Abstract
miRNAs (miRs) play an important role in cancer cell growth and migration; however, the potential roles of miRs in osteosarcoma remain largely uncharacterized. By applying a miR microarray platform and unsupervised hierarchical clustering analysis, we found that several miRs have altered expression levels in osteosarcoma cell lines and tumor tissues when compared to normal human osteoblasts. Three miRs, miR-199a-3p, miR-127-3p, and miR-376c were significantly decreased in osteosarcoma cell lines, while miR-151-3p and miR-191 were increased in osteosarcoma cell lines in comparison to osteoblasts. Transfection of precursor miR-199a-3p into osteosarcoma cell lines significantly decreased cell growth and migration, thus indicating that the inhibition effect is associated with an increase in the G1-phase and a decrease of the S-phase cell population. Additionally, we observed decreased mTOR and Stat3 expression in miR-199a-3p transfected cells. This study provides new insights for miRs in osteosarcoma and suggests that miR-199a-3p may play a functional role in osteosarcoma cell growth and proliferation. Restoring miR-199a-3p’s function may provide therapeutic benefits in osteosarcoma.
Keywords: miR-199a-3p, osteosarcoma
Introduction
Although osteosarcomas have been treated with chemotherapy for over 30 years, patients with recurrent or metastatic osteosarcomas still have very poor prognosis (1-3). Finding new strategies to treat recurrent or and metastatic osteosarcoma remains an important but unmet clinical need. Recently, several important studies have focused on the impact of microRNAs (miRNAs, miRs) on tumorigenesis and cancer progression (4-6). miRs are a class of small noncoding, single-stranded endogenous RNA fragments containing 19 to 25 nucleotides (nt) in length that repress translation and cleaves mRNA by base-pairing to the 3′ untranslated region (UTR) of the target gene. In a variety of cancers, miR expression is significantly altered, and this has potential to be a prominent diagnostic and prognostic tool (7). Elucidating the function of miRs in tumor pathogenesis and progression is important as they may play critical roles in the regulation of genes involved in controlling the development, proliferation/differentiation, apoptosis and drug resistance of tumor cells (8). Several studies have found that specific miR expression contribute to tumor growth, progression, metastasis, and drug resistance (7-11). However, not much is known about the expression and deregulation of miRs in osteosarcoma.
In this study, miR expression profiles in osteosarcoma cell lines were compared with osteoblast cell lines, leading us to identify a subset of miRs that are deregulated in osteosarcoma. In turn, these miRs may be involved in the pathogenesis of the tumor by possibly acting as tumor suppressor genes or oncogenes. Specifically, we demonstrate that miR-199a-3p expression is significantly decreased in human osteosarcoma cell lines and the overexpression of miR-199a-3p leads to inhibition of cell migration and cell growth, increase of G1-phase cell population, and down-regulation of a number of oncogenes such as Met, mTOR and Stat3.
Materials and Methods
Human osteoblasts cell lines culture
Human osteoblast cell lines HOB-c (OB1) were purchased from PromoCell GmbH (Heidelberg, Germany, purchased in 2009), osteoblast cell lines NHOST (OB2) were purchased from Lonza Wallkersville Inc. (Walkersville, MD, purchased in 2009), and osteoblast cell lines hFOB (OB3) were purchased from ATCC (Rockville, MD, purchased in 2009). These osteoblast cell lines were purchased with certificates of analysis and were not reauthenticated prior to use in this study. Osteoblast cell lines were cultured in osteoblast growth medium (PomoCell) with supplement Mix. The human normal skeletal muscle RNAs were purchased from Ambion (Applied Biosystems, Foster City, CA) and Invitrogen (Carlsbad, CA). Normal osteoblast cells and normal muscle tissues have been used previously as controls for genetic studies (mRNA and miRNA expression) in osteosarcoma cell lines and in sarcoma tumor tissues (11-13).
Human osteosarcoma cell lines culture
The human osteosarcoma cell line KHOS (OS1) was kindly provided by Dr. Efstathios Gonos (Institute of Biological Research & Biotechnology, Athens, Greece), and U-2OS (OS2) and Saos (OS3) were purchased from the ATCC (purchased in 2006) and these cell lines were not reauthenticated prior to use in these experiments. These cell lines were cultured in RPMI 1640 (Invitrogen) supplemented with 10% FBS, 100-units/ml penicillin and 100μg/ml streptomycin (Invitrogen). Cells were incubated at 37°C in 5% CO2-95% air atmosphere and passaged every 2 to 3 days.
Human sarcoma tissues
Twelve of the osteosarcoma tissue samples (OT1 to OT12) were obtained from Massachusetts General Hospital sarcoma tissue bank and were used in accordance with the policies of the institutional review board (IRB) of the hospital. All diagnoses were confirmed by light microscopy and immunohistochemistry.
Isolation of miRs
Total RNA was extracted from osteoblast and osteosarcoma cell lines and from frozen tissue samples using miRNANeasy Mini Kit (Qiagen GmbH, Hilden, Germany) by following the manufacturer’s instructions. The purity and quantity of the isolated small RNAs were assessed using 1% formaldehyde-agarose gel electrophoresis and by spectrophotometer measurement (Beckman). The RNA samples were submitted to LC Sciences (Houston, TX) for further analysis by Agilent Bioanalyzer (criteria, 28S/18S > 1 and RIN > 5).
