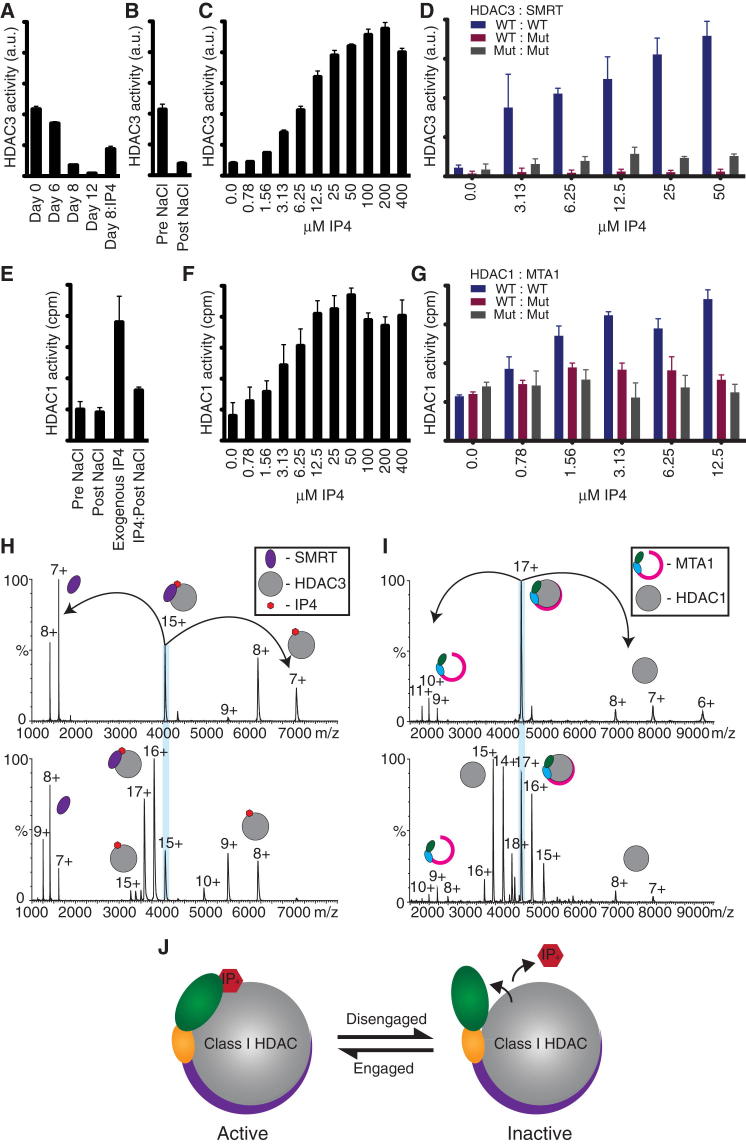
When characterizing this HDAC3 complex with the extended SMRT domain, we observed a gradual loss in HDAC activity with time (Figure 5A). HDAC activity could be partially recovered by addition of Ins(1,4,5,6)P4 to the “aged” complex (Figure 5A), suggesting that the aging is in part through loss of Ins(1,4,5,6)P4. Similarly, dialysis of fresh complex against high salt led to an immediate loss in HDAC activity (Figure 5B). Strikingly, activity can be fully restored (and indeed increased further) by the addition of exogenous Ins(1,4,5,6)P4 (Figure 5C). Mutations in HDAC3 and SMRT that would be expected to impair Ins(1,4,5,6)P4 binding to the complex abolished the ability of added Ins(1,4,5,6)P4, to enhance HDAC activity (Figure 5D). The activation characteristics suggest an apparent Kd of approximately 6 μM, which is similar to the concentration reported in mammalian cells (Barker et al., 2004). This strongly supports the hypothesis that Ins(1,4,5,6)P4 has a regulatory role in vivo.
Figure 5.
Regulation of HDAC Complexes by Ins(1,4,5,6)P4
(A) Time course showing that HDAC activity of the HDAC3:SMRT complex declines over 12 days. The “aged” complex can be partially reactivated upon addition of Ins(1,4,5,6)P4, suggesting that the “aging” is in part due to dissociation of Ins(1,4,5,6)P4.
(B) HDAC activity of HDAC3:SMRT is decreased after salt displacement of Ins(1,4,5,6)P4 (1 M NaCl).
(C) Activation of 1M NaCl-treated HDAC3:SMRT upon addition of Ins(1,4,5,6)P4.
(D) Wild-type HDAC3:SMRT is activated upon the addition of Ins(1,4,5,6)P4, whereas complexes with surface mutations within the Ins(1,4,5,6)P4 binding pocket do not respond to Ins(1,4,5,6)P4. Mutations in HDAC3, H17C, G21A, K25I, R265P, L266M, and R301A; mutations in SMRT, Y470A and Y471A.
(E) Ins(1,4,5,6)P4 is readily lost from the HDAC1:MTA1 complex during purification since the activity is unchanged after washing with 1 M NaCl. NaCl-treated HDAC1:MTA1 (1 M) can be activated by exogenous Ins(1,4,5,6)P4, but this activity is lost on further NaCl treatment.
(F) Ins(1,4,5,6)P4 stimulates the HDAC activity of HDAC1:MTA1.
(G) Wild-type HDAC1:MTA1 is activated upon the addition of Ins(1,4,5,6)P4, whereas complexes with surface mutations within the Ins(1,4,5,6)P4 binding pocket do not respond to Ins(1,4,5,6)P4.
