Abstract
Eukaryotic elongation factor 2 (eEF2) kinase is an unusual calcium- and calmodulin-dependent protein kinase that is regulated by insulin through the rapamycin-sensitive mTOR pathway. Here we show that insulin decreases the ability of eEF2 kinase to bind calmodulin in a rapamycin-sensitive manner. We identify a novel phosphorylation site in eEF2 kinase (Ser78) that is located immediately next to its calmodulin-binding motif. Phosphorylation of this site is increased by insulin in a rapamycin-sensitive fashion. Regulation of the phosphorylation of Ser78 also requires amino acids and the protein kinase phosphoinositide-dependent kinase 1. Mutation of this site to alanine strongly attenuates the effects of insulin and rapamycin both on the binding of calmodulin to eEF2 kinase and on eEF2 kinase activity. Phosphorylation of Ser78 is thus likely to link insulin and mTOR signaling to the control of eEF2 phosphorylation and chain elongation. This site is not a target for known kinases in the mTOR pathway, e.g., the S6 kinases, implying that it is phosphorylated by a novel mTOR-linked protein kinase that serves to couple hormones and amino acids to the control of translation elongation. eEF2 kinase is thus a target for mTOR signaling independently of previously known downstream components of the pathway.
mRNA translation is a key control point for gene expression and is regulated by diverse physiological stimuli. For example, it is activated by insulin, and this activation involves stimulation of a variety of components of the translational machinery, several of which are regulated through signaling involving the mammalian target of rapamycin, mTOR (21). mTOR is a large multidomain protein whose function is inhibited specifically by the immunosuppressant drug rapamycin. Translation components linked to mTOR include the translational repressor 4E-BP1 (eukaryotic initiation factor 4E-binding protein 1) (21) and the ribosomal protein (rp) S6 kinases (S6Ks) (4). mTOR signaling can also regulate the cell cycle, autophagy, and other processes (20, 39). The already high level of interest in mTOR signaling has been further increased by recent data showing that it plays important roles in the control of cell and organism size (20, 44) and in cell transformation (3, 31), as well as in certain benign tumors (30). There have recently been important advances in understanding how hormones such as insulin stimulate mTOR signaling. Insulin stimulates protein kinase B (PKB) via phosphatidylinositide (PI) 3-kinase, and PKB phosphorylates the product of the tuberous sclerosis complex TSC2 gene, also termed tuberin. TSC2 forms a heterodimer with TSC1 which represses mTOR activity apparently by acting as the GTPase-activator protein for the small G-protein Rheb (for reviews, see references 28 and 30). Phosphorylation of TSC2 by PKB alleviates this inhibitory restraint on mTOR signaling, resulting in its activation. However, the mechanisms that link mTOR to the control of these processes remain obscure. There is thus a pressing need to learn more about mTOR signaling, especially how it regulates its downstream effectors.
In addition to the S6Ks and 4E-BP1, both of which modulate translation initiation, mTOR signaling also regulates the translation elongation process through the phosphorylation of eukaryotic elongation factor 2 (eEF2) (40). eEF2 is a GTP-binding protein that mediates the translocation step of elongation (10). When phosphorylated at Thr56, eEF2 loses its ability to bind to ribosomes and is thus inactivated (12). Insulin and other stimuli induce the dephosphorylation of eEF2, and this effect is blocked by rapamycin (19, 40, 47, 48, 50). Where studied, this appears to involve decreases in the activity of the kinase that acts on eEF2, a highly specific enzyme called eEF2 kinase (40, 48, 48, 50). The ability of insulin to decrease eEF2 kinase activity is also blocked by rapamycin, implying that this effect is also mediated through mTOR.
eEF2 kinase is a highly unusual enzyme. The sequence of its catalytic domain differs substantially from that of other protein kinases, and it is not a member, e.g., of the main Ser-Thr-Tyr kinase superfamily (41). The C-terminal half of the eEF2 kinase polypeptide contains several sites of phosphorylation and, at the C-terminal tip, the binding site for the substrate eEF2 (16, 34). The activity of eEF2 kinase is normally completely dependent upon Ca2+ ions and calmodulin (CaM). The CaM-binding site has been identified as lying in the N terminus of the polypeptide, adjacent to the catalytic domain, in a region containing both hydrophobic and basic residues, as found for CaM-binding sites in other proteins (16, 34). We previously identified Ser366, which lies in the C terminus of the catalytic domain, as being phosphorylated by S6K and by p90RSK, which lies downstream of the classical mitogen-activated protein kinase pathway (50). The phosphorylation of eEF2 kinase at Ser359, which was first identified as a substrate for stress-activated protein kinase 4, is reported to be regulated in a rapamycin-sensitive manner in response to insulin-like growth factor 1 (IGF1) (26).
Here we identify a novel phosphorylation site in eEF2 kinase that is regulated markedly in response to insulin in an mTOR-dependent manner. This site (Ser78) is not phosphorylated by any known protein kinase in the mTOR pathway. eEF2 kinase is thus a target of signaling from mTOR independently of other known targets of this pathway, which implies the existence of a novel (probably mTOR-controlled) protein kinase that acts upon Ser78 in eEF2 kinase. Ser78 is located immediately adjacent to the CaM-binding site in eEF2 kinase. We show that insulin decreases the binding of eEF2 to CaM in vivo and that this effect is blocked by rapamycin. The mutually antagonistic effects of insulin and rapamycin on CaM binding are abolished by mutating Ser78 to a nonphosphorylatable residue, Ala. These data provide a molecular explanation for the mechanism by which insulin and mTOR control the activity of eEF2 kinase and thereby regulate peptide chain elongation.
MATERIALS AND METHODS
Materials.
[γ-32P]ATP and materials for protein purification were obtained from Amersham Pharmacia Biotech, United Kingdom. Unlabeled ATP was from Roche Molecular Biochemicals (Lewes, United Kingdom); cell culture media and human IGF1 were obtained from Gibco (Paisley, United Kingdom); microcystin-LR, wortmannin, rapamycin, PD98059, and U0126 were from Calbiochem (Nottingham, United Kingdom); and Immobilon P membranes were from Millipore (Bedford, United Kingdom). Other chemicals were of the highest purity available and were purchased from Merck (Poole, United Kingdom) or Sigma-Aldrich (Poole, United Kingdom). CaM was purified from bovine brain. [γ-32P]ATP and enhanced chemiluminescence reagents were purchased from Amersham Pharmacia Biotech; wortmannin and rapamycin were from Calbiochem. Bovine serum albumin (fatty acid-free) was from Roche Molecular Biochemicals. Cycloheximide (CHX) was from Sigma. Unless otherwise indicated, all other reagents were obtained from Sigma or Merck.
Antibodies for p70 S6k phosphorylated at Thr389, PKB-phosphorylated Thr308, phosphorylated ERK1/2, and PKB (total) were from Cell Signaling Technology (Hitchin, Hertsfordshire, United Kingdom). Antibodies for eEF2 kinase phosphorylated at Ser366 have been described previously (25, 26) and were provided by Jane Leitch, Division of Signal Transduction Therapy, University of Dundee. Antibodies against TSC2 were also provided by Leitch. Antibodies to the α1 and α2 subunits of AMP-activated protein kinase (AMPK) and to acetyl-coenzyme A carboxylase phosphorylated at Ser79 were kindly provided by D. G. Hardie, University of Dundee. Anti-FLAG antibody (M2) was from Sigma. Antisera for 4E-BP1, S6K1, eEF2 phosphorylated at Thr56, total eEF2, and phospho-Ser235 rpS6 were described previously (7). Rabbit anti-sheep immunoglobulin G and goat anti-rabbit immunoglobulin G antibodies, both conjugated to peroxidase, were obtained from Perbio Science Ltd. (Tattenhall, United Kingdom).
Cells and cell culture.
KB cells (human oral epidermoid carcinoma cells) were cultivated in Dulbecco's modified Eagle medium (DMEM) containing 10% (vol/vol) fetal calf serum, 100 U of penicillin per ml, and 0.1 mg of streptomycin per ml. Prior to hormone treatment, cells were grown to 90% confluence and were then starved of serum overnight. Mouse embryo fibroblasts (TSC2+/+ or TSC2−/−) (24, 52) were cultivated in DMEM containing 10% (vol/vol) fetal calf serum, 100 U of penicillin per ml, and 0.1 mg of streptomycin per ml. Prior to hormone treatment, cells were grown to 90% confluency and were then starved of serum for 3 h. CHO.K1 cells and embryonic stem (ES) cells were cultivated and treated as described previously (7, 50). Prior to hormone treatment, the ES cells were starved of serum for 4 h. Where used, signaling inhibitors were added 30 min before the addition of 100 nM insulin for 30 min. For amino acid starvation, CHO.K1 or KB cells were transferred to Dulbecco's phosphate-buffered saline or Earle's balanced salts solution, respectively. Both media contain d-glucose at a concentration of 1 g per liter.
Transfection of KB cells was carried out by using the GenePORTER 2 transfection system (Cambridge BioSciences) according to the manufacturer's instructions, with 10 μg of plasmid DNA per 10-cm cell culture dish
Preparation and analysis of cell extracts.