Quantitative μParaflo™ miR microarray assay
miR microarray assay was performed using a service provider (LC Sciences). The assay started from 5 μg total RNA sample, which was size fractionated using a YM-100 Microcon centrifugal filter (Millipore, Billerica, MA), and the small RNAs (< 300 nucleotides) isolated were 3′-extended with a poly(A) tail using poly(A) polymerase. An oligonucleotide tag was then ligated to the poly (A) tail for later fluorescent dye staining; two different tags were used for the two RNA samples in dual-sample experiments. Hybridization was performed overnight on a μParaflo microfluidic chip (miRHuman_13.0) using a micro-circulation pump (Atactic Technologies, Houston, TX). On the microfluidic chip, each detection probe consisted of a chemically modified nucleotide coding segment complementary to target miR (from miRNABase, http://microrna.sanger.ac.uk/sequences/) or other RNA (control sequences). The hybridization melting temperatures were balanced by chemical modifications of the detection probes. Hybridization used 100 μL 6×SSPE buffer (0.90 M NaCl, 60 mM Na2HPO4, 6 mM EDTA, pH 6.8) containing 25% formamide at 34 °C. After RNA hybridization, tag-conjugating Cy3 and Cy5 dyes were circulated through the microfluidic chip for dye staining. Fluorescence images were collected using a laser scanner (GenePix 4000B, Molecular Devices, Sunnyvale, CA) and digitized using Array-Pro image analysis software (Media Cybernetics, Silver Spring, MD).
Hierarchical cluster analysis
Multiple sample analysis involves normalization, data adjustment, t-Test, and clustering. Normalization is carried out using a cyclic LOWESS (Locally-weighted Regression) method. The normalization is to remove system-related variations, such as sample amount variations, different labeling dyes, and signal gain differences of scanners so that biological variations can be accurately revealed. The Log2 transformation converts intensity values into a Log2 scale. Gene centering and normalization transform the Log2 values using the mean and the standard deviation of individual genes across all samples using the following formula: Value = [(Value) − Mean (Gene)]/[Standard deviation(Gene)]. For hierarchical cluster analysis, the clustering was done using a hierarchical method and performed with average linkage and Euclidean distance metric. All data processes, except the clustering plot, were carried out using in-house (LC Sciences) developed computer programs. The clustering plot was generated using TIGR MeV (Multiple Experimental Viewer) software from The Institute for Genomic Research.
Statistical analysis
For statistical analysis of microarray data, a t-Test was performed between the “control” and “test” sample groups. T-values were calculated for each miR, and p-values were computed from the theoretical t-distribution. miRs with p-values below a critical p-value (typically 0.01) were selected for cluster analysis.
Taq Man reverse transcription-PCR (RT-PCR) for quantification of miR-199a-3p and miR-151-3p
Real-time RT-PCR was performed to validate differentially expressed miRNAs. For mature miR-199a-3p and miR-151-3p detection, cDNA reverse transcription was performed from total RNA samples using specific miR primers from the Taq Man MicroRNA Assays and reagents from the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems). The resulting cDNA was amplified by PCR using Taq Man miR-199a-3p and miR-151-3p MicroRNA Assay primers with the Taq Man Universal PCR Master Mix and analyzed with a StepOnePlus Real time PCR System (Applied Biosystems) according to the manufacturer’s instructions. RNU48 miRNA (RNU48 miR) was used as a control because RNU48 is one of the most highly abundant and relatively stable expression miRs across the human tissue, and is considered a good candidate for endogenous control. The relative levels of miR-199a-3p and miR-151-3p expression were calculated from the relevant signals by normalization with the signal for RNU48 miR expression. PCR reaction mixtures contained Taq Man human miR-199a-3p and miR-151-3p and Universal PCR Master Mix in a total volume of 20 μl. Cycling variables were as follows: 95°C for 10 min followed by 40 cycles at 95°C (15s) and annealing/extension at 60°C (1 min). All reactions were performed in triplicate.
miR-199a-3p precusor transfection
The miR-199a-3p precursor, miRNASelect™ pEP-miR-199A-1, was purchased from Cell Biolab,Inc (San Diego, CA). This precursor vector expresses miR-199a-3p precursor in its native context while preserving putative hairpin structures to ensure biologically relevant interactions with endogenous processing machinery and regulatory partners, which leads to properly cleaved miRs. The vector also contains a red fluorescence protein (RFP) for evaluating the transfection efficiency. The miR-199a-3p precursor is cloned between BamHI and Nhe I sites. A control miR vector, miRNASelect™ pEP-miR-Null (Cell Biolab,Inc), was used as a negative control. The miR-199a-3p precursor expression and control vectors were purchased as bacterial glycerol stocks. Individual colonies were obtained in cultured bacteria on LB ampicillin plates. The plasmid was isolated by EndoFree Plasmid Kit (QIAGEN, Valencia, CA). Transfections of miR-199a-3p precursor into KHOS, U-2OS and Saos cells were performed with Lipofectamine™ LTX and Plus™ Reagent (Invitrogen) according to the manufacturer’s instructions. Forty-eight hours post-transfection, the stable clones were selected in 2μg/ml of puromycin (Sigma-Aldrich, St. Louis, MO) containing medium.
Western blot analysis
Total protein from osteoblast and osteosarcoma cell lines and tissues was extracted by 1× RIPA lysis buffer (Upstate Biotechnology, Charlottesville, VA). Protein concentration was determined by the DC Protein Assay (Bio-Rad). The human Met (hepatocyte growth factor receptor), mTOR, Smad1, Stat3, MCL-1 and Bcl-XL antibodies were purchased from Cell Signaling (Dedham, MA). The mouse monoclonal antibody to human actin was purchased from Sigma-Aldrich. Western blot analysis was performed as previously described (14).
