Abstract
We isolated active mutants in Saccharomyces cerevisiae DNA polymerase α that were associated with a defect in error discrimination. Among them, L868F DNA polymerase α has a spontaneous error frequency of 3 in 100 nucleotides and 570-fold lower replication fidelity than wild-type (WT) polymerase α. In vivo, mutant DNA polymerases confer a mutator phenotype and are synergistic with msh2 or msh6, suggesting that DNA polymerase α-dependent replication errors are recognized and repaired by mismatch repair. In vitro, L868F DNA polymerase α catalyzes efficient bypass of a cis-syn cyclobutane pyrimidine dimer, extending the 3′ T 26,000-fold more efficiently than the WT. Phe34 is equivalent to residue Leu868 in translesion DNA polymerase η, and the F34L mutant of S. cerevisiae DNA polymerase η has reduced translesion DNA synthesis activity in vitro. These data suggest that high-fidelity DNA synthesis by DNA polymerase α is required for genomic stability in yeast. The data also suggest that the phenylalanine and leucine residues in translesion and replicative DNA polymerases, respectively, might have played a role in the functional evolution of these enzyme classes.
In all organisms with a DNA genome, the genome is replicated by DNA polymerases prior to cell division and one copy is transmitted to each daughter cell. In eukaryotic cells, the DNA polymerase α (pol α)-DNA primase complex initiates DNA replication; DNA primase synthesizes a short RNA chain, and pol α extends the RNA primer by ∼30 deoxyribonucleotides. pol ɛ and pol δ continue DNA synthesis in a processive manner and complete the elongation stage of DNA replication (60).
To ensure the survival of each daughter cell, DNA replication must be efficient and accurate. The insertion of the correct nucleotide depends in part on Watson-Crick base pair formation, but this mechanism is not sufficient to obtain high-fidelity DNA replication (23, 30). The observed high fidelity of chromosomal replication is achieved through mechanisms that enhance correct nucleotide insertion and that correct errors after they occur. Eukaryotic pol δ and pol ɛ remove misincorporated nucleotides with an intrinsic 3′-5′ exonuclease activity (37, 38). Some evidence supports the idea of a physical and functional interaction between mismatch repair (MMR) proteins and PCNA, which is an accessory protein of pol δ and pol ɛ (1, 6, 18, 59). Furthermore, polymerase-associated exonucleases may participate in the same pathway that involves MMR (38, 39, 56). Disruption of this proofreading-MMR pathway causes types of cancers in both humans and mice (13, 14, 17).
Little is known about error prevention and correction mechanisms that enhance the fidelity of DNA pol α. pol α lacks 3′-5′ exonuclease activity (36, 46), but it discriminates between correct and incorrect nucleotides during nucleotide incorporation and extension. Mutant polymerase characterization has provided some insight into the mechanism by which nucleotide insertion fidelity is achieved (7, 26, 41). However, it would be desirable to have a relevant experimental system with which one could ask whether error prevention by pol α has any significance at the cellular level. There have been a few pioneer works that addressed the relationship between genomic stability and pol α (20, 29, 45). Unlike the results seen with elongation DNA polymerases of pol δ and pol ɛ, however, a direct relationship between replication errors and genomic stability has not been established yet.
In this work, we have isolated pol α mutants with amino acid substitutions at the conserved palm residue Leu868. The mutants are viable and have highly active DNA polymerases but extremely low fidelity. In translesion DNA polymerases, unexpectedly, the residue equivalent to Leu868 is phenylalanine, suggesting that the intrinsic low fidelity and translesion activity of translesion DNA polymerases are related to the presence of phenylalanine at this amino acid position.
MATERIALS AND METHODS
Strain and plasmid construction.
A plasmid encoding Saccharomyces cerevisiae DNA pol α and a yeast strain (SIII pol1-17) were generously provided by Hiroyuki Araki at the National Institute of Genetics, Mishima, Japan. Human pol α cDNA was kindly provided by Heinz-Peter Nasheuer at Institut für Molekulare Biotechnologie, Jena, Germany. Yeast strain CG379 (MATα ade5 his7-2 leu2-3 leu2-112 trp1-289 ura3-52) was provided by Akihiko Kikuchi (Nagoya University) and was used to isolate pol α mutants. The coding region of S. cerevisiae pol η was isolated by PCR using genomic DNA and the following primers: rad30/NcoI/F (5′-GCATGTCAAAATTTACTTGGAAGGA) and rad30/HindIII/R (5′-TGAAGCCATATAATTGTCTATTTGG). The PCR product was digested with NcoI and HindIII and ligated into pFastBacHTb (Invitrogen, Carlsbad, Calif.).
Library construction.
pol α mutants were isolated from a library of palm mutants as described previously (21, 41, 53). Mutants were generated by randomization of the motif A region during synthesis of an oligonucleotide cassette. Mutant cassettes were inserted into the pol α gene through the use of a unique SacII restriction site downstream of the motif A region. The SacII restriction site was generated using a GeneEditor in vitro site-directed mutagenesis kit (Promega Inc., Madison, Wis.) and the following oligonucleotide (silent mutations introducing the SacII site are underlined): +SacII, 5′-T CAG GGT GTT TTA CCG CGG TTA TTA GCT CTG. Motif A cassettes were synthesized by PCR using two template DNA molecules, ypol13246-3337m/F and ypol13395-3321/R, whose DNA sequences are as follows: for ypol13246-3337m/F, 5′-CTTCATAAGAATTAC[GTTTTAGTCATGGACTTTAATTCTTTGTATCCATCTATTATCCAGGAATTTAATATATGT]TTTACCACCGTTGATAG); for ypol13395-3321/R, 5′-ACCCTGATCCACCTCTGAAGGAGGTACGCTTGGTAACTCATCGATATCTTCCTTATTTCTATCAACGGTGGTAAA). Nucleotides in brackets were randomized by including 94% wild-type (WT) nucleotide and 2% each of the other three nucleotides during synthesis. The PCR primers were ypol13216F and ypol13425R, with the sequences 5′-GGTGGTTTAGTTTTTGAACCTGAGAAAGGTCTTCATAAGAATTAC and 5′-CACCAGATTAGCTAATAACCGCGGTAAAACACCCTGATCCACCTC, respectively. The plasmid vector and PCR products were digested with EcoNI and SacII, purified, ligated into the vector, and transformed into DH5α cells (41). An aliquot of the library was plated on a Luria-Bertani agar plate containing 100 μg of ampicillin/ml, and the remainder was grown in 5 ml of liquid culture medium. Cells were cultured for 14 h and plated to estimate the library size (∼20,000 independent clones). Forty clones were analyzed to confirm proper construction. Plasmid DNA was purified from the liquid culture and transformed into SIII pol1-17 yeast. Transformants were grown in cultures on SD-URA plates (6.7 g of yeast nitrogen base w/o amino acids/liter, 5 g of Casamino Acids/liter, 20 g of glucose/liter, 20 mg of adenine sulfate/liter, 20 mg of tryptophan/liter, 15 g of Bacto agar/liter) at 27°C for 48 h. Colonies were replica plated to fresh plates and grown in cultures at 37°C for 72 h. Plasmids were isolated as described previously (41).
Construction of F34L pol η.
An S. cerevisiae pol η F34L mutant was constructed by PCR as follows: PCR was carried out with primers rad30/NcoI/F and rad30FL/R (5′-CTGCTCAACCTGTGCAAACAAGGCATTCATATCTATG) and with primers rad30/105F (5′-TGCACAGGTTGAGCAG) and rad30/HindIII/R. The two reaction products were purified, mixed, and reamplified using primers rad30/NcoI/F and rad30/HindIII/R. Amplified fragments were digested with NcoI and HindIII and ligated into pFastBacHTb.
Enzyme overexpression and purification.
His6-tagged WT and mutant pol α and pol η were expressed in insect cells and purified using a BAC-TO-BAC HT baculovirus expression system (Invitrogen) as described previously (41). The purity of the proteins was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Protein concentrations were determined using the Bradford dye-binding method (Bio-Rad, Hercules, Calif.). Specific activity was determined under the following conditions: 200 μg of activated calf thymus DNA/ml, 100 μM (each) dATP, dGTP, dCTP, and dTTP, 0.2 μCi of [3H]dTTP in 50 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 50 mM KCl, 2 mM dithiothreitol, and an aliquot of enzyme fraction were used in a final volume of 25 μl; reaction mixtures were incubated at 37°C for 30 min; and acid-insoluble radioactivity levels were measured. One unit of polymerase activity is the amount of protein that incorporates 1 nmol of dNMP per h.
Primer extension assay.
The primer extension assay was carried out using a previously described method (52). A 16-mer DNA primer (5′-CACTGACTGTATGATG) was 32P labeled at the 5′ end with T4 polynucleotide kinase and [γ-32P]ATP and annealed with a 1.2-fold molar excess of complementary 30-mer with or without a centrally located cyclobutane pyrimidine dimer (CPD) lesion. The 30-mer sequence is 5′-CTCGTCAGCATCTTCATCATACAGTCAGTG), where the underlined TT is a cis-syn CPD. For an abasic template, the 3′ T of the underlined TT is replaced with the abasic analog (32). The labeled template primer was used at a final concentration of 4 nM. Reaction mixtures contained 50 mM Tris-HCl, (pH 8.0), 2 mM dithiothreitol, 50 mM KCl, 5 mM MgCl2, 100 ng of bovine serum albumin/ml, and the indicated amount of enzyme in a final volume of 5 μl. The reaction was carried out at 37°C for 1 h and terminated by addition of an equal amount of 2× loading buffer containing 90% formamide, 20 mM EDTA, 0.05% xylene cyanol, and 0.05% bromophenol blue. Reaction products were separated using a 14% polyacrylamide gel containing 8 M urea and visualized by exposing the gel to X-ray film.
