Abstract
Polycomb repressive complex two (PRC2) has been implicated in embryonic stem (ES) cell pluripotency; however, the mechanistic roles of this complex are unclear. It was assumed that ES cells contain PRC2 with the same subunit composition as that identified in HeLa cells and Drosophila embryos. Here, we report that PRC2 in mouse ES cells contains at least three additional subunits: JARID2, MTF2, and a novel protein denoted esPRC2p48. JARID2, MTF2, and esPRC2p48 are highly expressed in mouse ES cells compared to differentiated cells. Importantly, knockdowns of JARID2, MTF2, or esPRC2p48 alter the level of PRC2-mediated H3K27 methylation and result in the expression of differentiation-associated genes in ES cells. Interestingly, expression of JARID2, MTF2, and esPRC2p48 together, but not individually, enhances Oct4/Sox2/Klf4-mediated reprograming of mouse embryonic fibroblasts (MEFs) into induced pluripotent stem cells, whereas knockdown or knockout of JARID2, MTF2, or esPRC2p48 significantly inhibits reprograming. JARID2, MTF2, and esPRC2p48 modulate H3K27 methylation and facilitate repression of lineage-associated gene expression when transduced into MEFs, and synergistically stimulate the histone methyl-transferase activity of PRC2 in vitro. Therefore, these studies identify JARID2, MTF2, and esPRC2p48 as important regulatory subunits of PRC2 in ES cells and reveal critical functions of these subunits in modulating PRC2’s activity and gene expression both in ES cells and during somatic cell reprograming.
Keywords: Epigenetics, Polycomb group protein, Embryonic stem cell, Histone modification
Introduction
Polycomb group proteins play an important role in the epigenetic regulation of development, cell fate determination, and maintenance, and have recently been implicated in embryonic stem (ES) cell pluripotency [1–4]. PcG proteins form multisubunit complexes to execute their repressive functions. Biochemical and genetic evidence indicates that PcG proteins exist in two major polycomb repressive complexes one and two (PRC1 and PRC2). [5–8]. PRC2 is composed of four core components EZH2, SUZ12, RaAp46/48, and EED; EZH2 is the catalytic subunit that mediates histone H3K27 methylation [9–12]. In addition, polycomb-like (PCL1) and EZH1 were also identified as additional subunits of PRC2 [13–16]. The core components of PRC1 include HPC, HPH, Bmi1/Mel18, and Ring1A/B; this complex condenses chromatin directly and also mediates ubiquitination of histone H2AK119 [17, 18]. Recent studies indicate that PRC1 and PRC2 function in a cooperative manner to maintain gene silencing. PRC2 initiates the repression process and PRC1 is involved in maintaining the repressed state [19, 20]. The histone modification activities are essential for PcG protein-mediated gene silencing [9–12, 17, 21].
Although initially identified as factors responsible for the heritable repression of Homeotic (Hox) genes, recent genome-wide studies reveal that PcG proteins associate with a large number of genes controlling development in murine and human ES cells [22, 23]. PcG proteins and histone modifications mediated by these proteins are proposed to maintain genes in a transcriptionally poised state and, therefore, contribute to ES cell pluripotency. Although the function of PcG proteins in maintaining ES cell pluripotency has been debated, defects in repression of lineage-specific gene expression have been documented in ES cells with knockouts of PRC2 subunits EZH2, SUZ12, and EED [15, 23–25]. These ES cells are also defective in reprograming of gene expression during lineage-commitment [15, 24, 25]. These studies suggest that PcG proteins are important for defining cellular gene expression programs and, consequently, cell fate determination [26, 27].
Despite extensive efforts devoted to defining the composition of PRCs, the majority of these studies have employed HeLa cells or Drosophila embryos as starting materials and assumed that all other cell types contain PRCs with the same subunit composition. Here, we report the purification and characterization of a PRC2 complex from mouse ES cells. This complex contains at least three additional regulatory subunits, and importantly, these subunits play critical roles in regulating the function of PRC2, and therefore, the pluripotency of ES cells and reprograming of somatic cells.
Materials and Methods
cDNAs, Recombinant Proteins, and Antibodies
cDNAs for JARID2, EZH1, MTF2, and esPRC2p48 were obtained from Open Biosystems (www.openbiosystems.com Huntsville, AL, US) and cloned into the lentiviral vector FG12 (Addgene, www.addgene.org Cambridge, MA, US) and verified by sequencing. The polycistronic Oct4, Sox2, and KLF4 lentiviral vector was described previously [28]. shRNA cassettes including human H1 promoter and targeting sequences were also cloned into lentiviral vector FG12. Targeting sequences for SUZ12, JARID2, MTF2, and esPRC2p48 are provided in Table S2. Recombinant proteins were purified from sf-9 cells with anti-Flag resin and Superose six gel filtration column as described previously [29]. Antibodies against SUZ12 and EZH2 were described in a previous publication [9]. Antibodies against JARID2, EZH1, H3K27me3, H3K27me1, H3K4me3, H3K4me2, and H3K4me1 were obtained from Abcam (www.abcam.com Cambridge, MA, US). Antibody against Anti-H3K27me2 was obtained from Millipore (www.millipore.com Billerica, MA, US). Antibodies against MTF2 and esPRC2p48 were generated with recombinant MTF2 (amino acids 44–155) and esPRC2p48 (151–327) as antigens.
AP, SSEA1 Staining, and Teratoma Formation
For alkaline phosphatase (AP) staining on original plates, cells were stained using the Vector Blue Alkaline Phosphatase Substrate Kit III (Vector Laboratories, www.vectorlabs.com Burlingame, CA, US) according to the manufacturer’s instructions. For immunostaining, induced pluripotent stem (iPS) cells were cultured on cover slips, fixed with 4% paraformaldehyde, and permeabilized with 0.5% Triton X-100. Cells were stained with primary antibodies against stage-specific embryonic antigen-1 (SSEA1) and Nanog (R&D Systems, www.rndsystems.com Minneapolis, MN, US) and then incubated with fluorophore-labeled secondary antibodies (Jackson Immunoresearch, www.jacksonimmuno.com West Grove, PA, US) before visualized under an Olympus microscope. For teratoma formation assay, 5 × 106 iPS cells in 100 μl of PBS were subcutaneously injected into NOD/SCID IL-2γR KO mice via a 21 G needle. Teratomas were surgically removed after 3–4 weeks and fixed in formalin at 4°C overnight, embedded in paraffin wax, and sectioned. Sections were stained with hematoxylin and eosin for pathological examination.
Immunoprecipitation, Western Blot, RT-PCR, and ChIP Assay
For immunopurification, affinity purified antibodies against EZH2, EZH1, SUZ12, JARID2, MTF2, and esPRC2p48 were cross-linked to protein A agarose beads as described [29] and were incubated at 4°C for 4 hours with an aliquot of the Superose six column fractions (Figs. 1B and S1B) or Hydroxyapatite fractions (Fig. 1D–1F) dialyzed against BC50. After washing with BC500 three times and BC50 two times, proteins bound to beads were used for silver staining, western blotting, and protein identification [29]. Parallel experiments were performed with equal amounts of rabbit IgG. Reverse transcription-polymerase chain reaction (RT-PCR) and chromatin immunoprecipitation (ChIP) were performed as described with primers listed in Table S2 [29].