Cell migration assay
Effects of over expression of miR-199a-3p on osteosarcoma cell migration were determined by Oris™Cell Migration Assay Kit (Platypus Technologies, LLC, Madison, WI) by following manufacturer’s instructions (15-17). In brief, osteosarcoma cell lines KHOS or U-2OS were transfected with either pER-miR-199a-3p precusor vector or pEP-miR-Null control vector as described above and seeded on Oris™ 96-well plate through one of the side ports of the Oris™ Cell Seeding Stopper. The assay was then incubated for 72 hours to permit cell migration. The Oris™ stoppers were removed and all wells received Calcein AM (Molecular Probes, Eugene, OR) Green to fluorescently stain the cells. Cell migration was visualized and examined by Nikon Eclipse Ti-U fluorescence microscope (Nikon Corp.), and images were captured with a SPOT RT digital camera (Diagnostic Instruments, Inc., Sterling Heights, MI). The cell fluorescence signals in the detection zones were measured with a SPECTRAmax® Microplate Spectrofluorometer (Molecular Devices, Sunnyvale, CA). Fluorescent signals that reflect the cell migration were evaluated using a two-sided Student’s t test (GraphPadPRISMH 4 software, GraphPad Software, San Diego, CA).
Cell proliferation assay
The miR-199a-3p precursor transfected cells (4,000 cells per well) were plated in 96-well plates and incubated in RPMI 1640 containing 10% FBS. After 24 h, 48 h, 72 h and 96 h of culture, 10 μl of MTT (5 mg/ml in PBS, purchased from Sigma) was added to each well and the plates were incubated for 4 h (18-21). The resulting formazan product was dissolved with acid-isopropanol, and the absorbance at a wavelength of 490 nm (A490) was read on a SPECTRAmax® Microplate Spectrophotometer. Experiments were performed in triplicate. Cell growth curves were fitted with use of GraphPad PRISM® 4 software.
Cell cycle flow cytometry assay
The miR-199a-3p transfected osteosarcoma cell lines KHOS, U-2OS and Saos stable clones were established as described above. Cells were cultured in a normal growth medium RPMI1640 with FBS for 48 h. The cells were collected from each flask and pelleted by spinning at 1200 rpm for 3 min. The cell pellets were then re-suspended in 1ml of cold phosphate buffer solution (PBS) and fixed in 70% ethanol at −20°C. Flow cytometry analysis of cell cycle was performed at Flow Cytometry Core Facility Center for Regenerative Medicine, Massachusetts General Hospital. Cell cycle analysis was performed on a Becton-Dickinson FACSCAN.
Results
Identification of altered expression of miRNAs between osteosarcoma cell lines and osteoblast cell lines
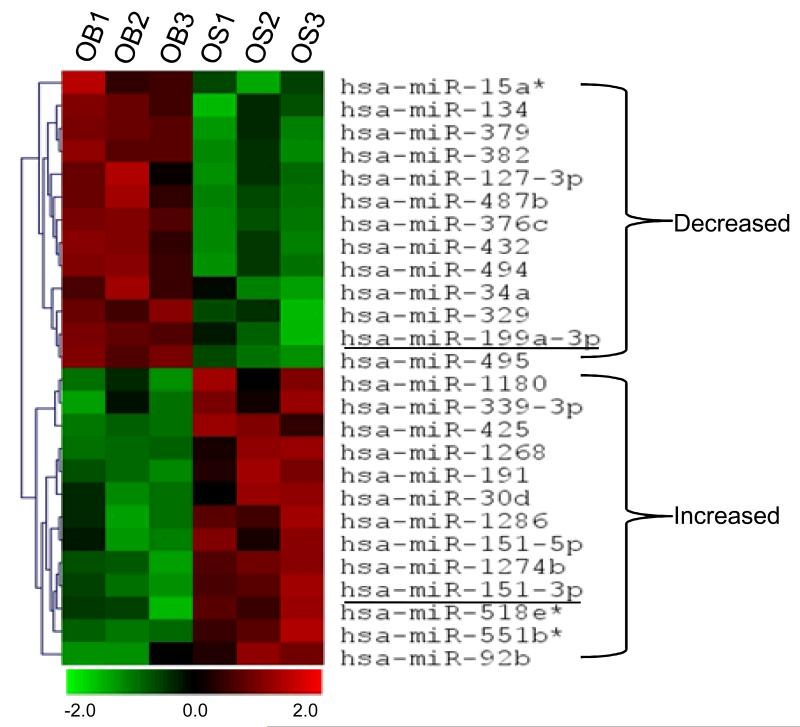
To investigate the expression profiles of miRs in osteosarcoma and osteoblast cell lines, global miR expression levels were measured using μParaflo™ miRNA microarray assay containing 875 unique mature miRs probes. We identified several miRs with expression levels that differed significantly between osteosarcoma and osteoblast cell lines. When we measured miR expression levels in osteosarcoma cell lines, the miRs that met the filtering criteria were analyzed by hierarchical clustering among the three osteoblast cell lines in an unsupervised manner. The clustering algorithm grouped both miRs and samples into clusters based on overall similarity in miR expression pattern without prior knowledge of sample identity. We found that osteoblast cell lines and osteosarcoma cell lines clustered separately as distinct miR expression profiles. We observed significantly reduced expression of miR-199a-3p, miR-127-3p, and miR-376c and significantly increased expression of miR-151-3p and miR-191 in osteosarcoma cell lines. In total, 26 miRs were differentially expressed between the two groups of cell lines at a level of p value<0.05 (Fig. 1). The top eight downregulated and up-regulated miRs are listed in Table 1 and Table 2.
