Abstract
The tail of histone H3 is an ideal medium for storing epigenetic information because displacement of histone H3 is heavily restricted during transcription. To maintain the locus-specific modifications of histone H3, histone molecules should be retained locally at the original position through multiple rounds of transcription. Here, we found that fission yeast Spt6, a highly conserved RNA polymerase II-interacting histone H3–H4 chaperone, is essential for the maintenance of Lys-4 and Lys-9 methylation of histone H3 in euchromatin and heterochromatin, respectively. In euchromatin, loss of Lys-4 methylated histone H3 and deposition of newly synthesized Lys-56 acetylated histone H3 induced by Spt6 inactivation were coupled with transcription. While in heterochromatin, Spt6 prevents histone turnover and cryptic transcription in parallel with Clr3 histone deacetylase. We propose that Spt6 retains posttranslationally modified histone H3 during transcription to maintain epigenome integrity.
Posttranslational modifications of histone molecules epigenetically control the state of the corresponding genomic loci in cis1. Transcription by RNA polymerase II (Pol II) can cause dissociation of pre-existing histone molecules in vitro2,3 and in vivo4,5,6,7,8,9. Therefore, factors that oppose this dissociation by serving as molecular liaisons between histones and DNA should exist to ensure the integrity of the epigenome. Here, we show that Spt6 (Suppressor of Ty 6), a highly conserved histone H3–H4 chaperone that interacts with elongating Pol II10,11,12,13,14, is one such molecular liaison candidate. Spt6 has been shown to play important roles in nucleosome restoration in highly transcribed regions11, in genetic alterations at immunoglobulin loci15,16, and in methylation of Lys-4 and Lys-9 in histone H316,17. However, the primary function of Spt6 in epigenetic regulation remains unclear.
We found that Spt6 prevents transcription-coupled nucleosome loss and loss of associated methylation of histone H3 at Lys-4 and Lys-9 in euchromatin and heterochromatin, respectively. The nucleosome loss caused by inactivation of Spt6 appears to be partially compensated for by Spt6-independent deposition of newly synthesized Lys-56 acetylated histone H3 molecules that do not carry locus-specific modifications. In other words, Spt6 prevents transcription-coupled histone turnover to maintain the posttranslational modifications of histone H3. We observed genetic interaction between Spt6 and Clr3 (cryptic loci regulator 3), a histone deacetylase that prevents histone turnover in heterochromatin18, indicating that Spt6 acts in parallel with Clr3HDAC to prevent histone turnover and thereby maintain heterochromatin. These results strongly suggest that the primary function of Spt6 is to maintain epigenomic integrity by retaining posttranslationally modified histone H3 during transcription.
Results
Spt6 is required for the maintenance of Lys-9 dimethylated histone H3 in heterochromatin
In a forward genetic screen for heterochromatin regulators that function in conjunction with Pol II19 in the fission yeast Schizosaccharomyces pombe, we isolated a hypomorphic allele of spt6, designated spt6-K20, in which a 246-bp region corresponding to the YqgFc RNase H-like domain14 of Spt6 has been deleted in-frame (Fig. 1a, Supplementary Figs. 1a–f and 2a–h, see Supplementary Methods for details). We found that spt6-K20 cells grew slower than wild-type cells (Supplementary Fig. 2a), whereas the complete deletion of spt6 (spt6Δ) caused a much more severe growth defect. Heterochromatin in fission yeast is marked by both Lys-9 methylation of histone H3 and the heterochromatin protein 1 (HP1) homolog Swi6HP1 (switch 6), which recognizes Lys-9 methylated histone H320. Insertion of ade6 in the pericentromere as a marker gene leads to passive construction of heterochromatin at the marker gene by spreading of heterochromatin marks from the native regions21. Chromatin immunoprecipitation (ChIP) followed by quantitative polymerase chain reaction (ChIP-qPCR) analysis for dimethylation of Lys-9 in histone H3 (K9me2) and Swi6HP1 demonstrated that placement of these heterochromatin-specific marks in the pericentromere (ade6 and dg) and subtelomere (tlh) depends solely on the Suv39h family Lys-9-specific methyltransferase Clr4Suv39h (cryptic loci regulator 4)20 (Fig. 1b, c).
