SUMMARY
The dynamic, posttranslational modification of proteins with a SUMO tag has been recognized as an important cellular regulatory mechanism relevant to a number of cancers as well as normal embryonic development. As part of a program aimed towards the identification of inhibitors of SUMO conjugating enzymes, we have developed a microfluidic electrophoretic mobility shift assay to monitor sumoylation events in real time. We disclose herein the use of this assay to discover the first cell permeable compound capable of blocking the transfer of SUMO-1 from the E2 enzyme UBC9 to the substrate. We screened a small collection of compounds and identified an oxygenated flavonoid derivative that inhibits sumoylation in vitro. Next, we carried out an in-depth mechanistic analysis that ruled out many common false positive mechanisms such as aggregation or alkylation. Furthermore, we report that this flavonoid inhibits a single step in the sumoylation cascade: the transfer of SUMO from the E2 enzyme (UBC9) thioester conjugate to the substrate. In addition to being the first example of a compound with this unique mechanism of action, this inhibitor has a discreet structure-activity relationship uncharacteristic of a promiscuous inhibitor. Cell-based studies showed that the flavonoid inhibits the sumoylation of topoisomerase-I in response to camptothecin treatment in two different breast cancer cell lines, while isomeric analogs are inactive. Importantly, this compound blocks sumoylation while not affecting ubiquitylation in cells. This work identifies a novel point of entry for pharmacological inhibition of the sumoylation cascade, and will serve as the basis for continued study of additional pharmacophores that modulate SUMO-conjugating enzymes such as UBC9.
INTRODUCTION
The Small Ubiquitin-like Modifier, SUMO, is a protein tag that is dynamically attached to and cleaved from lysines on protein substrates by a tightly regulated enzymatic cascade (Gareau and Lima, 2010; Ulrich, 2009; Zhao, 2007). The modification of proteins with a SUMO tag is a process that is critical to normal development (Van Nguyen et al., 2012), and has been implicated in a broad spectrum of disease states, most notably cancers such as breast, colon, ovarian (Wang and Banerjee, 2004), and multiple myeloma (Driscoll et al., 2010). High levels of SUMO-conjugating enzymes, in particular UBC9 (the sole SUMO E2 enzyme), have been associated with adverse clinical outcomes for cancer patients and correlate with decreased survival rates (Driscoll et al., 2010). Recently, sumoylation was also reported as a requirement for Myc-driven tumorigenesis (Kessler et al., 2012) and has been implicated as protective in the heat shock response (Golebiowski et al., 2009). Enzymes in the SUMO conjugation cascade are therefore of interest as drug targets. UBC9 has been discussed in the literature as a target (Duan et al., 2009; Mo and Moschos, 2005), but there are no inhibitors reported. In the case of the SUMO E1 enzyme, there are two reported inhibitors known: ginkgolic acid (Fukuda et al., 2009a) and kerriamycin B (Fukuda et al., 2009b).
The process of SUMO conjugation/deconjugation is often described as an equilibrium, and is biochemically analogous to the process of protein ubiquitylation/deubiquitylation (Bedford et al., 2011). The SUMO conjugation cascade involves E1 (activating), E2 (conjugating), and E3 (ligase) enzymes, while cleavage is modulated by isopeptidases (referred to as SENPs or sentrin-specific proteases). In this cascade, a SUMO conjugate is formed by the generation of an isopeptide bond between a lysine on the substrate and the C-terminal diglycine tail of SUMO. This process is highly substrate specific, and there is only one SUMO E2 enzyme (UBC9) and ~12 SUMO E3 enzymes in contrast to the ~40 ubiquitin E2 and ~700 ubiquitin E3 enzymes (Cohen and Tcherpakov, 2010). Given this disparity, it is not surprising that while ubiquitin is estimated to modify some 90% of the proteome, a far smaller percentage of the proteome has been definitively identified as a substrate for sumoylation (Golebiowski et al., 2009). Sumoylation enzymes are also evolutionarily conserved, with yeast and mammalian SUMO-conjugating enzymes displaying high homology. From a structural standpoint SUMO-1 shares a common fold with ubiquitin, however it only has 18% sequence homology. The other two SUMO homologues, SUMO-2 and -3, have roughly 50% sequence homology with SUMO-1 (but ~95% homology with each other) (Muller et al., 2001).
The consequences of sumoylation are associated with nuanced changes in protein structure, function, molecular recognition, and subcellular localization, while ubiquitination is primarily associated with induced protein degradation and DNA repair. Many discreet effects of sumoylation have been identified, with examples occurring in the areas of transcriptional activation/repression (Schmidt and Muller, 2003; Verger et al., 2003; Zhao, 2007), intracellular transport (Geoffroy et al., 2010; Majumdar et al., 2011; Muller et al., 1998), DNA damage (Andrews et al., 2005; Potts and Yu, 2005; Zhao and Blobel, 2005), chromosome assembly (Chung et al., 2004) tumorigenesis (Kessler et al., 2012; Wood et al., 2003), and stress response (Golebiowski et al., 2009) as well as several others (Zhao, 2007). Recent work suggests that the sumoylation response also acts synergistically on groups of proteins with similar functions. This is particularly evident in the DNA repair pathway, where sumoylation of several proteins in the pathway leads to efficient repair, with individual modifications having only a small effect (Psakhye and Jentsch, 2012). The specific effects of protein sumoylation remain an active area of investigation. This effort is hampered by the observation that endogenous levels of sumoylation are quite low: less than 1% of a substrate protein is estimated to be sumoylated at any given time. The exception to this is RanGAP1, largely considered to be an atypical sumoylation substrate.
Despite reports suggesting that SUMO-conjugating enzymes would likely be good drug targets (Mo and Moschos, 2005), there is a lack of chemical probes to monitor and manipulate the process of sumoylation. There are few biochemical assays available to identify and evaluate small molecule inhibitors in these pathways, and the kinetic study of sumoylation remains nontrivial (Alontaga et al., 2012). In the specific case of sumoylation there have only been two reports of small molecule inhibitors to date, both of which appear to affect the E1 SUMO activating enzyme (SAE) (Fukuda et al., 2009a; Fukuda et al., 2009b). However, inhibitors of other ubiquitin-like (UBL) signaling pathways have been identified as well. To date there have been a small number of prominent successes of small molecule inhibitors of ubiquitin and NEDD8 E1 conjugation (Brownell et al., 2010; Lei et al., 2003; Soucy et al., 2009; Ungermannova et al., 2012a; Ungermannova et al., 2012b; Yang et al., 2007; Zhong et al., 2012). There is also one notable example of a ubiquitin E2 inhibitor in the literature (Ceccarelli et al., 2011).
Existing high throughput sumoylation assay technologies rely on TR-FRET (Carlson et al., 2009) or electrochemiluminescence approaches (Rouleau et al., 2008), which although robust use heavily modified SUMO and substrate fusion proteins that are substantially different from endogenous proteins, and are not suitable for all screening scenarios (such as natural product extracts). An elegant method to identify natural product inhibitors of sumoylation has also been reported, using image-based analysis of a semi-intact cell system (Saitoh et al., 2006). This is a unique and powerful approach, but appears to be somewhat limited in throughput and is not appropriate to use for quantitative kinetic analysis of inhibitors. Our goal was to overcome the limitations of existing technologies and to develop an alternative, highly quantitative sumoylation assay that would be useful for screening large libraries (>100,000) of small molecules of diverse origins, including natural product extracts as well as pure compounds, enable the kinetic analysis of inhibitors, and minimize false positives such as autofluorescent compounds.
