Abstract
Sphingosine-1-phosphate is a potent sphingolipid mediator and the kinase that produces it, sphingosine kinase 1 (SphK1), has been implicated in cancer progression, inflammation, and cardiovascular diseases. In this issue of Structure, Wang and colleagues provide the scientific community with the long awaited structure of SphK1 (Wang et al., 2013).
Lipid-mediated signaling is rapidly emerging as a new frontier among cellular biologists, where an ever-increasing body of evidence implicates numerous types of lipids in the regulation of an incredibly diverse array of functions in metabolism and disease. At one of the epicenters of these new and exciting fields is the pleiotropic modulator sphingosine-1-phosphate (S1P), a simple lipid with a remarkable ability to modulate many complex cellular programs including cell proliferation, inflammation, mitogenesis, migration, angiogenesis, protection from apoptosis, cytokine and chemokine production, to name just a few (Maceyka et al., 2012). As a testament to S1P’s powerful biological effector properties, changes in its levels accompany and can exacerbate cancer progression, inflammation, and cardiovascular diseases (Maceyka et al., 2012). Although synthesis, catabolism and import-export systems tightly regulate intra- and extra-cellular S1P levels, at the heart of its cellular de novo biogenesis are two isoenzymes: sphingosine kinase type 1 and 2 (SphK1; SphK2) (Or Gandy and Obeid, 2013). Abnormal cellular SphK1 regulation has been implicated in pathological processes and linked to diseases such as cancer and inflammation. Hence, SphK1 has long been the focus of many studies in vivo and in vitro, and its elusive atomic structure highly sought by many.
In this issue of Structure, Walker and colleagues present the atomic structure of SphK1 in complexes that expand our mechanistic understanding of this classic pharmacological target including lipid, nucleotide, and inhibitor+nucleotide bound states (Wang et al., 2013). These structures provide a vital baseline from which to generate predictions and targeted modifications that further probe the many nuances of functional elements in SphK1 activity and regulation.
As expected from sequence-based prediction studies, core structural elements of SphK1 quaternary structure adopt a two-domain hinged fold common among members of the PF-like superfamily (Labesse et al., 2002). However, structurally distinct C-terminal components distinguish SphK1 from diacylglycerokinases (DGKs), NAD kinases, and other PF-like structural neighbors (Miller et al., 2008). How these structurally divergent features contribute to substrate specificity or function among this large group of proteins with highly diversified enzymatic functions remains to be determined.
At the interface region of SphK1 domains is a solvent-accessible cleft where nucleotide settles to await transfer of its γ-phosphate to the sphingosine substrate (Wang et al, 2013). Topologically, this is a remarkably similar nucleotide binding mode in DGK. Within this extended cavity is a strictly conserved S/G79-GDG82 motif, and SphK1 activity is intimately linked to these residues (Pitson et al., 2002). For example, S79D and G80D mutants are catalytically inactive, and G82D mutants show a ~44-fold weakening of ATP binding (Pitson et al., 2002). Wang et al. confirm that the last remaining uncharacterized residue in this motif, Asp 81, is also critical for catalysis as D81A mutants show significant losses in ATP hydrolysis rates. Moreover, their work provides an important structural correlation for the observed loss in activity among S/G79-GDG82 motif mutants, as modeling based on the lipid bound structure of SphK1 suggests that this conserved structure abuts both the nucleotide and the sphingosine head group, precisely positioning the 2-amino-1,3-diol moiety of sphingosine within a catalytically competent distance for ATP γ-phosphate transfer. However, as mutations at any position along this motif show such strong effects on SphK1 activity (Pitson et al., 2002), and the identity of the bound lipid could not be precisely identified (Wang et al., 2013), a complex between SphK1 and its cognate sphingosine substrate or S1P should help further our understanding of how structural elements of SphK1 contribute to its mechanism, and resolve how this important structural and catalytic motif controls substrate binding and turnover.
