Abstract
The cytoplasmic fate of mRNAs is dictated by the relative activities of the intimately connected mRNA decay and translation initiation pathways. In this study, we have found that yeast strains compromised for stages downstream of deadenylation in the major mRNA decay pathway are incapable of inhibiting global translation initiation in response to stress. In the past, the paradigm of the eIF2α kinase-dependent amino acid starvation pathway in yeast has been used to evaluate this highly conserved stress response in all eukaryotic cells. Using a similar approach we have found that even though the mRNA decay mutants maintain high levels of general translation, they exhibit many of the hallmarks of amino acid starvation, including increased eIF2α phosphorylation and activated GCN4 mRNA translation. Therefore, these mutants appear translationally oblivious to decreased ternary complex abundance, and we propose that this is due to higher rates of mRNA recruitment to the 40S ribosomal subunit.
Translation and mRNA decay are intimately linked to regulate the gene expression profile in response to a variety of nutritional and environmental stresses in Saccharomyces cerevisiae. Previously, a number of these yeast stresses have been found to inhibit translation at the initiation step, leading to a rapid loss of polysomes and an accumulation of inactive singlet ribosomes or monosomes. These stresses include glucose starvation (3, 23), salt stresses (28, 42), rapamycin addition (5), fusel alcohol addition (4), and severe amino acid starvation (41). In general, the inhibition of translation initiation is extremely rapid, and we rationalize these responses as follows. The inhibition of protein synthesis could allow the degradation of existing mRNAs and proteins, thus facilitating a switch in the genetic program that might be required to adapt to the particular stress. In addition, the inhibition of global translation initiation may allow specific specialized mRNAs to be translated and this activation could also support adaptation to the stress.
A precedent for this type of regulation exists in the general control pathway of yeast (17). Here global protein synthesis is rapidly inhibited upon severe amino acid starvation (41). It is thought that a specific protein kinase (Gcn2p) senses the levels of amino acids indirectly via the level of uncharged tRNA in the cell. An accumulation of uncharged tRNA stimulates the phosphorylation of the eIF2α subunit at serine 51 by Gcn2p, leading to the sequestration of a guanine nucleotide exchange factor, eIF2B. The eIF2B complex normally catalyzes the recycling of eIF2-GDP to the translationally active form, eIF2-GTP. eIF2-GTP is capable of interacting with the methionyl initiator tRNA (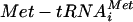 ) to form the ternary complex (eIF2-GTP
) to form the ternary complex (eIF2-GTP 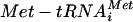 ). Therefore, inhibition of eIF2B (following severe starvation for amino acids and the subsequent phosphorylation of eIF2α) ultimately leads to decreased levels of the ternary complex (17). Recently it has been suggested that the ternary complex might interact with other translation initiation factors (eIFs 1, 5, and 3) to form a multifactor complex (MFC) prior to interaction with the 40S ribosomal subunit (43).
). Therefore, inhibition of eIF2B (following severe starvation for amino acids and the subsequent phosphorylation of eIF2α) ultimately leads to decreased levels of the ternary complex (17). Recently it has been suggested that the ternary complex might interact with other translation initiation factors (eIFs 1, 5, and 3) to form a multifactor complex (MFC) prior to interaction with the 40S ribosomal subunit (43).
As well as inhibiting global protein synthesis, starvation for amino acids induces the expression of a transcription factor, Gcn4p (17). The activation of this transcription factor at the translational level serves to overcome the amino acid stress by inducing the transcription of amino acid biosynthetic genes (18). The GCN4 mRNA contains four short upstream open reading frames (uORFs) that are involved in the translational activation via a complex mechanism involving translational reinitiation at downstream ORFs. The translation of this mRNA is exquisitely sensitive to the levels of ternary complex in the cell. Therefore, following translation of the first uORF (in the presence of high levels of ternary complex), reinitiation at downstream uORFs and subsequent translation termination is favored. In contrast, when ternary complex levels are low (for instance, following amino acid starvation), there is a greater chance that ternary complex will be acquired after ribosomes pass the uORFs, thus allowing GCN4 ORF translation (17).