In (A)–(G), error bars indicate the SEM. Mutations in HDAC1, R270A and R306P; mutations in MTA1, Y327A, Y328A, and K331A.
(H) Native MS spectra of HDAC3 (bottom) in complex with SMRT and MS/MS of the 15+ charge state (top) showing additional mass corresponding to one Ins(1,4,5,6)P4 molecule bound.
(I) Native MS spectra of HDAC1 (bottom) in complex with MTA1 and MS/MS of the 17+ charge state (top) confirming no additional mass in the complex.
(J) Model to show engagement of the SANT domain on the addition of Ins(1,4,5,6)P4.
Interestingly, the activity of HDAC1:MTA1 complex is not reduced by dialysis against high salt (Figure 5E). However, addition of Ins(1,4,5,6)P4 to the HDAC1:MTA1 results in a significant increase in HDAC activity in a similar fashion to that observed for the HDAC3:SMRT complex (Figure 5F). This Ins(1,4,5,6)P4-induced activity of the HDAC1:MTA1 complex is lost after dialysis against high salt (Figure 5E). These results suggest that the Ins(1,4,5,6)P4 is lost from the HDAC1:MTA1 complex during purification. Similarly to the HDAC3:SMRT complex, mutations in either HDAC1 or MTA1 that would be expected to be required for Ins(1,4,5,6)P4 binding abolished the enhancement in HDAC activity by Ins(1,4,5,6)P4 (Figure 5G). Interestingly, the apparent Kd for activation of HDAC1:MTA1 by Ins(1,4,5,6)P4 is 5 μM—similar to that of the HDAC3:SMRT complex.
To further characterize Ins(1,4,5,6)P4 binding to the two HDAC complexes, we used native and tandem mass spectrometry to explore the constitution of the complexes (Figures 5H and 5I). The experimental mass of the HDAC1:MTA1 complex matched the calculated mass for the proteins (with bound ions). In contrast, the HDAC3:SMRT complex was found to contain an additional mass of 496 Da, corresponding to bound Ins(1,4,5,6)P4. The difference in the Ins(1,4,5,6)P4 retention by the HDAC1 and HDAC3 complexes may result from MTA1 having one fewer Ins(1,4,5,6)P4-interacting lysine residues than SMRT. Taken together, our findings strongly support the hypothesis that Ins(1,4,5,6)P4 has a regulatory role in class I HDAC complexes in vivo (Figure 5J).
A Folding Transition Mediates Dimerization of HDAC1 in the NuRD Complex
The ELM2 dimerization domain in MTA1 is located immediately amino-terminal to the SANT domain and is largely helical in character. In the structure of the complex with HDAC1, this domain is sandwiched between its homodimeric partner and the deacetylase, making extensive interactions with both proteins. Interestingly, circular dichroism studies show that in isolation this domain has no cooperatively folded structure (Figure S3). Thus, the domain undergoes a dramatic folding transition on binding to HDAC1 so as to adopt a helical structure that mediates homodimerization of the whole complex.
The ELM2 dimerization domain is composed of four α helices. The interface with HDAC1 is mediated by helices H1 and H3 and excludes 1,278 Å2 from solvent exposure. In addition to important nonpolar interactions made by Trp199, Phe252, and Met255, there are a number of significant electrostatic interactions. Interestingly, mutation of the residues corresponding to either Trp199 or Phe252 in the related ELM2-SANT-containing corepressor protein MIER1 resulted in attenuated recruitment of HDAC1 (Ding et al., 2003).
Helices H1 and H4 of the ELM2 dimerization domain form the primary dimer interface with a smaller contribution from helix H2 (Figures 1D and 1E). In total, 28 nonpolar side chains are buried at the interface (14 from each monomer). The largely nonpolar and complementary nature of this dimer interface, as well as the extensive solvent excluded surface (2,332 Å2), indicates that this is a physiologically relevant interface and that the complete NuRD complex probably contains two HDAC enzymes. Given the similarity between HDAC1 and HDAC2, we envisage that the NuRD complex could contain either a homodimer of HDAC1 or HDAC2 or a heterodimer of the two enzymes. Consistent with this, HDAC1 and HDAC2 have often been reported to be both associated with the NuRD complex (e.g., Xue et al., 1998, Bantscheff et al., 2011). Importantly, the arrangement of the dimeric complex means that the active sites of the two HDACs are located on approximately the same face of the dimer, oriented approximately 50° away from the two-fold axis (Figure 1D). This may be functionally important, as such an orientation could potentially allow the complex to simultaneously target more than one nucleosome.
The overall sequence conservation for the ELM2 dimerization domain is low (Figure S4). However, the pattern of conservation suggests that the domain will adopt a similar fold in the related ELM2-containing corepressors—although helix H2 is lacking in the CoREST proteins (Figure S4). It remains to be seen whether all the related ELM2-containing corepressors form dimers and how many of the HDAC contacts are conserved.
Specificity of HDAC Complex Assembly
HDAC1 and HDAC2 are sister enzymes (83% identity) that are recruited to the same corepressor complexes (Laherty et al., 1997; Zhang et al., 1999; Humphrey et al., 2001). In contrast, HDAC3, which is 57% identical to HDAC1, is recruited uniquely to the SMRT and NCoR complexes (Guenther et al., 2000; Li et al., 2000; Yoon et al., 2003; Oberoi et al., 2011). Comparison of the structure of the HDAC3:SMRT complex with that of HDAC1:MTA1 reveals key differences between the two HDACs and their corepressors that dictate the specificity of complex assembly (Figure 6).