Figure 1. Spt6 is essential for the maintenance of Lys-9 dimethylated histone H3 in heterochromatin.
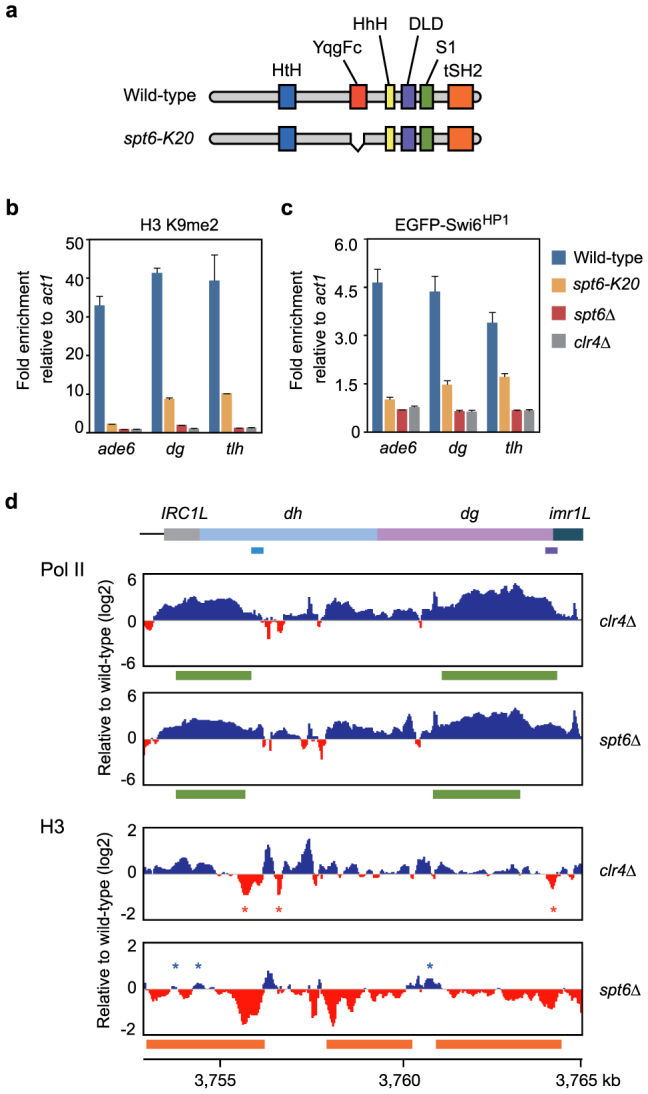
(a) Schematic representation of the spt6-K20 mutation. (b, c) ChIP-qPCR analysis of K9me2 (b) and EGFP-Swi6HP1 (c). Error bars, s.d. (d) ChIP-seq analysis of Pol II and histone H3 around the left side of centromere 1. Colored boxes at the top indicate the positions of pericentromeric repeat elements. The qPCR targets (dh in light blue and dg in purple) are shown. Positive (blue) and negative (red) values are filled with indicated colors. Red asterisks indicate the positions of nucleosome loss in clr4ΔSuv39h cells. Blue asterisks indicate the positions where nucleosome loss does not occur in spt6Δ cells. Regions with a significant increase in Pol II occupancy (green) and significant decrease in histone H3 occupancy (orange) as detected by MACS37 are shown.