Figure 1.
miR osteosarcoma and osteoblast cell lines profiles: Comparison of differentially expressed miRs genes in osteosarcoma cell lines (OS1,OS2 and OS3) as compared with osteoblast cell lines (OB1,OB2 and OB3): Hierarchical clusters of significantly altered miRs (as determined by ANOVA) across different samples. Red denotes high expression levels, whereas green depicts low expression levels. Each miR listed above is significantly differentially expressed (p<0.05) between the osteosarcoma cell lines (OS1,OS2 and OS3) and osteoblast cell lines (OB1, OB2 and OB3).
Table 1.
| A. Top decreased expression of miRs in osteosarcoma cell lines (OS) | |||
|---|---|---|---|
| Name of miRNA | OB (mean value) | OS (mean value) | Fold* |
| miR-199a-3p | 5,501** | 463 | −12 |
| miR-127-3p | 1,275 | 48 | −27 |
| miR-376c | 642 | 12 | −56 |
| miR-487b | 489 | 24 | −20 |
| miR-134 | 210 | 31 | −6.7 |
| miR-382 | 200 | 19 | −11 |
| miR-432 | 197 | 31 | −6.4 |
| miR-15a | 28 | 20 | −1.4 |
| B. Average values (>500) of decreased expression of miRs in OS | ||||||
|---|---|---|---|---|---|---|
| OB1 | OB2 | OB3 | OS1 | OS2 | OS3 | |
| miR-199a-3p | 7,003 | 5,294 | 4,204 | 921 | 360 | 107 |
| miR-127-3p | 853 | 2,750 | 220 | 21 | 89 | 35 |
| miR-376c | 733 | 829 | 365 | 7 | 19 | 10 |
p<0.05
The number is the raw data from miR microarray which reflects the relative abundance of miR in the cell line
Table 2.
| A. Top increased expression of miRNAs in osteosarcoma cell lines (OS) | |||
|---|---|---|---|
| Name of miRNA | OB (mean value) | OS (mean value) | Fold* |
| miR-191 | 1,334** | 3,317 | 2.5 |
| miR-151-3p | 232 | 986 | 4.3 |
| miR-425 | 149 | 319 | 2.1 |
| miR-1180 | 76 | 243 | 3.2 |
| miR-1274b | 28 | 66 | 2.4 |
| miR-551b | 26 | 46 | 1.8 |
| miR-518 | 25 | 32 | 1.3 |
| miR-1286 | 22 | 37 | 1.7 |
| B. Average values (>500) of increased expression of miRNAs in OS | ||||||
|---|---|---|---|---|---|---|
| OB1 | OB2 | OB3 | OS1 | OS2 | OS3 | |
| miR-191 | 1,485 | 1,339 | 1,178 | 2,382 | 4,143 | 3,427 |
| miR-151-3p | 296 | 222 | 178 | 773 | 841 | 1,343 |
p<0.05
The number is the raw data from miR microarray which reflects the relative abundance of miR in the cell line
Confirmatory studies with differentially expressed miRs by TaqMan Real-time PCR
We selected two of the most significant candidates for further confirmatory studies. miRs identified by miR microarray analysis were re-measured by real-time RT-PCR. First, the expression levels of miR-199a-3p and miR-151-3p in osteosarcoma and osteoblast cell lines were re-measured and we confirmed that miR-199a-3p is significantly decreased in osteosarcoma cell lines while miR-151-3p expression increased.
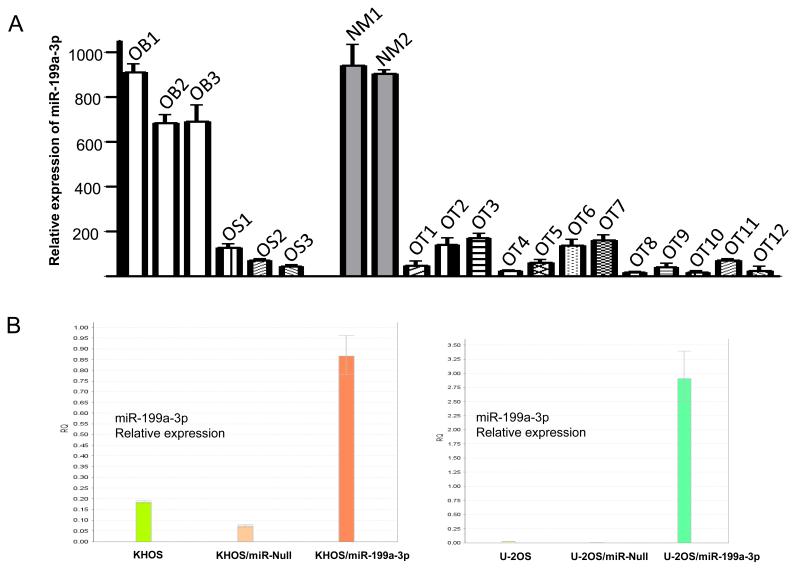
miR-199a-3p expression is decreased in osteosarcoma tumor tissues
We then focused on miR-199a-3p because it was decreased mostly in osteosarcoma cell lines and was ranked highest among miRs expressed in osteoblast cells (Table 1). To determine the expression of miR-199a-3p expression in osteosarcoma tissues, we measured miR-199a-3p expression levels in twelve cases of osteosarcoma tissue samples (OT1 to OT12) by TaqMan Real-time PCR. We observed that miR-199a-3p is not significantly decreased in these osteosarcoma tissues when compared with normal muscle tissues (NM1 and NM2) and osteoblast cells (Fig. 2A). These results are also consistent with previous studies that have shown decreased expression of miR-199a-3p in liver, bladder and ovarian cancer (22-24), suggesting that decreased miR-199a-3p levels are not tumor type specific.
