Abstract
Peripheral mechanisms preventing autoimmunity and maintaining tolerance to commensal microbiota involve CD4+Foxp3+ regulatory T cells1,2 generated in the thymus (tTregs) or extrathymically by induction of naive CD4+Foxp3− T cells (iTregs). Prior studies suggested that the T cell receptor (TCR) repertoires of tTregs and iTregs are biased towards self and non-self antigens, respectively 3–6 but their relative contribution in controlling immunopathology, e.g. colitis and other untoward inflammatory responses triggered by different types of antigens, remains unresolved 7. The intestine, and especially the colon, is a particularly suitable organ to study this question, given the variety of self-, microbiota- and food-derived antigens to which Tregs and other T cell populations are exposed. Intestinal environments can enhance conversion to a regulatory lineage 8,9 and favor tolerogenic presentation of antigens to naive CD4+ T cells 10,11, suggesting that intestinal homeostasis depends on microbiota-specific iTregs 12–15. Here, to identify the origin and antigen-specificity of intestinal Tregs, we performed single cell as well as high-throughput (HT) sequencing of the TCR repertoires of CD4+Foxp3+ and CD4+Foxp3− T cells and analyzed their reactivity against specific commensal species. We show that tTregs constitute the majority of Tregs in all lymphoid and intestinal organs, including colon, where their repertoire is heavily influenced by the composition of the microbiota. Our results suggest that tTregs, and not iTregs, dominantly mediate tolerance to antigens produced by intestinal commensals.
We used the TCRmini mice whose limited but diversified repertoire allows for a comprehensive comparison of TCRs in various organs and subpopulations 16. In these mice, thymocytes differentiate naturally as CD4+Foxp3+ and CD4+Foxp3− T cells, and efficiently repopulate peripheral lymphoid organs 16. Furthermore, the TCRβ chain is identical in all TCRmini T cells, enabling detection of TCR diversity through specific analysis of the TCRα chain 16. To identify Foxp3+ cells, we crossed TCRmini mice with Foxp3GFP reporter mice 17. The TCRminiFoxp3GFP and B6Foxp3GFP mice had very similar numbers of Foxp3GFP+ cells in different intestinal organs (Fig. S1) and CD4+ cells in both types of mice expressed comparable levels of α4β7 and CCR9 molecules that regulate homing to the intestine (Fig. S2). We also found that adoptive transfer of naive CD4+ T cells from TCRminiFoxp3GFP mice to lymphopenic, RAG-deficient mice caused inflammation in the colon and wasting disease. The disease could be prevented by co-transfer of TCRminiCD4+Foxp3+ thymocytes without affecting the colonization of the colon by CD4+Foxp3− T cells, indicating that tTregs can control intestinal inflammation at least in these experimental settings (Fig. S3).
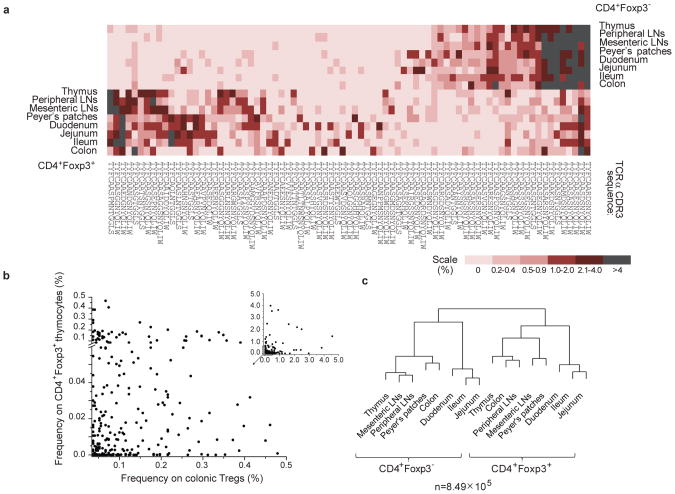
tTregs and iTregs have similar phenotypes, and overlapping but distinct TCR repertoires relative to their thymic or peripheral origin 16,18,19. To compare dominant TCRs on CD4+Foxp3− and Treg cells in lymphoid organs and in the intestine, we sorted individual cells, amplified the TCRα chains by RT-PCR and sequenced their CDR3 regions (the numbers of sequences analyzed by single-cell and HT sequencing, pertaining to Figs. 1-3, are shown in Tables S1-S3). As shown in Fig. 1a, the distribution of dominant TCRs from CD4+Foxp3− and Treg cells in all analyzed organs was asymmetrically skewed, and only a few TCRs were overrepresented in both CD4+ populations. Dissimilar allocation of abundant TCRs was previously observed in lymphoid organs and was attributed to separate thymic differentiation pathway for tTregs and limited conversion of CD4+Foxp3− cells 20. In addition, approximately half of the dominant TCRs found on intestinal Tregs (including colonic Tregs) were also found on CD4+Foxp3+ thymocytes, suggesting that the intestinal Treg repertoire includes a significant proportion of dominant clones of thymic origin (Fig. 1a).