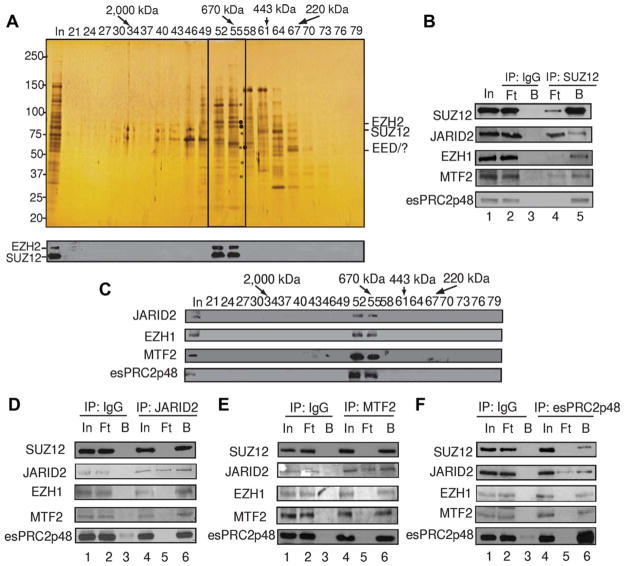
Figure 1.
Purification and identification of PRC2 from mouse ES cells. (A): Silver staining (top panel) and Western Blot assay (bottom panel) of an SDS-PAGE containing an aliquot of fractions derived from the Superose six column. Polypeptides co-purified with SUZ12 and EZH2 are indicated by asterisks. The elution profile of the protein markers is indicated on top of the panel. (B): Western blot analysis of an aliquot of Input (In), Flowthrough (Ft), and Bound (B) of samples derived from anti-SUZ12 and control IgG immunoprecipitation. (C): Western blot analysis of an aliquot of Superose six fractions. (D–F): Western blot analysis of an aliquot of Input (In), Flowthrough (Ft), and Bound (B) of samples derived from anti-JARID2 (D), anti-MTF2 (E), and anti-esPRC2p48 (F) and control IgG immunoprecipitation.
Information about protein purification, LC-MS/MS mass spectrometry identification, ES cell culture, histone methyl-transferase assay, histone demethylation assay, and peptide array binding assays can be found in Supporting information.
Results
Purification of PRC2 from Mouse ES Cell
To determine the subunit composition of PRC2 in ES cells, we undertook a biochemical purification from a large scale culture of mouse ES cells (≈3 × 1010 cells). Through a six-column scheme (Fig. S1A), we purified PRC2 to relative homogeneity (Fig. 1A). Silver staining of an SDS-PAGE gel containing fractions derived from the Superose six column revealed that unique polypeptides, which were not observed when PRC2 was purified from HeLa cells [9, 11, 13], co-purified with SUZ12 and EZH2 (Fig. 1A, top panel, marked with asterisks). To confirm that these polypeptides are indeed integral subunits of PRC2 in ES cells, we performed an affinity purification with anti-SUZ12 antibody. Silver staining revealed that anti-SUZ12 antibody, but not control IgG, specifically pulled down EZH2 and SUZ12 (Fig. S1B, labeled on the right side of the panel) as well as other subunits of PRC2, as revealed by western blot assay (Fig. 1B, top panel, compare lane three with five) and mass spectrometry analysis (Table S1). Interestingly, we observed that several of the poly-peptides, which co-purified with SUZ12 and EZH2 on the Superose six column, were also pulled down by anti-SUZ12 antibody (Fig. S1B, marked with arrow heads).
Because of limited amounts of sample, we subjected the anti-SUZ12 immunoprecipitates to LC-MS/MS identification. Analysis of mass spectrometry results revealed that, in addition to the previously reported PRC2 subunits SUZ12, EZH2, EED, Rbbp4, and Rbbp7 (mouse homologs for RbAP46 and RbAP48), JARID2, EZH1, MTF2, Hspa9, and a hypothetical protein E130012A19Rik, which we donated esPRC2p48 (ES cell-specific PRC2 subunit p48), were also specifically immunoprecipitated by anti-SUZ12 antibody (Table S1). Western blot analysis confirmed that anti-SUZ12 antibody not only specifically immunoprecipitated SUZ12 but also JARID2, EZH1, MTF2, and esPRC2p48 (Fig. 1B, second to fifth panels, compare lane three with 5), suggesting that JARID2, EZH1, MTF2, and esPRC2p48 might represent additional subunits of PRC2 in ES cells. Consistent with this notion, JARID2, EZH1, MTF2, and esPRC2p48 were found to co-purify with SUZ12 and EZH2 in the Superose six column and several other columns during the purification (Fig. 1C and data not shown).
To confirm that JARID2, EZH1, MTF2, and esPRC2p48 are bone fide subunits of PRC2 in ES cells, we performed a series of reciprocal immunoprecipitation assays with an aliquot of the hydroxyapatite column fraction (Fig. S1A). Compared to control IgG, antibodies against JARID2, MTF2, and esPRC2p48 not only efficiently immunoprecipitated the target proteins but also specifically immunoprecipitated PRC2 core component SUZ12 (Fig. 1D, top two panels; Fig. 1E, first and fourth panels; and Fig. 1F, top and bottom panels, compare lanes 1–3 with 4–6). The efficient immunoprecipitation depleted the target proteins as well as SUZ12 from the Flowthrough (Ft), suggesting that the interactions between JARID2, MTF2, esPRC2p48, and PRC2 are www.StemCells.com stable under stringent conditions (500 mM KCl with 0.05% NP40). In all immunoprecipitation assays, antibodies against JARID2, MTF2, and esPRC2p48 also efficiently immunoprecipitated the other newly identified subunits (Fig. 1D, bottom three panels; Fig. 1E, second, third, and bottom panels; Fig. 1F, second to fourth panels, compare lanes 1–3 with 4–6). These results suggest that JARID2, EZH1, MTF2, and esPRC2p48 are integral subunits of PRC2 in ES cells. However, we failed to detect interactions between EZH1 and EZH2 under our stringent conditions (Fig. S1C), suggesting that EZH2 and EZH1 might form mutually exclusive canonical and non-canonical PRC2 in ES cells. This observation is consistent with a recent report that EZH1 interacts with EZH2 in one step but not in tandem immunoprecipitation [15]. In summary, biochemical purification of PRC2 from mouse ES cells identified JARID2, EZH1, MTF2, and esPRC2p48 as additional subunits in ES cells.
Of the four subunits, JARID2 belongs to JmjC domain containing protein family, members of which mediate histone demethylation through an iron- and α-ketoglutarate (KG)-dependent oxidation mechanism [30, 31]. However, JARID2 is unlikely to have histone demethylation activity because key residues involved in the binding of Fe (ii) and α-KG are substituted (Fig. S2). MTF2 (also named PCL2), together with PHF1 (PCL1) and PHF19 (PCL3) are the three mammalian homologs of Drosophila Polycomb-like (Pcl) protein (Fig. S2). Pcl and PHF1 interact with PRC2 in Drosophila and HeLa cells, respectively, and facilitate the conversion of H3K27 from di-methyl to tri-methyl [13, 14, 32]. EZH1 and EZH2 are the two vertebrate paralogs of Drosophila Ez (Fig. S2), and EZH1 forms a non-canonical PRC2 complex that can partially compensate for the function of EZH2 in EZH2 knockout ES cells [15, 16]. esPRC2p48 is an uncharacterized protein with no predictable second structure (Fig. S2). In support of our biochemical purification results, JARID2 and MTF2 were recently identified as PRC2 interacting proteins with affinity purification and genome-wide screen approaches [33–38]; however, esPRC2p48 was not identified in these studies.