Cells were extracted into buffer containing 50 mM β-glycerophosphate (pH 7.5), 1 mM EGTA, 1 mM EDTA, 1% (vol/vol) Triton X-100, 1 mM Na3VO4, 100 nM microcystin-LR, 0.1% (vol/vol) β-mercaptoethanol, and protease inhibitors (leupeptin, pepstatin, and antipain, each at a concentration of 1 μg/ml, and 200 μM phenylmethylsulfonyl fluoride). Lysates were centrifuged at 13,000 rpm in an Eppendorf 5415D microcentrifuge to remove debris. Protein concentrations in the resulting supernatants were as previously described (8). Supernatants were removed to fresh tubes and then rotated at 4°C in the presence of protein G-Sepharose prebound with anti-FLAG or anti-total eEF2 kinase antibody, as appropriate. Immune complexes were washed four times with extraction buffer and resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer.
For CaM-eEF2 kinase coimmunoprecipitation experiments, the extraction and immunoprecipitation procedures were carried out by using a modified calcium-containing extraction buffer (50 mM HEPES, pH 7.5; 50 mM β-glycerophosphate; 50 mM NaCl; 1 mM CaCl2; 0.3%, wt/vol, 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS]), 1 mM Na3VO4, 100 nM microcystin-LR; 0.1% (vol/vol) β-mercaptoethanol, and protease inhibitors (leupeptin, pepstatin, and antipain, each at a concentration of 1 μg/ml, and 200 μM phenylmethylsulfonyl fluoride) in order to maintain the eEF2 kinase-CaM interaction.
Gel electrophoresis and Western blotting of lysates and immunoprecipitates were performed as described earlier (36) by using an Immobilon membrane, except that for the analysis of 4E-BP1, running gels containing 13.5% (wt/vol) acrylamide and 0.36% (wt/vol) methylene bis-acrylamide were employed. Blots were developed by enhanced chemiluminescence.
Phosphospecific antibodies for eEF2 kinase.
The antiserum for eEF2 kinase phosphorylated at Ser78 was raised in rabbits by using the peptides GSPANSpFHFKEC, where Sp indicates phosphoserine. The peptide was synthesized by Graham Bloomberg (University of Bristol, United Kingdom), coupled to keyhole limpet hemocyanin, and injected into rabbits at Diagnostics Scotland (Edinburgh, United Kingdom). Antibodies were affinity purified on phosphopeptide antigen-Sepharose columns and were used at a concentration of 0.5 μg/ml in the presence of the corresponding nonphosphorylated peptide (10 μg/ml). Antibodies against total eEF2 kinase were raised against bacterially expressed glutathione transferase (GST)-eEF2 kinase in rabbits at Diagnostics Scotland and then affinity purified on maltose binding protein-eEF2 kinase-Sepharose to eliminate anti-GST antibodies.
Mutagenesis of eEF2 kinase.
A modified cDNA encoding human eEF2 kinase with an N-terminal FLAG tag cloned between the BamHI sites of the vector pGEX-4T was kindly provided by Maria Deak of the Division of Signal Transduction Therapy, University of Dundee. Mutagenesis of Ser78 to alanine was performed by PCR using QuikChange (Stratagene). The forward primer was 5′-CGGCAAACGCCTTCCACTTCAAGGAAGCC-3′, and the reverse primer was 5′-GGCTTCCTTGAAGTGGAAGGCGTTTGCCG-3′. The template used was the pGEX-4T-eEF2 kinase vector. For transfection of KB cells, wild-type or mutant FLAG-tagged human eEF2 kinase was subcloned into the mammalian expression vector pcDNA3.1 by using the BamH1 sites either side of the insert.
Assays for eEF2 kinase.
eEF2 kinase was assayed as described earlier (40) with the following modifications. The cells were harvested in Ca2+-containing extraction buffer. The eEF2 kinase was immunoprecipitated with antibodies raised against either eEF2 kinase or the FLAG epitope, as appropriate, and the beads were washed four times in Ca2+-containing extraction buffer. The immunoprecipitated eEF2 kinase was then assayed for activity for 10 min at 30°C in the presence of 1 μg of eEF2 (purified from HeLa cells) and Mg-[γ-32P]ATP in a final volume of 30 μl of the Ca2+-containing buffer. The reaction was stopped by the addition of SDS-PAGE sample buffer, and the incorporation of 32P into eEF2 was determined by SDS-PAGE followed by staining with Coomassie brilliant blue and autoradiography.
Reproducibility.
All experiments were performed at least three times, with similar outcomes. In the case of Western blots, data from a typical experiment are shown.
RESULTS
Identification of a novel insulin-responsive phosphorylation site in eEF2 kinase.
Our laboratory has a long-standing interest in the mechanisms by which insulin and mTOR signaling regulate eEF2 kinase and the activity of eEF2. It has also previously been shown that conditions that deplete cellular ATP levels and/or stimulate the AMPK lead to increased phosphorylation of eEF2 and activate eEF2 kinase (23, 29). Other studies investigated whether eEF2 kinase was a substrate for AMPK in vitro (9). Recombinant human eEF2 kinase made as a GST fusion protein in Escherichia coli was incubated with a highly purified preparation of AMPK and [γ-32P]ATP. As described elsewhere (9), this led to the identification of three residues that can be phosphorylated by AMPK in vitro, Ser78, Ser366, and Ser398.
Ser366 and Ser398 lie C-terminal to the catalytic domain of the enzyme in a region that contains several sites of phosphorylation (Fig. 1A) (25, 26, 50). Ser398 has not previously been identified as a phosphorylation site in eEF2 kinase, and its phosphorylation is studied in detail elsewhere (9). It appears likely to be a physiological target for AMPK and is not regulated by insulin. Ser78 is also a novel site: it lies N terminal to the catalytic domain, immediately adjacent to the CaM-binding site (16, 34) (Fig. 1A). Given that the activity of eEF2 kinase is normally completely dependent upon Ca2+ or CaM, this is a critical region of the protein, and phosphorylation here could potentially affect its binding to CaM.
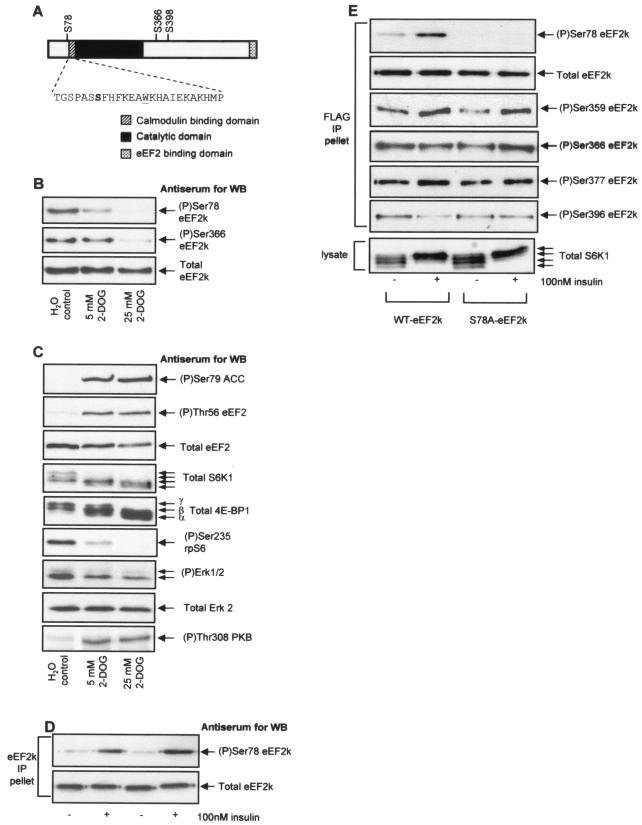
FIG. 1.
The treatment of cells with 2-DOG decreases the phosphorylation of several sites in eEF2 kinase. KB cells (70% confluent) were incubated in DMEM (containing 1 g of d-glucose per liter) plus fetal calf serum and treated with 5 or 25 mM 2-DOG for 30 min. (A) Ser78 is located adjacent to the CaM-binding site in eEF2 kinase. The CaM-binding, catalytic, and substrate-binding domains are indicated, as is the amino acid sequence (single-letter code) of the CaM-binding region. Also indicated are other relevant sites of phosphorylation within eEF2 kinase. (B) eEF2 kinase was immunoprecipitated from 100 μg of cell lysate protein and subjected to SDS-PAGE before Western blotting for total eEF2 kinase or phosphorylated eEF2 kinase by using the indicated phosphospecific antisera. (C) A total of 20 μg of cell lysate protein was subjected to SDS-PAGE and Western blotting for phosphorylated and total proteins as indicated. The α to γ species of 4E-BP1 and the different species of S6K1 (which reflect differing states of phosphorylation) are indicated. (D) KB cells (90% confluent) were starved of serum for 16 h and then treated with insulin for 30 min as indicated. eEF2 kinase was immunoprecipitated from 100 μg of cell lysate protein and subjected to SDS-PAGE before Western blotting for total eEF2 kinase or eEF2 kinase phosphorylated at Ser78. (E) KB cells were transfected with vectors encoding wild-type eEF2 kinase or a mutant in which Ser78 had been converted to alanine, each with an epitope (FLAG) tag. Cells were starved of serum and then in some cases treated with insulin (30 min) prior to lysis. The exogenous eEF2 kinase was then immunoprecipitated from the lysates by using anti-FLAG. Immunoprecipitates or cell lysates (as indicated) were then analyzed by SDS-PAGE and Western blotting for total eEF2 kinase, phosphorylated eEF2 kinase using the indicated phosphospecific antisera, or S6K1. eEF2k, eEF2 kinase; WB, Western blotting; WT, wild type; IP, immunoprecipitation.