Single-nucleotide-incorporation kinetics.
Incorporation efficiency levels for correct and incorrect nucleotides were measured essentially as described previously (15, 26). Each template position is indicated in Fig. 1. 32P-labeled 16-mer (5′-CACTGACTGTATGATG), 17-mer (5′-CACTGACTGTATGATGA), or 18-mer (5′-CACTGACTGTATGATGAA) primer was annealed to each 30-mer template with the sequences described above. The 32P-labeled template primer (40 nM) was incubated for 5 min at 37°C in a reaction mixture (5 μl) containing 100 mM Tris-HCl (pH 8.0), 2 mM dithiothreitol, 50 mM KCl, 5 mM MgCl2, 100 ng of bovine serum albumin/ml, various concentrations of dNTP, and an amount of enzyme that gave less than 20% template primer utilization. To obtain this, enzyme amounts were adjusted and monitored for the optimum concentrations, which depend on the primer template and substrate combinations (i.e., 0.2 to 6 nM S. cerevisiae pol η WT, 1 to 370 nM S. cerevisiae pol η F34L, 1 to 300 nM S. cerevisiae pol α WT, 0.2 to 2 nM S. cerevisiae pol α L868F, 1 to 200 nM human pol α WT, and 1 to 2 nM human pol α L864F). Data were analyzed on Hanes-Woolf plots, and apparent Km and kcat values (averaged from three measurements) were calculated from the graphs.
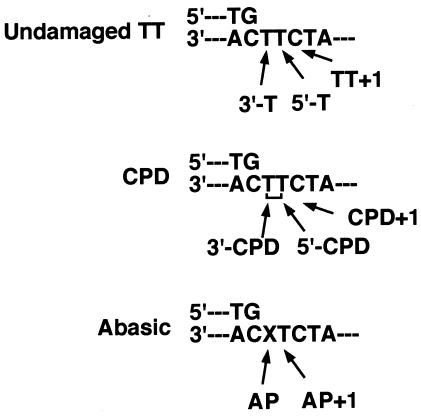
FIG. 1.
Each template site used for the kinetic analysis is schematically indicated.
In vitro forward mutation assay.
Replication errors were quantified using a M13mp2 forward mutation assay. Gapped M13mp2 substrate was constructed with a single-stranded gap in the lacZα complementation region. The reaction mixture contained 5 U of WT or mutant pol α, 400 ng of gapped M13mp2, 20 mM Tris-HCl (pH 8.0), 2 mM dithiothreitol, 10 mM MgCl2, 50 mM KCl, and 0.2 mM dNTPs. Reactions were incubated at 37°C for 10 min, and the reaction product (double-stranded phage DNA) was transfected into Escherichia coli. Mutant plaques were identified by their color. The complete gap-filling reaction was monitored by agarose gel electrophoresis of the reaction products. The background mutation rate (0.87 × 10−3) was subtracted prior to calculating error frequencies (54, 55). Nucleotide sequences of mutants were determined using an oligonucleotide primer with the DNA sequence 5′-CAGCCAGCTTTCCGGCA. The polymerase activity of pol η F34L was not sufficiently high to yield reliable data with this assay.
Generation of pol α mutant strains.
Plasmids carrying mutant alleles of S. cerevisiae pol α were digested with BamHI and ligated into Ycplac211. Ycplac211 derivatives were linearized with BspEI and transformed into CG379 yeast. Transformants were grown on SD-URA plates at 27°C for 72 h. Colonies were selected randomly and grown in yeast extract-peptone-dextrose (YEPD) liquid medium at 27°C overnight. Cells were collected and plated on SD containing 5′-fluoroorotic acid. Mutant strains were identified by PCR amplification and sequencing of the target region with primers confR2 (5′-ACCTTGACTAAACAGTTGAC) and SBR (5′-GTCGATCAACAGAACATAAG).
ΔMSH2, ΔMSH3, and ΔMSH6 strains were constructed by transformation with a PCR product in which one of the DNA sequence was interrupted by a URA3 marker gene, and the PCR product was generated using Ycplac33 plasmid as a URA3 source and forward and reverse primers of ΔMSH2 (5′-CAAGGTGGGAATGCTTGATATTGTCGATAATGAAGTGTATTCCAACCTAGTTAGTTTTGCTGGCCGCA and 5′-CGTGATGTCCTCATCGTCATGTTTTTGTTCTTTTAAATTTTTCTCGATATCCCTTTCGTCTTCAAGAA), ΔMSH3 (5′-GGTGAAGGACCTGAAAATGCATCATAGAGATAAAGTGCTTGTTATTAGAGTTAGTTTTGCTGGCCGCA and 5′-CGTTTGAGATATTTTCGTATTTCTGCCAATTCTTCCTTTAACTGTGACCGCCCTTTCGTCTTCAAGAA), and ΔMSH6 (5′-AAGCGATGAGGATGAATATTTACCAGATAAAAATGACGGCGATGAAGATGTTAGTTTTGCTGGCCGCA and 5′-AGGGAAAGAGGATATGTATTTTGAAACATCTCCCTTCAAATCGTTGTTCTCCCTTTCGTCTTCAAGAA), respectively. Transformed cells were grown on solid medium in the absence of uracil.
trp1 and his7 reversion assays.
The trp1 reversion assay and measurement of mutation rates were described previously (5, 31, 41). The mutation spectrum in the trp1 gene was analyzed using PCR-amplified DNA and primers Sctrp1(+)150 (5′-GAAAGAGAACAATTGACCCG) and Sctrp1(−)607 (5′-ACACCATTTGTCTCCACACC). The his7 reversion assay was performed in the same manner except that cells were plated on SD in the absence of histidine.
CAN1 forward mutation assay.
Strains were grown in YEPD liquid medium at 27°C for 24 h. Cells were collected and washed with water. An aliquot was plated on YEPD at approximately 103 cells/plate, and the remaining culture was plated on SD containing 60 mg of l-canavanine sulfate/liter. Plates were incubated for 4 days at 27°C, and the number of canavanine-resistant colonies was counted. The mutation spectrum in the can1 gene was analyzed using PCR-amplified DNA and primers can1−80F (5′-TGTCGTCAATCGAAAGTTTA) and can1+1840R (5′-TATGACTTATGAGGGTGAGA). Using most of the chromosome DNA from the mutant clones, we were able to recover the PCR fragment for sequencing. However, PCR products were not obtained from nine WT and eight L868F mutant clones and were not included in the Can1 forward mutation assay.
RESULTS
Screening pol α mutants.
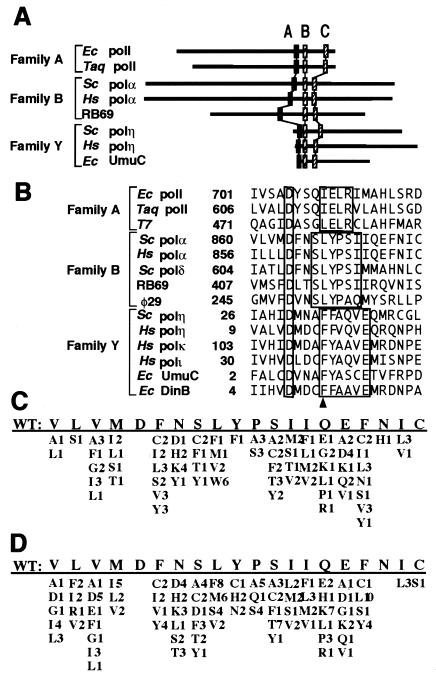
DNA polymerases from many different species have a structurally conserved sequence in the palm region (2, 4, 43, 50) (Fig. 2A). In this study, S. cerevisiae DNA pol α mutants with altered amino acids were isolated using a genetic complementation assay and the temperature-sensitive strain pol1-17. Active pol α mutants were identified with single or multiple amino acid substitutions (Fig. 2C and D). These mutants were rescreened by growing cells in the absence of tryptophan such that cell growth required reversion or suppression of a trp1 amber codon. The mutation rates for most of the mutants fell into a range of values 0.6- to 2.5-fold higher than those for the WT (data not shown). The experiments reported here were used to examine the properties of pol α L868F, whose spontaneous mutation rate for the pol1-17 strain was 8.7-fold higher than that of WT pol α.
FIG. 2.
DNA polymerase motif A. (A) Conserved motifs A, B, and C are illustrated schematically. (B) Amino acid sequences of family A, family B, and family Y DNA polymerases are compared. Conserved amino acids are boxed. Phe34 in S. cerevisiae pol η is indicated by an arrowhead. Sc, S. cerevisiae; Hs, Homo sapiens; RB69, bacteriophage RB69; φ29, bacteriophage φ29; Ec, E. coli; Taq, Thermus aquaticus; T7, bacteriophage T7. (C) Mutant pol α with single substitutions, which are shown under the WT sequence. Amino acid substitutions in active mutants are shown, and the number of isolates is indicated. Amino acid positions of the WT sequence are 860 to 879. Mutations at catalytically essential Asp864 were not obtained. (D) Mutant pol α with multiple substitutions is indicated.
Fidelity of S. cerevisiae L868F pol α.
In experiments designed to discover whether inaccurate DNA synthesis caused the mutator phenotype, WT pol α and Leu868 mutants carrying Phe, Met, Trp, or Val substitutions were purified and characterized. The specific activity of these mutants was similar to that of WT pol α (Table 1), but their in vitro mutation rate was significantly higher than that of the WT. In a lacZα forward mutation assay, WT pol α had a mutation frequency of 3.4 × 10−3 and L868F pol α had a 138-fold-higher mutation frequency (470 × 10−3) (Table 1).