Figure 6.
The Specificity of HDAC Corepressor Complex Assembly
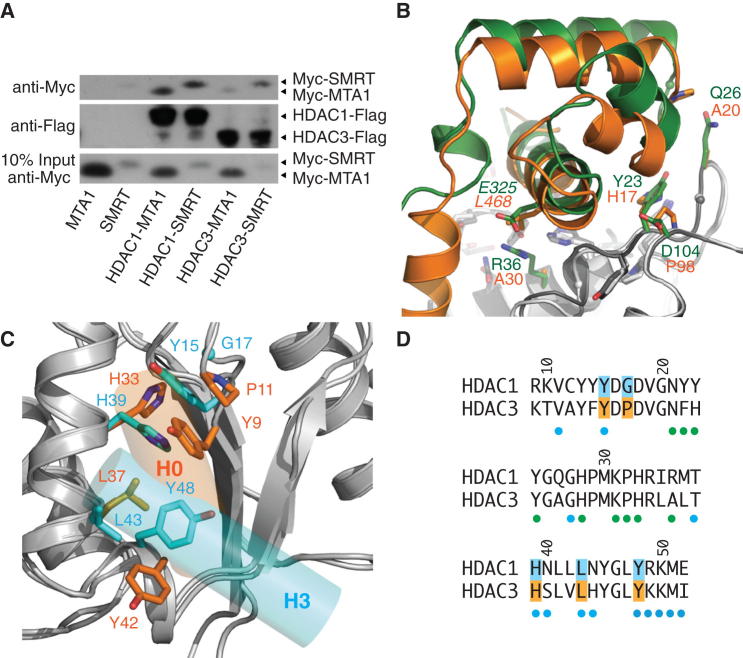
(A) Immunoblots showing that HDAC1 binds both MTA1-SANT and SMRT-SANT, whereas HDAC3 only binds SMRT-SANT efficiently.
(B) Comparison of MTA1-SANT (green) and SMRT-SANT (orange) bound to HDAC1 and HDAC3, respectively. The HDACs have been superposed. Key residues at the interface are labeled (MTA1 and HDAC1, orange; SMRT and HDAC3, green).
(C) Comparison of HDAC surfaces mediating interaction with the ELM2 dimerization domain. Key residues in HDAC1 and HDAC3 are shown in cyan and orange, respectively. The positions of the helix H3 of MTA1-ELM2 (cyan) and helix H0 of SMRT (orange) are indicated.
(D) HDAC sequence alignment highlighting residues shown in (C). The colored dots indicate residues in HDAC1 that bind to the SANT domain (green), the ELM2 domain (cyan). Residue numbers correspond to HDAC1.
Given the similarity of the SANT domains of MTA1 and SMRT, we sought to determine whether HDAC1 and HDAC3 can discriminate between the SANT domains. Coimmunoprecipitation assays show that HDAC1 appears to be able to bind the SANT domains from both MTA1 and SMRT. In contrast, HDAC3 appears to bind exclusively to the SMRT-SANT domain (Figure 6A). Examination of the structures suggests that in the HDAC1:MTA1 structure there is a key electrostatic interaction between Glu325 in MTA1 and Arg36 in HDAC1 (Figure 6B). In the HDAC3:SMRT complex, the equivalent residues are Leu468 and Ala30. Calculation of the surface charge of HDAC3 reveals a negatively charged surface that could not accommodate the negative charge of Glu325 in the MTA1-SANT domain and explains the specificity of HDAC3 for the SMRT-SANT. Interestingly, the bulkier side chains of Glu325 (MTA1) and Arg36 (HDAC1) cause helix H3 in MTA1-SANT to be slightly tipped away from the HDAC compared with the SMRT-SANT. This change is propagated to the supporting helices H1 and H2. This difference in SANT orientation matches with other amino acid differences (i.e., Gln26/Ala20, Tyr23/His17, and Asp104/Pro98 in HDAC1 and HDAC3, respectively).
The ELM2 dimerization domain of MTA1 forms a tight helical bundle that packs against HDAC1 in a region that is largely conserved between HDAC1 and HDAC3. However, four conserved side chains adopt very different configurations in the two HDACs, and this appears to be important for the specific interaction with the dimerization domain of MTA1 (Figures 6C and 6D). The critical difference between the two HDACs appears to be Gly17 in HDAC1 versus Pro11 in HDAC3. The lack of a side chain for Gly17 allows Tyr15 and His39 to make a coplanar stacking interaction in HDAC1. In HDAC3 Pro11 reorientates Tyr9, making an alternative stacking interaction resulting in the reorientation of His33. Importantly, the orientation of these residues in the HDAC1:MTA1 complex is identical to that in the free HDAC2 (Bressi et al., 2010), suggesting that this is a preformed interface that is specific for binding MTA1.
Two further conserved residues, Leu37 and Tyr42, also mediate interaction with MTA1 but adopt a very different conformation in the free HDAC2. It appears that these residues change conformation on binding the MTA1 dimerization domain so as to optimize the binding interface, but do not confer specificity.
Discussion
The HDAC1:MTA1 structure provides insights into how the combined ELM2-SANT domains are able to recruit histone deacetylase enzymes. Together, the two domains make very extensive interactions with HDAC1, wrapping completely around the catalytic domain such that both the carboxy- and amino-terminal ends are in close proximity with the active site of the enzyme.