A variety of other stresses have also been shown to activate GCN4 translation initiation. These include purine starvation, glucose starvation, fusel alcohol addition, salt stress, and rapamycin addition (4, 8, 15, 22, 30, 44, 48). Although the recently described effects of rapamycin on GCN4 do rely on increases in eIF2α phosphorylation (8, 22, 44), at least some of these stress responses do not appear to require an induction of eIF2α phosphorylation. Therefore, it seems plausible that these responses might decrease levels of ternary complex via alternative mechanisms. For example, we have proposed that fusel alcohols directly regulate the activity of eIF2B and thus lead to decreased ternary complex levels (4).
Starvation for glucose has a complex effect on translation initiation. Immediately following glucose removal (i.e., within minutes) there is a rapid inhibition of translation initiation (3, 23, 42). This inhibition still occurs in mutants that are incapable of phosphorylating eIF2α. In wild-type cells, however, activation of GCN4 translation at later time points following glucose starvation has been described and this activation is Gcn2p dependent (48). The precise mechanisms for these effects on global and GCN4 translation initiation remain to be elucidated.
Two general pathways of mRNA decay have been described for yeast. In both of these pathways, mRNAs are first deadenylated in a process thought to involve the Ccr4p/Pop2p deadenylase (40). In the major or 5′ to 3′ pathway, the 5′ cap structure is then removed by the Dcp1p/Dcp2p decapping complex and this facilitates the degradation of the body of the mRNA via Xrn1p, the 5′ to 3′ exoribonuclease (36, 39). In the exosome-mediated pathway, the deadenylated mRNA is degraded by the cytoplasmic exosome complex (which includes proteins with 3′ to 5′ exoribonuclease activity and putative RNA helicase activity) (26). As well as these general pathways for mRNA decay, pathways exist to degrade mRNAs containing premature stop codons. These nonsense-mediated decay pathways utilize many of the same components described for the general pathways of mRNA decay; however, these mRNAs are targeted for decay via a specific complex containing the Upf proteins (14, 27).
In this study we have analyzed a catalogue of mutants in these mRNA decay processes and assessed translational control in response to stress. As a result, we have found that major pathway mRNA decay mutants acting at stages downstream of deadenylation are globally resistant to stress-induced translational inhibition. At least in the case of severe amino acid starvation, these strains are translationally resistant to the decrease in ternary complex levels that occurs as a result of this stress. Thus, this study provides an example of the intimate relationship that exists between protein synthesis and mRNA turnover in the cytoplasm of cells and the interdependence of mechanisms involved in their regulation.
MATERIALS AND METHODS
Strains and growth conditions.
Table 1 lists the strains used in this study. In general the yMK116 (yRP840) strain background has been used throughout except where this proved impractical. In these cases isogenic strains were used across individual experiments. Gene disruptions were carried out as previously described (47). General yeast molecular genetic techniques and growth of cultures on standard yeast extract-peptone-dextrose (YPD) or synthetic complete dextrose (SCD) medium were performed as detailed previously (16). Glucose starvation was brought about by the removal of glucose for 5 or 10 min. Severe amino acid starvation was brought about by the removal of every amino acid for 10 min. Butanol was added at 1 or 0.75% (vol/vol) in liquid culture for 10 min. Rapamycin (Sigma-Aldrich, Poole, Dorset, United Kingdom) in 90% ethanol-10% Tween 20 was added to achieve a final concentration of 0.2 μg/ml for 1 h (an equal volume of drug vehicle was used as a control). Lithium acetate was added to galactose-grown cultures to achieve a final concentration of 30 mM for 15 min. Dilution series plate assays started with 3 μl of exponentially growing cultures at an optical density at 600 nm (OD600) of 0.3 and continued in fivefold dilutions.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| yMK23/24 (BUTR) | MATa or MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-P180 | 4 |