TABLE 1.
Error frequencies for WT and mutant pol α in a lacZα forward mutation assay
| DNA pol α | Specific activitya (%) | Plaque score
|
Mutation frequencyb (10−3) | |
|---|---|---|---|---|
| Total no. of plaques | No. of White plaques | |||
| S. cerevisiae WT | 100 | 31,289 | 135 | 3.44 |
| S. cerevisiae L868F | 113 | 7,412 | 3,489 | 470 |
| S. cerevisiae L868M | 95.0 | 10,115 | 1,298 | 127 |
| S. cerevisiae L868W | 85.9 | 9,995 | 271 | 26.2 |
| S. cerevisiae L868V | 40.1 | 10,103 | 173 | 16.3 |
| H. sapiens WT | 100 | 19,561 | 106 | 4.55 |
| H. sapiens L864F | 73.0 | 9,219 | 2,748 | 297 |
Specific activity of each polymerase was determined and is indicated as a value relative to that for WT pol α (Materials and Methods).
Mutation frequencies of WT and mutant enzymes were determined by plaque color. The background frequency (0.87 × 10−3) for this method was subtracted for each data point.
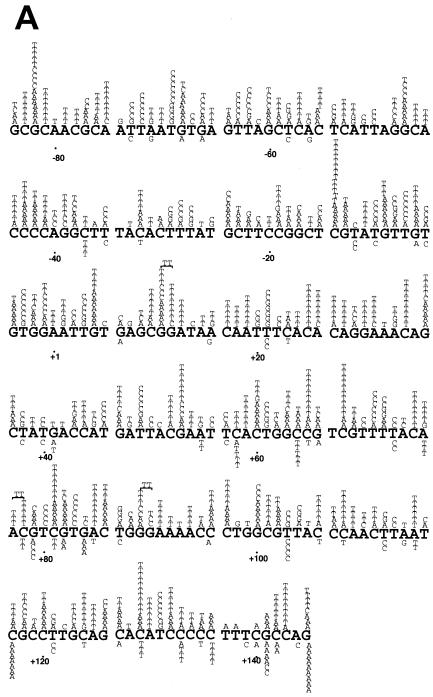
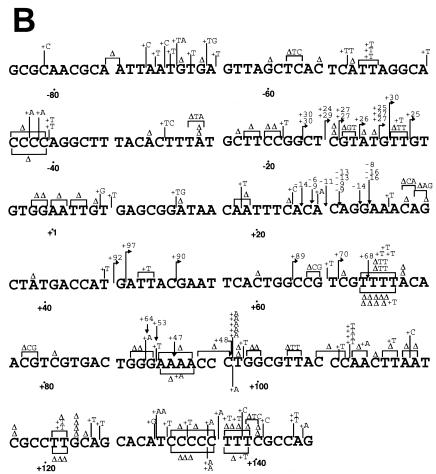
The mutation spectra of L868F mutant and WT pol α confirmed the observed difference in mutant frequencies. In the mutant phages, WT pol α made approximately one mutation in the target LacZα sequence; in contrast, L868F pol α made an average of nine errors in the same target sequence. Calculating on the basis of 1,234 mutations, the spontaneous error rate of L868F pol α was estimated to be 3 per 100 nucleotides, 570-fold higher than the results seen with the WT (Table 2). The spontaneous errors exhibited the following characteristics. (i) Base substitution errors made by L868F pol α were randomly distributed throughout the target DNA sequence and not clustered in “hot spots” as were those made by WT pol α (Fig. 3A). (ii) In vitro, L868F pol α generated 43% transversion mutations and 44% transition mutations whereas WT pol α generated 67% transitions and 13% transversions (Table 2). (iii) The relative frequencies of L868F pol α base substitution errors were C→T (21%) > A→T (12%) > G→T (7.9%), and the frequency of N→T substitutions was 41%. Thus, the rate of dAMP misincorporation was considerably higher than that of misincorporation of other nucleotides (Table 2). (iv) L868F pol α also generated frameshift errors, with a 930-fold-higher rate of insertions and a 320-fold-higher rate of deletions than WT pol α (Table 2). (v) WT pol α preferentially made single-base deletions at run sequences (18/18), while more than half (36/64) of the single-base deletions made by L868F pol α were in nonrun sequence. (vi) L868F pol α generated tandem base deletions as well as large deletions of 30 to 50 nucleotides.
TABLE 2.
Types of errors by S. cerevisiae WT and L868F pol α in a lacZα forward mutation assay
| Error type | WT
|
L868F
|
Fold increase (L868F/WT) | ||||
|---|---|---|---|---|---|---|---|
| n | % | Error ratea (10−5) | n | % | Error rateb (10−5) | ||
| All errors | 144 | 100 | 5.4 | 1,234 | 100 | 3,100 | 570 |
| Base substitutions | 115 | 80 | 4.6 | 1,073 | 87 | 2,700 | 590 |
| Transitions | 96 | 67 | 3.8 | 546 | 44 | 1,400 | 360 |
| T→C | 24 | 17 | 0.95 | 138 | 11 | 340 | 360 |
| C→T | 36 | 25 | 1.4 | 257 | 21 | 640 | 450 |
| A→G | 3 | 2.1 | 0.12 | 15 | 1.2 | 37 | 310 |
| G→A | 33 | 23 | 1.3 | 136 | 11 | 340 | 260 |
| Transversions | 19 | 13 | 0.75 | 527 | 43 | 1,300 | 1,700 |
| T→A | 0 | 0 | 0 | 33 | 2.7 | 82 | >2,100 |
| T→G | 1 | 0.69 | 0.040 | 48 | 3.9 | 120 | 3,000 |
| A→T | 5 | 3.5 | 0.20 | 152 | 12 | 380 | 1,900 |
| A→C | 1 | 0.69 | 0.040 | 10 | 0.81 | 25 | 630 |
| G→T | 1 | 0.69 | 0.040 | 98 | 7.9 | 240 | 6,200 |
| G→C | 5 | 3.5 | 0.20 | 66 | 5.3 | 160 | 830 |
| C→A | 6 | 4.2 | 0.24 | 116 | 9.4 | 290 | 1,200 |
| C→G | 0 | 0 | 0 | 4 | 0.32 | 10 | >250 |
| Frameshifts | 29 | 20 | 0.80 | 161 | 13 | 400 | 500 |
| 1-base deletions | 18 | 13 | 0.50 | 64 | 5.2 | 160 | 320 |
| 2-base deletions | 0 | 0 | 0 | 12 | 0.97 | 30 | >750 |
| ≥3-base deletions | 5 | 3.5 | 0.14 | 17 | 1.4 | 42 | 300 |
| 1-base insertions | 6 | 4.1 | 0.17 | 62 | 5.0 | 150 | 930 |
| 2-base insertions | 0 | 0 | 0 | 6 | 0.49 | 15 | >370 |
Error rates were determined as described previously (55). In the formula, rates were corrected by the probability of hitting silent sites. The mutational target includes 107 bases for base substitution and 154 for frameshift mutations.
M13 DNA contained multiple mutations (average, nine) that were found at both coding and silent sites. Thus, frequency values were not corrected by the probability of hitting silent sites and might be underestimated (55). Error frequencies were determined at 229 targets for both base substitution and frameshift mutations.
FIG. 3.
S. cerevisiae L868F DNA pol α mutation spectrum in M13mp2 lacZα. Mutation spectra were determined by sequencing 135 (WT pol α) and 137 (L868F pol α) mutant plaques. The M13mp2 DNA sequence is shown. Base substitutions (A) and frameshifts (B) produced by S. cerevisiae L868F are indicated above the M13mp2 sequence, and those produced by WT pol α are indicated below the sequence. The +1 and −1 frameshift mutations are indicated by Δ (deletion) and + (insertion), respectively. For a frameshift in a run sequence, the symbol is centered under the run. Endpoints of large deletions are indicated by one arrow pointing down and one arrow pointing up. The nucleotide position at the corresponding other endpoint is indicated at the arrow.
The overall mutation rates and the error specificities, but not the base substitution specificities, of pol α L868F and translesion DNA polymerase of pol η are remarkably similar (34, 35). Interestingly, WT pol η has a Phe at the position corresponding to L868 in pol α and Phe is conserved for Y family DNA polymerases whereas Leu/Ile is conserved in family A/B polymerases (Fig. 2A and B).
Translesion DNA synthesis activity.
Y family DNA polymerases are characterized by low-fidelity DNA synthesis and efficient bypass of template DNA damage (i.e., translesion DNA synthesis). This suggests that pol α Leu868 and pol η Phe34 in S. cerevisiae might influence both fidelity of replication and efficiency of lesion bypass. Thus, the lesion bypass efficiencies of F34L pol η and L868F pol α were compared on a DNA template with a CPD lesion. WT pol η synthesized DNA on damaged and undamaged template DNA with nearly equal levels of efficiency. In contrast, it seems that F34L pol η was less efficient on damaged than on undamaged template DNA (Fig. 4B). The opposite effect was observed for WT and L868F pol α: the mutant bypassed the CPD lesion more efficiently than the WT. In addition, the mutant bypassed an abasic site and 6-4 photoproduct with higher efficiency than the WT. Even at 10-fold excess, WT pol α incorporates nucleotides extremely poorly compared to the results seen with the 3′ T of CPD or 6-4 photoproduct (Fig. 4D). These results show that pol α L868F has an increased bypass ability and a different lesion bypass specificity compared to WT pol η, which efficiently bypasses the CPD lesion but not the 6-4 photoproduct or abasic site.
FIG. 4.