The importance of fission yeast Spt6 in heterochromatin organization was previously studied with the spt6-1 mutant allele17. Cells carrying the spt6-1 mutant allele have been shown not to affect K9me2 in the pericentromeric repeats. However, we found that both the spt6-K20 and spt6Δ mutations caused a significant decrease in the levels of K9me2 and Swi6HP1 (Fig. 1b, c). In spt6Δ cells, some K9me2 remained in the native regions (dg and tlh), whereas the modification was completely abrogated in the inserted marker gene (ade6) (Supplementary Fig. 3a, showing K9me2 level per histone H3 molecule). Therefore, Spt6 is essential for the maintenance of K9me2, especially in the inserted marker gene. Lys-9 methylation of histone H3 is coupled with generation of small interfering RNA (siRNA)22,23. Consistent with the substantial decrease in the K9me2 level, siRNA corresponding to the pericentromeric repeats was undetectable in spt6 mutant cells (Supplementary Fig. 4).
To examine the effect of spt6Δ on Pol II and histone H3 occupancy across the pericentromeric regions, we performed a ChIP analysis followed by high-throughput sequencing (ChIP-seq) of Pol II and histone H3. The Pol II occupancy in some pericentromeric cryptic regions was dramatically increased in spt6Δ cells, as was also shown in clr4ΔSuv39h cells24 (Fig. 1d). Transcriptional activation in clr4ΔSuv39h cells did not lead to an overall decrease in histone H3 occupancy, except in several sensitive regions, as previously reported (Fig. 1d, red asterisks)25. It has been shown that the spt6-1 mutation does not cause a decrease in nucleosome occupancy in the pericentromere17. In spt6Δ cells, however, a significant decrease in histone H3 occupancy over the cryptic transcribed regions was observed (Fig. 1d and Supplementary Fig. 3b). ChIP-qPCR and micrococcal nuclease treatment followed by qPCR (MNase-qPCR) analyses confirmed that nucleosome loss did occur in the cryptic transcribed regions (Supplementary Fig. 5a–c). Even in a region where Pol II occupancy was very low (Supplementary Fig. 3b, region dh-L), a 2-fold increase in Pol II occupancy was associated with a significant decrease in the levels of histone H3 occupancy (Supplementary Fig. 3c) and K9me2 (Supplementary Fig. 3a). Thus, in the absence of Spt6, nucleosome loss and a reduction in the level of K9me2 occur concomitantly in the cryptic transcribed regions in the pericentromere. Note that nucleosome loss did not appear to occur in some subregions within the cryptic transcribed regions in spt6Δ cells (Fig. 1d, blue asterisks). These subregions might be resistant to the cotranscriptional nucleosome loss caused by Spt6 inactivation.
Transcription-coupled nucleosome loss and compensatory deposition of Lys-56 acetylated histone H3 occur in spt6 mutant cells
Next, we studied the function of Spt6 in euchromatin. Partial inactivation of Spt6 causes nucleosome loss in actively transcribed genes11,17; therefore, we performed ChIP-seq and MNase-qPCR analyses to examine the effect of complete inactivation of Spt6 on nucleosome occupancy. As expected, significant nucleosome loss over the actively transcribed act1 gene was observed in spt6Δ cells (Fig. 2a and Supplementary Fig. 6). Genome-wide analysis of protein occupancy per gene revealed that the extent of the decrease in histone H3 occupancy in spt6Δ cells was greater in genes with higher Pol II occupancy (Fig. 2b), indicating that transcription induces nucleosome loss. In the naked regions from which pre-existing histone molecules have been dissociated, it is possible that nucleosomes could be reconstructed with other histone molecules. Newly synthesized histone H3, which does not carry “locus-specific” posttranslational modifications, is acetylated at Lys-56 (K56Ac) before deposition9,26,27. Therefore, this modification serves as a marker for newly deposited histone H328,29. As shown in Figure 2c, Western blotting analysis of bulk histones in the chromatin fraction revealed that the amounts of histones H3 and H4 do not change dramatically, and that the level of Lys-56 acetylated histone H3 (K56Ac) was apparently increased in spt6 mutant cells (Fig. 2c). These results strongly suggest that the cotranscriptional decrease in histone H3 occupancy is partially compensated for by Spt6-independent deposition of Lys-56 acetylated histone H3.