Fig. 1. TCR repertoires of intestinal Tregs are similar to the TCR repertoire of CD4+Foxp3+thymocytes.
a, The frequencies of fifteen dominant TCRs selected for each population from indicated organs in all organs (based on single cell TCR sequencing, see Table S1 for the number of TCR CDR3 sequences analysed). Color shades reflect the relative frequency with which a given TCR was found in each organ. b, The frequencies of dominant TCRs from colonic Tregs in the population of CD4+Foxp3+ thymocytes. c, The hierarchical diagrams depict similarity indices (MII) for TCR repertoires from CD4+Foxp3− and CD4+Foxp3+ populations ((b and c are based on HT sequencing, see Table S1 for the number of TCR CDR3 sequences analysed).
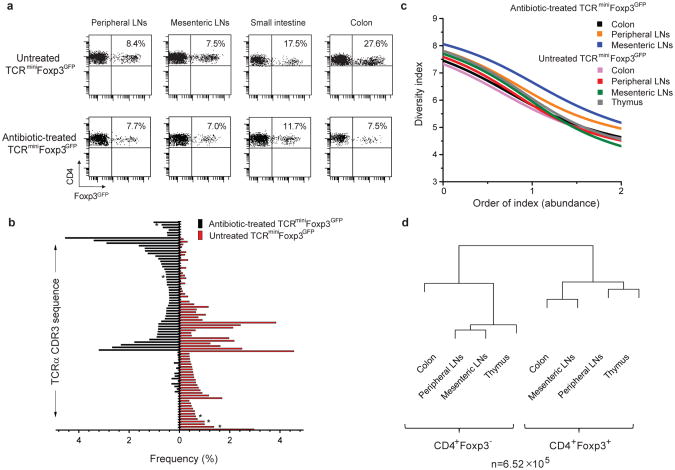
Fig. 3. Antibiotic-induced changes in colonic flora have profound influence on the TCR repertoire of colonic tTregs.
a, The effect of antibiotic treatment on the proportion of Tregs in indicated organs. Three mice per group were analyzed. b, Fifty dominant TCRs (Table S4) of colonic Tregs from untreated (red bars) or antibiotic-treated (black bars) mice and their frequencies in analyzed repertoires. TCRs not found on CD4+Foxp3+ thymocytes are star-marked. c, Diversity index (Renyi Entropy Function) of Tregs from indicated organs of untreated and antibiotic-treated mice. REF close to “0” corresponds to diversity of low abundant TCRs and close to “2” for high abundant TCRs. d, MII indices for TCR repertoires of CD4+Foxp3− and Treg populations from antibiotic-treated mice.
To comprehensively compare TCR repertoires on thymic, peripheral and intestinal CD4+Foxp3− and Treg clones we used HT sequencing (Fig. 1b, c), which also minimized the proportion of unique TCRs identified, i.e. found only in one organ. In the colon, unique Treg TCRs comprised just 9% of all TCR sequences retrieved from this organ, with overall 5% of TCRs found on CD4+Foxp3− T cells but not on CD4+Foxp3+ thymocytes, suggesting that the colonic population of iTregs expressing TCRs specific for the CD4+Foxp3− lineage is limited (data not shown). Accordingly, the remaining 86% of TCRs from colonic Tregs were expressed on CD4+Foxp3+ thymocytes, and Fig. 1b shows that a vast majority of dominant TCRs from colonic Tregs (found at least 10 times), were shared between both populations. These TCRs accounted for approximately half of all TCRs retrieved from CD4+Foxp3+ thymocytes, indicating that these thymocytes are not rare, recirculating mature iTregs (Fig. 1c and data not shown). As shown in Fig. 1c, the similarity indices (MII, depicted by the distance between branches of the dendrogram) calculated for the TCR repertoires from various intestinal Tregs and CD4+Foxp3− populations did not reveal higher similarity, which would be expected if a dominant portion of intestinal Tregs, including those from the colon, was represented by iTregs. In fact, not a single repertoire of Tregs clustered on the same branch of the dendrogram with CD4+Foxp3− repertoire(s), suggesting that these repertoires remained mostly dissimilar (as also shown for dominant TCRs in Fig. 1a and S4). Limited conversion in the mesenteric lymph nodes or colon of TCRminiFoxp3GFP mice was not a result of impaired recruitment of CD4+Foxp3− cells in this model because the conversion was apparent in the tumor environment, upon adoptive transfer of CD4+Foxp3− cells to lymphopenic hosts and in vitro (21 and data not shown).