The ELM2 and SANT domains are juxtaposed in at least 13 human corepressor proteins, most of which have been shown to recruit HDAC1 and/or HDAC2 (Toh et al., 2000; Ding et al., 2003; Lee et al., 2006; Wang et al., 2006; Bantscheff et al., 2011; Hao et al., 2011) (Figure S5). Sequence conservation strongly suggests that the mode of HDAC binding, as well as the involvement of an inositol phosphate cofactor, will be common to all these corepressor complexes. The conserved ELM2-specific motif binds in a conserved nonpolar groove on the surface of the HDAC. In MTA1, the region between the ELM2-specific motif and the SANT domain mediates dimerization of the complex, but sequence conservation suggests that this may not be the case for all ELM2-SANT-containing proteins.
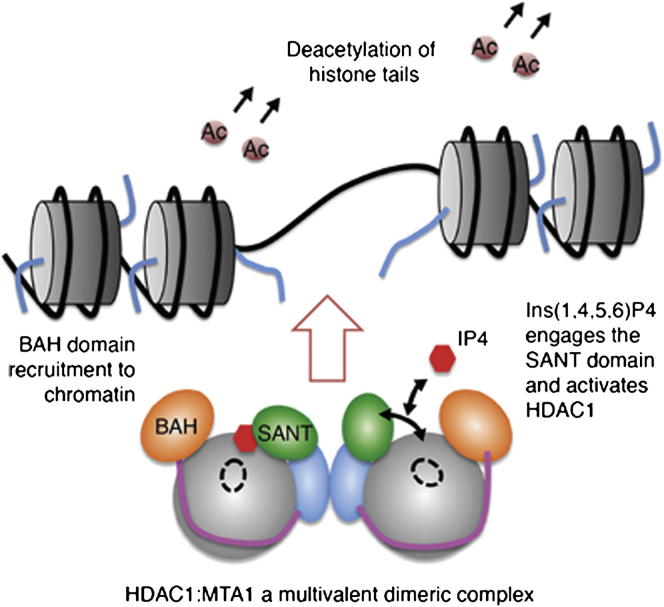
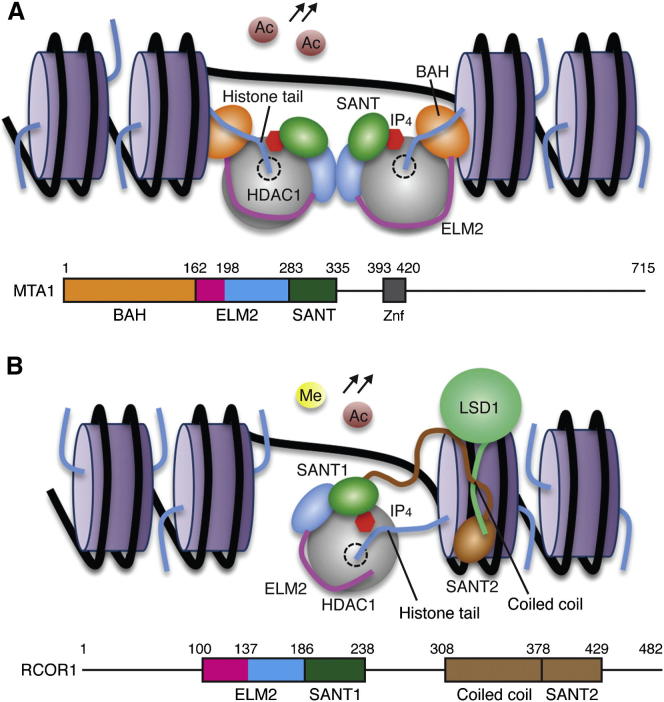
Intriguingly, within the different classes of corepressors, the protein context of the ELM2-SANT domain differs significantly (Figure S5). The MTA and RERE corepressors have an amino-terminal BAH (bromo-adjacent homology) domain immediately adjacent to the ELM2 domain. The BAH domain has been shown to mediate interaction with nucleosomes and histone tails, consistent with a role in targeting repression complexes to chromatin (Callebaut et al., 1999). A crystal structure of the SIR3 BAH domain in complex with a nucleosome revealed a mode of binding that involves recognition of the tail of histone H4 (Armache et al., 2011). The finding that the BAH domain from ORC1 also recognizes histone H4 tails supports the concept that BAH domains are chromatin recognition modules (Kuo et al., 2012). If we assume that the MTA1-BAH domain interacts with nucleosomes in a similar fashion to that of SIR3, then it is immediately clear that the BAH domain would serve as a substrate presentation module (Figure 7A) for HDAC1. Of course the BAH domains of MTA1 may well act in combination with other chromatin-targeting domains to contribute histone binding specificity to the NuRD complex. This model explains why HDAC enzymes themselves appear to lack substrate specificity since chromatin recognition modules within the specific corepressor complexes would control substrate selection.
Figure 7.
Mechanism of Substrate Presentation to Corepressor Complexes
(A) Schematic model for the interaction of the HDAC1:MTA1 dimeric complex with two adjacent nucleosomes. The MTA1-BAH domain (orange) is located immediately amino terminal to the ELM2-specific motif (magenta). An IP molecule (red) mediates the interface between HDAC1 and MTA1-SANT. Histone octamers (purple) and histone tails (blue) are illustrated schematically.