To study whether Ser78 is phosphorylated in vivo, we generated appropriate phosphospecific antisera, as described in Materials and Methods. A clear signal was observed with the anti-phospho-Ser78 antibody. However, this signal became weaker rather than stronger under conditions where AMPK is activated (i.e., when cells were treated with 2-deoxyglucose [2-DOG]) (Fig. 1B). Phosphorylation of Ser78 was abolished at the higher concentration of 2-DOG used here. Analysis with an antibody that recognizes Ser79 in acetyl-coenzyme A carboxylase, a known substrate for AMPK, confirmed that 2-DOG does indeed activate AMPK in KB cells (Fig. 1C). Thus, although Ser78 does appear to be phosphorylated in vivo, it seems unlikely to be a target for AMPK, and its phosphorylation by AMPK may only be an in vitro phenomenon. It was thus possible that the phosphorylation of this site might be regulated under other conditions in vivo, and we study this in the experiments described below.
Why should phosphorylation of Ser78 in eEF2 kinase be decreased by 2-DOG? Previous studies (15) have indicated that, in addition to activating AMPK, severe ATP depletion may impair mTOR signaling. In KB cells, increasing 2-DOG concentrations does indeed lead to a progressive decrease in the phosphorylation states of two targets of the mTOR pathway, S6K1 and 4E-BP1, and of rpS6, a physiological substrate of S6K1 (Fig. 1C). 2-DOG also caused a modest decrease in the phosphorylation (activation state) of Erk but actually increased the phosphorylation of an activating site in PKB. The fact that phosphorylation of Ser78 decreased under conditions of ATP depletion raised the possibility that it might be mediated via signaling through mTOR or perhaps Erk. We have previously shown that phosphorylation of Ser366 in eEF2 kinase can be mediated by signaling via mTOR and/or MEK; it is phosphorylated by kinases that lie downstream of these signaling proteins, i.e., S6K1 and p90RSK, respectively (50). The data in Fig. 1B show that Ser366 undergoes dephosphorylation in response to 2-DOG treatment consistent with the inactivation of mTOR and Erk signaling, although it was less sensitive to 2-DOG than the phosphorylation of Ser78.
Insulin activates mTOR signaling and thereby brings about the dephosphorylation of eEF2 (40, 48, 50). We therefore tested whether this hormone affected the state of phosphorylation of Ser78. As shown in Fig. 1D, insulin caused a marked increase in the signal observed with the anti-phospho-Ser78 antiserum, indicating that phosphorylation of this site is increased by insulin. However, it was important to verify that the anti-phospho-Ser78 antibody really detects only phosphorylation at this site in eEF2 kinase in vivo. To do this, we transfected KB cells with vectors encoding either wild-type eEF2 kinase or a mutant in which Ser78 has been altered to alanine. As shown in Fig. 1E, whereas a clear insulin-stimulated signal was observed for the wild-type eEF2 kinase by using the anti-phospho-Ser78 antibody, no signal whatsoever was observed for the Ser78Ala mutant. These data confirm that this reagent is indeed specific for eEF2 kinase phosphorylated at Ser78. We also show that mutation of Ser78 in eEF2 kinase to alanine does not affect the phosphorylation status of the other known phosphorylation sites in eEF2 kinase. This rules out Ser78 as a priming phosphorylation site for the sequential phosphorylation of the other known insulin-responsive phosphorylation sites in eEF2 kinase such as Ser359 and Ser366 (Fig. 1E).
The phosphorylation of Ser78 is regulated by insulin in an mTOR-dependent manner.
The above data show that Ser78 is thus an insulin-sensitive phosphorylation site in eEF2 kinase. To test whether the mTOR or extracellular signal-regulated kinase (ERK) signaling pathway(s) mediates its phosphorylation, we studied whether the phosphorylation of Ser78 was affected by agents that inhibit signaling through these pathways. As shown in Fig. 2A, insulin decreases the phosphorylation of eEF2 in KB cells, and this effect is blocked by rapamycin, indicating that it requires signaling through mTOR, as in other cell types (40, 48, 50). Examination of the phosphorylation of a key mTOR-sensitive site in S6K1 (Thr389) and of the S6K substrate rpS6 confirmed that rapamycin is completely effective at the concentration used here (Fig. 2B). As mentioned above, a basal signal is seen for the phosphorylation of Ser78 in serum-starved cells, and this signal is increased markedly in response to insulin. Rapamycin completely eliminated both the basal and insulin-stimulated phosphorylation of Ser78, demonstrating quite clearly that it is regulated via mTOR signaling (Fig. 2C). Wortmannin largely blocked the ability of insulin to bring about the dephosphorylation of eEF2 in KB cells (Fig. 2A), the phosphorylation of PKB, and the increases in phosphorylation of S6K1 and rpS6 seen in response to insulin, consistent with earlier data showing that PI 3-kinase signaling regulates the mTOR pathway in response to insulin (30) (Fig. 2B). This compound also prevented the increased phosphorylation of Ser78 that occurs in response to insulin, although it did not affect its basal phosphorylation (Fig. 2C).
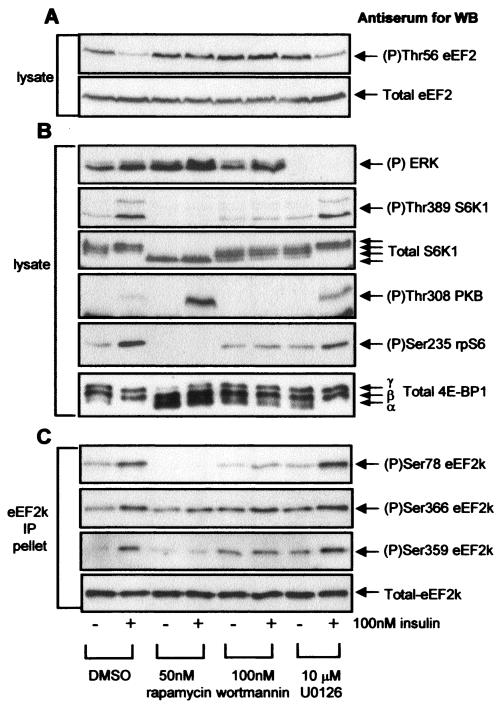
FIG. 2.
The phosphorylation of Ser78 in eEF2 kinase is regulated by insulin in a rapamycin-sensitive manner. KB cells (90% confluent) were starved of serum for 16 h, pretreated with signaling inhibitors for 30 min, and then treated with insulin for 30 min as indicated. (A) Cell lysate protein (20 μg) was subjected to SDS-PAGE followed by Western blotting for eEF2 phosphorylated at Thr56 or total eEF2. (B) Cell lysate protein (20 μg) was subjected to SDS-PAGE followed by Western blotting for phosphorylated or total proteins as indicated (see also legend to Fig. 1B). (C) Endogenous eEF2 kinase was immunoprecipitated from 100 μg of lysate protein and subjected to SDS-PAGE followed by Western blotting for phosphorylated or total eEF2 kinase, as indicated. eEF2k, eEF2 kinase; WB, Western blotting; DMSO, dimethyl sulfoxide; IP, immunoprecipitation.
In contrast, the ability of insulin to induce the phosphorylation of Ser78 in eEF2 kinase was not affected by an inhibitor of the MEK/ERK pathway, U0126 (Fig. 2C), even though this compound completely blocked basal and stimulated Erk phosphorylation in these cells (Fig. 2B). Insulin thus elicits dephosphorylation of eEF2 through signaling events that require PI 3-kinase and mTOR but not Erk signaling. Exactly the same pattern was seen for the insulin-induced phosphorylation of Ser78 in eEF2 kinase, suggesting that its phosphorylation might play an important role in the regulation of the activity of eEF2 kinase. In contrast, the phosphorylation state of Ser366, a target for S6K1 (50), does not correlate with the regulation of eEF2 phosphorylation. In particular, its phosphorylation is not affected by rapamycin (Fig. 2C) even though this drug abolishes the effects of insulin on eEF2 phosphorylation. This lack of effect of rapamycin on phosphorylation at Ser366 likely reflects an input from Erk signaling (e.g., via p90RSK) (50) which is not blocked by rapamycin (Fig. 2B).
An important point to note here is that whereas rapamycin blocks both the dephosphorylation of eEF2 and the phosphorylation of Ser78, it does not block the phosphorylation of Ser366. This may be because phosphorylation of Ser366 is already high in KB cells, presumably due to basal activity of S6K1 and p90RSK. These data suggest that Ser366 does not play a major role in the regulation of eEF2 kinase activity by insulin in these cells. In contrast, changes in the phosphorylation of Ser78 and of eEF2 do mirror one another.
The multisite phosphorylation of other targets of mTOR signaling—S6 kinases and 4E-BP1—is highly hierarchical (4, 21), and this could also apply to eEF2 kinase. However, these data show that phosphorylation of Ser78 is regulated independently of the phosphorylation of Ser366 and vice versa.
Ser78 is not a target for S6K1.
The data above show that the phosphorylation of Ser78 is regulated through mTOR signaling. The only protein kinases known to lie downstream of mTOR are the S6 kinases. To test whether they play a role in the phosphorylation of eEF2 kinase at Ser78, we made use of the compound Ro31-8220, which inhibits S6 kinase in vitro (1). As shown in Fig. 3, Ro31-8220 inhibits S6K1 within KB cells, i.e., it completely blocks the phosphorylation of rpS6, but does not affect the phosphorylation of S6K1 or 4E-BP1. This result indicates that Ro31-8220 completely blocks S6 kinase activity without affecting upstream mTOR signaling. Ro31-8220 had no effect on the basal phosphorylation of Ser78 in eEF2 kinase or on its regulation by insulin (Fig. 3). This result shows that Ser78 is not a direct substrate for the S6 kinases and also implies that the kinase that phosphorylates this site is not activated by the S6 kinases. Ser78 is, thus, presumably the target of an mTOR-regulated kinase other than the S6 kinases.