Figure 2. Cotranscriptional nucleosome loss and increased deposition of Lys-56 acetylated histone H3 in spt6Δ cells.
(a) Occupancy of histone H3 around the act1 locus. Normalized tag counts at positions from the left end of chromosome II are shown. Arrows indicate the direction and length of transcripts. (b) Comparison of histone H3 occupancy with respect to Pol II occupancy in wild-type cells. Genes with significant loss of histone H3 (<0.8-fold remaining) in spt6Δ cells are highlighted in red. FPKM: fragments per kilobase of transcribed region per million mapped reads. (c) Western blotting analysis of histones. Triton X-insoluble fractions extracted from equal numbers of cells were separated by SDS-PAGE and probed with indicated antibodies. The gels have been run under the same experimental conditions. Full-length blots are presented in Supplementary Figure 10. (d) Occupancy of histone H3 around TSS and TTS. Regions corresponding to the first few nucleosomes are highlighted in light blue. (e) Comparison of Pol II occupancy with respect to its level in wild-type cells. Genes with a significant change in Pol II occupancy (>2-fold change) in spt6Δ cells are highlighted in red.
To examine histone H3 occupancy with respect to transcription start sites (TSS) and termination sites (TTS), we analyzed four sets of genes transcribed at different levels (“Very high”, “High”, “Medium”, and “Low”) (Supplementary Fig. 7a, b, see Supplementary Methods for details). Median histone occupancy was slightly lower in the “Very high” and “Low” genes than in the “High” and “Medium” genes in wild-type cells (Fig. 2d and Supplementary Fig. 7c). In spt6Δ cells, the decrease in histone H3 occupancy was indeed greater in genes with higher transcription levels (Fig. 2d and Supplementary Fig. 8). In contrast to previous studies in which Spt6 was not completely depleted11, in our study a significant decrease in histone H3 occupancy was observed even in the “Low” genes (Fig. 2d and Supplementary Fig. 8). The weaker effect in the “Low” genes may reflect Spt6-independent deposition of K56Ac-marked histones that acts antagonistically to the cotranscriptional nucleosome loss. We noted that Pol II occupancy in genes with low expression levels was increased in spt6Δ cells (Fig. 2e). Consistently, the levels of both sense and antisense transcripts with low expression levels in wild-type cells were increased in spt6Δ cells (Supplementary Fig. 9). Therefore, despite the observation that Spt6-independent deposition can partially compensate for the decrease in nucleosome occupancy, such quantitative compensation may be insufficient for the maintenance of the epigenetically repressed state. Histone H3 occupancy was decreased predominantly in the region corresponding to the first few nucleosomes (Fig. 2d and Supplementary Fig. 8, highlighted in light blue), suggesting that either nucleosome loss predominantly occurs in this region or that compensatory deposition rarely occurs here.