| yMK36/37 (BUTS) | MATa or MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 | 4 |
| yMK75 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3 gcn2::URA3 | 3 |
| yMK116 | MATahis4-539 leu2-3,112 trp1-1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG | R. Parker (yRP840) |
| yMK117 | MATahis4-539 leu2-3,112 trp1-1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG dcp1::URA3 | R. Parker (yRP1070) |
| yMK157 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 p[TEF1-TIF4632 URA3 CEN] | This study |
| yMK158 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 p[URA3 CEN] | This study |
| yMK279 | MATahis4-539 leu2-3,112 trp1-1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG dcp1::URA3 p[DCP1 TRP1 CEN] | R. Parker (pRP783) |
| yMK283 | MATahis4-539 leu2-3,112 trp1-1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG dcp1::URA3 p[dcp1-4 TRP1 CEN] | R. Parker (pRP874) |
| yMK286 | MATahis4-539 leu2-3,112 trp1-1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG dcp1::URA3 p[dcp1-7 TRP1 CEN] | R. Parker (pRP877) |
| yMK290 | MATahis4-539 leu2-3,112 trp1-1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG dcp1::URA3 p[dcp1-13 TRP1 CEN] | R. Parker (pRP881) |
| yMK291 | MATahis4-539 leu2-3,112 trp1-1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG dcp1::URA3 p[dcp1-14 TRP1 CEN] | R. Parker (pRP882) |
| yMK296 | MATahis4-539 leu2-3,112 trp1-1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG dcp1::URA3 p[dcp1-21 TRP1 CEN] | R. Parker (pRP887) |
| yMK305 | MATahis4-539 leu2-3,112 trp1-1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG dcp1::URA3 p[dcp1-34 TRP1 CEN] | R. Parker (pRP896) |
| yMK321 | MATaade2 his3 leu2 trp1 ura3 lsm1::LEU2 | Sachs strain collection |
| yMK323 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 pbp1::TRP1 | Sachs strain collection |
| yMK324 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 pbp1::TRP1 pab1::HIS3 | Sachs strain collection |
| yMK329 | MATaHIS4 leu2-3,112 trp1-1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG | This study |
| yMK330 | MATaHIS4 leu2-3,112 trp1-1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG dcp1::URA3 | This study |
| yMK380 | MATα his3 leu2 trp1 ura3 xrn1::LEU2 | Sachs strain collection |
| yMK429 | MATaade2-1 HIS3 leu2-3,112 trp1-1 ura3-1 can1-100 GCD1-S180 | 3 |
| yMK431 | MATaade2-1 HIS3 leu2-3,112 trp1-1 ura3-1 can1-100 GCD1-P180 | 3 |
| yMK513 | MATα ade2-1 HIS3 leu2-3,112 trp1-1 ura3-1 can1-100 gcn2::URA3 | This study |
| yMK696 | MATα leu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG lsm1::TRP1 | R. Parker (yRP1365) |
| yMK697 | MATα his4-539 leu2-3,112 trp1-1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG p[AEM55 LSM2ts] | R. Parker (yRP1366) |
| yMK699 | MATα his4-539 leu2-3,112 trp1-1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG lsm6::HIS3 | R. Parker (yRP1369) |
| yMK700 | MATα his4-539 leu2-3,112 trp1-1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG lsm7::HIS3LYS2 | R. Parker (yRP1370) |
| yMK701 | MATaade2 his3 leu2 lys2 trp1 ura3 LHP1 | R. Parker (yRP1402) |
| yMK702 | MATaade2 his3 leu2 lys2 trp1 ura3 LHP1 lsm8-1 | R. Parker (yRP1405) |
| yMK703 | MATahis4-539 leu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG pat1::LEU2 | R. Parker (yRP1372) |
| yMK704 | MATahis4-539 leu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG dcp2::TRP1 | R. Parker (yRP1346) |