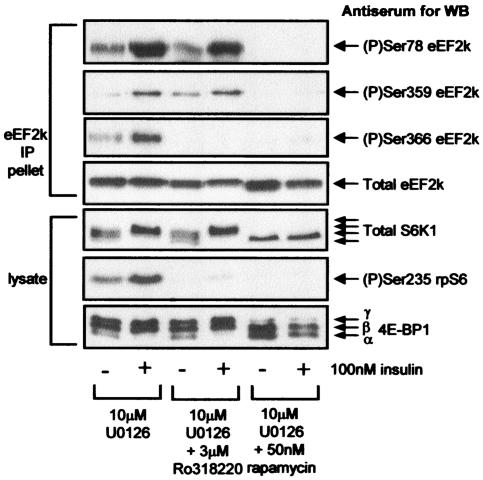
FIG. 3.
The phosphorylation of Ser78 in eEF2 kinase is insensitive to the broad specificity AGC kinase inhibitor Ro31-8220. Serum-starved (16 h) 90% confluent KB cells were pretreated with signaling inhibitors for 30 min and then with insulin for 30 min as indicated. eEF2 kinase was immunoprecipitated from 100 μg of lysate (IP pellet), or 20 μg of cell lysate protein was analyzed directly. Samples were subjected to SDS-PAGE followed by Western blotting (WB) for phosphorylated or total proteins, as indicated. eEF2k, eEF2 kinase.
Phosphorylation of eEF2 kinase at Ser366 was not markedly decreased either by rapamycin or U0126 when they were used individually (Fig. 2C). However, when U0126 was used in combination with either rapamycin or Ro31-8220, phosphorylation at this site was completely eliminated (Fig. 3). This finding is consistent with the idea that two separate pathways (S6K and p90RSK) provide input to the phosphorylation of Ser366.
The regulation of the phosphorylation of Ser78 depends on the amino acid status of the cells.
It is well established that in a range of cell types, amino acids positively regulate mTOR signaling. Thus, when cells are deprived of external amino acids, insulin fails to activate S6K1 or bring about the phosphorylation of 4E-BP1 (see, e.g., references 11, 22, and 49). Since the phosphorylation of Ser78 in eEF2 kinase is regulated in a rapamycin-sensitive manner, we anticipated that its phosphorylation would be influenced by the amino acid status of the cells. Previous work has shown that in CHO cells, amino acids are required for the basal phosphorylation of such targets of mTOR as 4E-BP1 and S6K1 and for their efficient regulation by insulin (7, 11, 49). When CHO cells were deprived of amino acids, we observed the expected dephosphorylation of 4E-BP1 and of S6K1 (a shift to more rapidly migrating forms), whereas dephosphorylation did not occur if the cells were kept in a medium containing amino acids (Fig. 4A). In amino acid-deprived cells, insulin fails to elicit the phosphorylation of 4E-BP1 or S6K1 (Fig. 4A) (7). Similarly, amino acid withdrawal caused the loss of the basal phosphorylation of Ser78 in eEF2 kinase and of the ability of insulin to increase phosphorylation at this site (Fig. 4A). It has recently been shown that treatment of amino acid-deprived CHO cells with protein synthesis inhibitors such as CHX restores basal and insulin-stimulated mTOR signaling (7). Consistent with this finding, treatment of amino acid-deprived CHO cells with CHX caused a slight rise in the basal phosphorylation of Ser78 and, much more strikingly, allowed insulin to elicit an increase in its phosphorylation to levels similar to those seen in cells kept in amino acids (Fig. 4A). These data are fully consistent with the earlier conclusion that phosphorylation of Ser78 is dependent upon intact mTOR signaling. They also demonstrate that Ser78 in eEF2 kinase is phosphorylated in CHO cells and indicate that, as in KB cells, its phosphorylation is controlled through mTOR signaling.
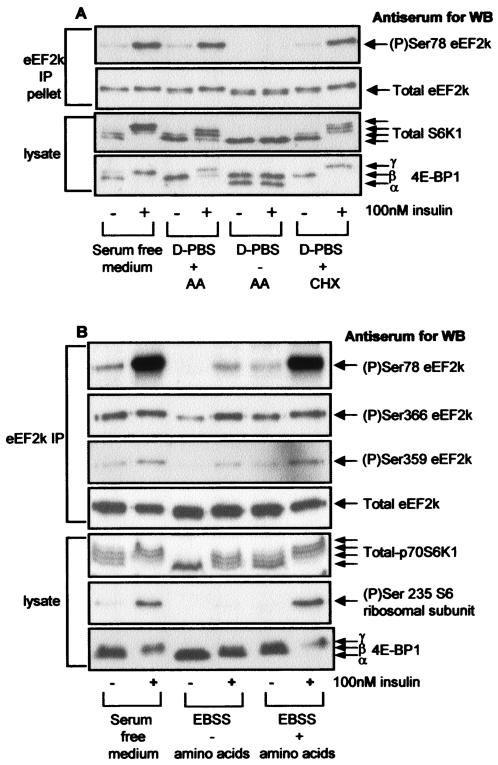
FIG. 4.
Phosphorylation of Ser78 in eEF2 kinase is sensitive to amino acid withdrawal. (A) CHO.K1 cells (90% confluent) were starved of serum for 16 h, preincubated for 1 h in serum-free Ham's F12 medium, Dulbecco's phosphate-buffered saline (D-PBS) containing 1 g of glucose per liter, or the latter containing 1 g of d-glucose per liter plus amino acids (AA) prior to treatment with insulin for 30 min as indicated. Where used, CHX was added 30 min prior to insulin stimulation. eEF2 kinase was immunoprecipitated from 100 μg of lysate protein or 20 μg of cell lysate protein and subjected to SDS-PAGE followed by Western blotting for phosphorylated and total proteins as indicated. (B) The method was the same as in panel A except that KB cells were used and were starved of amino acids for 4 h in Earle's balanced salts solution (EBSS); no treatments with CHX were included. Samples were analyzed as in panel A, except that Western blotting was also performed for phosphorylated rpS6 (Ser235) and for phosphorylation of Ser366 and Ser359 in eEF2 kinase. WB, Western blotting; IP, immunoprecipitation; eEF2k, eEF2 kinase.
Similar data were obtained for the effects of amino acid withdrawal in KB cells (Fig. 4B) except that amino acid deprivation does not completely prevent insulin-induced phosphorylation of S6K1. It nevertheless reduces basal phosphorylation of S6K1 and prevents its full activation by insulin, as shown by the failure of this hormone to increase the phosphorylation of rpS6 in cells deprived of amino acids (indeed, rpS6 phosphorylation was undetectable in the absence of amino acids). Most importantly, insulin had almost no effect on the phosphorylation of Ser78 in eEF2 kinase in amino acid-starved cells. In contrast, insulin still induced phosphorylation of Ser366 in the amino acid-deprived cells, likely because it still activates Erk signaling under these conditions (Fig. 4B).
Regulated, but not basal, phosphorylation of Ser78 requires PDK1.
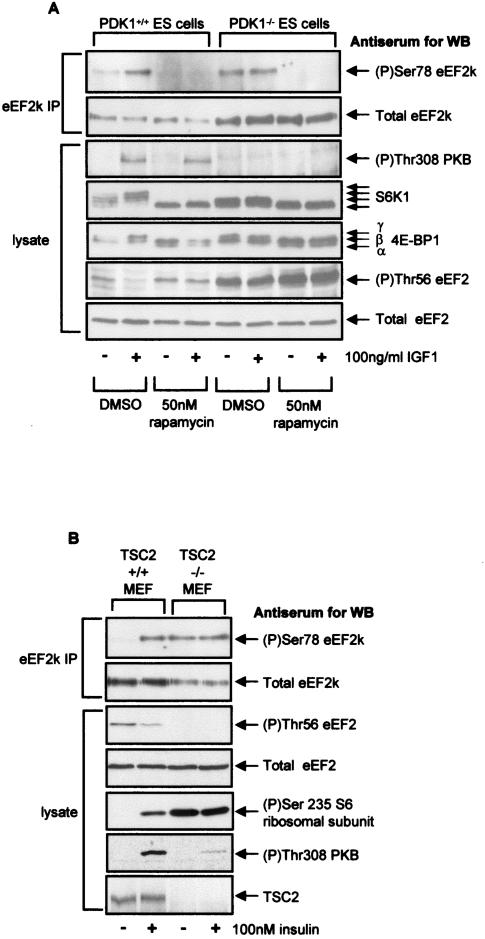
Phosphoinositide-dependent kinase 1 (PDK1) phosphorylates threonine residues in the T loop of a number of members of the AGC family of protein kinases (51). This modification is required for the activation of these protein kinases, which include PKB (Thr308) and S6K1 (Thr229) (2, 14, 38, 46). It has previously been shown that the ability of insulin to elicit dephosphorylation of eEF2 and inactivation of eEF2 kinase is lost in cells lacking PDK1, although basal mTOR signaling is still functional in these cells (50). If phosphorylation of eEF2 kinase at Ser78 plays a role in controlling its activity, we would anticipate that regulation of Ser78 phosphorylation would be lost in PDK1−/− cells. It was therefore important to test whether loss of PDK1 affected regulation of the phosphorylation of Ser78. To do this, we used mouse ES cells that express PDK1 or in which this gene has been disrupted and which thus lack functional PDK1 (51). The absence of PDK1 from these cells was confirmed by the lack of signal for the phosphorylation of Thr308 in PKB in the PDK1−/− cells in response to IGF1, whereas a signal is clearly seen in the PDK1+/+ cells (Fig. 5A). The overall level of eEF2 kinase is substantially higher in the PDK1−/− cells than in the PDK1+/+ controls, resulting in a higher basal level of phosphorylated eEF2 in the former (Fig. 5A).