In this study, we report the development of such a novel sumoylation assay that uses a microfluidic electrophoretic mobility shift system. This assay enables the real-time observation of the sumoylation of a fluorescent peptide substrate in a reconstituted biochemical cascade. Mobility shift assays are an example of a medium throughput separation-based assay that utilizes differences of electrophoretic mobility in a capillary between the substrate and product of an enzymatic reaction to directly monitor product formation. This approach has several benefits, including a highly quantitative kinetic or endpoint readout and a reduced number of false positives generated by autoflourescent compounds (Fanslau et al., 2010; Xiang et al., 2009; Xiang et al., 2010). We applied our assay to the kinetic screen of ~600 small molecules and natural product extracts, and report the identification of a flavonoid with potent SUMO inhibitory activity. A mechanistic study reveals that this molecule functions by blocking the transfer of SUMO-1 (and SUMO 2/3) from the E2-thioester complex to a variety of substrates. This inhibitor displays a discreet structure-activity relationship, and the ability to block a sumoylation event in the context of whole cells. It does not appear to function by aggregating, alkylating, or by generic mechanisms such as membrane disruption or reacting with oxygen. The discovery of this mechanistically unique small molecule represents the first example of a low molecular weight inhibitor of the function of UBC9, the sole SUMO E2 enzyme, and will provide the groundwork for future efforts into targeting UBC9 with small molecules.
RESULTS
Development of a Capillary Electrophoresis Assay to Study Sumoylation Events
Since there are currently no such high throughput assays that utilize single purified enzymes in the SUMO pathway (Alontaga et al., 2012; Carlson et al., 2009), we turned to a reconstituted biochemical cascade using recombinant SUMO-1, SUMO-activating enzyme 1/2 (SAE-1/2, the SUMO E1 enzyme) and UBC9 (the SUMO E2 enzyme) proteins with a peptide substrate. Importantly, this approach enables the assay to simultaneously probe for inhibitors of both the E1 and E2 enzymes. E3 enzymes, although important in vivo, are not necessary for in vitro sumoylation systems and were not included. We were motivated by the possibility that medium throughput electrophoretic mobility shift technology could serve as a flexible and quantitative assay. Furthermore, this approach has not been used for protein-based posttranslational modifications such as ubiquitylation or sumoylation previously.
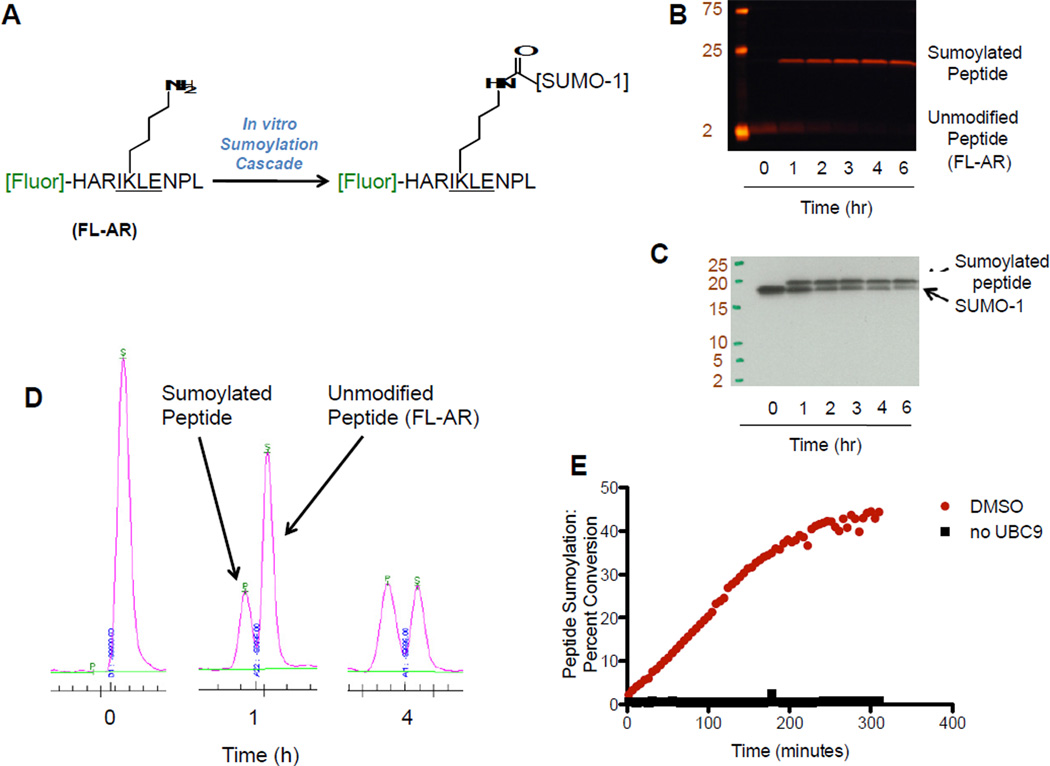
Although the identification of sumoylation substrates remains an active area of investigation, the majority of known substrates contain the tetrapeptide consensus sequence ψKxE/D, where ψ is a hydrophobic amino acid, K is the lysine where the incipient isopeptide bond is formed, x varies, and E/D is an acidic residue (Rodriguez et al., 2001). Interestingly, the consensus sequence is not an absolute requirement and discontinuous sumoylation epitopes have also been observed (Pichler et al., 2005). With this in mind, we synthesized a fluorescent 10-mer peptide derived from the androgen receptor that contained the SUMO consensus-sequence IKLE. This polypeptide was modified at the N-terminus with a fluorescent tag, 5-carboxyfluorescein (5-FAM), and is referred to as FL-AR (Figure 1A). We exposed FL-AR to a mixture of recombinant SUMO-1, SAE 1/2, UBC9, and ATP, and were able to observe a time dependent accumulation of a single, higher molecular weight fluorescent band as measured by in-gel fluorescence experiments (Figure 1B). The molecular weight of the band was consistent with a single SUMO-1 tag being attached to the fluorescent peptide. Furthermore, Western blot analysis with an anti-SUMO-1 antibody (Figure 1C) confirmed that a SUMO-1 tag had in fact been attached to the fluorescent substrate.
Figure 1.
Development of an Electrophoretic Mobility Shift Assay for Protein Sumoylation. (A) Sequence and reactivity of a fluorescent polypeptide substrate for the sumoylation assay. (B) In-gel fluorescence and (C) Western blot (with anti-SUMO-1 antibody) experiments showing the sumoylation of the fluorescent peptide. (D) Separation of the substrate peptide and sumoylated product using the LabChip EZ Reader II system. (E) Kinetic measurement of fluorescent peptide sumoylation. A sample from one 30 µL reaction mixture treated with 0.1% DMSO (either with or without UBC9) was analyzed using the LabChip EZ Reader II system every 4.88 minutes for 5 hours and percent conversion was monitored at each time point.
We next moved to analyze the reaction by a mobility shift protocol. We were pleased to find that under optimized separation conditions we could observe a near-baseline separation of FL-AR and the SUMO-1-FL-AR (Figure 1D). Furthermore, the accumulation of SUMO-1-FL-AR could be easily observed in a time dependent fashion, and the percent conversion could be quantified using a ratiometric measurement of peak height on an electropherogram (Figure 1D). Finally, miniaturization of the assay was straightforward, with the assay performing equally well in eppendorf tubes (250 µL total volume), 96-well (100 µL total volume) and 384-well (20 µL total volume) formats. Once optimized, we were able to obtain a separation-based readout of reaction progress for a complete 384 well plate in ~30 minutes by analyzing reactions that had been quenched with EDTA.