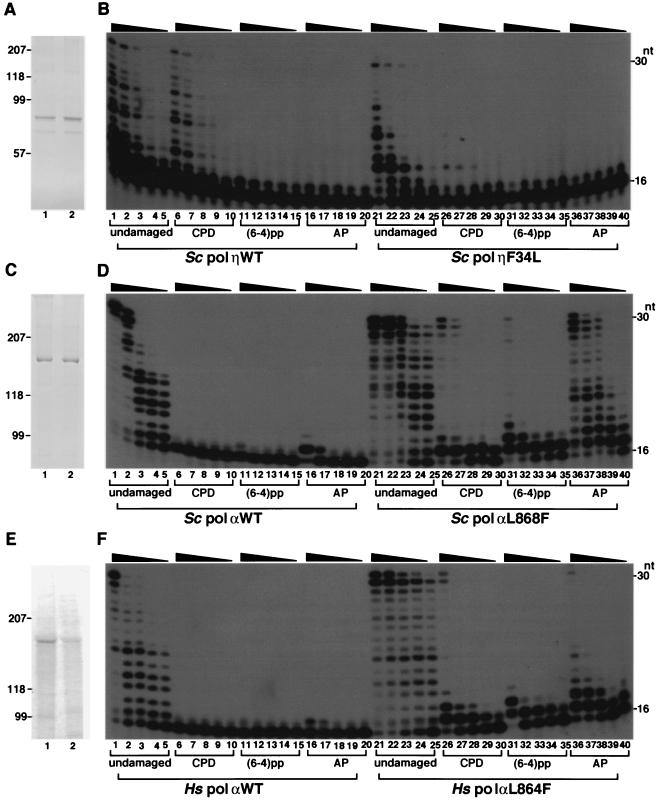
Translesion synthesis by pol η and α. Sodium dodecyl sulfate gel electrophoresis profiles for each enzyme preparation are shown in panels A (S. cerevisiae pol η WT and F34L), C (S. cerevisiae pol α WT and L868F), and E (human pol α WT and L864F). WT and mutant proteins were loaded onto lanes 1 and 2, respectively. Positions of protein standards (in kilodaltons) are indicated in the left. The second major bands shown in panel A were considered to be the degradation products of the major ones, because they were also stained by Western blotting (data not shown). For translesion DNA synthesis, enzymes were incubated with undamaged or damaged template as indicated. The primer terminus is properly base paired to the nucleotide adjacent to the first damaged base such that the damaged base is the first template site during primer extension. Enzymes were titrated to result in similar levels of primer extension efficiency for the WT and mutant strains on the undamaged template primer. (B) Reaction mixtures included pol η at the following concentrations in the indicated lanes: pol η WT at 3.5 (lanes 1, 6, 11, and 16), 1.8 (lanes 2, 7, 12, and 17), 0.9 (lanes 3, 8, 13, and 18), 0.44 (lanes 4, 9, 14, and 19), and 0.22 (lanes 5, 10, 15, and 20) nM; and pol η F34L at 440 (lanes 21, 26, 31, and 36), 220 (lanes 22, 27, 32, and 37), 110 (lanes 23, 28, 33, and 38), 52 (lanes 24, 29, 34, and 39), and 26 (lanes 25, 30, 35, and 40) nM. (D) Reactions were carried out in the presence of S. cerevisiae pol α at the following concentrations in the indicated lanes: WT pol α at 560 (lanes 1, 6, 11, and 16), 280 (lanes 2, 7, 12, and 17), 140 (lanes 3, 8, 13, and 18), 70 (lanes 4, 9, 14, and 19), and 35 (lanes 5, 10, 15, and 20) nM; and L868F pol α at 56 (lanes 21, 26, 31, and 36), 28 (lanes 22, 27, 32, and 37), 14 (lanes 23, 28, 33, and 38), 7 (lanes 24, 29, 34, and 39), and 3.5 (lanes 25, 30, 35, and 40) nM. (F) Reactions were carried out in the presence of human pol α at the following concentrations in the indicated lanes: WT and L864F pol α at 110 (lanes 1, 6, 11, 16, 21, 26, 31, and 36), 56 (lanes 2, 7, 12, 17, 22, 27, 32, and 37), 28 (lanes 3, 8, 13, 18, 23, 28, 33, and 38), 14 (lanes 4, 9, 14, 19, 24, 29, 34, and 39), and 7 (lanes 5, 10, 15, 20, 25, 30, 35, and 40) nM. Positions of primers and the full-length products are indicated as 16 and 30, respectively. Some reactions resulted in a 31-mer product that was 1 nucleotide longer than the template. This template-independent extra nucleotide forms a single-stranded DNA 3′ overhang, as observed for other proofreading-deficient polymerases.
To quantify how well mutant and WT polymerases utilize the CPD template, single-nucleotide-incorporation kinetics investigations were carried out. F34L pol η incorporated dAMP 770-fold less efficiently than WT pol η at an undamaged T and 3,900-fold less efficiently than the WT at the 3′ T of a CPD (Table 3). Thus, in pol η the mutation is primarily associated with the activity reduction, although F34L pol η discriminated between CPD and undamaged DNA 5.1-fold more efficiently than WT pol η, and it discriminated dGMP from dAMP 3.1-fold better at template T (Table 3).
TABLE 3.
Fidelity of WT and mutant pol η on TT dimer by steady-state kinetics
| Enzyme | Template:dNTPa | Km (μM) | kcat (min−1) | kcat/Km (μM−1 min−1) | fincb |
|---|---|---|---|---|---|
| S. cerevisiae pol η WT | 3′-T:dATP | 2.1 ± 1.3 | 7.5 ± 0.42 | 4.4 ± 1.9 | 1 |
| 3′-T:dGTP | 130 ± 14 | 2.3 ± 0.16 | 0.018 ± 0.0029 | 4.0 × 10−3 | |
| 3′-T:dCTP | 330 ± 160 | 0.37 ± 0.13 | (1.2 ± 0.15) × 10−3 | 2.6 × 10−4 | |
| 3′-T:dTTP | 210 ± 83 | 1.8 ± 0.16 | (9.8 ± 5.5) × 10−3 | 2.2 × 10−3 | |
| 3′-CPD:dATP | 6.0 ± 2.3 | 5.7 ± 0.76 | 1.1 ± 0.46 | 0.24 | |
| S. cerevisiae pol η F34L | 3′-T:dATP | 460 ± 43 | 2.6 ± 0.30 | (5.7 ± 0.14) × 10−3 | 1 |
| 3′-T:dGTP | 1,100 ± 150 | (7.5 ± 0.80) × 10−3 | (7.1 ± 0.53) × 10−6 | 1.3 × 10−3 | |
| 3′-T:dCTP | NDc | ND | ND | ND | |
| 3′-T:dTTP | ND | ND | ND | ND | |
| 3′-CPD:dATP | 600 ± 140 | 0.16 ± 0.023 | (2.8 ± 0.33) × 10−4 | 0.048 |
Template and dNTP represent template base and incoming dNTP, respectively. The name of each template site is schematically illustrated in Fig. 1.
finc, kcat/Km value relative to that for T:dATP (which was set at 1.0).
ND, incorporation was below the detection limit and thus was not detected.
The apparent kcat/Km values for L868F pol α were 30- and 26,000-fold higher at the undamaged T and 3′ T of the CPD, respectively, than those seen for WT pol α (Table 4). Therefore, the discrimination factors at a CPD differ by 870-fold between L868F and WT pol α. In pol α, the L868F mutation dramatically increases the efficiency of nucleotide incorporation at the CPD lesion but has relatively little effect on polymerase activity on undamaged DNA.
TABLE 4.