FIG. 5.
Phosphorylation of Ser78 in eEF2 kinase is dysregulated in cells lacking PDK1 or TSC2. (A) Ninety percent confluent ES cells (PDK1+/+ or PDK1−/− as indicated) were serum starved for 3 h and then pretreated with rapamycin (or dimethyl sulfide [DMSO] as a control) for 30 min prior to IGF1 treatment for 30 min. eEF2 kinase immunoprecipitated from 500 μg of cell lysate protein or 20 μg of total cell lysate protein was subjected to SDS-PAGE followed by Western blotting for phosphorylated and total proteins as indicated. The positions of the differently phosphorylated forms of S6K1 and 4E-BP1 are indicated by arrows. (B) Ninety percent confluent mouse embryo fibroblasts (TSC2+/+ or TSC2−/− as indicated) were serum starved for 3 h and then, where indicated, treated with 100 nM insulin for 30 min. eEF2 kinase that had been immunoprecipitated from 500 μg of cell lysate protein or 20 μg of total cell lysate protein was subjected to SDS-PAGE followed by Western blotting for phosphorylated and total proteins as indicated. eEF2k, eEF2 kinase; WB, Western blotting; IP, immunoprecipitation.
To examine the control of Ser78 phosphorylation in these two types of cells, some dishes were treated with IGF1 prior to extraction, and samples of lysates from IGF1-treated or control cells were subjected to immunoprecipitation with anti-eEF2 kinase antibody. Precipitates were analyzed by SDS-PAGE and Western blotting by using the anti-phospho-Ser78-specific antibody. Basal phosphorylation of Ser78 was observed in both cases, and this was blocked by rapamycin. This result shows that PDK1 is not required for the basal phosphorylation of Ser78, confirming that basal mTOR signaling is functional in PDK1−/− cells, in line with previously published data (50). While IGF1 treatment increases the phosphorylation of Ser78 in eEF2 kinase in PDK1+/+ cells, it fails to do so in PDK1−/− cells. A very similar situation is seen for the phosphorylation of S6K1 and 4E-BP1 (Fig. 5A). (The appearance of S6K as two higher-mobility forms in PDK1−/− cells probably reflects the fact that PDK1 directly acts on this enzyme [46].) These findings provide further evidence that S6K1 is not involved in the phosphorylation of Ser78 (since its phosphorylation would be completely lost in the knockout cells) and suggest that the activation of the phosphorylation of Ser78, but not its basal phosphorylation, requires PDK1. It is therefore likely that PDK1 is required for the activation of mTOR signaling by IGF1 (50). The loss of regulation of phosphorylation of Ser78 seen in PDK1−/− cells in response to IGF1 is consistent with an important role for this site in regulating eEF2 kinase activity as such regulation is also lost in these cells (50). Phosphorylation of Ser366 is completely absent in the PDK1−/− cells, consistent with the requirement of PDK1 for activation of both S6 kinase and p90RSK (51; G. J. Browne, unpublished data). The residual rapamycin sensitivity of eEF2 phosphorylation in the PDK1−/− cells, therefore, cannot be due to changes in the phosphorylation of Ser366 and likely reflects the modulation of the phosphorylation of eEF2 kinase at Ser78, which does remain sensitive to this drug in the knockout cells.
Ser78 in eEF2 kinase is constitutively phosphorylated in TSC2−/− cells.
To further confirm that Ser78 in eEF2 kinase is regulated through mTOR signaling, we made use of the availability of cells deficient in an upstream repressor of mTOR signaling, TSC2. We therefore expected mTOR signaling to be constitutively active in such cells, e.g., even in the absence of insulin (30, 52). To verify this assumption, we assessed the phosphorylation state of rpS6, which was already high in TSC2−/− cells and was not further increased by insulin but was low and enhanced by insulin in the corresponding TSC2+/+ cells (Fig. 5B). Activation of PKB by insulin was blunted in TSC2−/− cells, consistent with earlier data (52) and with the data shown in Fig. 1C and 2B, which indicate the operation of a feedback loop from activated mTOR signaling that represses signaling to PKB from the insulin receptor. Thus, basal mTOR signaling is indeed highly active in TSC2−/− cells.
In TSC2+/+ cells, basal phosphorylation of Ser78 in eEF2 kinase was undetectable, and phosphorylation of eEF2 was relatively high (Fig. 5B). In response to insulin, Ser78 became phosphorylated and eEF2 itself underwent dephosphorylation, as observed with other cell types elsewhere in this study. In contrast, in the TSC2−/− cells, the phosphorylation at Ser78 was already high in the absence of insulin and was not further increased by the addition of this hormone (Fig. 5B). Conversely, phosphorylation of eEF2 was low (indeed, undetectable) even without insulin treatment. These data provide further, independent evidence that the phosphorylation of Ser78 in eEF2 kinase is indeed regulated through mTOR signaling. Interestingly, the level of eEF2 kinase expression appears to be lower in the TSC2−/− cells than in the TSC2+/+ cells (Fig. 5B). These data agree with the previous finding that in PDK1−/− cells, eEF2 kinase expression is increased relative to the level of expression in wild-type cells (Fig. 5A). This fact suggests that the level of expression of eEF2 kinase may be negatively regulated by the mTOR pathway in addition to its activity.
Insulin reduces the ability of eEF2 kinase to bind CaM in an mTOR-dependent manner, and this effect requires Ser78.
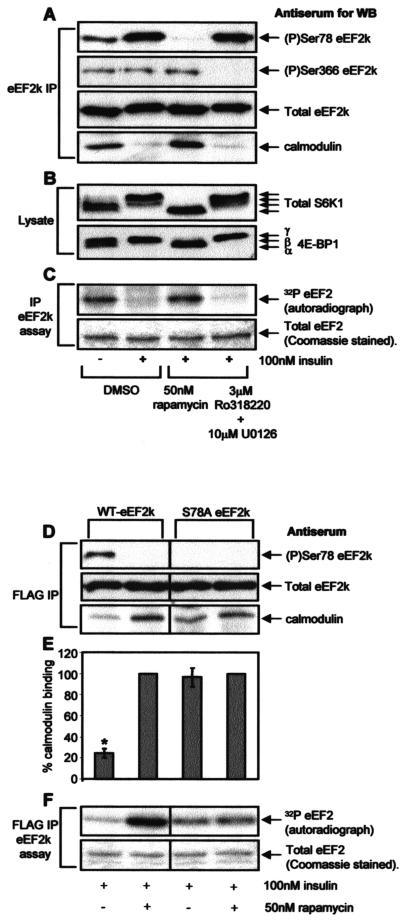
Ser78 lies very close to the CaM-binding motif in eEF2 kinase (Fig. 1A), and phosphorylation could, therefore, affect the binding of eEF2 kinase to CaM. To study whether insulin actually affected the ability of eEF2 kinase to bind CaM, eEF2 kinase was immunoprecipitated from lysates of cells that had been subjected to differing treatments. Immunoprecipitations were performed in the presence of Ca2+ ions in order to preserve the binding of eEF2 kinase to CaM. Insulin treatment of serum-starved KB cells resulted in a sharp reduction in the amount of CaM bound to the immunoprecipitated eEF2 kinase, and this reduction correlated with an increase in phosphorylation at Ser78 (Fig. 6A). The effect of insulin on both CaM binding and Ser78 phosphorylation was blocked by pretreatment with rapamycin but not by pretreatment with a combination of Ro31-8220 and U0126 (Fig. 6A). The data shown in Fig. 6B confirm that rapamycin fully blocks mTOR signaling (as evidenced by the mobility of S6K1 or 4E-BP1), while Ro31-8220 plus U0126 does not affect this signaling. These experiments demonstrate for the first time that insulin treatment causes a marked decrease in the binding of CaM to eEF2 kinase; this effect was blocked by rapamycin.
FIG. 6.