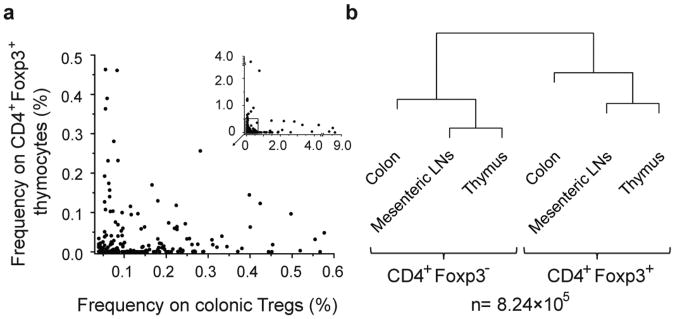
In view of reports analyzing mice with broader repertoire of TCRs than that of TCRmini mice 14, which suggested that iTregs are overwhelmingly abundant in the colon, we also examined if the extent of TCR diversity could play a role in determining the relative involvement of tTregs and iTregs in maintaining tolerance to colonic antigens. To address this question we analyzed TCRβFoxp3GFP transgenic mice in which the repertoire of TCRs is much larger than that of TCRmini mice due to the natural diversity of the TCRα chain. The HT sequencing of the TCRVα2+ chains of thymic, peripheral and colonic CD4+Foxp3+ and CD4+Foxp3− subpopulations from TCRβ transgenic mice revealed a pattern similar with the respective repertoires collected from TCRmini mice. Approximately 75% of all Vα2+ TCRs retrieved from colonic Tregs from TCRβFoxp3GFP transgenics were also expressed by CD4+Foxp3+ thymocytes, including many abundant colonic TCRs (found in the colonic Treg repertoire more than 20 times Fig. 2a), which accounted for approximately 20% of all TCRs retrieved from CD4+Foxp3+ thymocytes (data not shown). Furthermore, the MII indices calculated for the TCR repertoires from colonic, mesenteric and thymic CD4+Foxp3+ and CD4+Foxp3− cells retained the same hierarchical clustering as originally observed in TCRmini mice (Fig. 2b). Thus, we concluded that the extent of TCR diversity does not have significant influence on the predominance of tTregs in the colon.
Fig. 2. In TCRβFoxp3GFP transgenic mice the majority of colonic CD4+Foxp3+ T cells share TCRs with CD4+Foxp3+ thymocytes.
a, Dominant TCRs from colonic Tregs and their frequencies on CD4+Foxp3+ thymocytes b, The hierarchical dendrogram depicts MII indices between TCR repertoires from CD4+Foxp3− and CD4+Foxp3+ populations from the indicated organs. For calculation of MII, the dataset from CD4+Foxp3+ thymocytes was limited as described in Methods Summary.
To investigate whether changes in the composition of colonic microflora influence the repertoire of tTregs we treated the TCRminiFoxp3GFP mice with a cocktail of antibiotics. This treatment significantly altered the composition of the colonic microbiota and reduced the proportion of intestinal Tregs, particularly in the colon (Fig. 3a). As shown in Fig. S5, 6 out of 10 dominant commensal species cultured from the cecum of untreated mice, including members of Clostridiales, an abundant anaerobe known to induce colonic iTregs13, fell to undetectable levels in cultures from treated mice. Of the remaining 4 species cultured from the cecum, 2 significantly increased in biomass with antibiotic treatment and 2 remained unaffected. Antibiotic treatment also significantly altered the frequency of dominant TCRs identified in colonic Tregs, as some clones became undetectable, whereas others expanded, likely in response to the rebound growth of more antibiotic-resistant species (Fig. 3b). Most TCRs expressed by dominant Treg clones that contracted or expanded in antibiotic-treated mice were also found on CD4+Foxp3+ thymocytes, indicating that changes in the intestinal flora influence the repertoire of colonic tTregs (Fig. 3b). As shown in Fig. 3c, the diversity of the TCRs on colonic Tregs from antibiotic-treated and untreated mice (calculated from HT sequencing) was not significantly affected, despite their strong numerical reduction (Fig.3a). Calculation of the MII index between Tregs and CD4+Foxp3− populations from different organs of antibiotic-treated mice (Fig. 3d) showed that the repertoire of colonic Tregs remained similar (see Fig. 1c) to the rest of CD4+Foxp3+ repertoires, which argues against the significant recruitment of iTregs in response to the changing composition of bacterial antigens.