One of the openings to the ligand cavity of SphK1, like those of DGK and NAD kinase, is lined by the N-terminal S/G79-GDG82 motif and the C-terminal domain residues (Wang et al., 2013). Within the cavity, sphingosine is proposed to be guided into position by a tunneling mechanism that is driven by energetically favorable interactions between non-polar cavity residues and aliphatic carbons along the sphingosine tail group. The pocket itself is revealed not as a simple tunnel but is rather J-shaped, allowing bulky groups like those of the SKI-II inhibitor to nestle within the cavity and competitively occlude substrate binding, or, for example, for non-natural sphingosines with large cyclic fluorogenic tail groups like NBD or fluorescein to be modest substrates of SphK1. However, simple induced-fit interactions are not all that govern SphK1 catalytic activity as this enzyme displays high regioselectivity towards the phosphorylation of lipid substrates. For example, whereas D-erythro-dihydrosphingosine is a weak SphK1 substrate, D,L-threo-dihydrosphingosine is competitive inhibitor (Kohama et al., 1998).
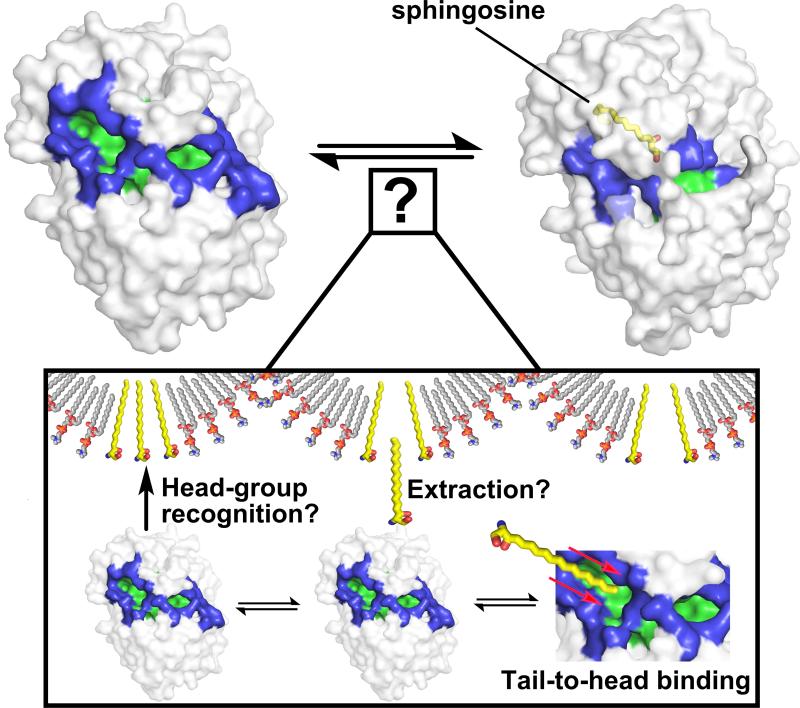
The structure of SphK1 provides a structural framework to understand how many unrelated residues are involved in catalysis and resolve the effects of distal mutations on SphK1 activity. However, outside of the magnifying glass of an atomic structure, a number of macroscopically important questions arise that relate to the biology of SphK1 in cells. For instance, a tail-to-head tunneling of lipid substrates to the SphK1 cavity implies that sphingosines are bound in some fashion by their aliphatic tail, a region devoid of any identifying features. At physiological concentrations, sphingosines are likely embedded in cellular membranes or intracellular vesicles, making the availability of “free” sphingosine in the cell extremely low, effectively hiding the tail group and only exposing the head group to the environment. As is exemplified by the translocationally regulated activity of SphK1, it may associate with lipid rafts (Hengst, 2009), where sphingosine might be enriched, and effectively “extract” it for catalysis. However, as a tail-to-head binding mechanism is most evident from the SphK1 structure, how does SphK1 bind to, identify, and extract lipids from their cognate loci imbedded in plasma, endosomal, or lysosomal membranes while retaining mechanistic consistency observed in the crystal structure (Fig. 1). These are important questions that remain to be answered and will likely require advanced biophysical methods to probe SphK1 lipid binding.
Figure 1.