JARID2, MTF2, and esPRC2p48 are Highly Expressed in ES Cells
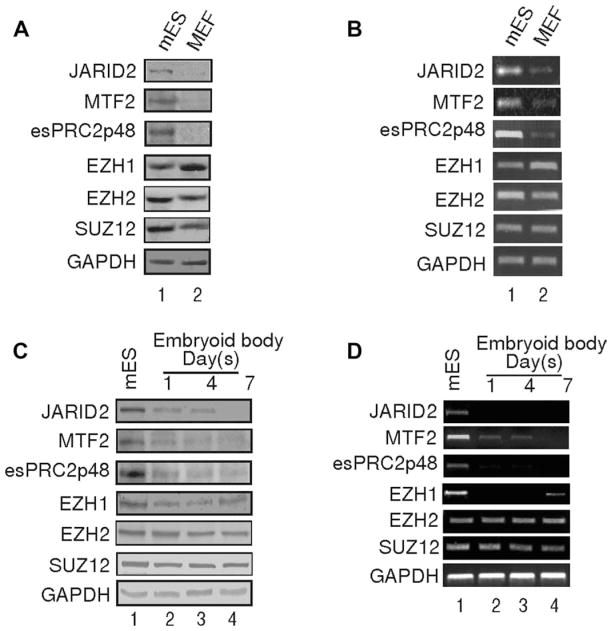
As previous biochemical purification of PRC2 from HeLa cells and Drosophila embryos did not identify JARID2, EZH1, MTF2, and esPRC2p48 [9–12], we reasoned that these subunits might be expressed at high levels in ES cells. To test this hypothesis, we compared the expressions of these proteins in mouse ES cells and mouse embryonic fibroblasts (MEFs). As shown in Figure 2A, western blot analysis revealed that JARID2, MTF2, and esPRC2p48 are expressed at high levels in mouse ES cells but are barely detectable in MEFs (top three panels). EZH1 is expressed in both ES cells and MEFs, but surprisingly, is present at higher levels in MEFs (Fig. 2A, fourth panel). EZH2 and SUZ12 are expressed in both cell types and have higher expression levels in ES cells (Fig. 2A, fifth and sixth panels). The western blot results were further confirmed by semi-quantitative RT-PCR assays. As shown in Figure 2B, semi-quantitative RT-PCR revealed that the levels of mRNA for JARID2, MTF2, and esPRC2p48 are significantly higher in ES cells than in MEFs, whereas the levels of mRNA for EZH1 is slightly higher in MEFs than in ES cells. To further determine whether JARID2, MTF2, and esPRC2p48 are exclusively expressed in ES cells, we examined the expression of these proteins during ES cell differentiation. We induced differentiation of ES cells in vitro by the formation of embryoid body (EB). During EB formation, the levels of ES cell markers Oct4 and Nanog decreased, and the levels of differentiation-associated genes Pax-3, Pax-7, and Brachyury increased, indicating that ES cells were successfully differentiated (Fig. S3, compare lane one with 2–4). Western blot analysis revealed that during ES cells differentiation, the levels of JARID2, MTF2, and esPRC2p48 decreased dramatically (Fig. 2C, top three panels, compare lane 1 with 2–4). The level of EZH1 also decreased as ES cells began to differentiate; however, the levels were restored at a later stage of differentiation (Fig. 2C, fourth panel, compare one with 2–4). In contrast to the dramatic downregulation of JARID2, MTF2, and esPRC2p48, changes in the levels of EZH2 and SUZ12 were modest (Fig. 2C, fifth and sixth panels). Western blot results were confirmed by semi-quantitative RT-PCR assays, suggesting that the regulation of JARID2, MTF2, and esPRC2p48 during ES cell differentiation occurs at the transcriptional level (Fig. 2D, compare lane one with 2–4). Taken together, these data suggest that PRC2 containing JARID2, MTF2, and esPRC2p48 as subunits might be specifically related to a pluripotent gene expression program. Consistent with this notion, JARID2 has been identified in a gene expression signature shared by human and mouse ES cells and MTF2 was shown to be uniquely expressed in mouse ES cells and during early development [38–40].
Figure 2.
JARID2, MTF2, and esPRC2p48 are highly expressed in ES cells. Western blot analysis (A, C) and semi-quantitative Reverse Transcription Polymerase Chain Reaction assay (B, D) of selected PRC2 subunits in mouse ES cells and MEFs (A, B) and during ES cell differentiation (C, D). GAPDH serves as a loading control. Parallel experiments, during which the Reverse Transcription reaction was omitted, did not give any specific signal and thus were not shown.
JARID2, MTF2, and esPRC2p48 Regulate Gene Expression in ES Cells
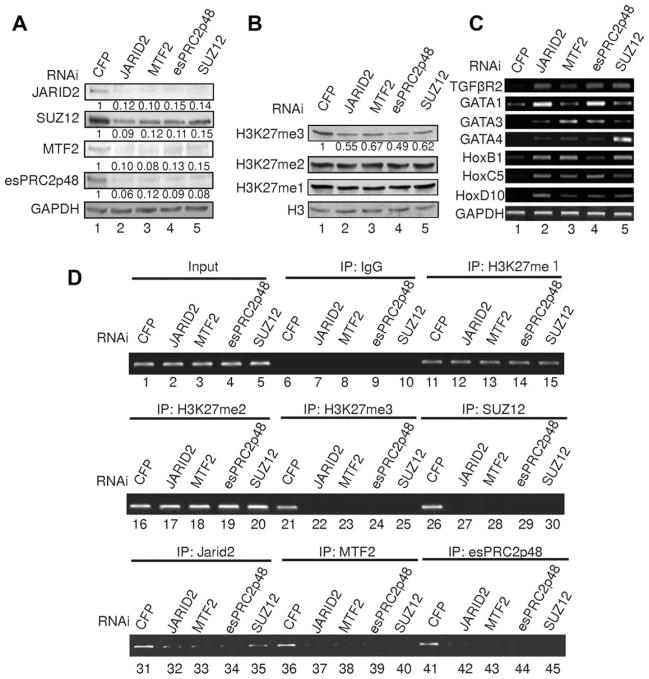
Since JARID2, MTF2, and esPRC2p48 represent additional subunits of PRC2 in ES cells, we investigated whether these subunits regulate PRC2 mediated H3K27 methylation and gene expression. For this purpose, we used shRNAs to specifically knockdown the expressions of these proteins in ES cells. Western blotting of total population of infected cells on 3 days post infection revealed that when JARID2 was knocked down by shRNA, the protein levels of MTF2, esPRC2p48, and SUZ12 were also significantly reduced (Fig. 3A, lane 2). Similarly, when MTF2, esPRC2p48, or SUZ12 were knocked down, the protein levels of other subunits were also reduced (Fig. 3A, lanes 3–5). RT-PCR analysis confirmed that the mRNA levels of other subunits were not affected by knocking down a specific subunit, suggesting that these shRNAs function through blocking protein translation. Token together, these data suggest that the stability of individual subunit of PRC2 depends on its integration into the complex. Similar findings were also reported for EED and SUZ12, in which deletion of these two proteins resulted in a significant reduction of EZH2 protein levels [41, 42].