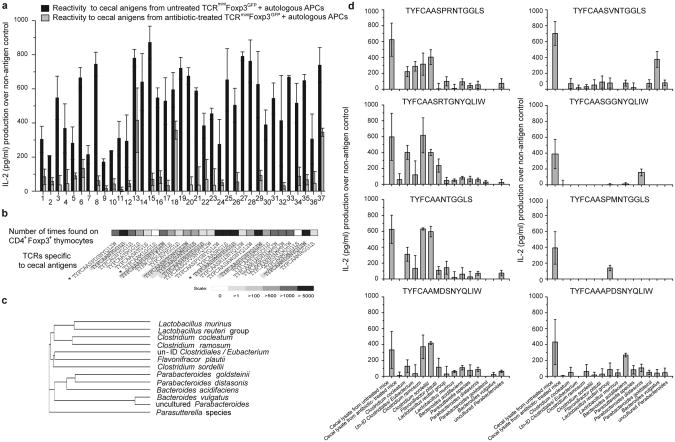
To identify Treg clones specific to antigens produced by commensal species, we created hybridomas from colonic Tregs 22, and sorted hybridomas that responded to sterile filtrates of cecal contents from untreated TCRminiFoxp3GFP mice (Fig. S6). Of these sub-cloned hybridomas, majority that responded to cecal filtrates from untreated mice did not respond to filtrates from antibiotic-treated mice, suggesting that most responding hybridomas expressed TCRs specific for microbial antigens present in untreated mice (Fig. 4a). We then identified 26 TCRs from hybridomas that responded to cecal filtrates from untreated mice and examined their expression on CD4+Foxp3+ thymocytes. Fig. 4b shows that over 90% of sequenced TCRs derived from colonic Tregs were also expressed by CD4+Foxp3+ thymocytes. Next, we tested the reactivity of hybridomas, against bacterial sonicates prepared from thirteen cultures of individual species identified in the ceca of TCRminiFoxp3GFP mice (Fig. 4c). Fig. 4d, (left column) shows that four hybridomas responded to isolates from Clostridiales (one of these also responded to phylogenetically related Falvonifractor), and other four hybridomas (Fig. 4d, right column) responded to sonicates from Bacteroides or Lactobacillus, suggesting that these responses were elicited by unidentified bacterial antigen(s). Overall, these results demonstrated that colonic Tregs and CD4+Foxp3+ thymocytes share multiple TCRs that recognize microbial antigens.
Fig. 4. TCRs from colonic tTregs recognize microbial antigens.
a. The response (± s.e.m from three experiments) of cloned colonic Treg hybridomas, which responded to cecal lysate from untreated TCRminiFoxp3GFP mice (Fig. S6), to re-stimulation with cecal lysate from untreated or antibiotic-treated mice. b, The abundance of Treg TCRs from hybridomas that responded to cecal lysates from untreated mice (shown in a) on CD4+Foxp3+ thymocytes. Hybridomas marked by asterisk also responded to cecal lysate from antibiotic-treated mice. c, Phylogenetic distance of bacterial strains tested here. d. The response of hybridomas highlighted in panel 4b to indicated bacterial sonicates.
Our study provides evidence for interactions between the host tTreg population and the complex communities of microbes present in the gut lumen. In-depth analysis of the TCR repertoire of colonic tTregs demonstrated that it is sufficiently broad to recognize microflora-derived antigens and that conversion of naive CD4+Foxp3− cells does not appreciably modify its diversity. These conclusions challenge the recent report suggesting that iTregs constitute the vast majority of colonic Tregs 14. In that study, the authors sampled TCR repertoires of colonic and peripheral CD4+ cells from a TCRβ transgenic line and used retrogenic mice to determine if TCRs derived from dominant colonic Treg clones support thymic selection of tTregs. None of the colonic TCRs examined supported tTreg development, but constitutive expression of TCR in retrogenic mice can compromise tTregs selection and skew thymocyte commitment to the Foxp3− lineage, irrespective of TCR origin 23. The number of TCRs examined in that study was about one order of magnitude smaller than that sequenced here, which would preclude detection of colonic TCRs on low-abundant clones in other organs. Nevertheless, half of the most abundant colonic TCRs were found on Tregs in lymphoid organs 14, where 93% of these clones were estimated to represent tTregs 20. Therefore, in both TCRβ transgenic lines 14,16, a large proportion of colonic Tregs can be of thymic origin.