Ser78 is required for the regulation of the binding of CaM to eEF2 kinase by insulin. (A to C) KB cells (90% confluent) were serum starved for 16 h and then pretreated with the indicated signaling inhibitors for 30 min prior to treatment with insulin for 30 min as indicated. The cells were lysed, and immunoprecipitations and eEF2 kinase assays were carried out in a modified extraction buffer containing 1 mM CaCl2 as described in Materials and Methods. In panel A, eEF2 kinase immunoprecipitated from 100 μg of lysate protein was subjected to SDS-PAGE followed by Western blotting for phosphorylated or total eEF2 kinase as indicated. In panel B, cell lysate protein (20 μg) was analyzed by SDS-PAGE and Western blotting using antisera for S6K1 or 4E-BP1, as indicated. Positions of the differently phosphorylated forms of these proteins are shown. In panel C, eEF2 kinase immunoprecipitated from 100 μg of cell lysate protein was incubated with 1 μg of purified eEF2 in the presence of [γ-32P]ATP. Samples were then subjected to SDS-PAGE. Gels were stained with Coomassie and analyzed by autoradiography to assess incorporation of radiolabel into eEF2 (i.e., eEF2 kinase activity, upper section). The lower section shows a portion of the stained gel to confirm equal loading of the substrate, eEF2. (D to F) KB cells were transfected with vectors encoding FLAG-tagged versions of wild-type eEF2 kinase or the Ser78Ala mutant. When cells had reached 90% confluence, they were starved of serum for 16 h and then pretreated with rapamycin for 30 min prior to treatment with insulin as indicated. The FLAG-tagged eEF2 kinase was immunoprecipitated from 100 μg of lysate protein by using immobilized FLAG antibody and subjected to SDS-PAGE followed by Western blotting for phosphorylated and total eEF2 kinase or bound CaM as indicated in panel D. The graph in panel E shows the binding of CaM to wild-type FLAG-eEF2 kinase or the Ser78Ala mutant immunoprecipitated from cells treated with insulin or insulin plus rapamycin. The amount of bound CaM, presented as a percentage of the CaM bound to the FLAG-eEF2 kinase immunoprecipitated from cells treated with insulin plus rapamycin, was determined by using Image/J software (available at rsb.info.nih.gov/ij/). The results are presented as the means ± standard errors of the means (n = 5). Alternatively, in panel F eEF2 kinase activity was determined by incubating the immunoprecipitated eEF2 kinase with 1 μg of purified eEF2 in the presence of [γ-32P]ATP to assay eEF2 kinase activity (see Materials and Methods) (40). Samples were then subjected to SDS-PAGE. Gels were stained with Coomassie brilliant blue and then analyzed by autoradiography to assess incorporation of the label into eEF2 (upper section). The lower section shows a portion of the stained gel to confirm equal loading of eEF2, the substrate. eEF2k, eEF2 kinase; WB, Western blotting; IP, immunoprecipitation; WT, wild type; *, P < 0.005.
Phosphorylation at Ser366 has previously been reported to decrease the sensitivity of eEF2 kinase to activation by Ca or CaM (50), and we were concerned that the effect on CaM binding might involve that site. However, Ro31-8220 plus U0126, a combination which completely blocks the phosphorylation of eEF2 kinase at Ser366 without affecting Ser78 (Fig. 3 and 6A), had no effect on the ability of insulin to decrease the binding of CaM to eEF2 kinase, thus ruling out a role for phosphorylation at Ser366 in the observed modulation of the binding of CaM to eEF2 kinase.
Given that the activity of eEF2 kinase is normally completely dependent upon Ca or CaM, one might anticipate that a decrease in the binding of CaM to eEF2 kinase should diminish its activity. To test this, we assayed the activity of eEF2 kinase in immunoprecipitates with purified eEF2 used as a substrate. No CaM was added to the assays, so that the activity measured reflects the CaM already bound to eEF2 kinase in the lysates or immunoprecipitates. As shown in Fig. 6C, treatment of KB cells with insulin markedly decreased the activity of eEF2 kinase, and this effect was completely abolished by prior treatment of the cells with rapamycin. In contrast, the combination of Ro31-8220 and U0126 had no effect on the activity of eEF2 kinase measured under this condition, ruling out roles for S6K1, p90RSK, or other kinases, such as protein kinase C (PKC), that are inhibited by this compound and therefore also phosphorylation at Ser366.
The data are consistent with the idea that phosphorylation at Ser78, next to the CaM-binding domain, impairs the binding of this essential activator to eEF2 kinase. To test this idea, we made use of a mutant of eEF2 kinase in which Ser78 is altered to alanine. Wild-type eEF2 kinase or the Ser78Ala mutant were expressed in KB cells, which were subsequently serum starved and then treated with insulin in the absence or presence of rapamycin. Lysates were prepared, and the exogenous (FLAG-tagged) eEF2 kinase was immunoprecipitated from them in the presence of Ca2+ ions to preserve binding to CaM. As shown in Fig. 6D, rapamycin greatly increased the binding of CaM to overexpressed wild-type eEF2 kinase in insulin-treated cells. In contrast, the amount of CaM bound to overexpressed eEF2 kinase (S78A) was very similar for the cells treated with insulin and those treated with insulin plus rapamycin (and also similar to the amount bound to the wild-type enzyme after treatment of the cells with rapamycin). The graph in Fig. 6E shows quantitative data for the effect of insulin or insulin plus rapamycin on CaM binding to wild-type or Ser78Ala eEF2 kinase from five independent experiments. As indicated, there is only a statistically significant difference in the amount of CaM bound to the wild-type eEF2 kinase under these different treatments. These data provide strong support for the idea that phosphorylation at Ser78 decreases CaM binding and that phosphorylation of this site is sufficient to elicit this effect. This phosphorylation is expected to decrease eEF2 kinase activity, an effect which is indeed observed in response to insulin (40, 48, 50).
As shown in Fig. 6C and F, the activity of wild-type eEF2 kinase was markedly increased by rapamycin treatment in cells treated with insulin, while this drug had little effect on the activity of the Ser78Ala mutant (Fig. 6F). These activity measurements closely parallel the effects reported above for CaM binding. Taken together, the data strongly suggest that phosphorylation at Ser78 plays a major role in the control of eEF2 kinase activity by insulin or mTOR signaling and that this regulation is linked to the role of this site in modulating the binding of eEF2 kinase to its essential activator, CaM.
DISCUSSION
The regulation of mRNA translation plays a key role in controlling gene expression, cell growth, and cell size in metazoan organisms, and the mTOR signaling pathway plays a central role in this regulation (40). A variety of components of the translational machinery are regulated by phosphorylation, and in several cases this regulation is linked to mTOR signaling. Most previous studies have focused on the control of translation initiation, and relatively little is known about the control of elongation.
In this study, we report three main findings. First, we identify a novel, physiological phosphorylation site in eEF2 kinase that is controlled through the mTOR signaling pathway (Ser78). Insulin increases its phosphorylation, and this effect is blocked by rapamycin. Second, we show that Ser78 is not a substrate for the S6 kinases, demonstrating that eEF2 kinase is regulated by mTOR signaling independently of known effectors of the pathway. The evidence for this conclusion is summarized below. eEF2 is thus the third well-characterized target for TOR signaling in mammalian cells (the others being the S6 kinases and 4E-BP1). Third, we provide evidence that phosphorylation at Ser78 plays a crucial role in regulating the binding of CaM to eEF2 kinase and thus in regulating its activity. Insulin decreases the binding of CaM to eEF2 kinase and also decreases eEF2 kinase activity. Both effects correlate with increased phosphorylation at Ser78, which lies immediately next to the CaM-binding site in eEF2 kinase, and both are blocked by rapamycin. The mutation of Ser78 to a residue which cannot be phosphorylated (Ala) almost completely eliminates the opposing effects of insulin and rapamycin on CaM binding and eEF2 kinase activity. These data therefore imply that phosphorylation at other sites in eEF2 kinase play little role in regulating its binding to CaM, at least in response to insulin.
In many cases, CaM-binding sites contain both positively charged (basic) and hydrophobic residues, and this is also the case in eEF2 kinase (Fig. 1A) (17, 34). It seems likely that by introducing a negative charge close to the CaM binding site in eEF2 kinase, phosphorylation at Ser78 weakens its affinity for CaM. Evidence for this idea includes our observations (i) that the effect of insulin on CaM binding is blocked by rapamycin, which completely eliminates phosphorylation at Ser78, and (ii) that the mutation of Ser78 to Ala almost completely abolishes the effects of rapamycin or insulin on CaM binding and eEF2 kinase activity.
Although Ser78 can be phosphorylated by AMPK in vitro, its phosphorylation in vivo changes in the opposite direction from what would be expected if it were a substrate for this kinase in vivo. Ser78 shows basal phosphorylation under conditions where AMPK is not activated, and its phosphorylation actually falls during ATP depletion. These observations and the facts that insulin increases the level of phosphorylation of this site and that this effect is blocked by rapamycin make it most unlikely that Ser78 is a true physiological target for AMPK. Rapamycin and insulin either do not affect AMPK activity or may do so in the opposite direction; for example, insulin has been reported to antagonize AMPK activation (6). We also show that the ability of insulin to bring about the phosphorylation of Ser78 is lost when cells are starved of amino acids. This finding is consistent with a large body of earlier work which showed that amino acids positively regulate mTOR signaling in mammalian cells (reviewed in reference 37) and makes particularly good sense in terms of the control of elongation. Almost all the amino acids used by translation are consumed during elongation. The loss of phosphorylation of Ser78 in amino acid-deprived cells will tend to result in higher activity of eEF2 kinase, leading to inhibition of eEF2, thereby reducing the consumption of amino acids.
It has previously been shown that the ability of IGF1 to induce the dephosphorylation of eEF2 and the inactivation of eEF2 kinase is abolished in cells that lack functional PDK1 (50). We show that phosphorylation at Ser78 is important in the regulation of eEF2 kinase activity, as the inability of IGF1 to induce the phosphorylation of Ser78 in PDK1−/− cells was associated with a failure of IGF1 to induce the dephosphorylation of eEF2. However, basal phosphorylation of Ser78 is still observed, indicating that the activity of the kinase that directly acts upon it is not dependent upon PDK1, whereas its activation by insulin probably is. Consistent with this model, we observed that the PI 3-kinase wortmannin inhibitor blocks the insulin-induced phosphorylation of Ser78 but not its basal phosphorylation. These events likely reflect the role of PI 3-kinase and PKB in signaling from the insulin or IGF1 receptors to the mTOR pathway, which involves the proteins TSC1 and TSC2 (reviewed in reference 30). Treatment of cells with 2-DOG or rapamycin (both of which impair mTOR signaling) led to an increase in the basal phosphorylation of Thr308 in PKB. This result may reflect the operation of a feedback loop from mTOR signaling which serves to impair the phosphorylation of PKB. Other evidence for such a loop has been reported (45).
