Summary
Histones H2a and H2b are stored in lipid droplets in Drosophila embryos complexed with the protein Jabba. Jabba mutant embryos degrade histones H2a and H2b but embryos survive by translating stored histone mRNA.
A critical feature of early embryogenesis in all metazoans is the need to provide the histone proteins in the oocyte required to assemble new chromatin during early embryogenesis. Immediately after fertilization the sperm chromatin is remodeled and maternal histone proteins replace the specialized sperm chromatin proteins. Subsequently both the maternal and paternal chromosomes replicate prior to the first cell division, followed by a series of zygotic divisions. In most embryos there is no transcription immediately after fertilization and in many species there are a substantial number of very rapid divisions that occur in the absence of zygotic transcription. These divisions result in a logarithmic increase in the amount of DNA and hence exponentially increasing demand for histone protein for assembly of newly synthesized chromatin. A number of distinct mechanisms have evolved to solve the problem of providing histones for early embryogenesis in different metazoans. In two well-studied systems large amounts of histone proteins are provided by maternal stores of histone proteins and histone mRNAs. In Xenopus there are over 14 cell divisions resulting in a 10,000 cell embryo in 8 hrs prior to initiation of zygotic transcription and in Drosophila, zygotic transcription of histone genes initiates in cycle 11 (about 1.5 hrs after fertilization), when the syncytial embryo contains over 1000 nuclei. In this issue Welte and coworkers show that in Drosophila (and likely many insects), histones H2a and H2b are stored in lipid droplets and that proper storage is essential for utilization of the stored histone for chromatin assembly [1].
Since most histone mRNAs are not polyadenylated but end instead in a stemloop sequence [2], translation of histone mRNAs requires unique factors. One of these is the stemloop binding protein (SLBP) that binds the stemloop at the 3′ end and is required not only for histone mRNA biosynthesis but also for translation of histone mRNA [3,4]. In mammals, activation of translation of the SLBP mRNA at oocyte maturation is required for translation of the maternal histone mRNAs for synthesis of histones during the initial cell cycle [5]. In Xenopus, the oocyte synthesizes both the histone mRNAs and proteins and stores them early in development [6]. These stored histones plus additional histones synthesized by activation of translation of the stored histone mRNA at oocyte maturation [7] provide the histone proteins necessary for early development. The Xenopus histones are stored in soluble complexes with histone chaperones [8]. In Drosophila stored histone mRNA is synthesized after the last cycle of DNA replication in the nurse cells [9,10]. The stored maternal histone protein is likely synthesized at this time and then transferred from the nurse cells to the oocyte with other proteins and mRNAs necessary for early embryogenesis. Thus, large amounts of histone mRNA and protein are stored in the unfertilized egg, but whether both are essential during development is not known.
The problem of histone storage is complicated by the fact that the two sets of core histones required to form a nucleosome are stored independently; the histone H3/H4 complex likely stored as a tetramer, and the histones H2a/H2b stored as a dimer. Ultimately, equal amounts of these two sets of histones are required for assembly of newly replicated chromatin. Excess histone proteins are toxic, and histone proteins are stored bound in complexes with histone chaperones. Histones are rapidly degraded unless they are present in complexes that sequester them within the cell. In yeast it is essential to degrade excess histones [11] and it is likely similar systems exist in all cells [12]. In growing mammalian cells there is only a small supply of “soluble” histone (not bound in chromatin) [13] sufficient to support about 5 minutes of DNA replication, but many embryos store sufficient histones for hundreds or thousands of cycles of DNA replication.
Previously Welte and coworkers showed that histones H2a and H2b were present in lipid droplets in early embryos of Drosophila and that these histones were transferred to nuclei during development [14]. In the current issue they show that storage in lipid droplets is essential for utilization of stored histones H2a and H2b during embryogenesis [1]. Starting with proteomic analysis of purified lipid droplets, they identified a novel protein, Jabba, that is necessary to sequester the H2a/H2b dimers on the surface of the lipid droplet. It seems likely that the histones are targeted to lipid droplets immediately after synthesis and enter the oocyte already bound to lipid droplets. Strikingly histones H3 and H4 are not present on lipid droplets but are stored in soluble form in a complex with a histone chaperone.