| yMK705 | MATahis4-539 leu2-3,112 trp1-1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ski2::LEU2 | R. Parker (yRP1195) |
| yMK706 | MATahis4-539 leu2-3,112 trp1-1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ski3::TRP1 | R. Parker (yRP1196) |
| yMK707 | MATahis4-539 leu2-3,112 lys2-201 trp1-1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ski8::URA3 | R. Parker (yRP1197) |
| yMK708 | MATα his4-539 leu2-3,112 lys2-201 trp1-Δ1 ura3-52(ski6-2/LYS2) ski6::URA3 | R. Parker (yRP1203) |
| yMK709 | MATα ade1-100 leu2-3,112 trp1-Δ1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG rrp4-1 | R. Parker (yRP1223) |
| yMK710 | MATahis4 leu2 lys2 trp1 ura3 cup1::LEU2/PGK1pG/MFA2pG upf1::URA3 | R. Parker (yRP1373) |
| yMK711 | MATahis4-539 leu2-3,112 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ccr4::NEO | R. Parker (yRP1616) |
| yMK712 | MATα leu2-3,112 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG pop2::URA3 | R. Parker (yRP1617) |
| yMK713 | MATahis4-539 leu2-3,112 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG ccr4::NEO pop2::URA3 | R. Parker (yRP1618) |
| yMK714 | MATaade2-101 his3-Δ200 leu2-3,112 lys2-201 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG edc1::HIS3 | R. Parker (yRP1503) |
| yMK769 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 dhh1::G418 | EUROSCARF |
| yMK793 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 pan2::LEU2 pan3::HIS3 | Sachs strain collection |
| yMK840 | MATaleu2-3,112 trp1-Δ63 ura3-52 | 9 |
| yMK841 | MATaleu2-3,112 trp1-Δ63 ura3-52 fun12::LEU2 | 9 |
| yMK861 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-P180 dcp1::G418 | This study |
| yMK863 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 dcp1::G418 | This study |
| yMK870 | MATaade1 ade2 gal1 lys2 ura3-52 | 29 |
| yMK871 | MATaade1 ade2 gal1 lys2 ura3-52 pap1-1 | 29 |
| yMK898 | MATaade3-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 | 10 |
| yMK899 | MATaade3-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 dob1-1 | 10 |
| yMK919 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 dcp1::G418 pan2::LEU2 pan3::HIS3 | This study |
| yMK921 | MATαade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 dcp1::G418 ccr4::G418 | This study |
| yMK926 | MATaade2-1 HIS3 leu2-3,112 trp1-1 ura3-1 GCD1-P180 p[GCN4-lacZ URA3 CEN] | This study |
| yMK927 | MATaade2-1 HIS3 leu2-3,112 trp1-1 ura3-1 GCD1-P180 p[ΔuORFs-GCN4-lacZ URA3 CEN] | This study |
| yMK936 | MATaade2-1 HIS3 leu2-3,112 trp1-1 ura3-1 GCD1-P180 dcp1::G418 p[GCN4-lacZ URA3 CEN] | This study |
| yMK937 | MATaade2-1 HIS3 leu2-3,112 trp1-1 ura3-1 GCD1-P180 dcp1::G418 p[ΔuORFs-GCN4-lacZ URA3 CEN] | This study/PICK> |
| yMK940 | MATaade2-1 HIS3 leu2-3,112 trp1-1 ura3-1 GCD1-P180 dcp1::G418 | This study |
| yMK1045 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 fal1::HIS3MX6 p[FAL1 LEU2 CEN] | 21 |
| yMK1046 | MATaade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 fal1::HIS3MX6 p[fal1-1 LEU2 CEN] | 21 |
Analysis of ribosome distribution on sucrose gradients.
Yeast cultures were grown to an OD600 of 0.7 and treated by the removal of glucose or amino acids or by the addition of butanol, rapamycin, or lithium as described above. Extracts were prepared in 100 μg of cycloheximide/ml, and these were layered onto 15 to 50% sucrose gradients. The gradients were sedimented via centrifugation at 40,000 rpm in a Beckman ultracentrifuge for 2.5 h, and the A254 was measured continuously to give the traces shown (3).
[35S]methionine incorporation assay.
Yeast strains were grown to an OD600 of 0.4 in SCD medium lacking methionine, and two 7.5-ml aliquots were pelleted and then resuspended in 20 ml of SCD-Met or SC-Met. Methionine was added to achieve a final concentration of 60 ng/ml, of which 0.5 ng/ml was [35S]methionine (New England Nuclear, Boston, Mass.) (cell-labeling grade; 1,175 Ci/mmol). Samples (1 ml) were taken at the indicated times and processed as described previously (3).