In some cases, phosphorylation at a serine residue may be mimicked by conversion of the serine residue to an acidic one (Asp or Glu). We prepared such mutants of eEF2 kinase (Ser78Asp and Ser78Glu); however, neither appeared to mimic phosphorylation. In both bases, the degree of CaM binding was high in rapamycin-treated cells and was not reduced by rapamycin. In this respect, these mutants resemble the Ser78Ala mutant (Fig. 6). A substantial number of other instances are known where Asp or Glu again fail to mimic phosphoserine.
We do not know the identity of the mTOR- and insulin-regulated kinase that phosphorylates Ser78 in vivo. The fact that the phosphorylation of Ser78 is completely blocked by rapamycin suggests that it is regulated by mTOR. However, the amino acid sequence around Ser78 differs from the sequences around mTOR-regulated sites in other proteins. Ser366 in eEF2 kinase and the sites in S6 (which are targets for the S6 kinases) lie in RXRXX(S/T) sequences characteristic of substrates for certain other AGC family kinases. The fact that Ser78 does not lie in such a motif likely accounts for the fact that it does not appear to be a direct target for S6 kinases (50). Thr389/412 in S6K1 (35) and the rapamycin-sensitive sites in certain PKC isoforms are in highly conserved ϕXXϕ(T/S)ϕ sequences (where ϕ denotes a hydrophobic residue) (33). Although the residue that follows Ser78 is hydrophobic (Phe), the residues N terminal to it do not conform to this pattern. Lastly, several mTOR-regulated sites in 4E-BP1 and the S6Ks are followed by prolines, which is not the case for Ser78. Both Thr389/412 in S6K1 and some sites in 4E-BP1 can be phosphorylated by mTOR in vitro. The fact that the local sequence around Ser78 does not resemble either of these motifs likely explains why it cannot be phosphorylated by mTOR in vitro. This inability may also reflect the fact that eEF2 kinase lacks the TOR-signaling motif found in proteins that can be phosphorylated directly by mTOR, i.e., the S6 kinases and 4E-BP1 (42). Recent work has shown that phosphorylation of several sites in 4E-BP1 and S6K1 is dependent upon this motif (13, 32, 32, 43) which acts to recruit mTOR to these substrates via its associated scaffold protein raptor. In a range of experiments, we have been unable to show that mTOR phosphorylates Ser78 in eEF2 kinase in vitro under conditions where it readily phosphorylates 4E-BP1 (E. Smith, unpublished data). Nonetheless, the effects of rapamycin on the phosphorylation of Ser78 and the requirement of amino acids for its control by insulin are entirely consistent with this residue being a target for control by the mTOR signaling pathway. Furthermore, the data from the TSC2−/− cells, in which Ser78 is constitutively highly phosphorylated, provide further strong evidence that phosphorylation of this site is controlled by mTOR signaling.
The only well-understood kinases that lie downstream of mTOR are the S6 kinases. However, as mentioned above, several lines of evidence indicate that they are not the enzymes that act at Ser78. First, as mentioned above, Ser78 does not lie in the RXRXXS consensus typical of other substrates for S6Ks (and other AGC family kinases). Second, S6K1 cannot phosphorylate Ser78 in vitro (reference 50 and data not shown). Third, the ability of S6K1 to phosphorylate eEF2 kinase is lost when Ser366 is mutated to Ala, indicating that Ser366 is the only S6K1 site in eEF2 kinase (50). Fourth, basal phosphorylation of Ser78 is still observed in PDK1−/− cells, which are devoid of S6K activity (51). Fifth, phosphorylation of Ser78 is not blocked by treatment of cells with Ro31-8220 at concentrations which completely inhibit S6K1. It has also been reported that certain PKC isoforms are regulated in an mTOR-dependent manner (33). However, these enzymes (PKCδ and PKCɛ) can be excluded as candidates for phosphorylating Ser78 in eEF2 kinase because (i) their expression is abolished, or almost abolished, in PDK1−/− cells (5) where phosphorylation of Ser78 is still observed and (ii) because phosphorylation of Ser78 is not blocked by the broad specificity PKC inhibitor Ro31-8220. Furthermore, it has also been demonstrated that the activity of PKC isoforms is dependent on phosphorylation within the T loop for activity and that PDK1 may be responsible for this phosphorylation in vivo. PKCδ and PKCɛ would not, therefore, be expected to be catalytically active in PDK1−/− cells (18, 27).
Thus, our data point to eEF2 kinase being the third independent target of mTOR signaling in mammalian cells. Identification of the kinase that directly phosphorylates Ser78 is now a high priority.
In summary, we have identified a novel, physiological phosphorylation site in eEF2 kinase that regulates its activity. This site is a target for regulation by the mTOR pathway, a key regulatory cascade which, despite its manifest importance for cell regulation, remains poorly understood. The phosphorylation of Ser78 serves to impair the binding of CaM to eEF2 kinase, and thus eEF2 kinase activity, in response to hormones such as insulin and to the availability of amino acids, key precursors for protein synthesis.
Acknowledgments
This work was supported by a project grant from the BBSRC.
We are grateful to Nick Morrice and David Campbell (University of Dundee) for valuable assistance with protein chemistry and mass spectrometry, and to Nick Redpath (formerly of Leicester University), Richard Lamb (Institute of Cancer Research, London, United Kingdom), Grahame Hardie, Dario Alessi, and Axel Knebel (all from the University of Dundee) for generous gifts of antisera and other reagents.
REFERENCES
- 1.Alessi, D. R. 1997. The protein kinase C inhibitors Ro31-8220 and GF109203X are equally potent inhibitors of MAPKAP kinase-1β and p70 S6 kinase. FEBS Lett. 402:121-123. [DOI] [PubMed] [Google Scholar]
- 2.Alessi, D. R., M. T. Kozlowski, Q.-P. Weng, N. Morrice, and J. Avruch. 1998. 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr. Biol. 8:69-81. [DOI] [PubMed] [Google Scholar]
- 3.Aoki, M., E. Blazek, and P. K. Vogt. 2001. A role for the kinase mTOR in cellular transformation induced by the oncoproteins P3K and Akt. Proc. Natl. Acad. Sci. USA 98:136-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avruch, J., C. Belham, Q. Weng, K. Hara, and K. Yonezawa. 2001. The p70 S6 kinase integrates nutrient and growth signals to control translational capacity. Prog. Mol. Subcell. Biol. 26:115-154. [DOI] [PubMed] [Google Scholar]
- 5.Balendran, A., G. R. Hare, A. Kieloch, M. R. Williams, and D. R. Alessi. 2000. Further evidence that 3-phosphoinositide-dependent protein kinase-1 (PDK1) is required for the stability and phosphorylation of protein kinase C (PKC) isoforms. FEBS Lett. 484:217-223. [DOI] [PubMed] [Google Scholar]
- 6.Beauloye, C., A. S. Marsin, L. Bertrand, U. Krause, D. G. Hardie, J. L. Vanoverschelde, and L. Hue. 2001. Insulin antagonizes AMP-activated protein kinase activation by ischemia or anoxia in rat hearts, without affecting total adenine nucleotides. FEBS Lett. 505:348-352. [DOI] [PubMed] [Google Scholar]
- 7.Beugnet, A., A. R. Tee, P. M. Taylor, and C. G. Proud. 2002. Evidence that intracellular amino acids regulate translation factor function in mammalian cells. Biochem. J. 372:555-566. [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 77:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Browne, G. J., S. G. Finn, and C. G. Proud. 5 January 2004. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. J. Biol. Chem. http://www.jbc.org/cgi/reprint/M309773200v1. [DOI] [PubMed]
- 10.Browne, G. J., and C. G. Proud. 2002. Regulation of peptide-chain elongation in mammalian cells. Eur. J. Biochem. 269:5360-5368. [DOI] [PubMed] [Google Scholar]
- 11.Campbell, L. E., X. Wang, and C. G. Proud. 1999. Nutrients differentially modulate multiple translation factors and their control by insulin. Biochem. J. 344:433-441. [PMC free article] [PubMed] [Google Scholar]
- 12.Carlberg, U., A. Nilsson, and O. Nygard. 1990. Functional properties of phosphorylated elongation factor 2. Eur. J. Biochem. 191:639-645. [DOI] [PubMed] [Google Scholar]
- 13.Choi, K.-M., L. P. McMahon, and J. C. Lawrence. 2003. Two motifs in the translational repressor PHAS-I required for efficient phosphorylation by mammalian target of rapamycin and for recognition by raptor. J. Biol. Chem. 278:19667-19673. [DOI] [PubMed] [Google Scholar]
- 14.Cohen, P., D. R. Alessi, and D. A. E. Cross. 1997. PDK1, one of the missing links in insulin signal transduction? FEBS Lett. 410:3-10. [DOI] [PubMed] [Google Scholar]
- 15.Dennis, P. B., A. Jaeschke, M. Saitoh, B. Fowler, S. C. Kozma, and G. Thomas. 2001. Mammalian TOR: a homeostatic ATP sensor. Science 294:1102-1105. [DOI] [PubMed] [Google Scholar]