Figure 3.
JARID2, MTF2, and esPRC2p48 regulate H3K27 methylation and gene expression in ES cells. (A): Western blot analysis of selected subunits of PRC2 in ES cells treated with shRNA as indicated on the top of the panels. GAPDH is used as a loading control. The numbers below the panels represent the relative proteins levels as compared to control (cells transduced with virus encoding non-targeting shRNAs CFP). (B): Western blot analysis of histone H3K27 methylation in ES cells treated with shRNA as indicated on the top of the panels. Histone H3 was used as a loading control. The numbers below the top panel represent the relative methylation levels. Quantification of top panel is shown under the image of western blot of H3K27me3. (C): Semi-quantitative Reverse Transcription Polymerase Chain Reaction assay of differentiation-associated genes in mouse ES cells treated with interference RNA as indicated on the top of the panels. GAPDH was used as a control. Parallel experiments, during which the Reverse Transcription reaction was omitted, did not give any specific signal and thus were not shown. (D): Chromatin immunoprecipitation (ChIP) assay of histone H3K27 methylation, JARID2, MTF2, esPRC2p48, and SUZ12 on TGFβR2 gene in ES cells treated with shRNAs as indicated on the top of the panels. Antibodies used are indicated in the top of the panels. A diagram of the TGFβR2 gene is illustrated in Figure 6C.
When JARID2, MTF2, or esPRC2p48 were knocked down in ES cells, although there was no apparent morphology of differentiation in these ES cell, we observed a decrease in H3K27 tri-methylation but not in di- and mono-methylation (Fig. 3B). The requirement of JARID2 for proper H3K27 methylation was also reported in recent studies at a global level or at specific loci [33, 36, 37], although conflicting results have been reported [34, 35]. Additionally, although loss of MTF2 does not affect global H3K27 methylation levels, MTF2 is required for proper H3K27 methylation at target genes [38]. Therefore, it is likely that JARID2, MTF2, and esPRC2p48 play important roles in regulating PRC2 catalyzed H3K27 methylation in vivo. Previous studies also revealed that ES cells express differentiation-associated genes when SUZ12 or EED was knocked out [23, 24]. To determine whether ES cells with knockdowns of JARID2, MTF2, or esPRC2p48 display a similar phenotype, we measured expression of differentiation-associated genes, which were reported to be expressed in SUZ12 and EED knockout ES cells, and Hox genes, which are classic targets of PRC2 and are repressed by PRC2 in ES cells (Fig. 3C). Compared to control, knockdown of JARID2, MTF2, or esPRC2p48 results in a significant increase of the expression of TGFβR2 gene expression (Fig. 3C, top panel), knockdown of JARID2 or esPRC2p48 resulted in a significant increase of GATA1 gene expression (Fig. 3C, second panel) and knockdowns of JARID2, MTF2, or esPRC2p48 resulted in significant increases in the expression of both GATA3 and GATA4 genes (Fig. 3C, third and fourth panels). Furthermore, knockdowns of JARID2, MTF2, or esPRC2p48 resulted in significant increases in the expression of all examined Hox genes (Fig. 3C, fifth to seventh panels). These results indicate that, similar to PRC2 core subunits, JARID2, MTF2, and esPRC2p48 are also required for the repressive function of PRC2 on differentiation-associated genes in ES cells. This conclusion is consistent with recent genome-wide studies indicating that JARID2 localizes to large number of PRC2 target genes and is required for targeting PRC2 in ES cells [33–37].
Given that histone H3K27 methylation catalyzed by PRC2 plays an important role in repressing lineage-specific gene expression in ES cells, we investigated whether the expression of lineage-specific genes in knockdown ES cells is related to the changes in H3K27 methylation. For this purpose, we used ChIP assays to measure the levels of degree-specific H3K27 methylation on selected genes in ES cells with or without knockdowns of JARID2, MTF2, or esPRC2p48. As shown in Figure 3D, knockdowns of JARID2, MTF2, esPRC2p48, or SUZ12 resulted in a specific reduction of H3K27 trimethylation (lanes 21–25) but had no effects on H3K27 mono- (lanes 11–15) and dimethylation (lanes 16–20) at the promoter of TGFβR2 gene. Interestingly, knockdown of any of JARID2, MTF2, esPRC2p48, or SUZ12 abolished the bindings of other PRC2 subunits to the target gene (Fig. 3D, middle panel, compare lanes 26–27–30; bottom panel, compare lanes 31–32–35, lanes 36–37–40, lanes 41–42–45). This observation further indicates that integral PRC2 in ES cells contains JARID2, MTF2, and esPRC2p48, and that localization of this complex to the target gene contributes to the repression of these genes in ES cells.
JARID2, MTF2, and esPRC2p48 Facilitate Somatic Cell Reprograming
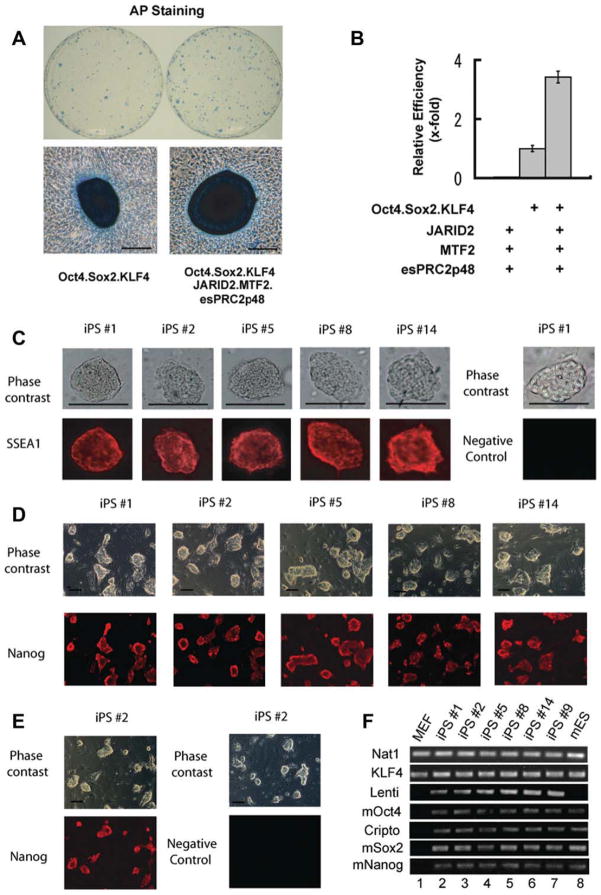
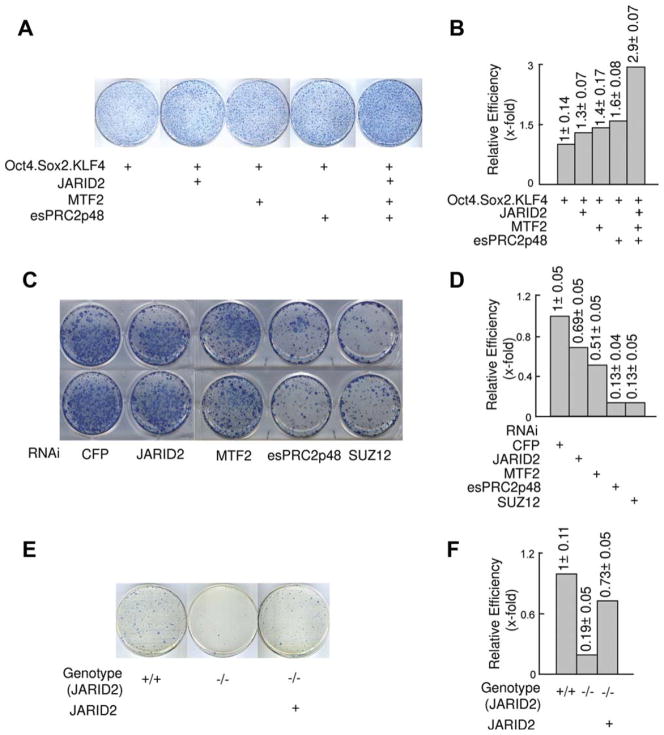