(B) Schematic model for the recruitment of HDAC1 to chromatin as part of the CoREST complex. A 70-residue linker (brown) bridges the RCOR1-SANT domain to the coiled-coil (LSD1 interaction) domain. Sequence alignments suggest that an IP molecule (red) will mediate the interface between HDAC1 and RCOR1-SANT.
See also Figure S5, which shows the domain context of HDAC corepressors.
The RCOR1, RCOR2, and RCOR3 corepressors also contain an ELM2-SANT domain that recruits HDAC1 and HDAC2 to the CoREST complex. RCOR1 does not contain an amino-terminal BAH domain; instead, there is a second SANT domain that has been shown to mediate nucleosome binding (Yang et al., 2006). Between the ELM2-SANT domain and this second SANT domain is a region that forms a coiled-coil with the lysine demethylase LSD1. This coiled-coil is separated from the ELM2-SANT domain by a 70-amino-acid linker. Combination of the LSD1:RCOR structure with the HDAC1:ELM2-SANT structure suggests that the demethylase and deacetylase enzymes are closely associated such that they can target histone tails on the same nucleosome (Figure 7B).
While it was previously thought that HDAC complexes are constitutively active, we recently showed that a potentially regulatory inositol tetraphosphate molecule [Ins(1,4,5,6)P4] is sandwiched between HDAC3 and its cognate corepressor, SMRT. This raises the important questions of whether the Ins(1,4,5,6)P4 actually regulates the activity of the complex and whether this regulation is unique to the HDAC3 complex.
Here we have demonstrated that Ins(1,4,5,6)P4 dramatically enhances the HDAC activity of both the HDAC3:SMRT and HDAC1:MTA1 complexes. On the basis of sequence conservation, we presume that inositol phosphates will activate all class I HDAC complexes that contain ELM2-SANT or equivalent domains. Interestingly, a major HDAC complex that does not contain an identifiable SANT domain is the ubiquitous Sin3A:HDAC1 complex. It remains to be established how HDAC1 is activated in this complex.
The apparent Kd of both HDAC3 and HDAC1 complexes for Ins(1,4,5,6)P4 is around 5 μM. If Ins(1,4,5,6)P4 were to act as a physiologically relevant regulator of these HDAC complexes, then Ins(1,4,5,6)P4 levels in the relevant cellular compartment would need to fluctuate around the this concentration. We could not identify any reports of Ins(1,4,5,6)P4 levels in the nucleus; however, the average concentration of InsP4 (both enantiomers) in the cell has been reported to vary between 3.6 and 10.5 μM, depending upon the stage of the cell cycle (Barker et al., 2004). On the basis of our in vitro assays, this would equate to at least a 2-fold change in HDAC activity. The match between the apparent Kd of binding to HDAC complexes with the cellular concentration of Ins(1,4,5,6)P4 would seem to strongly suggest that the Ins(1,4,5,6)P4 is regulatory in vivo.
It is interesting to compare the Ins(1,4,5,6)P4 regulation of HDACs with the well-established regulatory activity of Ins(1,4,5)P3 in the opening of calcium channels. The concentration of Ins(1,4,5)P3 needed to give maximal calcium release is about 2 μM (Marchant and Taylor, 1997). The reported average cellular concentration of Ins(1,4,5)P3 is 0.8–2.7 μM. The similarity in the ratio of these concentrations with those needed to activate the HDAC3 and HDAC1 complexes provides further support that Ins(1,4,5,6)P4 is a bona fide regulator of HDAC activity.
Since Ins(1,4,5,6)P4 and Ins(1,4,5)P3 are both biologically relevant signaling molecules and differ only in a single phosphate group, it is important to ask how these signaling pathways are kept distinct, i.e., what is the specificity of Ins(1,4,5)P3 receptors versus HDAC complexes. It is very clear from the structure of the Ins(1,4,5)P3 receptor that Ins(1,4,5,6)P4 would be unable to substitute for Ins(1,4,5)P3 since the –OH group at the 6 position is oriented toward the receptor and a phosphate group at this position could not be sterically tolerated (Bosanac et al., 2002). In contrast, the phosphate at the 6 position in the HDAC complex is clearly very important for binding since it is deeply buried and mediates multiple interactions with the complex. Thus, it appears that the two signaling pathways are kept distinct through differential specificity at the 6 position of the inositol phosphate.
An unexpected feature of the HDAC1:MTA1 complex is that MTA1 wraps completely around HDAC1 such that the Ins(1,4,5,6)P4-binding SANT domain is tethered to the corepressor via the ELM2 domain. We have shown that SMRT is tethered to HDAC3 in an analogous fashion. These findings suggest that these corepressor proteins are constitutively assembled into the class I HDAC complexes and that they are activated by freely dissociating inositol phosphates that bind at this interface. It is likely that the SANT domain is either “engaged” or “disengaged” from the catalytic domain of the HDAC depending upon the concentration of the inositol phosphate. Exactly the mechanism through which engaging the SANT domain activates the HDAC remains to be determined, but we and others have proposed that this involves a stabilization of the dynamics of the catalytic domain so as to open the channel to the active site of the enzyme (Arrar et al., 2012; Watson et al., 2012).
In conclusion, the structure of the HDAC1:MTA1 complex reveals that inositol phosphates are conserved regulators of class I HDAC complexes and offers further insight into the mechanism and specificity of the recruitment of HDACs to the NuRD repression complex. The structure also reveals the mechanism of complex dimerization and strongly suggests that substrate presentation and specificity is determined by the adjacent BAH domain serving as a chromatin recognition module.