The results of our study are consistent with the findings that tTregs recognize non-self antigens 22,24, become activated upon colonization of germ-free mice with standardized microbial flora (Schaedler flora)25, and prevent colitis in CNS1- deficient mice exclusively lacking iTregs 26 or in lymphopenic mice that received wild- type, naïve CD4+ cells 27. We conclude that iTregs can participate in maintaining tolerance to intestinal antigens, but that tTregs play the dominant role in this process.
Methods
Mice
TCRminiFoxp3GFP and TCRβFoxp3GFP (Vβ14Dβ2Jβ2.6) mice were obtained by mating B6Foxp3GFP 17 with TCRmini 16 and TCRβ 16 mice, respectively. The progeny was screened for the co-expression of Foxp3GFP reporter and TCRmini Vα2Vβ14+ dimer or TCRβ Vβ14+ chain, respectively. To eliminate expression of endogenous TCRα chains, all TCRmini mice were crossed with mice deficient in endogenous TCRα loci and were heterozygous for TCRα Vα2Jα26Jα2 minilocus to ensure expression of a single TCRα chain per T cell. All animals were housed in GRU animal facility in accordance to the Institutional regulations.
Purification of intestinal lamina propria T cells
Intestinal regions were opened longitudinally and contents were flushed with ice-cold Hanks balanced-salt solution (HBSS, Cellgro). Each region was cut into small pieces and washed with HBSS supplemented with 5% FCS (HyClone) and 2mM EDTA at 37°C. A single-cell suspension was obtained after treatment with Collagenase D (1.0 mg/ml) and DNase I (0.1 mg/ml) (both from Roche). A purified and concentrated suspension of lamina propria lymphocytes was obtained after centrifugation on Percoll (GE Healthcare) gradient (45% and 70%). The interface, enriched in leukocytes, was collected and used for experiments.
Isolation of thymocytes and T cells from lymphoid organs
Single-cell suspensions were prepared from the thymus, inguinal and mesenteric LNs by mechanical disruption. Peyer's patches were excised from the small intestine wall, and lymphocytes were isolated by enzymatic digestion for 20 min, using Collagenase D (1.0 mg/ml) and DNase I (0.1 mg/ml) at 37°C.
Flow cytometry, single-cell sorting and single cell RT PCR
Thymocytes and T cells were stained with antibodies against CD4, CD8, Vα2, Vβ14, CCR9 and α4β7 (BD Biosciences or eBioscience) and analyzed using BD FACS Canto (BD Biosciences). Single cells were sorted (MoFlo cell sorter, Beckman Coulter) into 96-well plates from a sorted (purity >99%) populations of CD4+Foxp3GFP+ and CD4+Foxp3GFP− T cells. cDNA was synthesized using MMLV reverse transcriptase (Promega) and random hexamers (IDT) followed by two rounds of PCR via Perfect Taq Polymerase (5 PRIME) 16. Products of CDR3 Vα chain obtained in the second PCR reaction were sequenced in the Genomic Core Facility at the University of Illinois (Urbana, IL). All necessary precautions were taken to prevent PCR contamination as previously described 16
High-throughput CDR3 sequencing
Cα specific primer used for cDNA synthesis: (5′-TCGGCACATTGATTTGGGAGTC-3′).
Primers with incorporated tags for Ion Torrent high-throughput sequencer (Vα2IT: 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGTCTCAGCCTGGAGACTCAGC-3′ and CαIT: 5′-CCTCTCTATGGGCAGTCGGTGATTGGTACACAGCAGGTTCTGGGT-3′).
CDR3 regions sequenced on the same chip and derived from different subsets were discriminated based on barcodes, which were validated for optimal performance with the Ion Torrent PGM. Data was analyzed using PACE (Parallel Algorithm for CDR3 Extraction) program and evaluated with statistical methods as described below and in ref 28.