Figure 2.
A, miR-199a-3p expression in osteosarcoma tumor tissues: Relative expression of miR-199a-3p was evaluated by Taq-Man Real-time RT-PCR as described in Materials and Methods. Human normal skeletal muscle RNAs and osteoblast cell line RNAs were used as controls. B, Establishment of miR-199a-3p stably overexpressed osteosarcoma cell lines and confirmation of overexpression of miR-199a-3p in transfected cell lines by real-time PCR. The miR-199a-3p precursor expression and pEP-miR-Null control vector were transfected with Lipofectamine™ LTX and Plus™ Reagent A to osteosarcoma cell lines and stable clones were selected with puromycin. Relative expression of miR-199a-3p in transfected osteosarcoma cell lines KHOS and U-2OS was assessed by real-time PCR with total RNA isolated from the indicated cell lines as described in methods.
Established miR-199-3p overexpressing stable cell lines
To determine the functional role of miR-199a-3p in osteosarcoma, we stably transfected either miR-199a-3p precursor in a vector containing red fluorescence protein (RFP) or control vector with RFP into KHOS and U-2OS osteosarcoma cell lines. Successful transfections were selected with puromycin, and then further selected with red fluorescence to obtain stable clones. We confirmed by Real-time RT-PCR that miR-199a-3p was highly expressed in puromycin selected clones as compared with untransfected cells, or cells transfected with the empty control vector miR-Null (Fig. 2B).
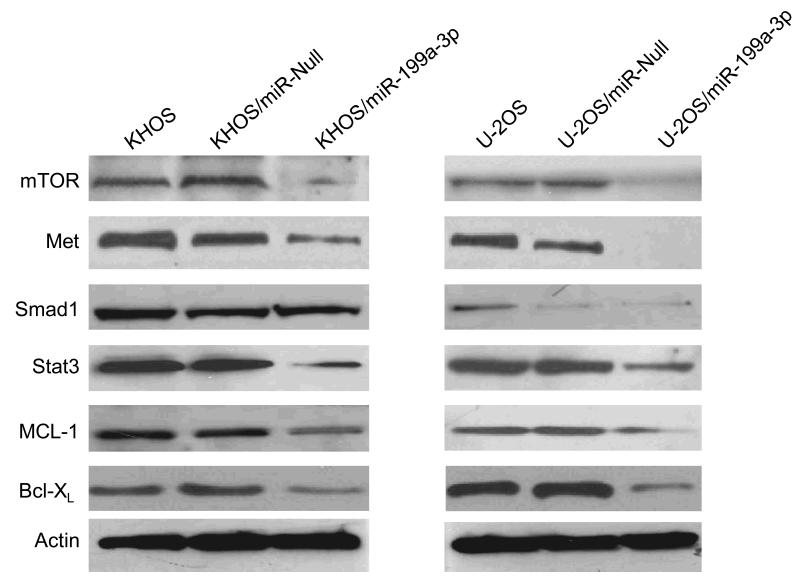
miR-199a-3p suppresses mTOR, Met and Stat3 expression
To establish the effects of reduced miR-199a-3p expression on target genes in osteosarcoma, we measured the protein levels of the miR-199a-3p targeted genes, Met and mTOR (25). First, we measured mTOR, Stat3 and Met expression levels in the normal osteoblast cell lines and compared them to levels in osteosarcoma cell lines. To assess whether miR-1999a-3p directly alters the expression of Met and mTOR in osteosarcoma cell lines, we measured Met and mTOR levels in osteosarcoma cell lines transfected with pre-miR-199a-3p. The Western blot results show that miR-199a-3p transfection decreases Met and mTOR expression levels in KHOS and U-2OS cells (Fig. 3). Furthermore, Stat3, MCL-1 and Bcl-XL protein levels were also decreased in miR-199-3p transfected cells (Fig. 3).
Figure 3.
Transfection of miR-199a-3p into osteosarcoma cells suppresses mTOR and Met expression. KHOS or U-2OS cells were transfected with either miR-199a-3p precursor or pEP-miR-Null control vector and stable clones were selected with puromycin. Expression mTOR, Met, Smad1, Stat3, MCL-1 and Bcl-XL were determined by Western blot as described in the Materials and Methods. β-actin was used as a control.