Experimental Procedures
Protein Expression and Purification
The ELM2-SANT domain of MTA1 (residues 162–335) and full-length HDAC1 were cloned into pcDNA3 vectors. The His10-Flag3-TEV-MTA1 construct was cotransfected with untagged-HDAC1 into suspension-grown HEK293F cells (Invitrogen) with polyethylenimine (PEI; Sigma) and harvested after 48 hr as described previously (Watson et al., 2012). Cells were lysed in 50 mM Tris-Cl (pH 7.5), 100 mM potassium acetate, 10% v/v glycerol, 0.5% v/v Triton X-100, and Roche Complete Protease Inhibitor (buffer A), and the insoluble material removed by centrifugation. The complex was purified on FLAG resin (Sigma) after incubation with Sepharose 4B (GE Healthcare). The resin was washed three times with buffer A, three times with 50 mM Tris-Cl (pH 7.5), 200 mM potassium acetate, and 5% v/v glycerol (buffer B), and three times with 50 mM Tris-Cl (pH 7.5), 50 mM potassium acetate, 5% v/v glycerol, 0.5 mM TCEP (buffer C) followed by an overnight TEV cleavage in buffer C. HDAC1:MTA1 was further purified by gel filtration chromatography on a Superdex S200 column (GE Healthcare) in buffer containing 25 mM Tris-Cl (pH 7.5), 25 mM potassium acetate, and 0.5 mM TCEP (buffer D). Fractions containing the complex were identified by SDS-PAGE, pooled, and concentrated to 5 mg/ml.
Various MTA1 constructs (residues 162–335, 196–277, and 277–335) were expressed in E. coli Rosetta (Novogen) with the pET30a vector. The polypeptides were expressed at 20°C as N-terminal His6 fusion proteins with a TEV cleavage site. Cells were lysed by sonication in 50 mM Tris-Cl (pH 8), 150 mM NaCl, 20 mM imidazole, 1 mM DTT, and Roche Complete Protease Inhibitor (buffer E), and the insoluble material was removed by centrifugation. The supernatant was applied to Ni-NTA resin (QIAGEN) and washed with buffer E containing 40 mM imidazole, and the protein was eluted with buffer E containing 200 mM imidazole. Proteins were dialyzed into 50 mM Tris-Cl (pH 8), 50 mM NaCl, and 1 mM DTT (buffer F), and the His tag was removed by TEV cleavage. Fragments were further purified in buffer F by gel filtration chromatography on a Superdex S75 column (GE Healthcare).
Crystallography
Diffracting crystals of 15 μm were obtained by sitting-drop vapor diffusion at 20°C against wells containing 0.1 M Na HEPES (pH 7.5), 2 M ammonium sulfate, and 5% PEG400. Single crystals were frozen in mother liquor with the addition of 15% glycerol (cryoprotectant), and data were collected at the Diamond synchrotron microfocus beamline I24, with use of a grid-scan tool to center the crystals (Aishima et al., 2010). Diffraction data from three crystals were processed with MOSFLM (Leslie, 2006) and combined with AIMLESS. The structure was solved by molecular replacement with HDAC3:SMRT (PDB code 4A69) as a search model using PHASER (McCoy et al., 2007). The HDAC1:MTA1 structure was built through multiple rounds of rebuilding and refinement with REFMAC (Collaborative Computational Project, Number 4, 1994) and COOT (Emsley et al., 2010).
The final model contains amino acids 165–333 of MTA1 (chain A), amino acids 8–376 of HDAC1 (chain B), one zinc ion, two potassium ions, four sulfate molecules, and one acetate molecule.
Immunoprecipitation
Myc-MTA1 constructs and HDAC1-Flag, or Myc-SMRT constructs and HDAC3-Flag, were transfected individually or cotransfected into HEK293F cells as described above. Cells were lysed by sonication in buffer containing 50 mM Tris-Cl (pH 7.5), 100 mM potassium acetate, 5% v/v glycerol, 0.3% v/v Triton X-100, and Roche Complete Protease Inhibitor, and the insoluble material was removed by centrifugation. The supernatant was applied to BSA-blocked FLAG resin for 2 hr before three washes with 50 mM Tris-Cl (pH 7.5), 100 mM potassium acetate, and 5% v/v glycerol. For the reconstitution experiments, Myc-MTA1 supernatant was incubated with the resin bound HDAC1-Flag for 2 hr before three washes with 50 mM Tris-Cl (pH 7.5), 100 mM potassium acetate, and 5% v/v glycerol.
Circular Dichroism
MTA1 (residues 205–310) was expressed in E. coli Rosetta (Novogen). Initial purification was carried out with NiNTA and followed by ion-exchange chromatography. Protein was dialyzed into 50 mM Tris-Cl (pH 8), 100 mM NaCl, and 1 mM DTT and concentrated to 1 mg/ml. A circular dichroism spectrum was measured from 200–250 nm at 20°C with a spectropolarimeter (Jasco J-715). A melting curve was obtained by monitoring of ellipticity CD at 210 nm as the temperature was increased from 5°C to 95°C.
HDAC Activity Assays
HDAC3 and SMRT (residues 350–480, 428–480, 350–427, 350–411) were coexpressed in HEK293 cells and purified on FLAG resin. HDAC3 activity was measured with the HDAC Assay Kit (Active Motif) and read with a Victor X5 plate reader (Perkin Elmer) as described (Watson et al., 2012).