- 16.Diggle, T. A., C. K. Seehra, S. Hase, and N. T. Redpath. 1999. Analysis of the domain structure of elongation factor-2 kinase by mutagenesis. FEBS Lett. 457:189-192. [DOI] [PubMed] [Google Scholar]
- 17.Diggle, T. A., T. Subkhankulova, K. S. Lilley, N. Shikotra, A. E. Willis, and N. T. Redpath. 2001. Phosphorylation of elongation factor-2 kinase on serine 499 by cAMP-dependent protein kinase induces Ca2+/calmodulin-independent activity. Biochem. J. 353:621-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dutil, E. M., A. Toker, and A. C. Newton. 1998. Regulation of conventional protein kinase C isozymes by phosphoinositide-dependent kinase 1 (PDK-1). Curr. Biol. 8:1366-1375. [DOI] [PubMed] [Google Scholar]
- 19.Everett, A. D., T. D. Stoops, A. C. Nairn, and D. Brautigan. 2001. Angiotensin II regulates phosphorylation of translation elongation factor-2 in cardiac myocytes. Am. J. Physiol. 281:H161-H167. [DOI] [PubMed] [Google Scholar]
- 20.Fingar, D. C., S. Salama, C. Tsou, E. Harlow, and J. Blenis. 2002. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16:1472-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gingras, A.-C., B. Raught, and N. Sonenberg. 2001. Control of translation by the target of rapamycin proteins. Prog. Mol. Subcell. Biol. 27:143-174. [DOI] [PubMed] [Google Scholar]
- 22.Hara, K., K. Yonezawa, Q.-P. Weng, M. T. Kozlowski, C. Belham, and J. Avruch. 1998. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF4E BP1 through a common effector mechanism. J. Biol. Chem. 273:14484-14494. [DOI] [PubMed] [Google Scholar]
- 23.Horman, S., G. J. Browne, U. Krause, J. V. Patel, D. Vertommen, L. Bertrand, A. Lavoinne, L. Hue, C. G. Proud, and M. H. Rider. 2002. Activation of AMP-activated protein kinase leads to the phosphorylation of elongation factor 2 and an inhibition of protein synthesis. Curr. Biol. 12:1419-1423. [DOI] [PubMed] [Google Scholar]
- 24.Jaeschke, A., J. Hartkamp, M. Saitoh, W. Roworth, T. Nobukuni, A. Hodges, J. Sampson, G. Thomas, and R. Lamb. 2002. Tuberous sclerosis complex tumor suppressor-mediated S6 kinase inhibition by phosphatidylinositide-3-OH kinase is mTOR independent. J. Cell Biol. 159:217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knebel, A., C. E. Haydon, N. Morrice, and P. Cohen. 2002. Stress-induced regulation of eEF2 kinase by SB203580-sensitive and -insensitive pathways. Biochem. J. 367:525-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knebel, A., N. Morrice, and P. Cohen. 2001. A novel method to identify protein kinase substrates: eEF2 kinase is phosphorylated and inhibited by SAPK4/p38delta. EMBO J. 20:4360-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Good, J. A., W. H. Ziegler, D. B. Parekh, D. R. Alessi, P. Cohen, and P. J. Parker. 1998. Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science 281:2042-2045. [DOI] [PubMed] [Google Scholar]
- 28.Manning, B. D., and L. C. Cantley. 2003. Rheb fills a GAP between TSC and TOR. Trends Biochem. Sci. 28:573-576. [DOI] [PubMed] [Google Scholar]
- 29.McLeod, L. E., L. Wang, and C. G. Proud. 2001. β-Adrenergic agonists increase phosphorylation of elongation factor 2 in cardiomyocytes without eliciting calcium-independent eEF2 kinase activity. FEBS Lett. 489:225-228. [DOI] [PubMed] [Google Scholar]
- 30.McManus, E. J., and D. R. Alessi. 2002. TSC1-TSC2: a complex tale of PKB-mediated S6K regulation. Nat. Cell Biol. 4:E214-E216. [DOI] [PubMed] [Google Scholar]
- 31.Neshat, M. S., I. K. Mellinghoff, C. Tran, B. Stiles, G. Thomas, R. Petersen, P. Frost, J. J. Gibbons, H. Wu, and C. L. Sawyers. 2001. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc. Natl. Acad. Sci. USA 98:10314-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nojima, H., C. Tokunaga, S. Eguchi, N. Oshiro, S. Hidayat, K. Yoshino, K. Hara, J. Tanaka, J. Avruch, and K. Yonezawa. 2003. The mTOR partner, raptor, binds the mTOR substrates, p70 S6 kinase and 4E-BP1, through their TOS (TOR signaling) motifs. J. Biol. Chem. 278:15461-15464. [DOI] [PubMed] [Google Scholar]
- 33.Parekh, D., W. Ziegler, K. Yonezawa, K. Hara, and P. J. Parker. 1999. Mammalian TOR controls one of two kinase pathways acting upon PKCdelta and epsilon. J. Biol. Chem. 274:34758-34764. [DOI] [PubMed] [Google Scholar]
- 34.Pavur, K. S., A. N. Petrov, and A. G. Ryazanov. 2000. Mapping the functional domains of elongation factor-2 kinase. Biochemistry 39:12216-12224. [DOI] [PubMed] [Google Scholar]
- 35.Pearson, R. B., P. B. Dennis, J. W. Han, N. A. Williamson, S. C. Kozma, R. E. H. Wettenhall, and G. Thomas. 1995. The principal target of rapamycin-induced p70S6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J. 14:5279-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price, N. T., S. F. Nakielny, S. J. Clark, and C. G. Proud. 1989. The two forms of the beta-subunit of initiation factor-2 from reticulocyte lysates arise from proteolytic degradation. Biochim. Biophys. Acta 1008:177-182. [DOI] [PubMed] [Google Scholar]
- 37.Proud, C. G. 2002. Regulation of mammalian translation factors by nutrients. Eur. J. Biochem. 269:5338-5349. [DOI] [PubMed] [Google Scholar]
- 38.Pullen, N., P. B. Dennis, M. Andjelkovic, A. Dufner, S. C. Kozma, B. A. Hemmings, and G. Thomas. 1998. Phosphorylation and activation of p70S6k by PDK1. Science 279:707-710. [DOI] [PubMed] [Google Scholar]
- 39.Ravikumar, B., A. Stewart, H. Kita, K. Kato, R. Duden, and D. C. Rubinsztein. 2003. Raised intracellular glucose concentrations reduce aggregation and cell death caused by mutant huntingtin exon 1 by decreasing mTOR phosphorylation and inducing autophagy. Hum. Mol. Genet. 12:985-994. [DOI] [PubMed] [Google Scholar]
- 40.Redpath, N. T., E. J. Foulstone, and C. G. Proud. 1996. Regulation of translation elongation factor-2 by insulin via a rapamycin-sensitive signalling pathway. EMBO J. 15:2291-2297. [PMC free article] [PubMed] [Google Scholar]
- 41.Ryazanov, A. G., M. D. Ward, C. E. Mendola, K. S. Pavur, M. V. Dorovkov, M. Wiedmann, H. Erdjument-Bromage, P. Tempst, T. G. Parmer, C. R. Prostko, F. J. Germino, and W. N. Hait. 1997. Identification of a new class of protein kinases represented by eukaryotic elongation factor-2 kinase. Proc. Natl. Acad. Sci. USA 94:4884-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schalm, S. S., and J. Blenis. 2002. Identification of a conserved motif required for mTOR signaling. Curr. Biol. 12:632-639. [DOI] [PubMed] [Google Scholar]
- 43.Schalm, S. S., D. C. Fingar, D. M. Sabatini, and J. Blenis. 2003. TOS motif-mediated raptor binding regulates 4E-BP1 multisite phosphorylation and function. Curr. Biol. 13:797-806. [DOI] [PubMed] [Google Scholar]
- 44.Shima, H., M. Pende, Y. Chen, S. Fumagalli, G. Thomas, and S. C. Kozma. 1998. Disruption of the p70S6k/p85S6k gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 17:6649-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takano, A., I. Usui, T. Haruta, J. Kawahara, T. Uno, M. Iwata, and M. Kobayashi. 2001. Mammalian target of rapamycin pathway regulates insulin signaling via subcellular redistribution of insulin receptors substrate 1 and integrates nutritional signals and metabolic signals of insulin. Mol. Cell. Biol. 21:5050-5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanhaesebroeck, B., and D. R. Alessi. 2000. The P13K-PDK1 connection: more than just a road to PKB. Biochem. J. 346:561-576. [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, L., and C. G. Proud. 2002. Regulation of the phosphorylation of elongation factor 2 by MEK-dependent signalling in adult rat cardiomyocytes. FEBS Lett. 531:285-289. [DOI] [PubMed] [Google Scholar]
- 48.Wang, L., X. Wang, and C. G. Proud. 2000. Activation of mRNA translation by insulin in rat cardiomyocytes involves multiple rapamycin-sensitive steps. Am. J. Physiol. 278:H1056-H1068. [DOI] [PubMed] [Google Scholar]
- 49.Wang, X., L. E. Campbell, C. M. Miller, and C. G. Proud. 1998. Amino acid availability regulates p70 S6 kinase and multiple translation factors. Biochem. J. 334:261-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, X., W. Li, M. Williams, N. Terada, D. R. Alessi, and C. G. Proud. 2001. Regulation of elongation factor 2 kinase by p90RSK1 and p70 S6 kinase. EMBO J. 20:4370-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams, M. R., J. S. C. Arthur, A. Balendran, J. van der Kaay, V. Poli, P. Cohen, and D. R. Alessi. 2000. Role of PDK1 in mediating the activation of AGC kinases in embryonic stem cells defined by targeted disruption of the PDK1 gene. Curr. Biol. 20:439-448. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, H., G. Cicchetti, H. Onda, H. B. Koon, K. Asrican, N. Bajraszewski, F. Vazquez, C. L. Carpenter, and D. J. Kwiatkowski. 2003. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J. Clin. Investig. 112:1223-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]