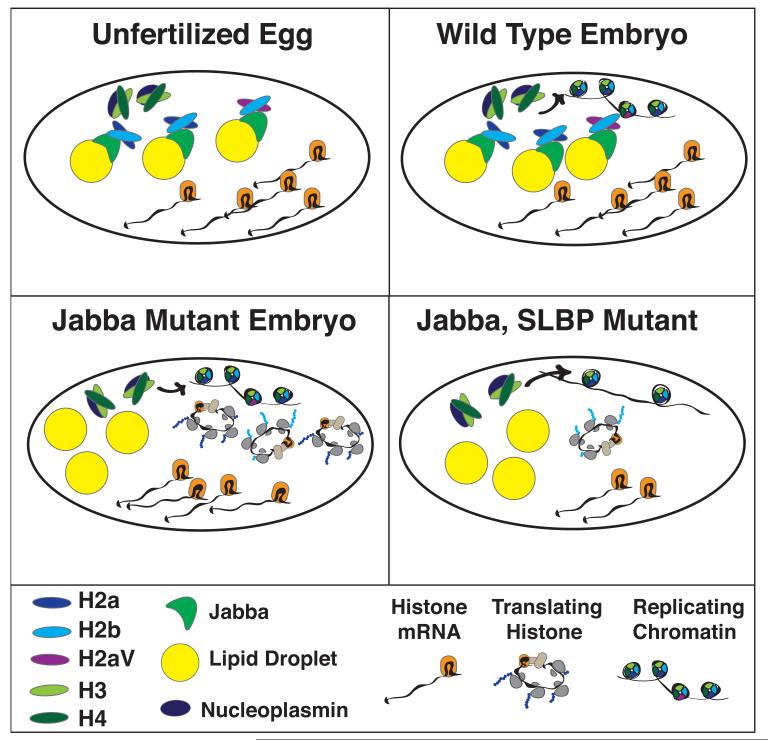
Analysis of mutants in Jabba, together with SLBP mutants [15], reveals a sophisticated interplay of storage and synthesis of histones to ensure the proper amount of histone protein for chromatin assembly until the zygotic histone genes are activated (Fig. 1). Strikingly, in the absence of Jabba there is no stored histone H2a/H2b protein in the eggs or early embryos. This is almost certainly due to degradation of the H2a and H2b proteins in the absence of Jabba. Because null mutants in Jabba are viable and fertile, the Jabba mutants must be able to synthesize sufficient histone H2a and H2b protein from maternal stores of histone mRNA. Genetic analysis reports a synthetic lethality between mutants in which the stored histone mRNA is mildly reduced and Jabba null mutants. Genetically reducing SLBP function ~50% in the Jabba mutant is lethal, as a consequence of reducd storage of histone mRNA and possibly also reduced translation of stored histone mRNA. Similar results were obtained with a reduction in the dosage of the essential histone variant H2aV, which is also present in lipid droplets in early embryos and dependent on Jabba vor storage in these organelles, presumably by forming H2b/H2aV heterodimers.
Fig. 1.
Utilization of stored histone proteins and mRNAs in Drosophila embryogenesis. A. Unfertilized eggs there are stored histone proteins - H3 and H4 bound to a nucleoplasmin-like histone chaperone, and H2a/H2b bound with Jabba in lipid droplets, as well as stored histone mRNAs. B. In the wild-type embryos most of the histone incorporated into chromatin come from the stored histone proteins. C. In Jabba mutant embryos, there is no stored histone H2a/H2b and these proteins are synthesized from the stored maternal histone mRNAs. D. In Jabba, SLBP mutant embryos where there is a slightly reduced (~50%) amount of stored histone mRNAs there are not sufficient histone H2a and H2b synthesized for appropriate assembly of chromatin and the embryos die.
Thus, Jabba is critical for the maintenance of the pool of H2a/H2b proteins for assembly into chromatin during embryogenesis. The amount of stored histone protein in the normal egg suggests that there is little, if any, need for the synthesis of histone proteins from maternal mRNAs. However the fact that Jabba mutants are viable and contain no stored histone H2a and H2b, demonstrates that there is sufficient maternal histone mRNA, including H2aV histone mRNA, to provide the histone protein necessary during embryonic development, and that these mRNAs are translated at a high rate in the absence of Jabba. It seems unlikely, although possible, that these histone mRNAs are normally translated at high rates and the histone protein rapidly degraded. More likely there is cross-talk between the stored histone pool and the demand for histones in chromatin to ensure that the proper amounts of histone protein are provided, since the amount of stored histone mRNA remains constant and the demand for new histone is increasing exponentially. In the Jabba mutants with a slightly reduced capacity for histone synthesis, this system cannot keep up with the demand for histones H2a and H2b, resulting in death of the embryo before activation of transcription from the zygotic genome can provide additional histone mRNA.