HDAC1 and MTA1 (residues 162–335) were coexpressed in HEK293 cells, and HDAC1 activity was measured with the HDAC Assay Kit (Millipore). In brief, purified HDAC1:MTA1 was incubated with 25,000 cpm [3H]-acetyl biotinylated-histone H4 peptide captured on streptavidin agarose in buffer containing 50 mM Tris-Cl (pH 7.5), 50 mM NaCl, and 5% v/v glycerol for 2 hr at 37°C. The reaction was quenched with acid before the beads were pelleted by centrifugation for 2 min at 14,000 g. Released [3H]-acetate was measured with a scintillation counter (Beckman LS 6500).
HDAC Activity Assay with Ins(1,4,5,6)P4 Titration
HDAC3:SMRT (residues 350–480) and HDAC1:MTA1 (residues 162–335) were expressed in HEK293 cells and purified on FLAG resin. For removal of intrinsically bound inositol phosphate, 1 μM protein was incubated in a buffer containing 50 mM Tris-Cl (pH 7.5), 1 M NaCl, and 5% v/v glycerol for 4 hr at room temperature. The protein was then dialyzed for 14 hr against 50 mM Tris-Cl (pH 7.5), 50 mM NaCl, and 5% v/v glycerol. To perform the titration, 62 nM HDAC3:SMRT or 50 nM HDAC1:MTA1 was incubated with varying concentrations of exogenous Ins(1,4,5,6)P4 (Cayman Chemical) for 30 min at 37°C before the HDAC activity was measured. Experiments were performed in triplicate, and data were analyzed with (GraphPad Prism version 5.0, GraphPad Software); Kd values were calculated by nonlinear curve fitting with a one-site binding (hyperbola) model [y = bmax · x / (Kd + x)].
Mass Spectrometry
Nano electrospray ionization (NanoESI) mass spectrometry (MS) and tandem MS (MS/MS) experiments were performed on a QSTAR XL instrument. Prior to MS analysis, 20 μl of a 1 mg/ml HDAC3:SMRT or 2.8 mg/ml HDAC1:MTA1 in 50 mM Tris-Cl, 50 mM potassium acetate, and 5% v/v glycerol was buffer exchanged once into 300 mM ammonium acetate solution (pH 7.4) with Bio-Rad Biospin columns. After appropriate dilution, the protein complexes were analyzed by native MS at 19 μM for the HDAC1:MTA1 and 4 μM for the HDAC3:SMRT complex. Protein solution was typically loaded for sampling via gold-plated borosilicate glass capillaries made in house as described previously (Nettleton et al., 1998). The following experimental parameters were used: capillary voltage, 1.3 kV; declustering potential, 100 V; focusing potential, 150 V; and second declustering potential, 15 V. Focusing rod offset varied from 60 to 100 V, MCP 2550. Argon was used as a collision gas at maximum pressure. All spectra were calibrated externally with a solution of cesium iodide (100 mg/ml).
Acknowledgments
We are grateful to Xiaowen Yang (PROTEX cloning service) for preparing all the expression constructs used in this work. We thank the staff at Beamline I24 at the Diamond Light Source, especially Robin Owen, for assistance with data collection. This work was supported by grant WT085408 from the Wellcome Trust.
Published: June 20, 2013
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplemental Information includes five figures and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2013.05.020.
Contributor Information
Louise Fairall, Email: louise.fairall@le.ac.uk.
John W.R. Schwabe, Email: john.schwabe@le.ac.uk.
Accession Numbers
The coordinates and structure factors for the HDAC1:MTA1 complex crystal structure reported in this paper have been deposited in the Protein Data Bank under accession code 4bkx.
Supplemental Information
References
- Aishima J., Owen R.L., Axford D., Shepherd E., Winter G., Levik K., Gibbons P., Ashton A., Evans G. High-speed crystal detection and characterization using a fast-readout detector. Acta Crystallogr. D Biol. Crystallogr. 2010;66:1032–1035. doi: 10.1107/S0907444910028192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache K.-J., Garlick J.D., Canzio D., Narlikar G.J., Kingston R.E. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science. 2011;334:977–982. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrar M., Turnham R., Pierce L., de Oliveira C.A.F., McCammon J.A. Structural insight into the separate roles of inositol tetraphosphate and deacetylase-activating domain in activation of histone deacetylase 3. Protein Sci. 2012;22:83–92. doi: 10.1002/pro.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantscheff M., Hopf C., Savitski M.M., Dittmann A., Grandi P., Michon A.-M., Schlegl J., Abraham Y., Becher I., Bergamini G. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat. Biotechnol. 2011;29:255–265. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]
- Barker C.J., Wright J., Hughes P.J., Kirk C.J., Michell R.H. Complex changes in cellular inositol phosphate complement accompany transit through the cell cycle. Biochem. J. 2004;380:465–473. doi: 10.1042/BJ20031872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosanac I., Alattia J.-R., Mal T.K., Chan J., Talarico S., Tong F.K., Tong K.I., Yoshikawa F., Furuichi T., Iwai M. Structure of the inositol 1,4,5-trisphosphate receptor binding core in complex with its ligand. Nature. 2002;420:696–700. doi: 10.1038/nature01268. [DOI] [PubMed] [Google Scholar]
- Bressi J.C., Jennings A.J., Skene R., Wu Y., Melkus R., De Jong R., O’Connell S., Grimshaw C.E., Navre M., Gangloff A.R. Exploration of the HDAC2 foot pocket: Synthesis and SAR of substituted N-(2-aminophenyl)benzamides. Bioorg. Med. Chem. Lett. 2010;20:3142–3145. doi: 10.1016/j.bmcl.2010.03.091. [DOI] [PubMed] [Google Scholar]