Spt6 prevents transcription-coupled loss of Lys-4 methylated histone H3
Newly deposited histone H3 molecules may not carry locus-specific posttranslational modifications. Therefore, the next question we sought to answer was whether or not inactivation of Spt6 facilitates cotranscriptional elimination of histone methylation in coding genes. We focused on four housekeeping genes in which Pol II occupancy was not dramatically altered by Spt6 inactivation (Fig. 3a). ChIP-qPCR analysis demonstrated that the extent of the decrease in histone H3 occupancy in spt6 mutant cells correlated with Pol II occupancy (Fig. 3a, b). In fission yeast, the Lys-4 residue of histone H3 is methylated solely by the Set1-family methyltransferase Set130,31. Western blotting analysis of bulk histones demonstrated the necessity of Set1 for di- and trimethylation at Lys-4 of histone H3 (K4me2 and K4me3, respectively) (Fig. 3c). In spt6 mutant cells, K4me2 and K4me3 could not be detected (Fig. 3c). ChIP-qPCR analyses consistently revealed that the level of K4me2 decreased significantly in spt6-K20 cells and was undetectable in spt6Δ cells (Fig. 3d; Note: the y-axis is magnified). The significant decrease in the level of K4me2 suggests that either Spt6 mediates Set1-dependent placement of Lys-4 methylation in histone H3, or that Spt6 retains Lys-4 methylated histone H3 during transcription. If the former is true, partial inactivation of Spt6 should attenuate Set1 activity, leading to a uniform reduction in the level of K4me2 in each gene. Importantly, K4me2 levels in spt6-K20 cells were lower in genes with higher Pol II occupancy (Fig. 3a, d). In spt6Δ cells, the levels of K56Ac and acetylation of histone H3 at Lys-9 and Lys-14 (AcH3) were higher in genes with higher Pol II occupancy (Fig. 3a, e, f). The level of histone H4 acetylation (AcH4) was also altered in spt6Δ cells (Fig. 3g). In contrast, these histone acetylation marks were not altered in the heterochromatin-defective clr4ΔSuv39h cells (Fig. 3e–g). These data suggest that inactivation of Spt6 causes cotranscriptional histone turnover, which in turn leads to impaired histone acetylation. Both the negative correlation between the levels of K4me2 and Pol II occupancy and the cotranscriptional histone turnover in spt6 mutant cells strongly suggest that transcription per se induces the loss of histones with locus-specific modifications when Spt6 does not function appropriately.
Figure 3. Spt6 is essential for the maintenance of Lys-4 methylated histone H3.
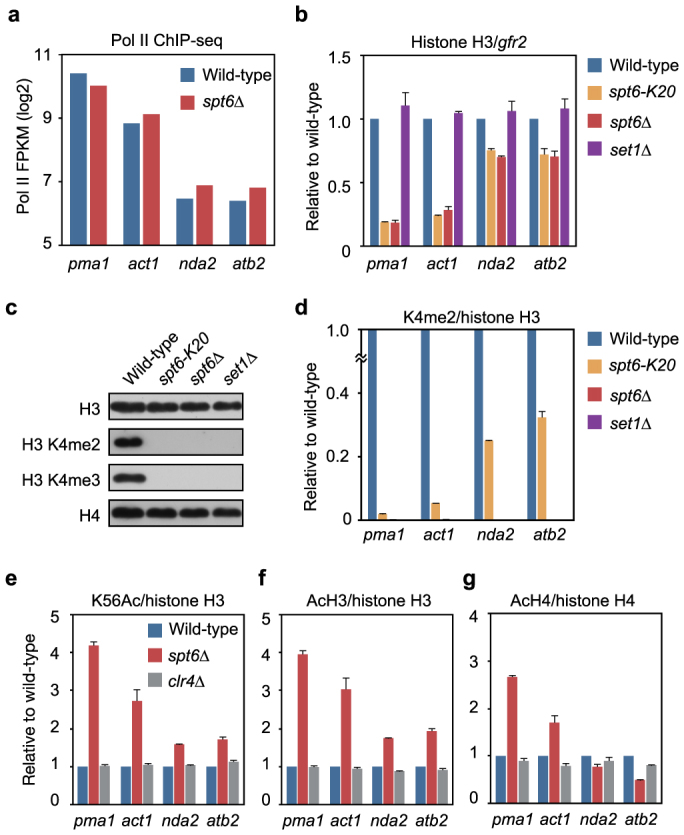
(a) Pol II occupancy of indicated genes is shown as FPKM. (b) ChIP-qPCR analysis of histone H3. Histone H3 levels in the indicated genes are normalized to the gene-free region gfr2. Fold-enrichment relative to wild-type cells is shown. (c) Western blotting analysis of histones and histone H3 modifications. Samples were prepared as described in Fig. 2c and probed with the antibodies indicated. The gels have been run under the same experimental conditions. Full-length blots are presented in Supplementary Figure 10. (d–g) ChIP-qPCR analysis of the level of K4me2 (d), K56Ac (e), AcH3 (f), and AcH4 (g) in the indicated genes. The y-axis in (d) is magnified for comparison. Error bars, s.d.