Assays of GCN4-lacZ reporter expression.
Standard methods for measuring the β-galactosidase activity for strains bearing GCN4-lacZ fusions were used (24). β-Galactosidase levels are expressed as nanomoles of o-nitrophenol-β-d-galactopyranoside (ONPG) hydrolyzed per minute per microgram of total protein.
Measurement of capped mRNA with the Xrn1p assay.
Yeast strains were grown to an OD600 of 0.7 in YPD and divided into three aliquots, and cells were pelleted. One of the cell pellets was immediately frozen in liquid nitrogen. The other two were resuspended in YPD or YP, incubated, pelleted, and frozen in liquid nitrogen such that the total incubation time was 10 min. RNA was extracted from the frozen pellets and used for Northern blot analysis (7). The Xrn1p assay was performed as described previously (7). Total RNA (5 μg) was incubated with 400 ng of purified Xrn1p (a gift from A. Johnson, University of Texas, Austin) in a final volume of 5 μl in 33 mM Tris-HCl (pH 8.0)-50 mM NaCl-2.5 mM MgCl2-0.2 mM dithiothreitol in the presence or absence of 5 mM EDTA. Reactions were incubated at 37°C for 30 min and stopped using RNA loading buffer.
Immunoblotting of eIF2α and phosphoserine 51 eIF2α.
Yeast strains were grown to an OD600 of 0.7 in SCD-His. Three 15-ml aliquots were taken per strain, the cells were pelleted, and SCD-His medium, SCD minus all amino acids, or SCD-His plus 50 mM 3-aminotriazole (3-AT) was added to each of the three aliquots, respectively. The cells were incubated, pelleted at 30°C, and rapidly frozen in liquid N2 such that the total incubation time was 15 min. The cells were lysed and protein samples were prepared, electrophoretically separated, and subjected to immunoblot analysis as described previously (3). The phosphospecific eIF2α (Biosource International) and eIF2α (a gift from G. Pavitt, University of Manchester Institute of Science and Technology, Manchester, United Kingdom) antibodies were used for the detection.
RESULTS
dcp1Δ and lsm1Δ mutants maintain translation following glucose removal.
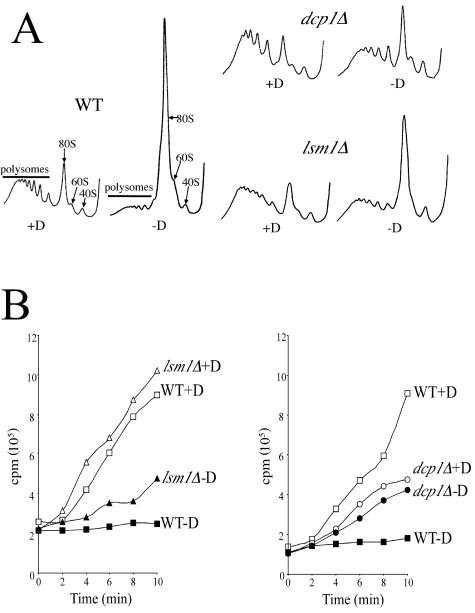
Previously we have found that the removal of glucose from yeast growth medium results in a rapid inhibition of translation initiation (3). This inhibition is characterized by a loss of polysomes and a concomitant increase in 80S monosomes (Fig. 1A). A candidate gene strategy was adopted to test a whole range of strains (either mutated in or overexpressing translation initiation factor and mRNA decay genes) (data not shown). Intriguingly, strains lacking either Dcp1p, a key component of the mRNA decapping enzyme, or Lsm1p, a member of the mRNA decay Lsm complex, maintain polysomal levels following glucose removal (Fig. 1A).