- Callebaut I., Courvalin J.C., Mornon J.P. The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 1999;446:189–193. doi: 10.1016/s0014-5793(99)00132-5. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Ding Z., Gillespie L.L., Paterno G.D. Human MI-ER1 alpha and beta function as transcriptional repressors by recruitment of histone deacetylase 1 to their conserved ELM2 domain. Mol. Cell. Biol. 2003;23:250–258. doi: 10.1128/MCB.23.1.250-258.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Guenther M.G., Lane W.S., Fischle W., Verdin E., Lazar M.A., Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Xu N., Box A.C., Schaefer L., Kannan K., Zhang Y., Florens L., Seidel C., Washburn M.P., Wiegraebe W., Mak H.Y. Nuclear cGMP-dependent kinase regulates gene expression via activity-dependent recruitment of a conserved histone deacetylase complex. PLoS Genet. 2011;7:e1002065. doi: 10.1371/journal.pgen.1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey G.W., Wang Y., Russanova V.R., Hirai T., Qin J., Nakatani Y., Howard B.H. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J. Biol. Chem. 2001;276:6817–6824. doi: 10.1074/jbc.M007372200. [DOI] [PubMed] [Google Scholar]
- Kuo A.J., Song J., Cheung P., Ishibe-Murakami S., Yamazoe S., Chen J.K., Patel D.J., Gozani O. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature. 2012;484:115–119. doi: 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laherty C.D., Yang W.M., Sun J.M., Davie J.R., Seto E., Eisenman R.N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Lee M.G., Wynder C., Bochar D.A., Hakimi M.-A., Cooch N., Shiekhattar R. Functional interplay between histone demethylase and deacetylase enzymes. Mol. Cell. Biol. 2006;26:6395–6402. doi: 10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A.G.W. The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- Li J., Wang J., Wang J., Nawaz Z., Liu J.M., Qin J., Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavathi B., Kumar R. Metastasis tumor antigens, an emerging family of multifaceted master coregulators. J. Biol. Chem. 2007;282:1529–1533. doi: 10.1074/jbc.R600029200. [DOI] [PubMed] [Google Scholar]
- Marchant J.S., Taylor C.W. Cooperative activation of IP3 receptors by sequential binding of IP3 and Ca2+ safeguards against spontaneous activity. Curr. Biol. 1997;7:510–518. doi: 10.1016/s0960-9822(06)00222-3. [DOI] [PubMed] [Google Scholar]
- Marks P.A., Xu W.-S. Histone deacetylase inhibitors: Potential in cancer therapy. J. Cell. Biochem. 2009;107:600–608. doi: 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettleton E.J., Sunde M., Lai Z., Kelly J.W., Dobson C.M., Robinson C.V. Protein subunit interactions and structural integrity of amyloidogenic transthyretins: evidence from electrospray mass spectrometry. J. Mol. Biol. 1998;281:553–564. doi: 10.1006/jmbi.1998.1937. [DOI] [PubMed] [Google Scholar]
- Oberoi J., Fairall L., Watson P.J., Yang J.-C., Czimmerer Z., Kampmann T., Goult B.T., Greenwood J.A., Gooch J.T., Kallenberger B.C. Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery. Nat. Struct. Mol. Biol. 2011;18:177–184. doi: 10.1038/nsmb.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren-Knaak M., Ishii H., Sun J.-M., Pazin M.J., Davie J.R., Peterson C.L. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- Toh Y., Kuninaka S., Endo K., Oshiro T., Ikeda Y., Nakashima H., Baba H., Kohnoe S., Okamura T., Nicolson G.L., Sugimachi K. Molecular analysis of a candidate metastasis-associated gene, MTA1: possible interaction with histone deacetylase 1. J. Exp. Clin. Cancer Res. 2000;19:105–111. [PubMed] [Google Scholar]
- Wang L., Rajan H., Pitman J.L., McKeown M., Tsai C.-C. Histone deacetylase-associating Atrophin proteins are nuclear receptor corepressors. Genes Dev. 2006;20:525–530. doi: 10.1101/gad.1393506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P.J., Fairall L., Santos G.M., Schwabe J.W.R. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012;481:335–340. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y.D., Perissi V., Staszewski L.M., Yang W.M., Krones A., Glass C.K., Rosenfeld M.G., Seto E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc. Natl. Acad. Sci. USA. 2000;97:7202–7207. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Dai X.-L., Huang H.-C., Jiang Z.-F. Targeting HDACs: a promising therapy for Alzheimer’s disease. Oxid. Med. Cell. Longev. 2011;2011:143269. doi: 10.1155/2011/143269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Wong J., Moreno G.T., Young M.K., Côté J., Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Yang M., Gocke C.B., Luo X., Borek D., Tomchick D.R., Machius M., Otwinowski Z., Yu H. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol. Cell. 2006;23:377–387. doi: 10.1016/j.molcel.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Yoon H.-G., Chan D.W., Huang Z.-Q., Li J., Fondell J.D., Qin J., Wong J. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. EMBO J. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ng H.H., Erdjument-Bromage H., Tempst P., Bird A., Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.