Spt6 acts in parallel with Clr3HDAC to maintain heterochromatin in the pericentromere
Recent studies have shown that heterochromatic silencing in fission yeast is maintained in part by a Clr3HDAC-mediated prevention of histone turnover18,25,32,33. In spt6 mutant cells, the majority of residual histone H3 in the pericentromere was not methylated at Lys-9 (Supplementary Fig. 3a), suggesting that the loss of Lys-9 methylation occurs as a result of cotranscriptional histone turnover. Therefore, we examined the relationship between Spt6 and Clr3HDAC genetically. ChIP-qPCR analysis showed that the levels of K56Ac in the pericentromere in both clr3ΔHDAC and clr4ΔSuv39h single mutant cells were higher than in wild-type cells (Fig. 4a), demonstrating that histone turnover is prevented by these factors18. The K56Ac level in the pericentromere was also increased in spt6-K20 single mutant cells (Fig. 4a).
Figure 4. Spt6 acts in parallel with Clr3 histone deacetylase to maintain heterochromatin in the pericentromere.

(a–f) ChIP-qPCR analysis of K56Ac (a), AcH3 (b), AcH4 (c), K9me2 (d), EGFP-Swi6HP1 (e), and Pol II (f) in the pericentromeric dg repeat. Error bars, s.d.
In order to determine whether Spt6 acts in the Clr3HDAC-dependent prevention of histone turnover, we combined the spt6-K20 mutation with clr3ΔHDAC. In the spt6-K20 clr3ΔHDAC double mutant cells, the K56Ac level in the pericentromere was higher than that in each of the single mutant cells (Fig. 4a), suggesting that Spt6 and Clr3HDAC have redundant mechanisms to prevent histone turnover. The increase in the K56Ac level was associated with an increase in the acetylation level of the N-terminal tail of both histone H3 (AcH3) and histone H4 (AcH4) (Fig. 4b, c). In addition, the levels of K9me2 and Swi6HP1 were further decreased in the double mutant cells to levels comparable to those in clr4ΔSuv39h cells (Fig. 4d, e). Consistently, Pol II occupancy, which was higher in each of the single mutant cells, was increased further in the double mutant cells (Fig. 4f). The additive effect observed here suggests that Spt6 acts in parallel with Clr3HDAC to prevent histone turnover and the associated subsequent loss of histone H3 Lys-9 methylation and derepression of cryptic transcription.
Discussion
Our data indicate that when Pol II lacking a functional Spt6 transcribes through the nucleosome template, nucleosome loss occurs in the wake of Pol II and eventually leads to Spt6-independent deposition of histones without locus-specific modifications. Therefore, we propose that during transcription, the histone H3–H4 chaperone Spt6 retains histone H3 to keep the rate of transcription-coupled histone turnover to a minimum in order to maintain locus-specific posttranslational modifications (Fig. 5). Alternatively, the transcription-coupled loss of histone modifications could result from an error in the recruitment of histone modification enzymes to the transcribed regions that is potentially mediated by Spt6. However, transcription-coupled nucleosome loss and increased acetylation of histone H3 at Lys-56 in the spt6 mutant cells support the idea that elimination of locus-specific histone modifications occurs as a result of histone turnover. The precise mechanism of Spt6-dependent maintenance of histone modifications should be studied at the molecular level in the future.
Figure 5. Model describing Spt6-dependent retention of epigenetically modified histones during transcription.