Since JARID2, MTF2, and esPRC2p48 are highly expressed in ES cells and contribute to the repression of differentiation-associated genes, we postulated that these proteins may contribute to cell pluripotency. To explore this possibility, we transduced JARID2, MTF2, and esPRC2p48 into MEFs, which express very low levels of these proteins, and investigated the production of iPS cells [43]. As EZH1 is expressed in MEFs, we did not include EZH1. After 2–4 days of infection, changes in fibroblast morphology were observed with the formation of large, flat colonies, which were mostly composed of rounded cells (Fig. S4A). Those colonies failed to proliferate during subsequent culture. However, we found that coinfection of MEFs with JARID2, MTF2, and esPRC2p48, and a polycistronic lentiviral vector encoding Oct4/Sox2/KLF4 (referred to as OSK) [28] resulted in small colonies emerging on top of the large colonies at days 6–7 post-infection. These newly emerging colonies proliferated rapidly and gradually adopted the morphology of typical mouse ES cell colonies (Fig. S4B), and stained positively for AP, a marker expressed in the early stages of somatic cell reprograming (Fig. 4A). Compared to OSK infection alone, co-infection with JARID2, MTF2, and esPRC2p48 increased the number of AP-positive colonies over three-folds, as assessed on day 14 post infection (Fig. 4B). Since AP-positive staining is not a definitive criterion for iPS cell identification, we undertook a series of experiments to functionally characterize these colonies.
Figure 4.
JARID2, MTF2, and esPRC2p48 facilitate somatic cell reprograming. (A): MEFs were infected with the polycistronic OSK vector (left) or in combination with JARID2/MTF2/esPRC2p48 lentiviral vectors (right). Emerging colonies of iPS cells were visualized by alkaline phosphatase (AP) staining on day 14 post infection. Representative plates from three independent experiments are shown. Scale bars: 200 μm. (B): Quantification of iPS cell colony number shown in A. Reprograming efficiency was shown as relative fold changes. Data correspond to the average and SD of three independent experiments. (C): Morphology (top rows) and SSEA1 staining (bottom rows) of selected iPS colonies derived from MEFs infected with the polycistronic OSK vector plus JARID2, MTF2, and esPRC2p48 lentiviral vectors. Original magnification is ×100. (D, E): Morphology (top rows) and Nanog staining (bottom rows) of selected iPS colonies derived from MEFs infected with the polycistronic OSK vector plus JARID2, MTF2, and esPRC2p48 lentiviral vectors. Staining omitted primary antibody was shown as a negative control. (F): RT-PCR analysis of MEFs, mouse ES cells, and iPS colonies derived from MEFs infected with the polycistronic OSK vector in combination with JARID2, MTF2, and esPRC2p48 lentiviral vectors. Nat1 was used as a control. Parallel experiments, during which the Reverse Transcription reaction was omitted, did not give any specific signal and thus were not shown.
To determine whether these AP-positive colonies indeed represent iPS cells, we randomly picked 20 colonies from MEFs infected with OSK alone or coinfected with JARID2, MTF2, and esPRC2p48. All colonies displayed typical mouse ES cell morphology during subsequent culturing (Fig. 4C, top row), and stained positively for pluripotent marker SSEA1 (Fig. 4C, bottom row). In addition, we measured the expression of endogenous Oct4, Sox2, Nanog, and Cripto by RT-PCR assay in five colonies randomly picked from coinfection of OSK and JARID2, MTF2, esPRC2p48 (lanes 2–6), and one colony from OSK infection alone (lane 7) (Fig. 4F). Oct4, Sox2, Nanog, and Cripto were expressed in these cells at levels similar to the levels in mouse ES cells, but distinct from the parental MEFs (Fig. 4F, compare lanes 2–7 with lanes 1, 8). The expression of Nanog protein was further confirmed by immunostaining (Fig. 4D, 4E). To further validate the pluripotency of these colonies, we injected the five cell lines into SCID mice and examined the resulting teratomas by HE staining of histological sections. As shown in Figure S5, tissues derived from all three germ layers (ectoderm, endoderm, and mesoderm) are present in the teratomas. As all these assays revealed that these AP-positive colonies indeed represent iPS cells; we conclude that JARID2, MTF2, and esPRC2p48 increase the efficiency of somatic cell reprograming.
To further determine the role of individual subunit in somatic cell reprograming, we transduced MEFs with individual lentiviruses encoding JARID2, MTF2, and esPRC2p48 together with OSK. In contrast to the three-fold enhancement upon coinfection, no major effects were observed for individual JARID2, MTF2, and esPRC2p48 (Fig. 5A, 5B). These results suggest that JARID2, MTF2, and esPRC2p48 must function together to facilitate somatic cell reprograming. These data are consistent with our biochemical studies that JARID2, MTF2, and esPRC2p48 represent integral subunits of PRC2 in ES cells and that all three subunits are required to maximally stimulate the histone methyltransferase activity of PRC2 in vitro (see below).
Figure 5.
Role of JARID2, MTF2, and esPRC2p48 in somatic cell reprograming. (A, B): Effects of individual JARID2, MTF2, and esPRC2p48 on somatic cell reprograming. iPS cell colonies were visualized by alkaline phosphatase (AP) staining. Quantification (average and SD) of two independent experiments is shown. (C, D): Effects of knockdown of JARID2, MTF2, esPRC2p48, and SUZ12 on somatic cell reprograming. iPS cell colonies were visualized by AP staining at day 18. Quantification (average and SD) of iPS cell colonies from two independent experiments at day 12 after transduction of MEFs is shown. (E, F): Effects of JARID2 knockout on somatic cell reprograming. iPS cell colonies were visualized by AP staining at day 32. Quantification (average and SD) of iPS cell colonies from two independent experiments at day 32 is shown.