FIG. 1.
mRNA decapping mutants maintain protein synthesis following glucose starvation. (A) Polysome traces from yMK116 (WT), yMK117 (dcp1Δ), and yMK321 (lsm1Δ). Yeast were grown in SCD medium, harvested, and then resuspended for 10 min in medium containing (+D) or lacking (−D) glucose. Polysomes were analyzed as described in Materials and Methods. The 40S (small ribosomal subunit), 60S (large ribosomal subunit), 80S (monosome), and polysome peaks are labeled. Even though the mutant strains were derived from different backgrounds, only a single wild type is shown for simplicity, as the inhibition of translation is identical in the different wild-type strains (data not shown). (B) [35S]methionine incorporation into proteins over time in the presence (+D) or absence (−D) of glucose for strains yMK23 (WT) and yMK321 (lsm1Δ) (left panel) and strains yMK116 (WT) and yMK117 (dcp1Δ) (right panel). Yeast strains were grown in SCD medium without methionine, harvested, and resuspended in a labeling mixture in the presence or absence of glucose. Aliquots were taken at the indicated times, and the levels of [35S]methionine incorporation into proteins were determined.
That polysomes were maintained in these mutants after glucose removal could be explained either by a continued synthesis of proteins or by decreased rates of downstream translational steps such as elongation and termination. To distinguish between these possibilities, the overall rate of protein synthesis was measured by [35S]methionine incorporation assays in either the presence or the absence of glucose (Fig. 1B). Strains lacking Lsm1p do not show a significant difference in growth rates in rich medium; hence, the rate of protein synthesis for this strain is similar to that of the isogenic background. In rich medium, however, the dcp1Δ mutant strain has a doubling time approximately twofold higher than its isogenic wild type (data not shown). In consistency with this observation, the rate of protein synthesis in the presence of glucose for the dcp1Δ mutant is lower than that seen with the wild type. For both the lsm1Δ and dcp1Δ strains, however, it is clear that (following a switch to medium with no glucose) the level of [35S]methionine incorporation is significantly higher than that seen with the appropriate wild-type strains. Indeed the dcp1Δ mutant strain continues to translate at a rate typical of that seen with glucose-containing medium. This demonstrates that the observed maintenance of polysomes following glucose removal from these mutants is due to sustained protein synthesis and does not occur due to slowed rates of elongation or termination in these strains.
Loss of decapping activity in DCP1 mutants allows the maintenance of translation initiation following glucose removal.
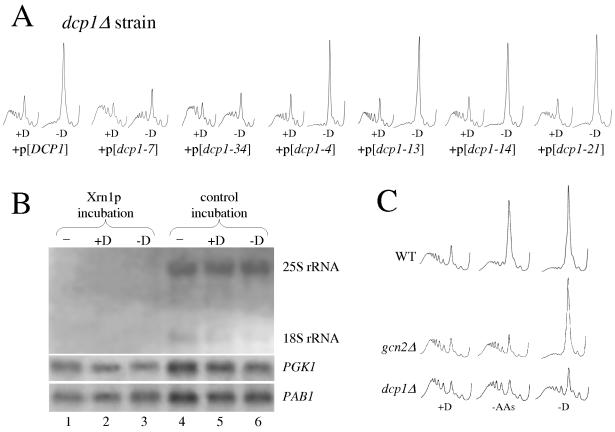
Strains lacking the decapping component Dcp1p have been characterized by their lack of decapping function and a resulting inability to decay mRNAs in a 5′ to 3′ direction (6). In addition, the original loss-of-function mutation in LSM1 (SPB8) was shown to be partially inhibited for mRNA decapping (7). This is consistent with the partial maintenance of protein synthesis observed following glucose removal in an lsm1Δ mutant (Fig. 1). To investigate whether a decrease in decapping function was responsible for continued translation following glucose removal, a well-characterized series of DCP1 mutants was assessed (37). Strains with mutations in DCP1 that either have little effect (dcp1-13, dcp1-14, and dcp1-21) or are partial-loss-of-function mutations (dcp1-4) in terms of mRNA decapping (37) show a redistribution of polysomes upon glucose withdrawal typical of that observed for wild-type strains (Fig. 2A). However, dcp1-7 and dcp1-34 mutants, which are known to have a strong loss of mRNA decapping function (37), maintain protein synthesis following glucose withdrawal (Fig. 2A). Hence, where decapping activity is low in the DCP1 mutants, translation is maintained following glucose removal.
FIG. 2.