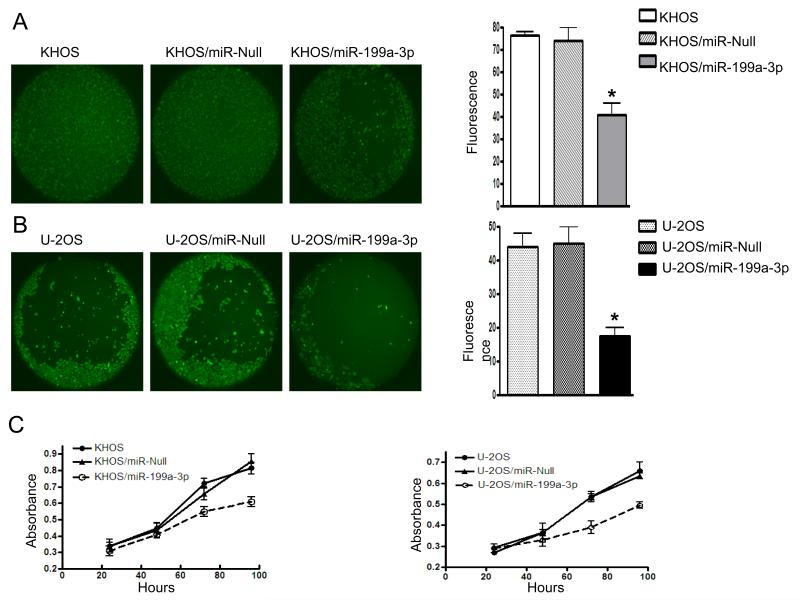
miR-199a-3p inhibits osteosarcoma cell migration and proliferation
To assess the phenotype of miR-199a-3p expression in the growth of osteosarcoma cell lines, cell migration and proliferation in miR-199a-3p stable transfected cells were compared with untransfected cells and cells transfected with empty control vector. We observed a significant decrease in the number of cell migrations in both KHOS and U-2OS cell lines transfected with miR-199a-3p (Fig.4A and Fig. 4B). Furthermore, overexpression of miR-199a-3p also decreased cell viability and inhibited the growth and proliferation of osteosarcoma cell lines (Fig. 4C).
Figure 4.
Transfection of miR-199a-3p into osteosarcoma cells decreases cell migration and proliferation. Cell migration was determined by Oris™Cell Migration Assay Kit as described in the Materials and Methods. A and B, Effects of overexpression of miR-199a-3p on osteosarcoma cell line KHOS and U-2OS. Cell migration were visualized and examined by fluorescence microscope and images were captured with a SPOT RT digital camera (left panel of A and B). The cell fluorescence signals in the detection zones were measured with a SPECTRAmax® Microplate Spectrofluorometer. The bar graph depicts the fluorescent signal that reflect the cell migration (right panel of A and B). The data were representative of one of three independent experiments. *, paired t test, P<0.01. C, The growth and proliferation of osteosarcoma cells were determined by MTT after 24 h, 48 h, 72 h and 96 h after transfection of miR-199a-3p precursor into KHOS (left panel of C) and U-2OS (right panel of C) as described in “Materials and Methods”.
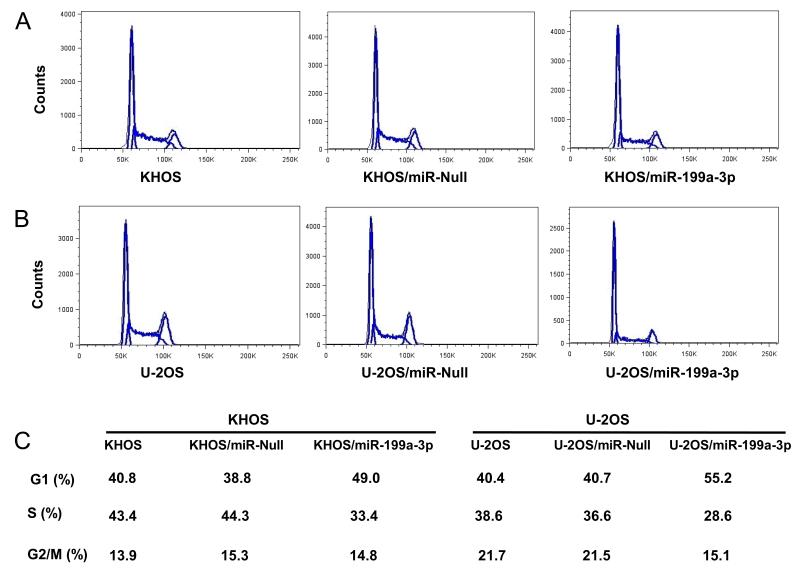
Effect of miR-199a-3p expression on cell cycle distribution in osteosarcoma cell lines
To characterize the effects of miR-199a-3p expression in cell cycle regulation, miR-199a-3p transfected osteosarcoma cell lines were analyzed by flow cytometry. We observed a significant increase in the G1-phase cell population (40.8 % vs. 49.0 % in KHOS and 40.4 % vs. 55.2 % in U-2OS); and a decrease of S-phase (43.4 % vs. 33.4 % in KHOS and 38.6 % vs. 28.6 % in U-2OS) after transfection of miR-199a-3p in both KHOS and U-2OS, whereas, the empty vector miR-Null transfectants exhibited no cell cycle changes (Figure 5).
Figure 5.
Effect of miR-199a-3p expression on cell cycle distribution in osteosarcoma cell lines. A, Cell cycle analysis of osteosarcoma cell line KHOS transfected with miR-199a-3p precursor. B, Cell cycle analysis of osteosarcoma cell line U-2OS transfected with miR-199a-3p precursor. C, Percentages of cells in cell cycle of G1, S and G2/M phases of each group. KHOS and U-2OS cell lines were transfected with miR-199a 3p precursor or control vector (miR-Null), stable clones were selected in 2μg/ml of puromycin and flow cytometry analysis of cell cycle was performed as described in “Materials and Methods”.