We next determined whether endogenous JARID2, MTF2, and esPRC2p48 are required for somatic cell reprograming. In these experiments, we used Oct4, Sox2, KLF4, and c-myc encoded by individual viruses to achieve a higher reprograming efficiency [43] and knocked-down JARID2, MTF2, and esPRC2p48 individually. As shown in Figure 5C, 5D, the number of AP-positive colonies was reduced when JARID2 was knocked down by shRNA (at least 30%). More profound effects were observed when MTF2, esPRC2p48, and PRC2 core subunit SUZ12 was knocked down (>50%). These observations were further confirmed with MEFs derived from JARID2 knockout mice; reprograming was nearly abolished in JARID2 knockout MEFs (Fig. 5E, 5F). Importantly, reprograming of JARID2 knockout MEFs could be rescued by transduction of JARID2 (Fig. 5E, 5F). The requirement for PRC2 core component EED and SUZ12 in somatic cell reprograming was also reported in recent studies using a cell fusion approach [44]. Taken together, these studies reveal that endogenous PRC2 subunits including these regulatory subunits identified in this study are required for somatic cell reprograming.
JARID2, MTF2, and esPRC2p48 Repress Lineage-Specific Gene Expression During Somatic Cell Reprograming
As JARID2, MTF2, and esPRC2p48 repress lineage-specific gene expression in ES cells, we investigated whether these subunits also contribute to the repression of lineage-specific gene expression during somatic cell reprograming, an important “de-differentiation” step [45]. Transduction of MEFs with either JARID2, MTF2, and esPRC2p48 or OSK resulted in a time-dependent decrease of the expression of TGFβR2, Snail1, and TGFβ1 genes, which are normally expressed in MEFs (Fig. 6A, top three panels, lanes 1–4). More prominent repression was noted when cells were co-infected (Fig. 6A, top three panels, lanes 5–6). Similar observations were also made for HOX A9, C5, and D10 genes, which are also expressed in MEFs (Fig. 6A, fourth to sixth panels), and in ZEB2 gene, which is not expressed in MEFs but exhibits increased expression during somatic cell reprograming (Fig. 6A, seventh panel). These data suggest that Oct4/Sox2/KLF4 and JARID2/MTF2/esPRC2p48 act synergistically to repress lineage-specific gene expression during somatic cell reprograming.
Figure 6.
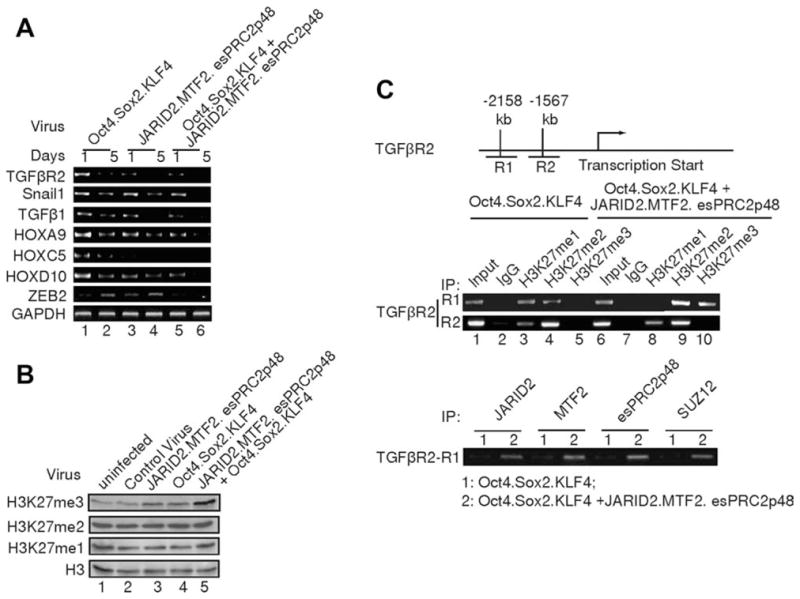
JARID2, MTF2, and esPRC2p48 facilitate lineage-specific gene repression during somatic cell reprograming. (A): Semi-quantitative Reverse Transcription Polymerase Chain Reaction assay of fibroblast-associated genes in MEFs infected with lentiviral vectors as indicated on the top of the panels. GAPDH was used as a loading control. Parallel experiments, during which the Reverse Transcription reaction was omitted, did not give any specific signal and thus were not shown. (B): Western blot analysis of H3K27 methylation in MEFs infected with lentiviral vectors as indicated on the top of the panels. Histone H3 was used as a loading control. (C): Chromatin immunoprecipitation (ChIP) assay of histone H3K27 methylation on TGFβR2 gene in MEFs infected with the indicated lentiviral vectors. Antibodies used for ChIP are indicated on the top of the images. A diagram of the TGFβR2 gene is illustrated on the top of the panels.
Given that JARID2, MTF2, and esPRC2p48 form a complex with PRC2, we investigated whether exogenous expression of JARID2, MTF2, and esPRC2p48 in MEFs affects H3K27 methylation. As shown in Figure 6B, infection of MEFs with JARID2/MTF2/esPRC2p48 or OSK both resulted in an increase of H3K27 trimethylation without obvious effects on di- and mono-methylation. These data indicate that (1) exogenous expression of JARID2, MTF2, and esPRC2p48 indeed affect the H3K27 methylation status in MEFs and (2) reprograming factors Oct4/Sox2/KLF4 also regulate PRC2 mediated H3K27 methylation. The latter regulation might be important for somatic cell reprograming, as knockdowns of PRC2 subunits significantly reduce reprograming efficiency (Fig. 5C–5F). More interestingly, we observed a dramatic increase of tri-, but not mono- and di-methylation, of H3K27 when MEFs were coinfected with JARID2/MTF2/esPRC2p48 and OSK (Fig. 6B, top panel, compare lane 5 with 3, 4). These data suggest that Oct4/Sox2/KLF4 and JARID2/MTF2/esPRC2p48 have a synergistic stimulatory effect on H3K27 tri-methylation. To further determine whether the changes of H3K27 methylation contribute to the repression of lineage-specific gene expression during somatic cell reprograming, we measured the levels of H3K27 methylation at the TGFβR2 gene, which exhibits the most dramatic change upon co-infection. In control uninfected MEFs, the levels of H3K27 methylation at the TGFβR2 gene promoter were undetectable (Fig. S7), consistent with the high expression of TGFβR2 gene in MEFs. However, co-infection resulted in a dramatic increase in the levels of H3K27 di- and tri-methylation at the promoter region of TGFβR2 gene, compared to OSK alone, as assessed on day 5 post infection (Fig. 6C, top panel, compare lanes 4, 5 with 9, 10). Consistent with the increase of H3K27 di- and tri- methylation, a reduction of H3K27 monomethylation was observed upon coinfection (Fig. 6C, top panel, compare lane 3 with 8). ChIP assay also revealed that JARID2, MTF2, and esPRC2p48 colocalize with H3K27 trimethylation (Fig. 6C, bottom panel), suggesting that the increase of H3K27 trimethylation results from the binding of JARID2, MTF2, and esPRC2p48. No significant changes were observed in H3K27 methylation levels at other regions of TGFβR2 gene (Fig. 6C, middle panel). These results suggest that the increase in H3K27 tri-methylation triggered by JARID2, MTF2, and esPRC2p48 binding contributes to the repression of lineage-specific genes.