In the unbound state (top left), SphK1 has two openings (colored blue) through which the sphingosine substrate can enter the ligand cavity (colored green). In the sphingosine (colored yellow) bound state (top right), conformational rearrangements of the opening occlude access to cavity residues. The J-shaped ligand cavity (not shown) is lined with non-polar residues that help guide sphingosine by a head-to-tail tunneling mechanism (Wang et al., 2013; inset, bottom right). However, in cells, sphingosine (inset, colored yellow) is likely embedded with other membrane lipids such as phosphatydylcholine (inset, colored cpk), and little is known regarding how SphK1 recognizes, binds, and extracts sphingosine from membranes.
Ultimately, the crystal structure of SphK1 is a stepping stone to new perspectives in our quest to resolve the cellular function and regulation of this key enzyme. However, as is often the case, although new structures answer a number of existing questions, many other exciting new unknowns arise. SphK1 is no exception. Further structural studies might explain the seemingly contradictory observation that some SphK1 inhibitors (Paugh et al., 2008, Orr Gandy and Obeid, 2013) markedly affect cell growth and survival, while a potent and selective inhibitor did not (Schnute et al., 2012). It will also help determine the conformational regulatory sites on SphK1 (Lim et al., 2012). Elucidation of the structural basis of SphK1 substrate recognition and catalysis will no doubt lead to better understanding of the regulation of this important enzyme in health and diseases and hopefully to the development of therapeutic molecules that target it.
ACKNOWLEDGMENTS
Work in the authors’ laboratory is supported by grants from the NIH to S.S.: R37GM043880 and RO1CA61774. S.L. was supported by NIH T32 HL094290.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Hengst JA, Guilford JM, Fox TE, Wang X, Conroy EJ, Yun JK. Arch. Biochem. Biophys. 2009;492:62–73. doi: 10.1016/j.abb.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohama T, Olivera A, Edsall L, Nagiec MM, Dickson R, Spiegel S. J. Biol. Chem. 1998;273:23722–23728. doi: 10.1074/jbc.273.37.23722. [DOI] [PubMed] [Google Scholar]
- Labesse G, Douguet D, Assairi L, Gilles AM. Trends Biochem. Sci. 2002;27:273–275. doi: 10.1016/s0968-0004(02)02093-5. [DOI] [PubMed] [Google Scholar]
- Lim KG, Tonelli F, Li Z, Lu X, Bittman R, Pyne S, Pyne NJ. J. Biol. Chem. 2011;286:18633–18640. doi: 10.1074/jbc.M111.220756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maceyka M, Harikumar KB, Milstien S, Spiegel S. Trends Cell Biol. 2012;22:50–60. doi: 10.1016/j.tcb.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DJ, Jerga A, Rock CO, White SW. Structure. 2008;16:1036–1046. doi: 10.1016/j.str.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr Gandy KA, Obeid LM. Biochim. Biophys. Acta. 2013;1831:157–166. doi: 10.1016/j.bbalip.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paugh SW, Paugh BS, Rahmani M, Kapitonov D, Almenara JA, Kordula T, Milstien S, Adams JK, Zipkin RE, Grant S, Spiegel S. Blood. 2008;112:1382–1391. doi: 10.1182/blood-2008-02-138958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitson SM, Moretti PA, Zebol JR, Zareie R, Derian CK, Darrow AL, Qi J, D’Andrea RJ, Bagley CJ, Vadas MA, Wattenberg BW. J. Biol. Chem. 2002;277:49545–49553. doi: 10.1074/jbc.M206687200. [DOI] [PubMed] [Google Scholar]
- Schnute ME, McReynolds MD, Kasten T, Yates M, Jerome G, Rains JW, Hall T, Chrencik J, Kraus M, Cronin CN, et al. Biochem. J. 2012;444:79–88. doi: 10.1042/BJ20111929. [DOI] [PubMed] [Google Scholar]
- Wang Z, Min M, Xiao S-H, Johnstone S, Romanow W, Meininger D, Xu H, Liu J, Dai J, An S, Thibault S, Walker N. Structure. 2013 doi: 10.1016/j.str.2013.02.025. this issue. [DOI] [PubMed] [Google Scholar]