Discussion
The current study identified 26 miRs with expression levels that were decreased or increased in osteosarcoma cell lines as compared to osteoblast cell lines. Among them, miR-199a-3p, miR-127-3p, and miR-376c were continuously decreased in osteosarcoma cell lines, while miR-151-3p and miR-191 were increased in osteosarcoma cell lines. Real-time RT-PCR confirmed that these miRs were differentially expressed in osteosarcoma cell lines and osteosarcoma tissues when compared to osteoblast cell lines. Specifically, miR-199a-3p, a miR previously reported to be decreased in liver, bladder, and ovarian cancer (22-24), decreased significantly in osteosarcoma tissue samples in this study. These studies suggest that miR-199a-3p may be part play a role in the pathogenesis of a variety of cancers, including osteosarcoma.
A functional analysis of these miRs may lead to better understanding of the mechanisms by which miRs mediate proliferation and transformation. We show that miR-199a-3p decreases the expression of several oncogenes and antiapoptotic genes, including Met and mTOR, as well as Stat3, MCL-1 and Bcl-XL. These results are consistent with several studies that have suggested that miR-199a-3p is a potential tumor suppressor (24-27). First, the expression of miR-199a-3p is decreased in all proliferating cell lines tested except for fibroblasts. Second, introduction of the miR-199a-3p precursor induced apoptosis in cancer cells. Third, miR-199a-3p downregulates both Met proto-oncogenes and ERK2 (25, 28). In support of our finding that miR-199a-3p functions as a tumor suppressor, it was recently reported that expression of miR-199a-3p was significantly reduced in ovarian, liver, breast, bladder and liver cancer when compared with normal tissues (22-24, 26, 29). We also found that the level of anti-apoptotic protein MCL-1 and Bcl-XL decreased significantly in cells transfected with miR-199a-3p. In fact, anti-apoptotic factors such as MCL-1 and BCL-XL are overexpressed in a variety of human tumors, including osteosarcoma, and down-regulation by siRNA does indeed inhibit cell growth and induced apoptosis (30, 31).
We found that miR-199a-3p expression had a significant effect on osteosarcoma cell growth in vitro. Overexpression of miR-199a-3p by transfection significantly decreased osteosarcoma cell growth and migration. This growth suppressive effect is associated with an increase in G1-phase population and a decrease of the S-phase followed by restoration in miR-199a-3p expression. Similar effects on cellular proliferation rate and cell cycle distribution after miR-199a-3p overexpression has previously been described for liver cancer and bladder cancer cells (22, 24, 26). A decreased level of Met, mTOR and Stat3 upon miR-199a-3p overexpression is in concordance with a reduced cellular proliferation rate. Interestingly, miR-199a-3p was reported to regulate mTOR and Met to influence the doxorubicin sensitivity in liver cancer cells (26). This study also demonstrated an inverse correlation between miR-199a-3p and mTOR levels in human liver cancer tissues (26). These results suggest that deregulation of miR-199a-3p expression is not tumor type specific.
In summary, our study identified that there is decreased expression of miR-199a-3p in osteosarcoma and that miR-199a-3p showed tumor suppressive abilities in vitro by affecting proliferation, migration and cell cycle. These results suggest that miR-199a-3p may have a tumor suppressor function in human osteosarcoma. A recent study showed that miR-199a-3p may target oncogenes such as Met and mTOR as well as Stat3 in osteosarcoma. These results provide support for restoring miR-199a-3p as gene therapy, as they may turn out to be promising candidates for biomarkers and gene therapy targets for treating human osteosarcoma.
Supplementary Material
Acknowledgements
This project was supported, in part, by grants from the Gattegno and Wechsler funds. Support has also been provided by the Kenneth Stanton Fund. Dr. Duan is supported, in part, through a grant from Sarcoma Foundation of America (SFA) and a grant from an Academic Enrichment Fund of MGH Orthopaedics. Dr. Choy is supported by the Harvard Catalyst | The Harvard Clinical and Translational Science Center (Award #UL1 RR 025758 and financial contributions from Harvard University and its affiliated academic health care centers). In addition, we would like to acknowledge Dr. Christoph Eicken at LC Sciences, LLC for providing useful advices during the analyzing of miRNA expression data.