Hybridoma assays
Colonic CD4+Foxp3GFP+ T cells were expanded in vitro for 7 days as described 29 and fused with BW thymoma stably transfected with the NFATGFP. It should be noted that as a result of fusion, Tregs lose expression of Foxp3GFP, and therefore expression of NFATGFP in hybridomas can be used as a marker of their activation. After 10 days, the heterogeneous pool of hybrids was incubated overnight with splenocytes or bone marrow derived dendritic cells from TCRα deficient mice preincubated overnight with sterile lysate obtained from cecum of TCRminiFoxp3GFP mice. Responding hybridomas were sorted based on the NFATGFP expression (Fig.S6,) cloned into 96-well plates, and two weeks later were restimulated with sterile cecum lysates from untreated or antibiotic-treated TCRminiFoxp3GFP mice in the presence of autologous APCs.
Response of cloned hybridomas toward cecal lysates and microbial sonicates was measured using HT-2 assay 22. In brief, 105 hybridoma cells were incubated with 105 bone marrow-derived dendritic cells (or splenocytes from TCRα deficient mice) alone (no antigen control) and lysate or the indicated bacterial sonicates each in the non-toxic range of different concentrations. After 24 hr, the amount of secreted IL-2 was measured with the detector HT-2 cell line and compared to values from a standard curve derived from recombinant IL-2 (Peprotech). The proliferation of HT-2 cells in response to IL-2 was measured with MTT (Sigma) assay 22 and the response at optimal concentration of bacterial sonicate is shown in Fig. 4. The TCRα CDR3 regions from responding hybridomas were amplified, sequenced and cross-referenced to our database of TCRα CDR3 collected from various subpopulations of CD4+ T cells.
Adoptive transfer
RAG-2 deficient mice were injected i.v. with sorted CD4+ subpopulations from TCRminiFoxp3GFP mice. Recipients received either naive, pLN CD4+ cells, or in addition thymic or peripheral CD4+Foxp3GFP+ cells. After adoptive transfer, mice weight was monitored on a weekly basis. After 5 weeks all recipients were sacrificed and their colons were examined for the signs of inflammation. The proportions of Foxp3GFP− to Foxp3GFP+ of CD4+ T cells in the colon were similar in all recipients (data not shown).
Parallel Algorithm for CDR3 Extraction (PACE)
Parallel Algorithm for CDR3 Extraction (PACE) is an application to obtain sequences of TCR CDR3 regions generated by high-throughput sequencers. This algorithm employs BLAST as a core component for sequence comparison to locate known V and J regions in high volumes of sequencing data. Extracted sequences were managed in a centralized SQL database and reported in FASTA format.
Accuracy of CDR3 sequencing
To control for possible contaminations during sorting or PCR, cells from multiple sorts from different CD4+ cell subsets were individually processed, sequenced and data from respective subsets were compared. In addition, it was verified that TCR repertoires obtained from the same cell populations by single-cell and high-throughput TCR sequencing were similar, demonstrating that both sequencing approaches yield comparable data (Fig. 1c and Fig. S4a,b). To ensure that dominant CDR3 regions are not contaminants, it was checked whether in each dataset these regions were encoded by multiple different nucleotide sequences (indicative of selection at the protein level and not the amplification of a single clonotype due to artificial contamination). Examples of this analysis are shown in Fig. S7 and Table S5).
The accuracy of the high-throughput CDR3 sequencing was ensured by the use of high fidelity DNA polymerase with a low intrinsic error rate (AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen). In addition, Ion Torrent Suite software filters were used during data processing to exclude low quality reads and erroneous sequences derived from mixed DNA templates. Most common Ion Torrent sequencer errors are base insertions and deletions occurring in homopolymers, which result in frame shifts and stop codons. To identify errors within Vα2 and Jα2 (or Jα26) segments, all sequences were aligned to constant regions. To estimate the application-specific error within the CDR3 region, two monoclonal CDR3 regions from TCRmini mice were amplified (approximately 1×105 reads were collected) and the reads that differed from the original template were counted. This approach estimated that less than 3% of CDR3 regions may contain errors (Table S6), which is significantly below the threshold adversely affecting the statistical similarity and overlap analysis 28.
Microbiology
Ceca with content were dissected under sterile conditions, placed in cryovials and immediately snap frozen in liquid nitrogen. Samples were further processed and analyzed in the Harvard Digestive Disease Center's (HDDC) Microbiome Core facility. Phylogenetic identification and subtyping was based on 16S rRNA classification. Molecular speciation of bacteria was performed with 16S rRNA gene analyses. Assembled sequences were loaded into the Ribosomal Database Project's (RDP) SEQMATCH tool to identify the taxonomic assignment (http://rdp.cme.msu.edu/).