Fidelity of WT and mutant pol α on TT dimer and abasic sites by steady-state kinetics
| Enzyme | Template:dNTPa | Km (μM) | kcat (min−1) | kcat/Km (μM−1 min−1) | fincb |
|---|---|---|---|---|---|
| S. cerevisiae pol α WT | 3′-T:dATP | 6.0 ± 0.14 | 0.97 ± 0.16 | 0.16 ± 0.24 | 1 |
| 3′-T:dGTP | 2,200 ± 360 | 0.015 ± 0.0021 | (6.8 ± 0.83) × 10−6 | 4.3 × 10−5 | |
| 3′-T:dCTP | 2,500 ± 410 | (6.9 ± 0.96) × 10−3 | (2.8 ± 0.84) × 10−6 | 1.7 × 10−5 | |
| 3′-T:dTTP | 1,700 ± 550 | (4.7 ± 0.30) × 10−3 | (2.6 ± 0.81) × 10−6 | 1.6 × 10−5 | |
| 3′-CPD:dATP | 1,100 ± 420 | (1.1 ± 0.75) × 10−3 | (1.1 ± 0.63) × 10−6 | 6.8 × 10−6 | |
| 3′-CPD:dGTP | 2,900 ± 420 | (1.0 ± 0.14) × 10−3 | (3.3 ± 0.0) × 10−7 | 2.1 × 10−6 | |
| 3′-CPD:dCTP | NDc | ND | ND | ND | |
| 3′-CPD:dTTP | 1,400 ± 450 | (6.0 ± 0.43) × 10−4 | (4.4 ± 0.96) × 10−7 | 2.8 × 10−6 | |
| AP:dATP | 76 ± 9.0 | 2.1 ± 0.28 | 0.028 ± 0.001 | 0.17 | |
| S. cerevisiae pol α L868F | 3′-T:dATP | 1.3 ± 0.55 | 6.5 ± 3.0 | 4.8 ± 0.39 | 1 |
| 3′-T:dGTP | 16 ± 3.0 | 2.3 ± 1.1 | 0.15 ± 0.077 | 0.031 | |
| 3′-T:dCTP | 59 ± 2.7 | 3.0 ± 0.022 | 0.050 ± 0.0019 | 0.011 | |
| 3′-T:dTTP | 52 ± 9.6 | 5.8 ± 1.4 | 0.12 ± 0.031 | 0.024 | |
| 3′-CPD:dATP | 160 ± 37 | 4.4 ± 1.1 | 0.029 ± 0.0092 | 6.1 × 10−3 | |
| 3′-CPD:dGTP | 280 ± 110 | 0.80 ± 0.17 | (3.0 ± 0.56) × 10−3 | 6.3 × 10−4 | |
| 3′-CPD:dCTP | 670 ± 160 | 0.30 ± 0.042 | (4.7 ± 0.56) × 10−4 | 9.7 × 10−5 | |
| 3′-CPD:dTTP | 1,400 ± 290 | 1.4 ± 0.51 | (1.0 ± 0.50) × 10−3 | 2.2 × 10−4 | |
| 3′-CPD:dPTP | 0.22 ± 0.011 | 1.4 ± 0.067 | 6.2 ± 0.26 | 1.3 | |
| AP:dATP | 1.7 ± 0.57 | 8.4 ± 2.4 | 5.2 ± 0.95 | 1.1 | |
| AP:dGTP | 8.7 ± 3.2 | 3.5 ± 0.85 | 0.41 ± 0.049 | 0.086 | |
| AP:dCTP | 68 ± 19 | 2.8 ± 0.32 | 0.043 ± 0.0080 | 9.0 × 10−3 | |
| AP:dTTP | 18 ± 2.0 | 2.6 ± 0.57 | 0.15 ± 0.024 | 0.031 | |
| AP:dPTP | 0.066 ± 0.0059 | 2.9 ± 0.23 | 45 ± 6.6 | 9.4 | |
| H. sapiens pol α WT | 3′-T:dATP | 4.4 ± 0.33 | 1.4 ± 0.23 | 0.32 ± 0.031 | 1 |
| 3′-CPD:dATP | 170 ± 50 | (5.0 ± 0.75) × 10−3 | (3.2 ± 1.3) × 10−5 | 1.0 × 10−4 | |
| H. sapiens pol α L864F | 3′-T:dATP | 2.5 ± 0.65 | 1.5 ± 0.30 | 0.60 ± 0.13 | 1 |
| 3′-CPD:dATP | 200 ± 43 | 0.87 ± 0.15 | (4.6 ± 1.3) × 10−3 | 7.6 × 10−3 |
Kinetic analysis was also carried out for L868F pol α at other damaged and undamaged template sites. L868F pol α incorporated dAMP with similar levels of efficiency at the 5′ T and 3′ T of CPD (Table 4 and Table 5). The 5′-adjacent template C neighboring the CPD (designated CPD + 1) was utilized as efficiently as other undamaged template residues (Table 5). Surprisingly, L868F incorporated dAMP opposite an abasic site or an undamaged template nucleotide with similar levels of efficiency (Table 4). Furthermore, L868F pol α incorporated the pyrene nucleotide dPTP (33, 51) at the 3′ T of CPD as efficiently as it incorporated dAMP at an undamaged T (Table 4).
TABLE 5.
Efficiency of S. cerevisiae pol α L868F at replicating through TT dimer and abasic sites by steady-state kinetics
| Template:dNTPa | Km (μM) | kcat (min−1) | kcat/Km (μM−1 min−1) | fincb |
|---|---|---|---|---|
| 5′-T:dATP | 0.59 ± 0.17 | 14 ± 3.2 | 24 ± 3.9 | 1 |
| 5′-CPD:dATP | 73 ± 25 | 4.1 ± 1.5 | 0.062 ± 0.031 | 2.6 × 10−3 |
| AP+1:dATP | 3.2 ± 1.4 | 3.5 ± 0.59 | 1.2 ± 0.44 | 0.050 |
| TT+1:dGTP | 0.95 ± 0.60 | 3.8 ± 1.7 | 4.4 ± 1.1 | 1 |
| CPD+1:dGTP | 1.1 ± 0.28 | 3.0 ± 0.37 | 2.7 ± 0.37 | 0.60 |
Template and dNTP represent template base and incoming dNTP, respectively. The name of each template site is schematically illustrated in Fig. 1.
finc, kcat/Km value relative to that for T:dATP (which was set at 1.0).
pol α Leu868 is functionally and evolutionarily conserved.
Because the Leu/Ile residue is found in replicative DNA polymerases, it seems likely that it would be functionally and evolutionarily conserved. This idea is supported by the characteristics of WT and L864F human pol α. Primer extension analysis showed that human L864F pol α incorporated dAMP at the 3′ T of CPD 77-fold more efficiently than the WT (Fig. 4F and Table 4). The mutation frequency of human L864F pol α was 65-fold higher than that seen with the WT (Table 1). Sequencing data showed that the overall error rate for human L864F was 0.012, or 1 in 80 nucleotides, which was 180-fold higher than seen with the WT (data not shown). Thus, WT and mutant human pol α are similar to their S. cerevisiae counterparts, strongly suggesting that Leu868/864 has a conserved function in yeast and human pol α. Like yeast pol η, the Leu mutant of human DNA pol η also decreased the translesion activity (data not shown).
Phenotype of Leu868 mutant pol α.
The in vitro data discussed above suggest that Leu868 in S. cerevisiae pol α is, at least in a part, a determinant of polymerase fidelity and that this residue might have played a role in the evolution of family B and Y DNA polymerases. If this hypothesis is correct, predictable phenotypic effects are to be expected in mutant yeast strains. This idea was tested by replacing the WT chromosomal pol α allele with an allele expressing L868F, L868M, or L868W pol α. The replication fidelity of these strains was measured using trp1 and his7 reversion assays and a CAN1 forward mutation assay (Table 6). The trp1+, his7+, and CAN1r mutation rates in pol1-L868F yeast cells were 15-, 7.1-, and 8.1-fold higher, respectively, than those seen with the WT (Table 6). In contrast, pol1-L868M and pol1-L868W yeast strains had trp1+ and his7+ reversion rates similar to those of the WT yeast and pol1-L868M had a moderate increase in the CAN1r forward mutation rate (2.9-fold higher than that of the WT). The base substitution specificity of these yeast strains was examined by limited sequencing of the trp1 gene in the revertants. In WT yeast, 82% of the revertants carried the original amber codon, indicating that the amber codon was suppressed by some other mechanism (e.g., mutation at distal site like a suppressor tRNA). The remaining revertants in WT yeast had mutations at the target codon including 27% transitions and 73% transversions (Table 7). In pol1-L868F cells, 17% of the revertants retained the amber codon and the rest contained 8% transitions and 92% transversions at the target codon. In pol1-L868F yeast, the overall reversion rate at trp1-289 was 71 ×10−8, which is 42-fold higher than the trp1-289 reversion rate in WT yeast (1.7 × 10−8). At the can1 locus in WT and pol1-L868F cells, transitions were more frequent than transversions (Table 8). Interestingly, N→T substitutions, which are characteristic of L868F pol α, also increased in the mutant strain.
TABLE 6.
Mutation rate of S. cerevisiae strains that carry either WT or mutant pol1a
| pol1 | MMR mutation rate forb:
|
|||||
|---|---|---|---|---|---|---|
| CAN1r WT (10−7) | his+ WT (10−8) |
trp+ (10−8)
|
||||
| WT | ΔMSH2 | ΔMSH3 | ΔMSH6 | |||
| WT | 4.8 (1)b | 2.1 (1) | 5.7 (1) | 27 (1) | 2.1 (1) | 36 (1) |
| L868F | 39 (8.1)d | 15 (7.1)d | 83 (15)d | -c | 24 (11) | -c |
| L868M | 14 (2.9)d | 2.6 (1.2) | 8.4 (1.5) | 1,400 (52)d | 4.4 (2.1) | 2,200 (61)d |
| L868W | 4.8 (1.0) | 3.2 (1.5) | 5.0 (0.9) | 400 (15) | 2.0 (1.0) | 170 (4.7)d |
Experiments were carried out (using independent clones) more than five times.
The numbers in parentheses represent severalfold induction values relative to the WT value.
-, double mutant was not successfully created.
Statistically relevant (Student t test; P < 0.05).
TABLE 7.
Types of base substitutions observed with S. cerevisiae WT and pol1-L868F in trp1 reversion assay
| Error type | WT
|
pol1-L868F
|
Fold increase (L868F/WT) | ||||
|---|---|---|---|---|---|---|---|
| n | % | Reversion ratea (10−8) | n | % | Reversion ratea (10−8) | ||
| All errors | 22 | 100 | 1.7 | 26 | 100 | 71 | 42 |
| Transitions | 6 | 27 | 2 | 7.7 | |||
| T→C(A→G) | 6 | 27 | 2 | 7.7 | |||
| Transversions | 16 | 73 | 24 | 92 | |||
| T→G(A→C) | 6 | 27 | 0 | 0 | |||
| T→A(A→T) | 0 | 0 | 1 | 3.8 | |||
| G→T(C→A) | 4 | 18 | 13 | 50 | |||
| G→C(C→G) | 6 | 27 | 10 | 39 | |||
Reversion rates were determined as described for Table 6.
TABLE 8.