Financial support: The Gattegno and Wechsler funds, Kenneth Stanton Fund and Sarcoma Foundation of America
Abbreviations
- miR-199a-3p
microRNA-199a-3p
- mTOR
mammalian target of rapamycin
- Stat3
signal transducer and activator of transcription 3
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Bielack SS, Marina N, Ferrari S, Helman LJ, Smeland S, Whelan JS, et al. Osteosarcoma: the same old drugs or more? J Clin Oncol. 2008;26:3102–3. doi: 10.1200/JCO.2008.17.1108. author reply 4-5. [DOI] [PubMed] [Google Scholar]
- 2.Chou AJ, Geller DS, Gorlick R. Therapy for osteosarcoma: where do we go from here? Paediatr Drugs. 2008;10:315–27. doi: 10.2165/00148581-200810050-00005. [DOI] [PubMed] [Google Scholar]
- 3.O’Day K, Gorlick R. Novel therapeutic agents for osteosarcoma. Expert Rev Anticancer Ther. 2009;9:511–23. doi: 10.1586/era.09.7. [DOI] [PubMed] [Google Scholar]
- 4.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–5. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–15. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs--the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 7.Mishra PJ, Merlino G. MicroRNA reexpression as differentiation therapy in cancer. J Clin Invest. 2009;119:2119–23. doi: 10.1172/JCI40107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan D, Dong Xda E, Chen X, Wang L, Lu C, Wang J, et al. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem. 2009;284:29596–604. doi: 10.1074/jbc.M109.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan Z, Choy E, Nielsen GP, Rosenberg A, Iafrate J, Yang C, et al. Differential expression of microRNA (miRNA) in chordoma reveals a role for miRNA-1 in Met expression. J Orthop Res. 2010;28:746–52. doi: 10.1002/jor.21055. [DOI] [PubMed] [Google Scholar]
- 11.Subramanian S, Lui WO, Lee CH, Espinosa I, Nielsen TO, Heinrich MC, et al. MicroRNA expression signature of human sarcomas. Oncogene. 2008;27:2015–26. doi: 10.1038/sj.onc.1210836. [DOI] [PubMed] [Google Scholar]
- 12.Palmieri A, Pezzetti F, Brunelli G, Ilaria Z, Carinci F. A comparison between genetic portraits of normal osteoblasts and osteosarcoma cell lines. Indian J Dent Res. 2009;20:52–9. doi: 10.4103/0970-9290.49069. [DOI] [PubMed] [Google Scholar]
- 13.Palmieri A, Pezzetti F, Graziano A, Riccardo D, Zollino I, Brunelli G, et al. Comparison between osteoblasts derived from human dental pulp stem cells and osteosarcoma cell lines. Cell Biol Int. 2008;32:733–8. doi: 10.1016/j.cellbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Duan Z, Bradner JE, Greenberg E, Levine R, Foster R, Mahoney J, et al. SD-1029 inhibits signal transducer and activator of transcription 3 nuclear translocation. Clin Cancer Res. 2006;12:6844–52. doi: 10.1158/1078-0432.CCR-06-1330. [DOI] [PubMed] [Google Scholar]
- 15.Gough W, Hulkower KI, Lynch R, McGlynn P, Uhlik M, Yan L, et al. A quantitative, facile, and high-throughput image-based cell migration method is a robust alternative to the scratch assay. J Biomol Screen. 2011;16:155–63. doi: 10.1177/1087057110393340. [DOI] [PubMed] [Google Scholar]
- 16.Jiang L, Liu X, Kolokythas A, Yu J, Wang A, Heidbreder CE, et al. Downregulation of the Rho GTPase signaling pathway is involved in the microRNA-138-mediated inhibition of cell migration and invasion in tongue squamous cell carcinoma. Int J Cancer. 2010;127:505–12. doi: 10.1002/ijc.25320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li G, Luna C, Qiu J, Epstein DL, Gonzalez P. Targeting of integrin beta1 and kinesin 2alpha by microRNA 183. J Biol Chem. 2010;285:5461–71. doi: 10.1074/jbc.M109.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 19.Rastinejad F, Conboy MJ, Rando TA, Blau HM. Tumor suppression by RNA from the 3′ untranslated region of alpha-tropomyosin. Cell. 1993;75:1107–17. doi: 10.1016/0092-8674(93)90320-p. [DOI] [PubMed] [Google Scholar]
- 20.Denizot F, Lang R. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J Immunol Methods. 1986;89:271–7. doi: 10.1016/0022-1759(86)90368-6. [DOI] [PubMed] [Google Scholar]
- 21.Carmichael J, DeGraff WG, Gazdar AF, Minna JD, Mitchell JB. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res. 1987;47:936–42. [PubMed] [Google Scholar]
- 22.Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, et al. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–27. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 24.Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345–52. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 25.Kim S, Lee UJ, Kim MN, Lee EJ, Kim JY, Lee MY, et al. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2) J Biol Chem. 2008;283:18158–66. doi: 10.1074/jbc.M800186200. [DOI] [PubMed] [Google Scholar]
- 26.Fornari F, Milazzo M, Chieco P, Negrini M, Calin GA, Grazi GL, et al. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70:5184–93. doi: 10.1158/0008-5472.CAN-10-0145. [DOI] [PubMed] [Google Scholar]
- 27.Migliore C, Petrelli A, Ghiso E, Corso S, Capparuccia L, Eramo A, et al. MicroRNAs impair MET-mediated invasive growth. Cancer Res. 2008;68:10128–36. doi: 10.1158/0008-5472.CAN-08-2148. [DOI] [PubMed] [Google Scholar]
- 28.Lee YB, Bantounas I, Lee DY, Phylactou L, Caldwell MA, Uney JB. Twist-1 regulates the miR-199a/214 cluster during development. Nucleic Acids Res. 2009;37:123–8. doi: 10.1093/nar/gkn920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang F, Zheng Z, Guo J, Ding X. Correlation and quantitation of microRNA aberrant expression in tissues and sera from patients with breast tumor. Gynecol Oncol. 2010;119:586–93. doi: 10.1016/j.ygyno.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 30.Dai Y, Grant S. Targeting multiple arms of the apoptotic regulatory machinery. Cancer Res. 2007;67:2908–11. doi: 10.1158/0008-5472.CAN-07-0082. [DOI] [PubMed] [Google Scholar]
- 31.Wang ZX, Yang JS, Pan X, Wang JR, Li J, Yin YM, et al. Functional and biological analysis of Bcl-xL expression in human osteosarcoma. Bone. 2010;47:445–54. doi: 10.1016/j.bone.2010.05.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.