Lipid droplets were initially thought to interact primarily with proteins involved in lipid metabolism but they also are storage depots for a number of other proteins in the embryo [14], and they are bound to different sets of proteins in different cells [16]. There are reports of lipid droplets containing histones in other some proteomic studies [17] although the function, if any, of these extranuclear histones is not known. They could potentially form an additional pool of histone proteins in the cytoplasm that could other than lipid metabolism, and more of these are likely to be discovered in the future.
References
- 1.Li Z, Thiel K, Thul PJ, Beller M, Kuhnlein RP, Welte MA. Lipid droplets control the maternal histone supply of Drosophila embryos. 2012. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanchez R, Marzluff WF. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell. Biol. 2002;22:7093–7104. doi: 10.1128/MCB.22.20.7093-7104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorgoni B, Andrews S, Schaller A, Schumperli D, Gray NK, Muller B. The stem-loop binding protein stimulates histone translation at an early step in the initiation pathway. RNA. 2005;11:1030–1042. doi: 10.1261/rna.7281305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnold DR, Francon P, Zhang J, Martin K, Clarke HJ. Stem-loop binding protein expressed in growing oocytes is required for accumulation of mRNAs encoding histones H3 and H4 and for early embryonic development in the mouse. Dev. Biol. 2008;313:347–358. doi: 10.1016/j.ydbio.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodland HR, Adamson ED. The synthesis and storage of histones during oogenesis of Xenopus laevis. Dev. Biol. 1977;57:118–135. doi: 10.1016/0012-1606(77)90359-1. [DOI] [PubMed] [Google Scholar]
- 7.Sánchez R, Marzluff WF. The oligoA tail on histone mRNA plays an active role in translational silencing of histone mRNA during Xenopus oogenesis. Mol. Cell. Biol. 2004:2513–2525. doi: 10.1128/MCB.24.6.2513-2525.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dilworth SM, Black SJ, Laskey RA. Two complexes that contain histones are required for nucleosome assembly in vitro: role of nucleoplasmin and N1 in Xenopus egg extracts. Cell. 1987;51:1009–1018. doi: 10.1016/0092-8674(87)90587-3. [DOI] [PubMed] [Google Scholar]
- 9.Ruddell A, Jacobs-Lorena M. Biphasic pattern of histone gene expression during Drosophila oogenesis. Proc. Natl. Acad. Sci. USA. 1985;82:3316–3319. doi: 10.1073/pnas.82.10.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ambrosio L, Schedl P. Two discrete modes of histone gene expression during oogenesis in Drosophila melanogaster. Dev. Biol. 1985;111:220–231. doi: 10.1016/0012-1606(85)90447-6. [DOI] [PubMed] [Google Scholar]
- 11.Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- 12.Singh RK, Paik J, Gunjan A. Generation and management of excess histones during the cell cycle. Front Biosci. 2009;14:3145–3158. doi: 10.2741/3441. [DOI] [PubMed] [Google Scholar]
- 13.Bonner WM, Wu RS, Panusz HT, Muneses C. Kinetics of accumulation and depletion of soluble newly synthesized histone in the reciprocal regulation of histone and DNA synthesis. Biochemistry. 1988;27:6542–6550. doi: 10.1021/bi00417a052. [DOI] [PubMed] [Google Scholar]
- 14.Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- 15.Sullivan E, Santiago C, Parker ED, Dominski Z, Yang X, Lanzotti DJ, Ingledue TC, Marzluff WF, Duronio RJ. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 2001;15:173–187. doi: 10.1101/gad.862801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodges BD, Wu CC. Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J. Lipid Res. 2010;51:262–273. doi: 10.1194/jlr.R003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welte MA. Proteins under new management: lipid droplets deliver. Trends Cell Biol. 2007;17:363–369. doi: 10.1016/j.tcb.2007.06.004. [DOI] [PubMed] [Google Scholar]