Types of errors by S. cerevisiae WT and pol1-L868F in CAN1r forward mutation assaya
| Error type | WT
|
pol1-L868F
|
||
|---|---|---|---|---|
| n | % | n | % | |
| All errors | 19 | 100 | 22 | 100 |
| Base substitutions | 13 | 68 | 14 | 64 |
| Transitions | 4 | 21 | 9 | 41 |
| T→C | 1 | 5.3 | 1 | 4.5 |
| C→T | 1 | 5.3 | 4 | 18 |
| A→G | 0 | 0 | 1 | 4.5 |
| G→A | 2 | 11 | 3 | 14 |
| Transversions | 9 | 47 | 5 | 23 |
| A→T | 0 | 0 | 4 | 18 |
| A→C | 2 | 11 | 0 | 0 |
| G→T | 0 | 0 | 1 | 4.5 |
| G→C | 2 | 11 | 0 | 0 |
| C→A | 2 | 11 | 0 | 0 |
| C→G | 3 | 16 | 0 | 0 |
| Frameshifts | 4 | 21 | 7 | 32 |
| 1-base deletions | 4 | 21 | 4 | 18 |
| 1-base insertions | 0 | 0 | 3 | 14 |
| Others | 2b | 11 | 1c | 4.5 |
Lagging strand synthesis was determined on the basis of the findings of Wyrick et al. (61).
Two errors represent a duplication of a CAN1 sequence (284 to 310) and a 5′-CAAAA→5′-AAAAAA mutation. The duplication may be directed by a direct repeat sequence (5′-TACCAATA).
An error represents a duplication of a CAN1 sequence (252 to 268) with no apparent repeat sequence.
MMR recognition and repair of DNA synthesis errors introduced by L868M/W pol α.
Mutagenesis was examined in MMR-defective strains carrying L868M/W pol α. Targeted disruptions were introduced into MSH2, MSH3, or MSH6 in WT and mutant pol α strains, and the trp1 reversion rate was measured (Table 6). In the double mutants msh2::pol1“msh2:: pol1-L868M/W and msh6::pol1-L868M/W, mutation rates increased synergistically. In contrast, disruption of msh3 did not significantly increase the mutation rate of pol1-L868M, pol1-L868W, or pol1-L868F. Similar results were observed using the CAN1 forward mutation assay (data not shown). These results suggest that DNA synthesis errors introduced by L868 pol α are recognized and repaired by MMR.
DISCUSSION
Errors of S. cerevisiae pol α L868F mutations in vitro.
In the elongation polymerases of pol δ and pol ɛ, error prevention pathways have been identified by knocking out the proofreading exonuclease activity (8, 38, 39). In the absence of the exonuclease activity in pol α, we have taken an alternative strategy to isolate active but inaccurate mutant DNA polymerases. After an in vivo screening, we characterized mutations in the conserved Leu residue of pol α and a comparable mutation of pol η. The spontaneous error rate of L868F pol α is 3 per 100 nucleotides (570-fold higher than that of the WT) (Table 2) and is consistent with the low-fidelity character of the corresponding mutations in prokaryotic DNA polymerases (42, 47, 48). Moreover, the error rate, random distribution of errors, and deletion spectrum introduced by this mutation resemble results for WT human pol η (35) (Fig. 3A), although the base substitution spectrums of pol η and L868F pol α are not identical.
L868F pol α generates more frameshift errors at run sequences than WT pol α, but it also generates many frameshift mutations at nonrun sequences. More than 50% of single-base deletion errors occurred at nonrun sequences. L868F pol α also generated large deletions of 30 to 50 nucleotides (Table 2 and Fig. 3B). Large deletions involve DNA rearrangement and relocation of the primer terminus. These template regions may form stable secondary structures. For example, one deletion hot spot (−15 to +30) contains the palindromic sequence 5′-TGTGTG-3′ , 5′-CACACA-3 at one end of the deleted region, and a partial repeat of this sequence at a distal site. pol η appears to make a similar mutation at the same site (35). L868F pol α also generates tandem base deletions, which are characteristic of the mutation spectrum of pol η (35).
Structural implications of low-fidelity DNA synthesis.
The three-dimensional structures of replicative and Y family DNA polymerases have shown that significant structural differences are present in the fingers subdomain. Replicative DNA polymerases have a narrow catalytic pocket with limited space for the nucleotide substrate and the template primer. In contrast, because the fingers are truncated in translesion DNA polymerases (62), the catalytic pocket is relatively wide and large enough to fit two nucleotides during bypass of CPD (57). The functional importance of the fingers subdomain is confirmed by the properties of fingers subdomain mutants (11, 12). Another structural study suggests that the catalytic pocket of Y family polymerases might favor frequent template misalignment errors (28).
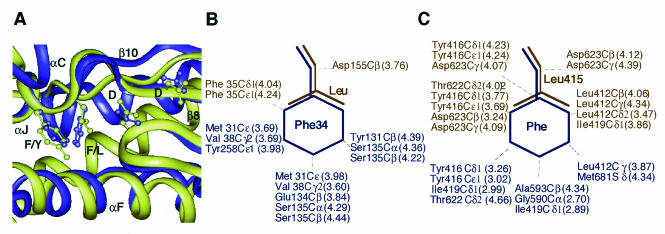
In contrast, Y and conventional A and B DNA polymerases share very similar structures in the palm subdomain (10, 19, 25, 44, 50). In accord with the previous studies, ternary structures of the motif A regions of a family B (RB69) polymerase and a family Y (S. cerevisiae pol η) polymerase superimpose fairly well (Fig. 5). The side chains of Leu/Phe do not face toward the catalytic pocket or DNA. Therefore, fidelity reduction may not be due to the altered interaction with the template, primer, or incoming nucleotide. However, it is possible that substitution of Phe for Leu868 in S. cerevisiae promotes novel interactions between the Phe aromatic ring and proximate side chains that might stabilize the catalytic complex (Fig. 5); an analogous manner has been reported in Taq pol I (52, 54). This stabilization might allow more time for chemistry to take place and increase the probability of phosphoryl transfer involving incorrectly paired nucleotides or nucleotides paired with damaged template bases. On the other hand, stabilization may have little effect on the rate of catalysis involving correctly paired nucleotides because this reaction is intrinsically very fast. This would be an interpretation explaining why the mutant pol α particularly increased both misincorporation efficiency and insertion efficiency at damaged sites while it gained relatively little activity enhancement for the correct base pairs.
FIG. 5.
Structural models of S. cerevisiae pol η and RB69 pol. (A) The representations of the crystal structures of S. cerevisiae pol η (blue) and RB69 pol (dark yellow) are superimposed. Side chains are indicated as F/Y (Phe35 in pol η and Tyr416 in RB69 are indicated with blue and dark yellow colors), F/L (Phe34 in pol η and Leu415 in RB69), and D (Asp30 and Asp155 in pol η and Asp623 and Asp411 in RB69). The α helix C, F, J, and β sheets 8 and 10 in pol η are also indicated. (B) Van der Waals contacts of Phe34 in S. cerevisiae pol η are shown in purple. A putative interaction with the Leu substitution (dark yellow) is also simulated. Distances between atoms are shown in parentheses. (C) Van der Waals contacts of Leu415 in RB69 pol are shown in blue. A putative interaction with the Phe substitution (blue) is also simulated. These structure models are also relevant to the recently crystallized Dpo4 pol catalytic complex with CPD (27).
It should be noted that biochemical characterization of WT and the mutant polymerases in pol α and η showed that the functions of the Leu and Phe residues are not really identical. L868F pol α shows a significant loss of fidelity and a gain in translesion activity. In contrast, F34L pol η and F10L E. coli UmuC (3, 49), presumably, are associated with a large loss in catalytic activity. This difference may be attributed to the other part of structure differences between family B and Y DNA polymerases. Translesion polymerases interact weakly with DNA and have a structurally simpler catalytic pocket than replicative polymerases (28, 57, 62); thus, they might be associated with the structural fragility upon catalysis.
Translesion DNA synthesis.
L868F pol α bypasses the 3′ T and 5′ T of CPD lesions ∼160- and ∼380-fold less efficiently, respectively, compared to the results seen with nondamaged TT (Table 4 and Table 5), while WT pol η incorporates dAMP with similar levels of efficiency on damaged and undamaged DNA (Table 3) (24). In addition, L868F pol α is equally efficient with templates containing CPD and 6-4 photoproduct (Fig. 4D) and inserts a nucleotide opposite an abasic site as efficiently as opposite a template base (Table 4). These characteristics are not observed for pol η (Fig. 4B) and other Y family DNA polymerases.
L868F pol preferentially incorporated dAMP at several DNA lesions (Table 4 and data not shown). At an abasic site or the 3′ T of CPD, dAMP and dGMP had the highest and next highest incorporation rates. dPMP was also incorporated opposite these lesions 9.4- and 1.3-fold more efficiently compared to the results seen with dAMP incorporation opposite template T. These results suggest that like T7 DNA polymerase (51), L868F pol α transiently reorients the DNA lesion with respect to the DNA backbone. In accord with this hypothesis it appears that when the structure of RB69 pol is superimposed on the Dpo4/CPD ternary complex, a displacement of the CPD template by a few angstroms would allow enzyme not to crash into the fingers subdomain (data not shown). This way of template-polymerase interaction would mimic the abasic-like structure and facilitate the incorporation of dAMP and dPMP.
Effects of Leu868 mutations in vivo.
L868F pol α is a unique mutant of pol α in that it has impaired fidelity without a loss of overall catalytic function. It may be important that the biochemical properties of this enzyme have a significant biological impact on genomic stability. For example, yeast strains with pol1-L868F have a 42-fold-higher base substitution rate at trp1-289 than WT yeast; that base substitution rate is comparable to or higher than the base substitution rate in exonuclease-deficient mutants of pol3-01 or pol2-04 (8, 37). This result may be somewhat surprising, because the primary role of pol α is thought to be initiation of lagging strand DNA synthesis during chromosomal DNA replication. The 3′ region of trp1 overlaps with ARS1 on chromosome IV (58, 61) such that pol α may be active on both DNA strands in this region. The mutation frequency was also significantly higher than that of the WT at his7 and CAN1, however, so the effect of pol1-L868F on genomic stability was not locus specific (Table 6).