In wild-type cells, Pol II transcribes the nucleosome templates, keeping histones H3 and H4 in its wake. The histone H3–H4 chaperone Spt6 interacts directly with the phosphorylated C-terminal domain of elongating Pol II. Spt6 retains posttranslationally modified histone H3 during transcription so that the state of the chromatin is maintained. In the absence of Spt6 activity (spt6Δ), modified histones are displaced during transcription. Spt6-independent deposition of K56Ac-marked histones partially compensates for the loss of nucleosome occupancy. However, as the newly deposited histones do not carry appropriate locus-specific modifications, epigenetic control of the transcribed region is impaired.
The essentiality of Spt6 for the maintenance of histone positioning and associated modifications suggests that intracellular mechanisms exist that inactivate Spt6 in cis to reset the chromatin state or create accessible chromatin34. As almost all of the genome is transcribed to some extent35, inactivation of Spt6 could cause a catastrophic breakdown in epigenomic integrity. Furthermore, Spt6 breakdown in human cells could be involved in the epigenetic alterations frequently observed in cancer cells36.
Methods
Genetic manipulations
A fission yeast wild-type strain was mutagenized with 254-nm ultraviolet light to obtain mutants defective in pericentromeric silencing. The corresponding mutation was identified by whole-genome sequencing.
Chromatin immunoprecipitation
Exponentially growing cells were subjected to chromatin immunoprecipitation (ChIP). For ChIP-qPCR analysis of histone modifications, the immunoprecipitation efficiency of K4me2, K9me2, K56Ac, AcH3 and AcH4 at each locus was first normalized to that of the C-terminal region of histone H3 or histone H4 (for AcH4) in order to evaluate the level of modification per histone molecule. The modification levels were then compared to those in the wild-type control (y-axis: relative to wild-type) or the internal control regions indicated. For ChIP-qPCR analysis of K9me2 and Swi6 in Figures 1 and 4, the immunopreciptation efficiency at each locus was compared to those in the act1 region. For ChIP-seq analysis, approximately 10 million single-end 49-base reads were mapped onto the 972 reference genome. Pileup data generated by MACS37 were analyzed using the R-statistical environment (http://www.R-project.org). The R-scripts used in this study are available upon request.
Full Methods and any associated references are available as Supplementary Information.
Accession codes
High-throughput sequencing data in this publication have been deposited in DRA (http://trace.ddbj.nig.ac.jp/dra) under accession numbers DRA000544, DRA000855, DRA000856, and DRA000857. Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-1373. Contig sequences have been deposited in the DDBJ (http://www.ddbj.nig.ac.jp/) under accession numbers AB762285 and AB762286.
Author Contributions
T.I. performed the northern blotting analysis; J.N. generated the egfp-swi6+ strain; K.O. and H.K. performed experiments and analyzed the data; Y.M. and T.U. were involved in study design; and H.K. designed the study, wrote the R-scripts used to analyze the data, and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Supplementary Material
Supplementary Information
Acknowledgments
We thank R. C. Allshire for providing strains, and we would like to acknowledge the technical expertise of the Center for Integrated Research in Science, Shimane University. We also thank Ms. Yuko Fukuma for technical assistance. This study was supported by a Grant-in-Aid for Scientific Research (to H. K., T. I., J. N., Y. M. and T. U.) and a grant from the JST PRESTO program (to H. K. and T. I.).