JARID2, MTF2, and esPRC2p48 Stimulate PRC2’s Histone Methyltransferase Activity
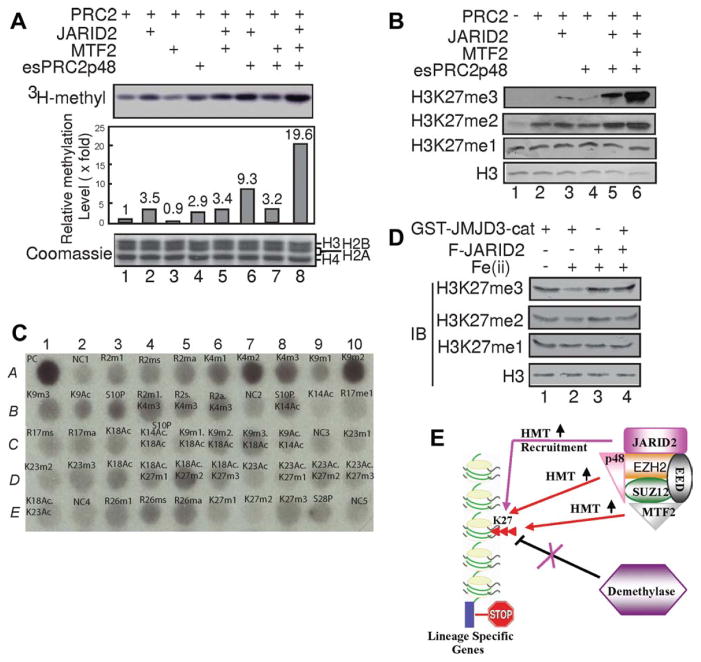
As JARID2, MTF2, and esPRC2p48 are integral subunits of PRC2 in ES cells, and regulate PRC2-mediated H3K27 methylation in vivo, we investigated whether these subunits directly regulate the histone methyltransferase activity of PRC2 in vitro. As shown in Figure 7A, JARID2 and esPRC2p48 stimulate the histone methyltransferase activity of PRC2 core complex (EZH2, SUZ12, EED, RbAP46/68) by 3.5- and 2.9-fold, respectively (compare lane 1 with 2, 4). In contrast, no obvious effect was observed for MTF2 (Fig. 7A, compare lane 1 with 3). We also did not observe any additive effects of MTF2 on JARID2 and MTF2 (Fig. 7A, compare lanes 1–4 with 5, 7). However, we did observe an additive effect of JARID2 and esPRC2p48 (Fig. 7A, compare lanes 2, 4 with 6). Intriguingly, we observed a dramatic stimulation of PRC2’s histone methyltransferase activity when all three subunits were present (2.2-fold of the effect of JARID2 plus esPRC2p48 and 19.6-fold of PRC2 alone; Fig. 7A, compare lane 1, 6 with 8). This synergistic stimulation suggests that JARID2 and esPRC2p48 are required for potentiating the effects of MTF2. The stimulatory effect of JARID2 on PRC2’s histone methyltransferase activity was also reported by a recent publication [33]; however, an inhibitory effect was reported for JARID2 in two other publications [34, 35]. The discrepancy might result from an altered GST-JARID2 conformation, differences in the purity of reconstituted complexes, or an altered conformation of JARID2 in the absence of MTF2 and esPRC2p48. With nucleosomes purified from EZH2 knockout ES cells as substrates [15], we further found that JARID2, MTF2, and esPRC2p48 together strongly stimulate H3K27 di- and tri-methylation by PRC2 (Fig. 7B, top two panels, compare 6 with 1–5). The synergistic stimulation of PRC2’s histone methyltransferase activity is consistent with our in vivo studies demonstrating that JARID2, MTF2, and esPRC2p48 are all required for somatic cell reprograming.
Figure 7.
JARID2, MTF2, and esPRC2p48 regulate the function of PRC2. (A): JARID2, MTF2, and esPRC2p48 modulate the histone methyl-transferase activity of PRC2 core complex (EZH2, SUZ12, EED, and RbAP46/68). PRC2 was incubated with recombinant proteins as indicated on the top of the panels on ice for 40 min before histone methyltransferase reaction was started. The top panel is an autoradiograph of the bottom panel. Quantification of two independent experiments is shown in the middle panel. (B): Western blot assay of the histone methyltransferase reaction products with antibodies as indicated at the left side of the panels. Histone methyltransferase assay were performed as above except cold SAM was used in the assay. (C): Peptide array binding assay of JARID2. Histone peptides containing indicated modifications (Table S3) were incubated with recombinant JARID2 and the binding was revealed by anti-His antibody. (D): JARID2 inhibits histone demethylation. Histone demethylation reactions were performed in the presence or absence of JARID2 and reaction product was analyzed by western blot assay with antibodies as indicated at the left side of the panels. (E): Model for JARID2, MTF2, and esPRC2p48 in the regulation of PRC2 function and gene expression. JARID2 recruits PRC2 to target loci. JARID2, MTF2, and esPRC2p48 then synergistically stimulate the histone methyltransferase activity of PRC2. JARID2 could also protect nucleosomes from demethylation by binding to methylated histone tails. All these mechanisms contribute to the high levels of H3K27 methylation and gene repression.
To further delineate the mechanisms by which JARID2 regulates PRC2’s function, we investigated whether JARID2 interacts with modified histone tails. As shown in Figure 7C, JARID2 did not interact with histone peptides without any modifications (A2, B7, C9, E2, and E10, peptide sequences in Table S3); however, JARID2 exhibited strong interactions with peptides containing H3K4me2 and H3K9me2 (A7 and A10). To provide functional insights into the interaction, we examined whether JARID2 could protect histones from demethylation. As shown in Figure 7D, JARID2 can efficiently protect H3K27 from demethylation by JMJD3 (compare lane two with 4). These results, in conjunction with several recent publications, suggest that JARID2 regulates histone methylation through at least three mechanisms (Fig. 7E). First, JARID2 might recruit PRC2 to target loci [33–36]; second, JARID2 stimulates the histone methyltransferase activity of PRC2 [33] and, more importantly, cooperates with esPRC2p48 to potentiate the effects of MTF2 (Fig. 7E); and third, JARID2 interacts with nucleosomes and protects histones from demethylation (Fig. 7C, 7D). Whether similar mechanisms are used by other Jumonji domain-containing proteins remains to be determined.