The in vivo mutation spectrum at trp1-289 in pol1-L868F yeast includes 50% of G→T (C→A) transversions (Table 7), in accordance with the result that dAMP misincorporations were the predominant form of base substitution errors during in vitro DNA synthesis by L868F pol α. The mutation spectrum in the can1 gene provides a way to further understanding of the relationship between in vitro and in vivo mutations. This gene locates in 26-kb proximity to the active ARS503 (61), allowing us to speculate that lagging strand DNA synthesis occurs with the Watson strand of this locus as a template. In accord with the findings regarding the trp1 spectrum, sequencing results showed the unique increase of C→T and A→T missense mutations (misincorporation of dAMP; Table 8). These results implicate the pol α mutant as one mechanism causing a high rate of spontaneous mutagenesis; the results also suggest that error discrimination by pol α is required for maintenance of genomic stability in yeast.
We also used the pol1-L868F yeast strain carrying a WT or ΔRAD30 yeast pol η allele for the study of translesion DNA synthesis activity in vivo. Preliminary experiments showed that methyl methanesulfonate and UV sensitivity characteristics were not significantly affected by the presence of the pol1-L868F allele (data not shown). However, further experiments must be carried out to justify the idea that pol1-L868F has a limited effect on methyl methanesulfonate and UV damage response in vivo; we have not excluded the possibility that the results simply reflect the limited involvement of pol α in the chromosomal DNA replication.
The results of this study suggest that there is a direct link between misincorporation by pol α L868F and genomic instability. Although an earlier study showed that pol α pol1-Y867A increased the mutation rate in a MMR-deficient strain (45). In our hands, pol α Tyr867 mutants had reduced activity but did not have reduced fidelity (data not shown). In another case, aberrant DNA replication (associated with large deletions) was observed when temperature-sensitive strains were grown at the semipermissive temperature (20, 29). In contrast, pol α L868F is an active DNA polymerase but a strong mutator in vitro and primarily makes base substitutions in CAN1r forward mutation assays (Table 8). In the mutated can1 locus of pol1-L868F yeast, furthermore, the direct-repeat-directed deletion was not observed (Table 8). This is in agreement with the observation that direct-repeat-directed deletions are an indication of low levels of activity or low copy numbers for DNA polymerases (22, 29, 41). These data suggest that base substitution errors by pol α L868F directly lead to the mutator phenotype in pol1-L868F cells. However, other mutagenic mechanisms such as oxidative damage (9, 40) or check point-dependent mutagenesis (8, 20) as factors contributing to the decrease in genomic stability in pol1-L868F yeast have not yet been examined.
An error prevention pathway for pol α.
The results presented here show that the fidelity of pol α has an impact on overall genomic stability in yeast. Furthermore, subsequent genetic study showed MSH2 and MSH6 alleles play a role in correcting pol α-dependent replication errors. These data are consistent with the finding that the presence of the MSH2-MSH6 protein complex prevents missense and frameshift mutations (17). In contrast, we did not observe the apparent link between pol α and the MSH2-MSH3 complex, which weakly binds to mismatched and frameshifted DNA and strongly binds to small (4- to 14-nucleotide) DNA loops (16). In our in vitro assay, L868F pol α primarily generated missense and frameshift mutations (Table 2), with less than 1.5% consisting of ≥2-base loops. Given these data, it seems likely that the MSH2-MSH3 complex, at least in the pol1-L868F strain, plays a minor role in the error-correction pathway.
Previous reports have suggested that MMR might function in the presence of PCNA, although no interaction between pol α and PCNA has been reported. Therefore, it is unlikely that MMR proteins encounter the pol α errors during primer DNA synthesis. After polymerase switching takes place, PCNA may be loaded and travel along the DNA (at least until the Okazaki fragments are ligated). Therefore, we prefer the explanation suggesting a mechanism in which a PCNA-MMR complex repairs mismatches in primer DNA after RNA primers have been removed.
In this article, we demonstrated that the mutation of Leu868 affects polymerase activity and DNA replication fidelity in pol α. We also suggest that fidelity DNA replication by pol α, like that by pol δ and pol ɛ, is also required to maintain genome integrity and probably to prevent cancers in higher eukaryotes. It seems that this specific residue and the corresponding Phe34 in pol η are also involved in the evolution of replicative and translesion DNA polymerases.
Acknowledgments
We thank Tazuko Tomita for DNA sequencing and Yasutomo Ito for model construction. We are also grateful to Akio Sugino, Yasuo Kawasaki, Akihiko Kikuchi, and Katsunori Sugimoto for critical advice and consistent encouragement.
This work was supported in part by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan, by the Hori Foundation, and in part by the U.S. National Institutes of Health (grant EB002059 to E.T.K.).
REFERENCES
- 1.Ayyagari, R., K. J. Impellizzeri, B. L. Yoder, S. L. Gary, and P. M. Burgers. 1995. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol. Cell. Biol. 15:4420-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasco, M. A., J. M. Lazaro, L. Blanco, and M. Salas. 1993. Phi 29 DNA polymerase active site. The conserved amino acid motif “Kx3NSxYG” is involved in template-primer binding and dNTP selection. J. Biol. Chem. 268:16763-16770. [PubMed] [Google Scholar]
- 3.Boudsocq, F., H. Ling, W. Yang, and R. Woodgate. 2002. Structure-based interpretation of missense mutations in Y-family DNA polymerases and their implications for polymerase function and lesion bypass. DNA Repair (Amsterdam) 1:343-358. [DOI] [PubMed] [Google Scholar]
- 4.Braithwaite, D. K., and J. Ito. 1993. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 21:787-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capizzi, R. L., and J. W. Jameson. 1973. A table for the estimation of the spontaneous mutation rate of cells in culture. Mutat. Res. 17:147-148. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C., B. J. Merrill, P. J. Lau, C. Holm, and R. D. Kolodner. 1999. Saccharomyces cerevisiae pol30 (proliferating cell nuclear antigen) mutations impair replication fidelity and mismatch repair. Mol. Cell. Biol. 19:7801-7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Copeland, W. C., N. K. Lam, and T. S. Wang. 1993. Fidelity studies of the human DNA polymerase alpha. The most conserved region among alpha-like DNA polymerases is responsible for metal-induced infidelity in DNA synthesis. J. Biol. Chem. 268:11041-11049. [PubMed] [Google Scholar]
- 8.Datta, A., J. L. Schmeits, N. S. Amin, P. J. Lau, K. Myung, and R. D. Kolodner. 2000. Checkpoint-dependent activation of mutagenic repair in Saccharomyces cerevisiae pol3-01 mutants. Mol. Cell 6:593-603. [DOI] [PubMed] [Google Scholar]
- 9.Earley, M. C., and G. F. Crouse. 1998. The role of mismatch repair in the prevention of base pair mutations in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95:15487-15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franklin, M. C., J. Wang, and T. A. Steitz. 2001. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell 105:657-667. [DOI] [PubMed] [Google Scholar]
- 11.Glick, E., J. S. Chau, K. L. Vigna, S. D. McCulloch, E. T. Adman, T. A. Kunkel, and L. A. Loeb. 2003. Amino acid substitutions at conserved tyrosine 52 alter fidelity and bypass efficiency of human DNA polymerase eta. J. Biol. Chem. 278:19341-19346. [DOI] [PubMed] [Google Scholar]
- 12.Glick, E., K. L. Vigna, and L. A. Loeb. 2001. Mutations in human DNA polymerase eta motif II alter bypass of DNA lesions. EMBO J. 20:7303-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldsby, R. E., L. E. Hays, X. Chen, E. A. Olmsted, W. B. Slayton, G. J. Spangrude, and B. D. Preston. 2002. High incidence of epithelial cancers in mice deficient for DNA polymerase delta proofreading. Proc. Natl. Acad. Sci. USA 99:15560-15565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldsby, R. E., N. A. Lawrence, L. E. Hays, E. A. Olmsted, X. Chen, M. Singh, and B. D. Preston. 2001. Defective DNA polymerase-delta proofreading causes cancer susceptibility in mice. Nat. Med. 7:638-639. [DOI] [PubMed] [Google Scholar]
- 15.Goodman, M. F., S. Creighton, L. B. Bloom, and J. Petruska. 1993. Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. Biol. 28:83-126. [DOI] [PubMed] [Google Scholar]
- 16.Habraken, Y., P. Sung, L. Prakash, and S. Prakash. 1997. Enhancement of MSH2-MSH3-mediated mismatch recognition by the yeast MLH1-PMS1 complex. Curr. Biol. 7:790-793. [DOI] [PubMed] [Google Scholar]