References
- Smolle M. & Workman J. L. Transcription-associated histone modifications and cryptic transcription. Biochim. Biophys. Acta 1829, 84–97 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges C. et al. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science 325, 626–628 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaeva O. I., Hsieh F. K. & Studitsky V. M. RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones. Proc. Natl Acad. Sci. USA 107, 11325–11330 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirbelauer C., Bell O. & Schubeler D. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 19, 1761–1766 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz B. E. & Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 19, 804–814 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. K. et al. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 36, 900–905 (2004). [DOI] [PubMed] [Google Scholar]
- Jamai A., Imoberdorf R. M. & Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol. Cell 25, 345–355 (2007). [DOI] [PubMed] [Google Scholar]
- Dion M. F. et al. Dynamics of replication-independent histone turnover in budding yeast. Science 315, 1405–1408 (2007). [DOI] [PubMed] [Google Scholar]
- Rufiange A. et al. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell 27, 393–405 (2007). [DOI] [PubMed] [Google Scholar]
- Mayer A. et al. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 17, 1272–1278 (2010). [DOI] [PubMed] [Google Scholar]
- Ivanovska I. et al. Control of chromatin structure by spt6: different consequences in coding and regulatory regions. Mol. Cell. Biol. 31, 531–541 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. et al. Solution structure of tandem SH2 domains from Spt6 protein and their binding to the phosphorylated RNA polymerase II C-terminal domain. J. Biol. Chem. 286, 29218–29226 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortvin A. & Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272, 1473–1476 (1996). [DOI] [PubMed] [Google Scholar]
- Close D. et al. Crystal structures of the S. cerevisiae Spt6 core and C-terminal tandem SH2 domain. J. Mol. Biol. 408, 697–713 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki I. M. et al. Histone chaperone Spt6 is required for class switch recombination but not somatic hypermutation. Proc. Natl Acad. Sci. USA 108, 7920–7925 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum N. A. et al. The Histone Chaperone Spt6 Is Required for Activation-induced Cytidine Deaminase Target Determination through H3K4me3 Regulation. J. Biol. Chem. 287, 32415–32429 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiely C. M. et al. Spt6 is required for heterochromatic silencing in the fission yeast Schizosaccharomyces pombe. Mol. Cell. Biol. 31, 4193–4204 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aygun O., Mehta S. & Grewal S. I. HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat. Struct. Mol. Biol. 20, 547–554 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H. et al. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science 309, 467–469 (2005). [DOI] [PubMed] [Google Scholar]
- Nakayama J. et al. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113 (2001). [DOI] [PubMed] [Google Scholar]
- Ekwall K. et al. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91, 1021–1032 (1997). [DOI] [PubMed] [Google Scholar]
- Motamedi M. R. et al. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119, 789–802 (2004). [DOI] [PubMed] [Google Scholar]
- Volpe T. A. et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837 (2002). [DOI] [PubMed] [Google Scholar]
- Chen E. S. et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451, 734–737 (2008). [DOI] [PubMed] [Google Scholar]
- Garcia J. F. et al. Combinatorial, site-specific requirement for heterochromatic silencing factors in the elimination of nucleosome-free regions. Genes Dev. 24, 1758–1771 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan T. et al. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 4, e1000270 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubota T. et al. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell 25, 703–712 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S. et al. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489, 452–455 (2012). [DOI] [PubMed] [Google Scholar]
- Rufiange A. et al. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell 27, 393–405 (2007). [DOI] [PubMed] [Google Scholar]
- Roguev A. et al. High conservation of the Set1/Rad6 axis of histone 3 lysine 4 methylation in budding and fission yeasts. J. Biol. Chem. 278, 8487–8493 (2003). [DOI] [PubMed] [Google Scholar]
- Noma K. & Grewal S. I. Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc. Natl Acad. Sci. USA 99, 16438–16445 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T. et al. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell 128, 491–504 (2007). [DOI] [PubMed] [Google Scholar]
- Yamane K. et al. Asf1/HIRA facilitate global histone deacetylation and associate with HP1 to promote nucleosome occupancy at heterochromatic loci. Mol. Cell 41, 56–66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman R. E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm B. T. et al. Dynamic repertorie of a eukaryotic transcriptome surveyed at single-nucleosome resolution. Nature 453, 1239–1243 (2008). [DOI] [PubMed] [Google Scholar]
- Berdasco M. & Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev. Cell 19, 698–711 (2010). [DOI] [PubMed] [Google Scholar]
- Feng J. et al. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7, 1728–1740 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information