Statistical analysis
The comparison of various TCR repertoires was conducted by means of the assessment of their respective diversities as well as overlap between populations (Fig. 1c, 2b, 3c, d and S4a). In the current context, under the term ‘diversity’ we understand both the richness as well as the abundance patterns of the repertoires.
For the sake of quantifying the TCR diversity for a single repertoire, we have adopted the information-theoretic approach based on the notion of the Renyi entropy function of order α denoted Hα30. This function quantifies diversity by means of the formula
| (1) |
where the pi is the observed frequency of the i-th species, and the order α is a nonnegative exponential weight parameter. The value of α below unity gives more weight to the less abundant (and thus possibly under-sampled) species, whereas the values above unity give more weight to the more abundant species. For α=1 the above formula is not well-defined but may be obtained by taking the limiting expression as α approaches unity. In this case
| (2) |
is the usual Shannon entropy function known in the information theory, which weights equally the contributions of all observed species to the repertoire diversity.
By plotting the values of the Renyi entropy Hα against its index, we are able to analyze diversity of TCR populations graphically in terms of their diversity, weighted towards rare (α less than one) and abundant (α greater than one) species (Fig. 3c). Since the quantities pi are the empirical counts of the observed species, for the sake of obtaining bounds on the sampling error in the values Hα we apply the computational methods based on the non-parametric bootstrap as described e.g., 28. In order to improve the robustness of Hα against unseen species (i.e., the possible under–sampling of the species richness) we have also considered a version of the analysis where the so-called Chao-Shen correction was applied to compute the Shannon entropy, as described 28. In our particular case, it turned out that the results of this alternative analysis differed only marginally from the original ones.
The pairwise overlap analysis as described in Figure 1c, 2b, 3d and S4a was conducted on the basis of the hierarchical clustering of the repertoires with an appropriately chosen dissimilarity function. Similarly as for the analysis of the single repertoire diversity, we have adopted an information- theoretical approach in order to pairwise compare TCR repertoires. Since the pairs of observed frequencies of different TCR species may be arranged in a two-way contingency table, the entropy based index known as the mutual information index (MII) can be applied to measure the association between observed TCR frequencies and the corresponding class (repertoire) labels. If MI is the usual mutual information statistic in two-way contingency table and n1, n2are the proportions of species observed in different TCR populations (i.e., n1+n2 =1), the MII is given by the formula
| (3) |
The MII may be shown to take values between zero and unity, with the zero value admitted only for repertoires with linearly dependent vectors of TCR frequencies. This property makes MII an appropriate measure of dissimilarity for the current purpose of TCR populations clustering. For the practical purpose of computing the sampling errors in values of MII for all pairs of repertoires, the statistical computational bootstrap methods were applied as explained above. With the MII as the dissimilarity measure we used a canonical clustering procedure based on agglomerative clustering with Ward linkage method. The outcome of the algorithm is presented as a dendrogram or a tree diagram with its leaves representing TCR populations. The leaves are located at the tree-distances from each other computed according to their MII values.
Supplementary Material
Acknowledgments
This work was supported by basic research grants from NIH (AI 5R01AI079277 to L.I., and DMS1106485 and R01CA152158 to G.A.R.). The HDDC Microbiome core is supported by P30-DK034854 and Brigham & Women's Hospital in Boston, MA. Microbiological analyses were performed by M. Delaney, A. Dubois and Q. Liu in the HDDC Microbiome Core, with additional review of findings by Dr. A.B. Onderdonk. We thank J. Pihkala and H. Ignatowicz for technical assistance, M. Kuczma, L. Wojciech, E. Szurek, A. Miazek and P. Muranski for the discussion and R. Markowitz for editing the manuscript.
Footnotes
The authors declare competing financial interest: details accompany the full-text HTML version of the paper at www.nature.com/nature.
Supplemental Information is linked to the online version of this paper at www.nature.com/nature.
Author Contributions A.C. performed most experiments and analyzed the data; M.S and G.A.R. performed statistical analyses; S.S.P and R.A.M. designed the PACE program; T.L.D. provided expertise in colonic T cells isolation; L.B. performed microbiological study; P.Kr. established TCRmini and Foxp3GFP mice models; P.K. and L.I. designed the study, analyzed the data and wrote the paper.