Once it was clear that an electrophoretic mobility shift assay would be suitable for the detection of SUMO-1-FL-AR, we monitored product formation in kinetic mode. Use of the mobility shift assay to measure sumoylation in real time was accomplished by repeated analysis of a single 30 µL reaction mixture over the course of 300 minutes. In this experiment, sumoylated product was produced in a roughly linear scale over the first ~100 minutes of the reaction. In the absence of Ubc9, no conversion was observed (Figure 1E). We also measured the IC50 of ginkgolic acid, a previously reported inhibitor of SAE (Fukuda et al., 2009a), by analyzing reactions that were quenched with EDTA at the 90 minute time point. The IC50 of ginkgolic acid was 9.1 µM, comparable to the literature value of 3.0 µM (not shown).
A Kinetic Screen for Inhibitors of Sumoylation
As part of our interest in screening natural products, we initially evaluated a well characterized plate of 80 extracts. This plate was assembled to include commonly problematic extracts with autofluorescence, high salt, polyphenols/tannins, and high viscosity. Of the 80 samples on this plate, nine showed inhibitory activity in our assay. We were pleased to find that none of the nine were autofluorescent. However, taxonomic investigation of the active extracts indicated that seven extracts were from known producers of polyphenols/tannins. As we were concerned that this type of molecule might be broadly interfering with our assay system, we next performed a kinetic screen of 500 flavonoids, chalcones, and related polyphenolic compounds in the electrophoretic mobility shift assay. For this kinetic screen, 20 µL reaction mixtures in 384 well plates were dosed with inhibitors at 30 µM (roughly comparable to the concentration expected in extracts) and initiated with ATP. After incubating for 30 miniutes at room temperature, samples from each well were analyzed every ~15 minutes. Conversion to the sumoylated product was monitored using the LabChip EZ Reader II system for 4–5 time points for each sample, for a total reaction time of 90–120 minutes. In this method (using a 12-sipper chip), 12 samples including a DMSO control, ginkgolic acid (a positive control of inhibition), and 10 test compounds were monitored simultaneously (see Supporting Information). By slightly offsetting analysis times, 80 compounds were kinetically screened in each experiment and a total of 500 compounds were screened in 7 separate experiments.
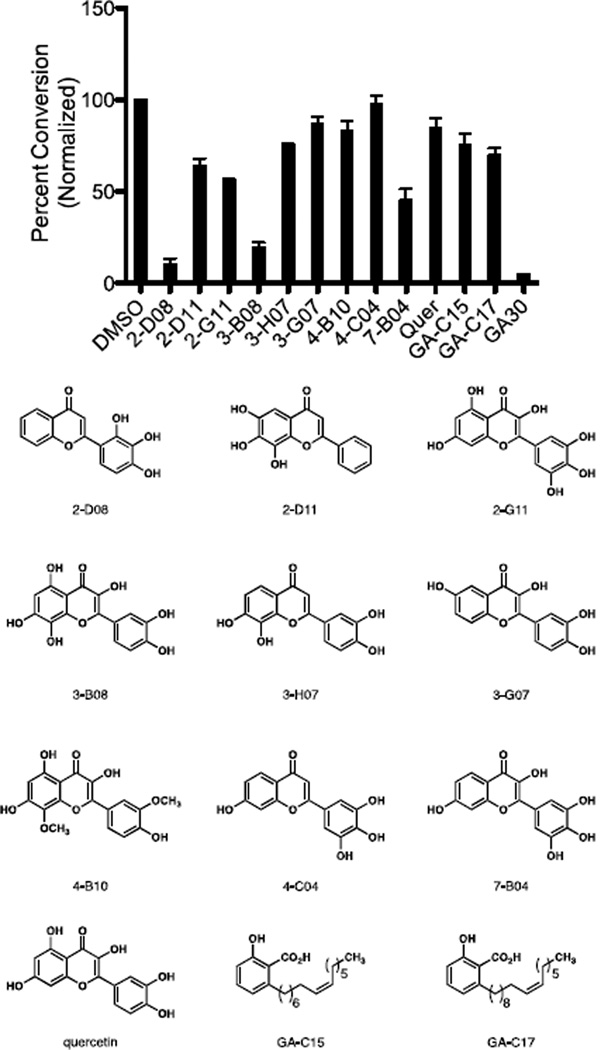
We were encouraged to find that of the 500 pure compounds screened, only ten compounds showed greater than 90% inhibition after 100 minutes. We re-assayed all ten compounds at a lower concentration of 5 µM in an effort to distinguish more potent compounds. At this concentration, a clear distinction could be made among the different compounds, with the most potent compound being 2-D08 (2’,3’,4’-trihydroxyflavone) (Figure 2). A number of other synthetic and natural polyhydroxylated flavonoids proved to be slightly less active. Both the C15 and C17 analogs of ginkgolic acid were evaluated as well, and proved to be slightly less potent.
Figure 2.
Ten Most Active Compounds from the Kinetic Screen. GA30 = Ginkgolic acid C15, 30 µM. All other compounds are assayed at 5 µM. The sumoylation reaction was performed for 90 minutes at room temperature, and the reaction was quenched by the addition of EDTA. Percent conversion was measured using the LabChip EZ Reader II system as described in the Experimental Procedures. Percent conversion is normalized to a DMSO-treated control sample in which percent conversion is 100. Values represent the mean of three replicates.
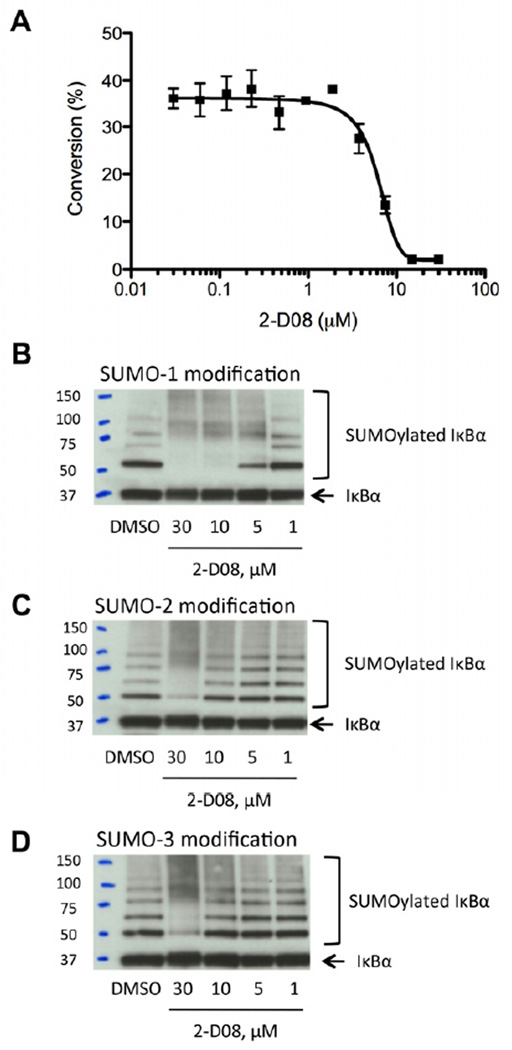
In an effort to further evaluate the properties of 2-D08 we next performed a more in-depth biochemical analysis. The IC50 for 2-D08 in this assay was measured to be 6.0 ± 1.3 µM (Figure 3A). We then evaluated the ability of 2-D08 to inhibit the sumoylation of fully intact recombinant IκBα, a well-studied sumoylation substrate (Desterro et al., 1998). Western blot analysis of in vitro sumoylation experiments showed that 2-D08 exhibited a dose-dependent inhibition of the SUMO-1 modification of recombinant IκBα (Figure 3B). This inhibition occurred in a concentration range consistent with the IC50 measured in the screening assay. Furthermore, 2-D08 also inhibited the conjugation of SUMO-2 (Figure 3C) and SUMO-3 (Figure 3D) to IκBα within a similar concentration range.