Discussion
PcG proteins play important roles in regulating gene expression program and determining cell fate [26, 27]. Although the core components of PRC2 have been well-defined, critical regulatory subunits, such as Pcl (PHF1), which are required for generating the trimethylation status of H3K27 and therefore the repression function, are continuously being identified [13, 14, 32]. We report here that PRC2 in mouse ES cells contains additional subunits JARID2, MTF2, and esPRC2p48. This study, together with recent reports describing JARID2 and MTF2 as PRC2 interacting proteins [33–38], revealed that these additional subunits play crucial regulatory roles in PRC2 mediated H3K27 methylation and gene expression in ES cells. Although PcG proteins may not be essential for the establishment and maintenance of pluripotency, PRC2 provides an additional layer of regulation to safeguard ES cell pluripotency by preventing aberrant gene expression. JARID2 and MTF2, as well as esPRC2p48 reported in this study, are required for PRC2 to efficiently repress lineage-specific gene expression by facilitating the generation and maintenance of high levels of H3K27 trimethylation in ES cells. With all three subunits identified in one study, we provide evidence that these regulatory subunits can modulate the function of PRC2 and facilitate somatic cell reprograming. Exogenous expression of JARID2, MTF2, and esPRC2p48 might contribute to the repression of lineage-specific gene expression and thus somatic cell reprograming. This result, together with the recent demonstration that ES cell specific chromatin remodeling BAF complex subunits facilitate somatic reprograming, suggest that epigenetic regulators specific for ES cells can be used to increase the efficiency of somatic cell reprograming [46]. Furthermore, this study also indicates that PRC2 might act as a cofactor for reprograming factors to repress lineage-specific gene expression during somatic cell reprograming. The co-localization of PRC2 and pluripotency factors in ES cells is consistent with this possibility [22]. The observation that JARID2/MTF2/esPRC2p48 and Oct4/Sox2/KLF4 have a synergistic effect on H3K27 methylation provides direct experimental evidences for this hypothesis. The significant reduction of somatic cell reprograming efficiency upon knockdown or knockout of JARID2, MTF2, and esPRC2p48 also support this notion. Further definition of the roles of PcG proteins in the induction and maintenance of pluripotency may suggest new strategies for somatic cell reprograming and lineage-specific differentiation.
Supplementary Material
Acknowledgments
We thank Stuart H. Orkin for EZH2 knockout ES cells, Danny Reinberg for anti-EZH1 antibody, Yang Shi for JMJD3 plasmid, Ying Huang and Jinbiao Ma for MTF2 and esPRC2p48 antigens, and Laura A. Fabrizio for help with mass spectrometric analysis. H.B.W. is a Sidney Kimmel Scholar and is supported by NIH grant (GM081489). Work in the laboratory of T.T. was supported by NIH grants (DK073391 and HL057619). Work in the laboratory of Y.L. is supported by NIH grant (HL67050) and American Heart Association grant (0615633Z). Work in the laboratory of P.T. was supported by NCI Cancer Center Support Grant P30 CA08748. H.B.W. is a Sidney Kimmel Scholar and is supported by NIH grant (GM081489). Work in the laboratory of T.T. was supported by NIH grants (DK073391 and HL057619). Work in the laboratory of Y.L. is supported by NIH grant (HL67050) and American Heart Association grant (0615633Z).Work in the laboratory of P.T. was supported by NCI Cancer Center Support Grant P30 CA08748.
Footnotes
Author contributions: H.W., T.T.: experimental strategy designing, manuscript writing; H.J., Z.Z.: contributing for purification; Z.Z.: determining the expression of PRC2 subunits in MEFs and ES cells, during ES cell differentiation with help from A.J., W.Y.; Z.Z.: designing and performing the knockdown experiments in ES cells and somatic cell reprograming experiments with help from C.S., C.L., C.C., L.W., K.P.; A.J.: performing the immunoprecipitation, histone methylation, and demethylation assays; C.S., L.W., K.L.: contributing teratoma assays and OSK lentivirus production; Q.D.: analyzing teratomas; Y.G., J.M.: performing the peptide array binding assay; M.M., Y. L.: deriving MEF from JARID2 knockout mice and wild-type littermates; H.E.-B., P.T.: performing mass spectrometric analysis.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
See www.StemCells.com for supporting information available online.
References
- 1.Orlando V. Polycomb, epigenomes, and control of cell identity. Cell. 2003;112:599–606. doi: 10.1016/s0092-8674(03)00157-0. [DOI] [PubMed] [Google Scholar]
- 2.Mohn F, Schübeler D. Genetics and epigenetics: Stability and plasticity during cellular differentiation. Trends Genetics. 2009;25:129–136. doi: 10.1016/j.tig.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: Digging Waddington’s canal. Nat Biotechnol. 2009;10:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- 4.Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 5.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Rajasekhar VK, Begemann M. Concise review: Roles of polycomb group proteins in development and disease: A stem cell perspective. Stem Cells. 2007;25:2498–2510. doi: 10.1634/stemcells.2006-0608. [DOI] [PubMed] [Google Scholar]
- 7.Valk-Lingbeek ME, Bruggeman SWM, van Lohuizen M. Stem cells and cancer: The polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 9.Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 10.Czermin B, Melfi R, McCabe D, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 11.Kuzmichev A, Nishioka K, Erdjument-Bromage H, et al. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller J, Hart CM, Francis NJ, et al. Histone methyltransferase activity of a drosophila polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 13.Cao R, Wang H, He J, et al. Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol Cell Biol. 2008;28:1862–1872. doi: 10.1128/MCB.01589-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarma K, Margueron R, Ivanov A, et al. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol. 2008;28:2718–2731. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen X, Liu Y, Hsu Y-J, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margueron R, Li G, Sarma K, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Wang L, Erdjument-Bromage H, et al. Role of histone H2A ubiquitination in polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 18.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 19.Levine SS, King IFG, Kingston RE. Division of labor in polycomb group repression. Trends Biochem Sci. 2004;29:478–485. doi: 10.1016/j.tibs.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Simon JA, Tamkun JW. Programming off and on states in chromatin: Mechanisms of polycomb and trithorax group complexes. Curr Opin Genet Dev. 2002;12:210–218. doi: 10.1016/s0959-437x(02)00288-5. [DOI] [PubMed] [Google Scholar]
- 21.de Napoles M, Mermoud JE, Wakao R, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 24.Pasini D, Bracken AP, Hansen JB, et al. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 27.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 28.Chang C-W, Lai Y-S, Pawlik KM, et al. Polycistronic lentiviral vector for hit and run reprogramming of adult skin fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27:1042–1049. doi: 10.1002/stem.39. [DOI] [PubMed] [Google Scholar]
- 29.Joo H-Y, Zhai L, Yang C, et al. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- 30.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 31.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 32.Nekrasov M, Klymenko T, Fraterman S, et al. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at polycomb target genes. EMBO J. 2007;26:4078–4088. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li G, Margueron R, Ku M, et al. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen X, Kim W, Fujiwara Y, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng JC, Valouev A, Swigut T, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasini D, Cloos P, Walfridsson J, et al. JARID2 regulates binding of the polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 37.Landeira D, Sauer S, Poot R, et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA polymerase II to developmental regulators. Nat Cell Biol. 2010;12:618–624. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker E, Chang WY, Hunkapiller J, et al. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2010;6:153–166. doi: 10.1016/j.stem.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Y, Li H, Liu Y, et al. Evolutionarily conserved transcriptional co-expression guiding embryonic stem cell differentiation. Plos One. 2008;3:e3406. doi: 10.1371/journal.pone.0003406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Assou S, Cerecedo D, Tondeur S, et al. A gene expression signature shared by human mature oocytes and embryonic stem cells. BMC Genomics. 2009;10:10. doi: 10.1186/1471-2164-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pasini D, Bracken AP, Jensen MR, et al. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montgomery ND, Yee D, Chen A, et al. The murine polycomb group protein EED is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 44.Pereira CF, Piccolo FM, Tsubouchi T, et al. ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell. 2010;6:547–556. doi: 10.1016/j.stem.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 45.Mikkelsen TS, Hanna J, Zhang X, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singhal N, Graumann J, Wu G, et al. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.