Reference List
- 1.Belkaid Y, Rouse BT. Natural regulatory T cells in infectious disease. Nat Immunol. 2005;6(4):353. doi: 10.1038/ni1181. [DOI] [PubMed] [Google Scholar]
- 2.Sakaguchi S, Powrie F, Ransohoff RM. Re-establishing immunological self-tolerance in autoimmune disease. Nat Med. 2012;18(1):54. doi: 10.1038/nm.2622. [DOI] [PubMed] [Google Scholar]
- 3.Jordan MS, et al. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol. 2001;2(4):301. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- 4.Ribot J, Romagnoli P, van Meerwijk JP. Agonist ligands expressed by thymic epithelium enhance positive selection of regulatory T lymphocytes from precursors with a normally diverse TCR repertoire. J Immunol. 2006;177(2):1101. doi: 10.4049/jimmunol.177.2.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coutinho A, et al. Thymic commitment of regulatory T cells is a pathway of TCR-dependent selection that isolates repertoires undergoing positive or negative selection. Curr Top Microbiol Immunol. 2005;293:43. doi: 10.1007/3-540-27702-1_3. [DOI] [PubMed] [Google Scholar]
- 6.Hsieh CS, et al. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat Immunol. 2006;7(4):401. doi: 10.1038/ni1318. [DOI] [PubMed] [Google Scholar]
- 7.Curotto de Lafaille MA, Lafaille JJ. Natural and adaptive Foxp3+ regulatory T cells: more of the same or a division of labor? Immunity. 2009;30(5):626. doi: 10.1016/j.immuni.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Barnes MJ, Powrie F. Regulatory T cells reinforce intestinal homeostasis. Immunity. 2009;31(3):401. doi: 10.1016/j.immuni.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 9.Mucida D, et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317(5835):256. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 10.Coombes JL, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204(8):1757. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denning TL, et al. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat Immunol. 2007;8(10):1086. doi: 10.1038/ni1511. [DOI] [PubMed] [Google Scholar]
- 12.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107(27):12204. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lathrop SK, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478(7368):250. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haribhai D, et al. A Requisite Role for Induced Regulatory T Cells in Tolerance Based on Expanding Antigen Receptor Diversity. Immunity. 2011;35(1):109. doi: 10.1016/j.immuni.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pacholczyk R, et al. Origin and T cell receptor diversity of Foxp3+CD4+CD25+ T cells. Immunity. 2006;25(2):249. doi: 10.1016/j.immuni.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 17.Kuczma M. Foxp3-deficient regulatory T cells do not revert into conventional effector CD4+ T cells but constitute a unique cell subset. J Immunol. 2009;183(6):3731. doi: 10.4049/jimmunol.0800601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh CS, et al. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 2004;21(2):267. doi: 10.1016/j.immuni.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Wong J, Mathis D, Benoist C. TCR-based lineage tracing: no evidence for conversion of conventional into regulatory T cells in response to a natural self-antigen in pancreatic islets. J Exp Med. 2007;204(9):2039. doi: 10.1084/jem.20070822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lathrop SK, et al. Antigen-specific peripheral shaping of the natural regulatory T cell population. J Exp Med. 2008;205(13):3105. doi: 10.1084/jem.20081359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuczma M, et al. Intratumoral convergence of the TCR repertoires of effector and Foxp3+ CD4+ T cells. PLoS One. 2010;5(10):e13623. doi: 10.1371/journal.pone.0013623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pacholczyk R, et al. Nonself-antigens are the cognate specificities of Foxp3+ regulatory T cells. Immunity. 2007;27(3):493. doi: 10.1016/j.immuni.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bautista JL, et al. Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nat Immunol. 2009;10(6):610. doi: 10.1038/ni.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suffia IJ, et al. Infected site-restricted Foxp3+ natural regulatory T cells are specific for microbial antigens. J Exp Med. 2006;203(3):777. doi: 10.1084/jem.20052056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34(5):794. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 26.Josefowicz SZ, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482(7385):395. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fahlen L, et al. T cells that cannot respond to TGF-beta escape control by CD4+CD25+ regulatory T cells. J Exp Med. 2005;201(5):737. doi: 10.1084/jem.20040685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rempala GA, Seweryn M. Methods for diversity and overlap analysis in T-cell receptor populations. J Math Biol. 2012 doi: 10.1007/s00285-012-0589-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh N, et al. Generation of T cell hybridomas from naturally occurring FoxP3+ regulatory T cells. Meth Mol Biol. 2011;707:39. doi: 10.1007/978-1-61737-979-6_3. [DOI] [PubMed] [Google Scholar]
- 30.Renyi A. On Measures of Information and Entropy. Proceedingds of the 4th Berkley Symposium on Mathematics, Statistics and Probability. 1961:547–561. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.