Figure 3.
Potency and Selectivity of 2-D08. (A) IC50 for 2-D08 is measured to be 6.0 µM using the mobility shift assay. Reactions were performed at varying concentrations of 2-D08 and quenched with EDTA after 90 minutes. Conversion was measured using the Caliper EZ Reader II system. Values represent the mean of three replicates. (B) 2-D08 dose-dependently inhibits the sumoylation of IκBα with SUMO-1, SUMO-2 (C), and SUMO-3 (D) as observed by Western blot using anti-IκBα (C-21) antibody. Sumoylation of full-length protein IκBα (human, recombinant) was performed with recombinant SUMO-1, SUMO-2, or SUMO-3 in the presence/absence of 2-D08 at room temperature for 90 minutes and sumoylated IκBα level was measured by Western blot analysis with an anti-IκBα.
Structure-Activity Relationship and Mechanism of Action
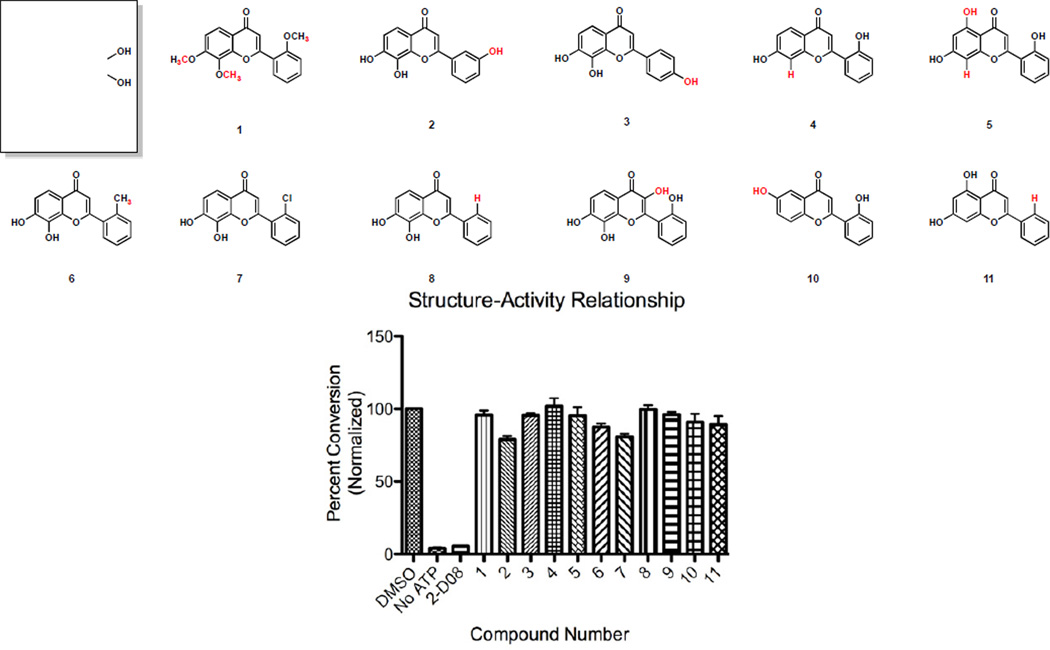
We next evaluated a series of compounds closely related to 2-D08 and evaluated them in the sumoylation assay. Interestingly, a discreet SAR could be observed (Figure 4). Small deviations from the core structure of 2-D08 resulted in a substantial loss of inhibitory activity. While 2-D08 displayed 90% inhibition at 30 µM, the permethylated derivative 1 exhibited only 4% inhibition. The meta- and para- isomers 2 and 3 were substantially less active at 21% and 4% inhibition, respectively. Since a strict requirement for an ortho- substituent could be indicative of a non-planar pharmacophore, we synthesized two other analogs containing chloro (7) and methyl (6) substituents in the ortho- position and evaluated compounds without a phenol (8). These compounds displayed markedly decreased inhibition, indicating that the phenol functionality is a requirement. Similarly, other similar compounds (4, 5, 10, 11) also exhibited a substantial loss of activity, while other catechols retained some activity (Figure 2).
Figure 4.
Structure-Activity Relationship of 2-D08. Sumoylation reactions were performed for 2 hours at room temperature, and then were quenched by the addition of EDTA. Percent conversion was measured using the LabChip EZ Reader II system as described in Experimental Procedures. Percent conversion is normalized relative to a DMSO-treated control sample in which percent conversion is 100. Values represent the mean of three replicates.
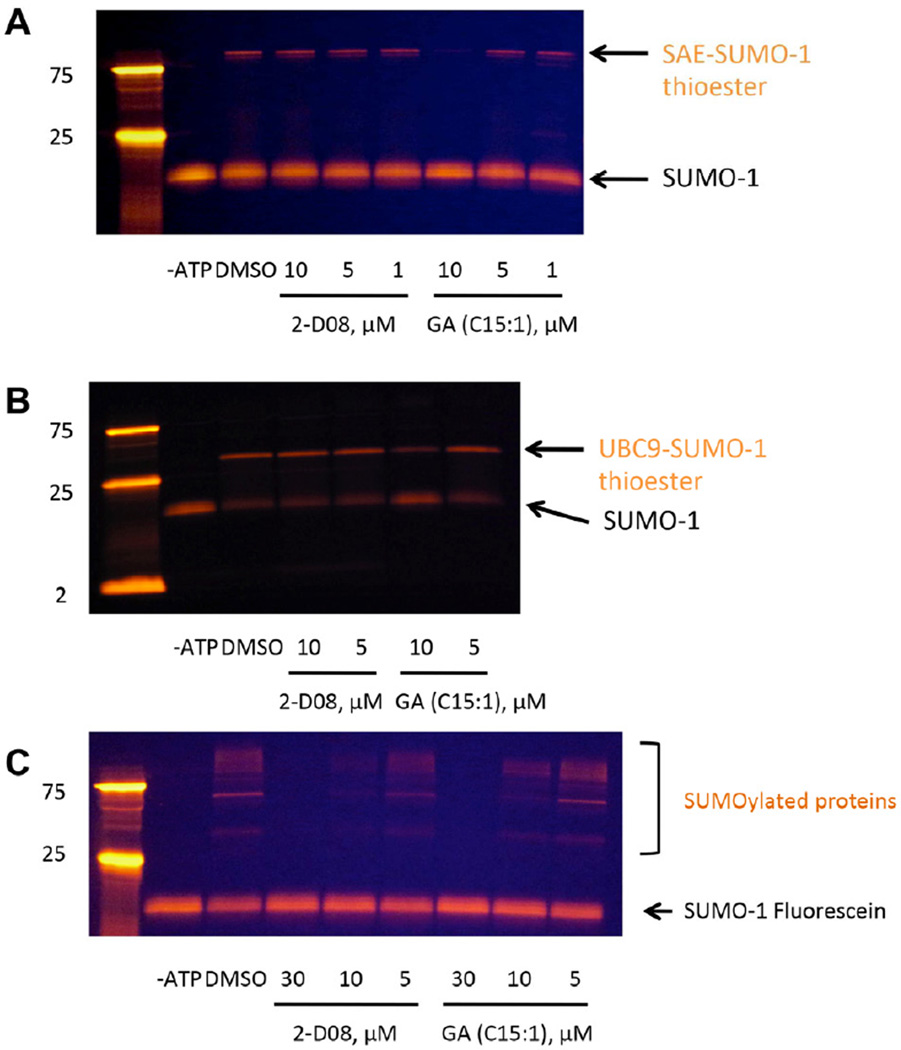
Having established an initial structure-activity relationship for 2-D08, we next moved to study its mechanism of action. Our initial screening assay involved a reconstituted sumoylation cascade, including recombinant SAE 1/2, UBC9, and SUMO-1. In order to evaluate whether 2-D08 inhibited individual steps in the cascade, we utilized a fluorescently labeled SUMO-1 (referred to as FL-SUMO-1) (Alontaga et al., 2012), rather than a fluorescent substrate. Straightforward gel shift experiments clearly indicated the activation and transfer of SUMO-1 between the enzymes. In the presence of FL-SUMO-1, SAE 1/2, and ATP, clean conversion to the SAE-(FL-SUMO-1) thioester complex was observed. Treatment with ginkgolic acid (an inhibitor of SAE) inhibited this process, while 2-D08 had no effect (Figure 5A). This indicated that while ginkgolic acid inhibited E1 activation, 2-D08 did not. In the presence of FL-SUMO-1, SAE 1/2, UBC9, and ATP, but no peptide substrate, formation of the UBC9-(FL-SUMO-1) thioester was also observed. Again, ginkgolic acid cleanly blocked this process while 2-D08 had no effect (Figure 5B). In a third experiment, where an intact cascade comprised of FL-SUMO-1, SAE 1/2, UBC9, and ATP were combined with an unlabeled protein substrate, 2-D08 cleanly inhibited formation of the product (Figure 5C). These data clearly indicate that 2-D08 inhibits the transfer of SUMO-1 from the E2 thioester to the SUMO substrate, without disrupting any of the other individual steps in the biochemical cascade.