- 17.Harfe, B. D., and S. Jinks-Robertson. 2000. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34:359-399. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, R. E., G. K. Kovvali, S. N. Guzder, N. S. Amin, C. Holm, Y. Habraken, P. Sung, L. Prakash, and S. Prakash. 1996. Evidence for involvement of yeast proliferating cell nuclear antigen in DNA mismatch repair. J. Biol. Chem. 271:27987-27990. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, R. E., S. Prakash, and L. Prakash. 1999. Requirement of DNA polymerase activity of yeast Rad30 protein for its biological function. J. Biol. Chem. 274:15975-15977. [DOI] [PubMed] [Google Scholar]
- 20.Kai, M., and T. S. Wang. 2003. Checkpoint activation regulates mutagenic translesion synthesis. Genes Dev. 17:64-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, B., T. R. Hathaway, and L. A. Loeb. 1996. Human immunodeficiency virus reverse transcriptase. Functional mutants obtained by random mutagenesis coupled with genetic selection in Escherichia coli. J. Biol. Chem. 271:4872-4878. [DOI] [PubMed] [Google Scholar]
- 22.Kokoska, R. J., L. Stefanovic, J. DeMai, and T. D. Petes. 2000. Increased rates of genomic deletions generated by mutations in the yeast gene encoding DNA polymerase δ or by decreases in the cellular levels of DNA polymerase δ. Mol. Cell. Biol. 20:7490-7504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunkel, T. A., and K. Bebenek. 2000. DNA replication fidelity. Annu. Rev. Biochem. 69:497-529. [DOI] [PubMed] [Google Scholar]
- 24.Kusumoto, R., C. Masutani, S. Iwai, and F. Hanaoka. 2002. Translesion synthesis by human DNA polymerase eta across thymine glycol lesions. Biochemistry 41:6090-6099. [DOI] [PubMed] [Google Scholar]
- 25.Li, Y., S. Korolev, and G. Waksman. 1998. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 17:7514-7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Limsirichaikul, S., M. Ogawa, A. Niimi, S. Iwai, T. Murate, S. Yoshida, and M. Suzuki. 2003. The Gly952 residue of Saccharomyces cerevisiae DNA polymerase alpha is important in discriminating correct deoxyribonucleotides from incorrect ones. J. Biol. Chem. 278:19079-19086. [DOI] [PubMed] [Google Scholar]
- 27.Ling, H., F. Boudsocq, B. S. Plosky, R. Woodgate, and W. Yang. 2003. Replication of a cis-syn thymine dimer at atomic resolution. Nature 424:1083-1087. [DOI] [PubMed] [Google Scholar]
- 28.Ling, H., F. Boudsocq, R. Woodgate, and W. Yang. 2001. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107:91-102. [DOI] [PubMed] [Google Scholar]
- 29.Liu, V. F., D. Bhaumik, and T. S. Wang. 1999. Mutator phenotype induced by aberrant replication. Mol. Cell. Biol. 19:1126-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loeb, L. A., and T. A. Kunkel. 1982. Fidelity of DNA synthesis. Annu. Rev. Biochem. 51:429-457. [DOI] [PubMed] [Google Scholar]
- 31.Luria, S. E., and M. Delbruck. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masutani, C., R. Kusumoto, S. Iwai, and F. Hanaoka. 2000. Mechanisms of accurate translesion synthesis by human DNA polymerase eta. EMBO J. 19:3100-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matray, T. J., and E. T. Kool. 1999. A specific partner for abasic damage in DNA. Nature 399:704-708. [DOI] [PubMed] [Google Scholar]
- 34.Matsuda, T., K. Bebenek, C. Masutani, F. Hanaoka, and T. A. Kunkel. 2000. Low fidelity DNA synthesis by human DNA polymerase-eta. Nature 404:1011-1013. [DOI] [PubMed] [Google Scholar]
- 35.Matsuda, T., K. Bebenek, C. Masutani, I. B. Rogozin, F. Hanaoka, and T. A. Kunkel. 2001. Error rate and specificity of human and murine DNA polymerase eta. J. Mol. Biol. 312:335-346. [DOI] [PubMed] [Google Scholar]
- 36.Mendelman, L. V., J. Petruska, and M. F. Goodman. 1990. Base mispair extension kinetics. Comparison of DNA polymerase alpha and reverse transcriptase. J. Biol. Chem. 265:2338-2346. [PubMed] [Google Scholar]
- 37.Morrison, A., J. B. Bell, T. A. Kunkel, and A. Sugino. 1991. Eukaryotic DNA polymerase amino acid sequence required for 3′-5′ exonuclease activity. Proc. Natl. Acad. Sci. USA 88:9473-9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrison, A., A. L. Johnson, L. H. Johnston, and A. Sugino. 1993. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 12:1467-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morrison, A., and A. Sugino. 1994. The 3′→5′ exonucleases of both DNA polymerases delta and epsilon participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol. Gen. Genet. 242:289-296. [DOI] [PubMed] [Google Scholar]
- 40.Ni, T. T., G. T. Marsischky, and R. D. Kolodner. 1999. MSH2 and MSH6 are required for removal of adenine misincorporated opposite 8-oxo-guanine in S. cerevisiae. Mol. Cell 4:439-444. [DOI] [PubMed] [Google Scholar]
- 41.Ogawa, M., S. Limsirichaikul, A. Niimi, S. Iwai, S. Yoshida, and M. Suzuki. 2003. Distinct function of conserved amino acids in the fingers of Saccharomyces cerevisiae DNA polymerase alpha. J. Biol. Chem. 278:19071-19078. [DOI] [PubMed] [Google Scholar]
- 42.Patel, P. H., H. Kawate, E. Adman, M. Ashbach, and L. A. Loeb. 2001. A single highly mutable catalytic site amino acid is critical for DNA polymerase fidelity. J. Biol. Chem. 276:5044-5051. [DOI] [PubMed] [Google Scholar]
- 43.Patel, P. H., and L. A. Loeb. 2001. Getting a grip on how DNA polymerases function. Nat. Struct. Biol. 8:656-659. [DOI] [PubMed] [Google Scholar]
- 44.Patel, P. H., M. Suzuki, E. Adman, A. Shinkai, and L. A. Loeb. 2001. Prokaryotic DNA polymerase I: evolution, structure, and “base flipping” mechanism for nucleotide selection. J. Mol. Biol. 308:823-837. [DOI] [PubMed] [Google Scholar]
- 45.Pavlov, Y. I., P. V. Shcherbakova, and T. A. Kunkel. 2001. In vivo consequences of putative active site mutations in yeast DNA polymerases alpha, epsilon, delta, and zeta. Genetics 159:47-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perrino, F. W., and L. A. Loeb. 1989. Differential extension of 3′ mispairs is a major contribution to the high fidelity of calf thymus DNA polymerase-alpha. J. Biol. Chem. 264:2898-2905. [PubMed] [Google Scholar]
- 47.Shinkai, A., and L. A. Loeb. 2001. In vivo mutagenesis by Escherichia coli DNA polymerase I. Ile(709) in motif A functions in base selection. J. Biol. Chem. 276:46759-46764. [DOI] [PubMed] [Google Scholar]
- 48.Shinkai, A., P. H. Patel, and L. A. Loeb. 2001. The conserved active site motif A of Escherichia coli DNA polymerase I is highly mutable. J. Biol. Chem. 276:18836-18842. [DOI] [PubMed] [Google Scholar]
- 49.Sommer, S., G. Coste, and A. Bailone. 2000. Specific amino acid changes enhance the anti-recombination activity of the UmuD'C complex. Mol. Microbiol. 35:1443-1453. [DOI] [PubMed] [Google Scholar]
- 50.Steitz, T. A. 1999. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 274:17395-17398. [DOI] [PubMed] [Google Scholar]
- 51.Sun, L., M. Wang, E. T. Kool, and J. S. Taylor. 2000. Pyrene nucleotide as a mechanistic probe: evidence for a transient abasic site-like intermediate in the bypass of dipyrimidine photoproducts by T7 DNA polymerase. Biochemistry 39:14603-14610. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki, M., A. K. Avicola, L. Hood, and L. A. Loeb. 1997. Low fidelity mutants in the O-helix of Thermus aquaticus DNA polymerase I. J. Biol. Chem. 272:11228-11235. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki, M., D. Baskin, L. Hood, and L. A. Loeb. 1996. Random mutagenesis of Thermus aquaticus DNA polymerase I: concordance of immutable sites in vivo with the crystal structure. Proc. Natl. Acad. Sci. USA 93:9670-9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki, M., S. Yoshida, E. T. Adman, A. Blank, and L. A. Loeb. 2000. Thermus aquaticus DNA polymerase I mutants with altered fidelity. Interacting mutations in the O-helix. J. Biol. Chem. 275:32728-32735. [DOI] [PubMed] [Google Scholar]
- 55.Tosaka, A., M. Ogawa, S. Yoshida, and M. Suzuki. 2001. O-helix mutant T664P of Thermus aquaticus DNA polymerase I: altered catalytic properties for incorporation of incorrect nucleotides but not correct nucleotides. J. Biol. Chem. 276:27562-27567. [DOI] [PubMed] [Google Scholar]
- 56.Tran, H. T., D. A. Gordenin, and M. A. Resnick. 1999. The 3′→5′ exonucleases of DNA polymerases δ and ε and the 5′→3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:2000-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trincao, J., R. E. Johnson, C. R. Escalante, S. Prakash, L. Prakash, and A. K. Aggarwal. 2001. Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis. Mol. Cell 8:417-426. [DOI] [PubMed] [Google Scholar]
- 58.Tschumper, G., and J. Carbon. 1980. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene 10:157-166. [DOI] [PubMed] [Google Scholar]
- 59.Umar, A., A. B. Buermeyer, J. A. Simon, D. C. Thomas, A. B. Clark, R. M. Liskay, and T. A. Kunkel. 1996. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell 87:65-73. [DOI] [PubMed] [Google Scholar]
- 60.Waga, S., and B. Stillman. 1998. The DNA replication fork in eukaryotic cells Annu. Rev. Biochem. 67:721-751. [DOI] [PubMed] [Google Scholar]
- 61.Wyrick, J. J., J. G. Aparicio, T. Chen, J. D. Barnett, E. G. Jennings, R. A. Young, S. P. Bell, and O. M. Aparicio. 2001. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294:2357-2360. [DOI] [PubMed] [Google Scholar]
- 62.Zhou, B. L., J. D. Pata, and T. A. Steitz. 2001. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol. Cell 8:427-437. [DOI] [PubMed] [Google Scholar]