Figure 5.
Mechanism of Action of 2-D08 Using a Fluorescently Labeled SUMO-1. (A) Ginkgolic acid, but not 2-D08, inhibits E1-SUMO-1 thioester formation. (B) Ginkgolic acid, but not 2-D08, inhibits UBC9-SUMO-1 thioester formation. For thioester bond formation assays under non-reducing condition, reaction mixtures were incubated for 37 °C for 20 minutes in the absence of DTT, and thioester bond formation was detected by in-gel fluorescence imaging. For thioester bond formation assay under reducing conditions, see Supporting Information. (C) Both ginkgolic acid and 2-D08 inhibit the sumoylation of IκBα. Sumoylation reactions were performed with full-length protein IκBα and SUMO-1-Fluorescein at room temperature for 90 min, and then sumoylated protein level was detected by in-gel fluorescence imaging.
Inhibition of Sumoylation in Cancer Cell Lines
Sumoylation has been shown to modulate a diverse variety of cellular phenomena. A particularly important role for sumoylation has been in DNA repair, whereby sumoylation of repair enzymes results in changes in their function and subcellular localization and other consequences. A specific example is the observation that treatment of cells with camptothecin (CPT) resulted in the rapid accumulation of sumoylated topoisomerase I (topo-I) (Desai et al., 2001; Mao et al., 2000). This effect has also been replicated in vitro. Though the consequences of this accumulation are complex and not well understood, high levels of sumoylation of topo-I appear to be at least one mechanism leading to CPT resistance. This observation is supported by experiments showing that cells overexpressing SUMO have an increased resistance to CPT (Yang et al., 2006).
In order to study the effects of 2-D08 on topo-I, we first confirmed that 2-D08 inhibited the sumoylation of a topo-I fragment in vitro (Figure 6A). 2-D08, but not the closely related meta- and para- analogs 2 and 3, inhibited the sumoylation of the topo-I fragment at a concentration of 30 µM using a reconstituted sumoylation cascade. We next moved to evaluate the sumoylation of topo-I in intact ZR-75-1 cells and in BT-474 cells, breast cancer cell lines where the sumoylation of topo-I has been observed previously (Desai et al., 2001). After 15 minutes of treatment with CPT, an accumulation of topo-I-SUMO conjugates was observed in both cell lines. However, when cells were pretreated with 2-D08 for several hours, this effect was inhibited. Critically, the meta- and para- analogs 2 and 3 were completely inactive in cells at the same concentration, again highlighting the specificity of this effect (Figure 6B, C).
Figure 6.
2-D08 Inhibits the Sumoylation of Topoisomerase-I in Response to Camptothecin Treatment. (A) 2-D08, but not the inactive meta- or para- isomers 2 or 3, inhibits the sumoylation of a Topo-I fragment. (B) 2-D08 but not the inactive meta- or para- isomers 2 or 3, inhibits Topo-I sumoylation in ZR-75-1 cells. (C) 2-D08, but not the inactive meta- or para-isomers 2 or 3, Inhibits Topo-I sumoylation in BT-474 cells. (D) Treatment of BT-474 cells with MG-132 results in an increase in high molecular weight ubiquitylated proteins relative to a DMSO control. In MG-132-treated BT-474 cells, PYR-41 inhibits global ubiquitylation, but 2-D08 does not. Concentration of compounds: 2-D08, 100 µM; compound 2, 100 µM; compound 3, 100 µM; CPT, 10 µM; PYR-41, 50 µM; MG-132, 10 µM.
Finally, we performed a ubiquitylation assay to see if 2-D08 affects protein ubiquitylation in cells. In BT-474 cells treated with MG-132, a proteasome inhibitor, accumulation of high molecular weight ubiquitylated adducts could be observed by Western blot. In the presence of 2-D08 and MG-132, no inhibition of this process was observed. In contrast, when cells were treated with both PYR-41 (a known ubiquitylation inhibitor) (Yang et al., 2007) and MG-132 (a proteasome inhibitor) (Tsubuki et al., 1996), the ubiquitin conjugates were substantially decreased, indicating that while PYR-41 inhibits ubiquitylation, 2-D08 does not (Figure 6D).
DISCUSSION
Protein sumoylation continues to raise interest as a key regulator of intracellular events and as a driver of several human cancers. The ability to perturb specific steps within the sumoylation cycle would be invaluable in developing our understanding of the roles of conjugating enzymes in cellular homeostasis as well as disease states, and could lead to a new approach to cancer chemotherapy. In this report, we describe the development of a novel sumoylation assay that relies on electrophoretic mobility shift technology. Furthermore, we show that this assay is useful in a kinetic screen to identify mechanistically novel, cell permeable small molecules capable of inhibiting single steps within the biochemical cascade. An active molecule identified from this screen (2-D08) does not block the formation of the UBC9-SUMO thioester, but prevents transfer of the SUMO tag to a number of substrates. Furthermore, we show that the newly identified inhibitor is capable of blocking the sumoylation of topo-I in response to CPT treatment in the context of two different breast cancer cell lines. Finally, it does not inhibit the global ubiquitylation increase in response to proteasome inhibition, indicating pathway selectivity. To our knowledge, 2-D08 is the first compound with this unique and selective mechanism of action. The discovery of this molecule provides a new inroad for the pharmacological perturbation of the SUMO pathway.
A variety of flavonoids closely related to 2-D08 have been studied in several biological contexts. 2-D08 is a synthetic flavone that has been reported in studies evaluating radical scavenging (Cotelle et al., 1992; Seyoum et al., 2006), antioxidant (Cotelle et al., 1996), and antimutagenic (Laget et al., 1995) effects. Although 2-D08 has been studied in several other systems, activity profiles in these reports do not correlate with observations made here. In these previous studies, 2-D08 is not the most potent compound and is often substantially less potent than other molecules studied. As one specific example, 2-D08 and the 7,8-dihydroxyflavone 8 display nearly identical radical formation activities (Seyoum et al., 2006), while they exhibit markedly different activities in the sumoylation assay with no detectable effect for 8 in the biochemical assay. The lack of correlation with other reported activities indicates that the effects we observe in both biochemical and cell-based assays are not a result of these other mechanisms.
Importantly, 2-D08 appears to act in the present context by a biochemical inhibition of protein function, rather than by a physiochemical function (for example, such as by nonspecific cysteine alkylation, reaction with oxygen, or aggregation). This observation is supported by our experiments showing that 2-D08 still inhibits SUMO conjugation in the presence of detergents such as Triton X-100 or CHAPS (aggregation is not present in either case as measured by dynamic light scattering), or in the presence of catalase to degrade hydroperoxide adducts (Tjernberg et al., 2004), (see supporting information for details). Additionally, modifications that make 2-D08 more hydrophobic, which should increase aggregation in aqueous medium, result in a decrease in activity. Furthermore, 2-D08 is active independent of thiol concentration: our assay is conducted at a relatively high concentration of DTT (1 mM), and 2-D08 is equally active this and lower thiol concentrations (data not shown), demonstrating that nonspecific thiol alkylation is not a mechanism for inhibition in this assay. We have observed that 2-D08, but not isomeric analogs, inhibits sumoylation events in response to stimuli in several breast cancer cell lines. Furthermore, 2-D08 inhibits SUMO conjugation but not global ubiquitylation, indicating that it is not a pan-E2 inhibitor and exhibits pathway specificity. It is clear that 2-D08 is a sumoylation inhibitor and not a ubiquitylation inhibitor. However, we can not rule out at this time that 2-D08 may have activity on pathways other than the sumoylation/ubiquitylation processes that may or may not be related in more complex cell-based contexts. This study shows that the electrophoretic mobility shift assay can be used to identify cell permeable sumoylation inhibitors that are mechanistically analyzed in a straightforward manner. Furthermore, this work indicates that the biochemical blockade of SUMO transfer from UBC9 to the substrate is a step that should be evaluated further for the pharmacological inhibition of sumoylation and the study of more druglike sumoylation inhibitors as anticancer compounds. Our future work includes the screening of a large and diverse library of compounds for the identification of more potent and/or structurally and mechanistically unique inhibitors of protein sumoylation.
SIGNIFICANCE
Despite the discovery that protein sumoylation plays a critical role in the growth of cancer cells (notably Myc-driven tumor cell lines), and is associated with adverse clinical outcomes, few small molecule inhibitors of this process exist. To address this, we have developed an electrophoretic mobility shift assay suitable for medium throughput analysis of compound libraries. Furthermore, we have identified a novel sumoylation inhibitor. This report represents, to the best of our knowledge, the first example of a cell permeable small molecule capable of blocking SUMO transfer from the UBC9-thioester complex to a variety of substrates. In addition to the consensus sequence-containing peptide initially used in the assay, sumoylation of a fully intact protein substrate (IκBα) and a fragment of topo-I were also inhibited. This activity was identified using an in vitro approach, however we were also able to observe blockade of an induced sumoylation event in intact cells as well. We were able to show the inhibition of topo-I sumoylation in cancer cells and that global ubiquitylation remained unaffected, indicating that 2-D08 is a pathway-specific inhibitor. UBC9, the sole SUMO E2 enzyme, is an important cancer target, and the results described herein demonstrate that further investigation of molecules that inhibit this step in the process of SUMO conjugation is warranted. Future work will focus on the continued use of the mobility shift assay for the identification, optimization, and in-depth study of more potent pharmacophores that have this unique mechanism of action.
EXPERIMENTAL PROCEDURES
Reagents, Proteins, and Cells
Adenosine 5′-triphosphate (ATP) magnesium salt, DL-dithiothreitol (DTT), and ginkgolic acid (C17:1) were purchased from Sigma-Aldrich. Ginkgolic acid (C15:1) was purchased from EMD Millipore. Library of 500 flavones was purchased from TimTec LLC and INDOFINE Chemical Company, Inc. The following recombinant proteins were purchased and used without further purification: SUMO E1 (E-315, Boston Biochem, Inc), GST-SUMO E1 (E-310, Boston Biochem, Inc), UBC9 (BML-UW9320, Enzo Life Sciences, Inc.), SUMO-1 (N-terminal His6-tag, BML-UW9195, Enzo Life Sciences, Inc.), SUMO-2 (1–93, ALX-201-089-C500, Enzo Life Sciences, Inc.), SUMO-3 (1–92, ALX-201-087-C500, Enzo Life Sciences, Inc), IκBα (untagged, BML-UW9975, Enzo Life Sciences, Inc.), Topoisomerase I fragment (1–200, B007, LAE Biotech), SUMO-1 Fluorescein (UL-735, Boston Biochem, Inc). ZR-75-1 and BT-474 breast cancer cells were obtained from American Type Culture Collection (ATCC).
Sumoylation Assay: General Procedure
The in vitro sumoylation assay was performed in 20 µL Tris buffer (50 mM Tris pH 9.0, 5 mM MgCl2, 1 mM DTT and 2 mM ATP) with SUMO E1 (0.1 µM), SUMO-1 (His-tag, 1.4 µM), UBC9 (0.25 µM), and a fluorescent peptide FL-AR (1.0 µM). The reaction was initiated by the addition of ATP (final concentration of ATP = 2 mM). After incubation for 90 min at room temperature, samples were mixed with NuPAGE LDS sample buffer (Invitrogen) or Laemmli sample buffer (Bio-Rad), the resulting mixtures were boiled for 5 min and then SUMOylation level was measured by in-gel fluorescence imaging or western blot. Alternatively, the sumoylation assay was performed using IκBα (0.35 µM) or topoisomerase I fragment (1–200) (0.25 µM) as SUMO substrates instead of the fluorescent peptide FL-AR and their sumoylation level was measured by Western blot.
Thioester Bond Formation Assays
The E1-SUMO1 thioester bond formation assay was performed in 20 µL Tris buffer (50 mM Tris pH 8, 5 mM MgCl2 and 2 mM ATP) with GST-SUMO E1 (E-310, 0.35 µM) and SUMO-1 Fluorescein (0.5 µM) in the absence or presence of 1 mM DTT. After incubation for 20 min at 37 °C1 samples were loaded on a Criterion Tris-HCl Pre cast Gel (Bio-Rad) and resolved by electrophoresis. E1-SUMO1 thioester bond formation was detected by in-gel fluorescence imaging.
UBC9-SUMO1 thioester bond formation assay was performed with E1 (E-315, 0.10 µM), Ubc9 (0.70 µM) and SUMO-1 Fluorescein (0.50 µM) in the absence of DTT. After incubation for 20 min at 37 °C, a set of samples were mixed with SDS sample buffer without DTT. Another set of samples were mixed with SDS sample buffer with DTT (150 mM) and boiled for 5 min. Samples were loaded on a Criterion Tris-HCl Precast Gel (Bio-Rad) and resolved by electrophoresis. UBC9-SUMO1 thioester bond formation was detected by in-gel fluorescence imaging.
Western Blots
Samples were loaded on Criterion Tris-HCl precast gels (Bio-Rad), resolved by electrophoresis and transferred to a nitrocellulose membrane (Whatman, Protran BA85). Membranes were blocked with 5% nonfat dry milk (Bio-Rad) for 1 h, and incubated with a primary antibody in Tween buffer (5 mM Tris, 2.5 mM EDTA, 50 mM NaCl, 0.1% Tween 20) for 1 h at room temperature. Membrane was washed with Tween buffer and incubated with an HRP-conjugated secondary antibody for 1 h at room temperature. After washing membrane, the blot was visualized using the ECL system on BioMax MR film (Eastman Kodak). The following antibodies were used for western blot: anti-SUMO-1 (CT, Enzo Life Sciences, Inc., 1:1500 dilution), anti-Topo I (H-5, Santa Cruz biotechnology, 1:1500 dilution), anti-Topo I (F14, LAE Biotech International, 1:1500 dilution), anti-IκBα (C-21, Santa Cruz biotechnology, 1:1000 dilution), anti-ubiquitin (P4D1, Santa Cruz biotechnology, 1:1000 dilution), anti-actin (C4, EMD Millipore Corporation, 1:3000 dilution), HRP-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, 1:5000 dilution), HRP-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, 1:5000 dilution).
Topo I Sumoylation in Cancer Cells
ZR-75-1 and BT-474 breast cancer cells were cultured at 37 °C in RPMI 1640 medium (Mediatech, Inc.) with 10% fetal bovine serum (FBS, Mediatech, Inc.) and 1% antimycotic-antibiotic (Mediatech, Inc.). Cells were treated with DMSO or the corresponding compound for 6 h, followed by treatment with camptothecin (Selleck Chemicals) for 15 min. The SUMOylated Topo I level was measured by western blot according to procedures reported in the literature (Desai et al., 2001; Mao et al., 2000).
Ubquitination in Cancer Cells
BT-474 cells were treated with DMSO, 2-D08 or PYR-41 (Sigma-Aldrich) in the absence/presence of MG-132 (Santa Cruz) for 6 h at 37 °C. Cells were washed with phosphate-buffered saline, lysed in SDS lysis buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol) on ice, and boiled for 5 min. Equal amounts of proteins from each sample were mixed with SDS sample buffer and boiled for 5 min. Western blot was carried out and the ubiquitinated protein level was measured using anti-ubiquitin (P4D1) antibody.
In-Gel Fluorescence Imaging
Samples were loaded on Criterion Tris-HCl precast gels (Bio-Rad) and resolved by electrophoresis. In-gel fluorescence images were obtained by UVP MultiDoc-It imaging systems (Ultra-Violet Products Ltd) and images were analyzed using UVP Doc-It LS image analysis software (Ultra-Violet Products Ltd).
Sumoylation Electrophoretic Mobility Shift Assay
The in vitro sumoylation assay was carried out as described in the general procedure above in 384-well plate format. After the desired reaction time, EDTA (0.25 M, 10 µL) was added to each well instead of sample buffer so as to quench reaction. Samples were analyzed using a LabChip EZ Reader II (Caliper Life Sciences, Inc.) and run conditions were the following: upstream voltage of −2500 V, downstream voltage of −500 V, and pressure of −1.0 psi. Percent conversion is defined as 100 × P/(P+S), where P and S are peak heights of sumoylated product SUMO-1-FL-AR and peptide substrate FL-AR, respectively.
Supplementary Material
Acknowledgments
The authors thank Dr. Jeffrey Gildersleeve and Dr. Barry O’Keefe for helpful discussions in the preparation of this manuscript. We thank the Molecular Targets Laboratory, National Cancer Institute for providing the plate of natural product extracts. The authors wish to acknowledge Laurel Provencher (Perkin Elmer) for help in initial assay development. This work was supported by the Intramural Research Program of the NIH, Center for Cancer Research, and the National Cancer Institute, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alontaga AY, Bobkova E, Chen Y. Biochemical analysis of protein SUMOylation. Chapter 10, Unit10 29. Curr Protoc Mol Biol. 2012 doi: 10.1002/0471142727.mb1029s99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews EA, Palecek J, Sergeant J, Taylor E, Lehmann AR, Watts FZ. Nse2, a component of the Smc5–6 complex, is a SUMO ligase required for the response to DNA damage. Mol Cell Biol. 2005;25:185–196. doi: 10.1128/MCB.25.1.185-196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov. 2011;10:29–46. doi: 10.1038/nrd3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- Carlson CB, Horton RA, Vogel KW. A Toolbox Approach to High-Throughput TR-FRET-Based SUMOylation and DeSUMOylation Assays. Assay Drug Dev Techn. 2009;7 doi: 10.1089/adt.2008.0188. [DOI] [PubMed] [Google Scholar]
- Ceccarelli DF, Tang X, Pelletier B, Orlicky S, Xie W, Plantevin V, Neculai D, Chou YC, Ogunjimi A, Al-Hakim A, et al. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell. 2011;145:1075–1087. doi: 10.1016/j.cell.2011.05.039. [DOI] [PubMed] [Google Scholar]
- Chung TL, Hsiao HH, Yeh YY, Shia HL, Chen YL, Liang PH, Wang AH, Khoo KH, Shoei-Lung Li S. In vitro modification of human centromere protein CENP-C fragments by small ubiquitin-like modifier (SUMO) protein: definitive identification of the modification sites by tandem mass spectrometry analysis of the isopeptides. The Journal of biological chemistry. 2004;279:39653–39662. doi: 10.1074/jbc.M405637200. [DOI] [PubMed] [Google Scholar]
- Cohen P, Tcherpakov M. Will the ubiquitin system furnish as many drug targets as protein kinases? Cell. 2010;143:686–693. doi: 10.1016/j.cell.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Cotelle N, Bernier JL, Catteau JP, Pommery J, Wallet JC, Gaydou EM. Antioxidant properties of hydroxy-flavones. Free Radical Bio Med. 1996;20:35–43. doi: 10.1016/0891-5849(95)02014-4. [DOI] [PubMed] [Google Scholar]
- Cotelle N, Bernier JL, Henichart JP, Catteau JP, Gaydou E, Wallet JC. Scavenger and Antioxidant Properties of 10 Synthetic Flavones. Free Radical Bio Med. 1992;13:211–219. doi: 10.1016/0891-5849(92)90017-b. [DOI] [PubMed] [Google Scholar]
- Desai SD, Li TK, Rodriguez-Bauman A, Rubin EH, Liu LF. Ubiquitin/26S proteasome-mediated degradation of topoisomerase I as a resistance mechanism to camptothecin in tumor cells. Cancer Res. 2001;61:5926–5932. [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- Driscoll JJ, Pelluru D, Lefkimmiatis K, Fulciniti M, Prabhala RH, Greipp PR, Barlogie B, Tai YT, Anderson KC, Shaughnessy JD, Jr, et al. The sumoylation pathway is dysregulated in multiple myeloma and is associated with adverse patient outcome. Blood. 2010;115:2827–2834. doi: 10.1182/blood-2009-03-211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Trent JO, Ye H. Targeting the SUMO E2 conjugating enzyme Ubc9 interaction for anti-cancer drug design. Anticancer Agents Med Chem. 2009;9:51–54. doi: 10.2174/187152009787047716. [DOI] [PubMed] [Google Scholar]
- Fanslau C, Pedicord D, Nagulapalli S, Gray H, Pang S, Jayaraman L, Lippy J, Blat Y. An electrophoretic mobility shift assay for the identification and kinetic analysis of acetyl transferase inhibitors. Anal Biochem. 2010;402:65–68. doi: 10.1016/j.ab.2010.03.025. [DOI] [PubMed] [Google Scholar]
- Fukuda I, Ito A, Hirai G, Nishimura S, Kawasaki H, Saitoh H, Kimura K, Sodeoka M, Yoshida M. Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem Biol. 2009a;16:133–140. doi: 10.1016/j.chembiol.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Fukuda I, Ito A, Uramoto M, Saitoh H, Kawasaki H, Osada H, Yoshida M. Kerriamycin B inhibits protein SUMOylation. J Antibiot (Tokyo) 2009b;62:221–224. doi: 10.1038/ja.2009.10. [DOI] [PubMed] [Google Scholar]
- Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy MC, Jaffray EG, Walker KJ, Hay RT. Arsenic-induced SUMO-dependent recruitment of RNF4 into PML nuclear bodies. Mol Biol Cell. 2010;21:4227–4239. doi: 10.1091/mbc.E10-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335:348–353. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laget M, DeMeo M, Wallet JC, Gaydou EM, Guiraud H, Dumenil G. Antimutagenic activities of 24 synthetic flavones with the Salmonella microsomal assay. Arch Pharm Res. 1995;18:415–422. [Google Scholar]
- Lei X, Johnson RP, Porco JA., Jr Total synthesis of the ubiquitin-activating enzyme inhibitor (+)-panepophenanthrin. Angew Chem Int Ed Engl. 2003;42:3913–3917. doi: 10.1002/anie.200351862. [DOI] [PubMed] [Google Scholar]
- Majumdar A, Petrescu AD, Xiong Y, Noy N. Nuclear Translocation of Cellular Retinoic Acid-binding Protein II Is Regulated by Retinoic Acid-controlled SUMOylation. J Biol Chem. 2011;286:42749–42757. doi: 10.1074/jbc.M111.293464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Sun M, Desai SD, Liu LF. SUMO-1 conjugation to topoisomerase I: A possible repair response to topoisomerase-mediated DNA damage. P Natl Acad Sci USA. 2000;97:4046–4051. doi: 10.1073/pnas.080536597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo YY, Moschos SJ. Targeting Ubc9 for cancer therapy. Expert Opin Ther Targets. 2005;9:1203–1216. doi: 10.1517/14728222.9.6.1203. [DOI] [PubMed] [Google Scholar]
- Muller S, Hoege C, Pyrowolakis G, Jentsch S. SUMO, ubiquitin's mysterious cousin. Nat Rev Mol Cell Biol. 2001;2:202–210. doi: 10.1038/35056591. [DOI] [PubMed] [Google Scholar]
- Muller S, Matunis MJ, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. Embo Journal. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler A, Knipscheer P, Oberhofer E, van Dijk WJ, Korner R, Olsen JV, Jentsch S, Melchior F, Sixma TK. SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nat Struct Mol Biol. 2005;12:264–269. doi: 10.1038/nsmb903. [DOI] [PubMed] [Google Scholar]
- Potts PR, Yu HT. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol. 2005;25:7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psakhye I, Jentsch S. Protein Group Modification and Synergy in the SUMO Pathway as Exemplified in DNA Repair. Cell. 2012 doi: 10.1016/j.cell.2012.10.021. [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. The Journal of biological chemistry. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- Rouleau N, Wang J, Karras L, Andrews E, Bielefeld-Sevigny M, Chen Y. Highly sensitive assays for SUMOylation and small ubiquitin-like modifier-dependent protein-protein interactions. Anal Biochem. 2008;375:364–366. doi: 10.1016/j.ab.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N, Uchimura Y, Tachibana T, Sugahara S, Saitoh H, Nakao M. In situ SUMOylation analysis reveals a modulatory role of RanBP2 in the nuclear rim and PML bodies. Exp Cell Res. 2006;312:1418–1430. doi: 10.1016/j.yexcr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Schmidt D, Muller S. PIAS/SUMO: new partners in transcriptional regulation. Cellular and Molecular Life Sciences. 2003;60:2561–2574. doi: 10.1007/s00018-003-3129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyoum A, Asres K, El-Fiky FK. Structure-radical scavenging activity relationships of flavonoids. Phytochemistry. 2006;67:2058–2070. doi: 10.1016/j.phytochem.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Tjernberg A, Hallen D, Schultz J, James S, Benkestock K, Bystrom S, Weigelt J. Mechanism of action of pyridazine analogues on protein tyrosine phosphatase 1B (PTP1B) Bioorg Med Chem Lett. 2004;14:891–895. doi: 10.1016/j.bmcl.2003.12.014. [DOI] [PubMed] [Google Scholar]
- Tsubuki S, Saito Y, Tomioka M, Ito H, Kawashima S. Differential inhibition of calpain and proteasome activities by peptidyl aldehydes of di-leucine and tri-leucine. J Biochem. 1996;119:572–576. doi: 10.1093/oxfordjournals.jbchem.a021280. [DOI] [PubMed] [Google Scholar]
- Ulrich HD. The SUMO system: an overview. Methods Mol Biol. 2009;497:3–16. doi: 10.1007/978-1-59745-566-4_1. [DOI] [PubMed] [Google Scholar]
- Ungermannova D, Parker SJ, Nasveschuk CG, Chapnick DA, Phillips AJ, Kuchta RD, Liu X. Identification and mechanistic studies of a novel ubiquitin E1 inhibitor. J Biomol Screen. 2012a;17:421–434. doi: 10.1177/1087057111433843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermannova D, Parker SJ, Nasveschuk CG, Wang W, Quade B, Zhang G, Kuchta RD, Phillips AJ, Liu X. Largazole and its derivatives selectively inhibit ubiquitin activating enzyme (e1) Plos One. 2012b;7:e29208. doi: 10.1371/journal.pone.0029208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nguyen T, Angkasekwinai P, Dou H, Lin FM, Lu LS, Cheng J, Chin YE, Dong C, Yeh ET. SUMO-specific protease 1 is critical for early lymphoid development through regulation of STAT5 activation. Mol Cell. 2012;45:210–221. doi: 10.1016/j.molcel.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verger A, Perdomo J, Crossley M. Modification with SUMO - A role in transcriptional regulation. Embo Rep. 2003;4:137–142. doi: 10.1038/sj.embor.embor738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Banerjee S. Differential PIAS3 expression in human malignancy. Oncol Rep. 2004;11:1319–1324. [PubMed] [Google Scholar]
- Wood LD, Irvin BJ, Nucifora G, Luce KS, Hiebert SW. Small ubiquitin-like modifier conjugation regulates nuclear export of TEL, a putative tumor suppressor. P Natl Acad Sci USA. 2003;100:3257–3262. doi: 10.1073/pnas.0637114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Asmussen G, Booker M, Hirth B, Kane JL, Jr, Liao J, Noson KD, Yee C. Discovery of novel sphingosine kinase 1 inhibitors. Bioorg Med Chem Lett. 2009;19:6119–6121. doi: 10.1016/j.bmcl.2009.09.022. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Hirth B, Kane JL, Jr, Liao J, Noson KD, Yee C, Asmussen G, Fitzgerald M, Klaus C, Booker M. Discovery of novel sphingosine kinase-1 inhibitors. Part 2. Bioorg Med Chem Lett. 2010;20:4550–4554. doi: 10.1016/j.bmcl.2010.06.019. [DOI] [PubMed] [Google Scholar]
- Yang M, Hsu CT, Ting CY, Liu LF, Hwang J. Assembly of a polymeric chain of SUMO1 on human topoisomerase I in vitro. The Journal of biological chemistry. 2006;281:8264–8274. doi: 10.1074/jbc.M510364200. [DOI] [PubMed] [Google Scholar]
- Yang Y, Kitagaki J, Dai RM, Tsai YC, Lorick KL, Ludwig RL, Pierre SA, Jensen JP, Davydov IV, Oberoi P, et al. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 2007;67:9472–9481. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- Zhao J. Sumoylation regulates diverse biological processes. Cell Mol Life Sci. 2007;64:3017–3033. doi: 10.1007/s00018-007-7137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XL, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. P Natl Acad Sci USA. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong HJ, Ma VP, Cheng Z, Chan DS, He HZ, Leung KH, Ma DL, Leung CH. Discovery of a natural product inhibitor targeting protein neddylation by structure-based virtual screening. Biochimie. 2012;94:2457–2460. doi: 10.1016/j.biochi.2012.06.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.