Decapping activity is required for translational inhibition, but this is not via a global decapping activation in response to stress. (A) Polysome traces from the dcp1Δ strain (yMK117) containing various plasmid-borne (+p) alleles of DCP1. Wild-type DCP1 (yMK279) or mutant alleles of DCP1 (yMK286 [dcp1-7], yMK305 [dcp1-34], yMK283 [dcp1-4], yMK290 [dcp1-13], yMK291 [dcp1-14], and yMK296 [dcp1-21]) were grown in SCD-TRP, harvested, and then resuspended in the same medium in the presence (+D) or absence (−D) of glucose for 10 min. (B) Northern blot stained with methylene blue to visualize rRNA levels (upper panel). The same blot was probed for either PAB1 mRNA or PGK1 mRNA (lower panels). A wild-type strain (yMK23) was grown in rich medium, and RNA was prepared either instantly (−) or following a 10-min incubation in rich medium with (+D) or without (−D) glucose. RNA samples were incubated in vitro with Xrn1p either in the presence (control incubation) or absence (Xrn1p incubation) of EDTA (which inhibits the 5′ to 3′ exoribonuclease). (C) Polysome traces from yMK116 (WT), yMK75 (gcn2Δ), and yMK117 (dcp1Δ) strains. Strains were grown in SCD medium, harvested, and then resuspended in the same medium (+D), medium without amino acids (−AAs), or medium without glucose (−D). Even though the mutant strains are derived from different backgrounds, only a single wild type is shown for simplicity, as the levels of inhibition of translation in the different wild-type strains were identical (data not shown).
An activation of decapping does not explain the inhibition of translation initiation following glucose removal.
Previously we showed that for wild-type strains, there is no major change in the abundance of several mRNA species after 10 min in the absence of glucose (3). As decapping mutants are translationally resistant to glucose removal, it is formally possible that in wild-type strains rapid global mRNA decapping might follow glucose removal and lead to the observed inhibition of protein synthesis. To explore whether there is a global decapping of mRNAs following glucose removal, an assay was used to assess the level of capped mRNA. Here RNA was prepared from cells without incubation (control) or incubated for short periods in either rich medium or medium lacking glucose. The sensitivity of these RNA samples to in vitro incubation in the presence of active Xrn1p was assessed by Northern blotting (Fig. 2B). Xrn1p degrades noncapped RNAs in a 5′ to 3′ direction; hence, the rRNA (which is not modified by a 5′ cap structure) is completely degraded during this treatment (Fig. 2B; cf. lanes 4 to 6 versus lanes 1 to 3). The presence of a cap structure protects the body of mRNAs from this in vitro Xrn1p exonucleolytic decay treatment (7). The levels of either the PGK1 or PAB1 mRNAs do not significantly differ between the samples prepared from yeast incubated in the presence or absence of glucose (Fig. 2B; cf. lanes 2 and 3). This suggests that the 5′ cap is present on these transcripts and inhibits degradation by the exogenously added Xrn1p regardless of the starvation for glucose. Hence, it appears that there is no global decapping of mRNA following glucose removal from wild-type strains.
As the inhibition mechanism following glucose removal is as yet unknown, polysome profiles of a dcp1Δ mutant were examined following amino acid removal. Severe amino acid starvation results in polysomal runoff indicative of an inhibition of translation initiation (41). The translational inhibition following amino acid starvation occurs via a well-described pathway that has not been shown to involve mRNA decay (17). Therefore, polysomal runoff in response to severe amino acid starvation involves eIF2α phosphorylation via Gcn2p kinase; thus, the gcn2Δ mutant is translationally resistant to this stress (Fig. 2C). Surprisingly, polysomal profiling of a dcp1Δ mutant shows that in contrast to the results seen with the wild-type strain, active translation is maintained following amino acid removal as well as glucose removal (Fig. 2C). Therefore, rather than a global decapping of mRNA upon either amino acid starvation or glucose removal accounting for the polysomal runoff and hence the translational resistance of the dcp1Δ strain, it seems more likely that the dcp1Δ mutant is generally resistant to translational control.
An analysis of mRNA decay mutants following either glucose or amino acid starvation.
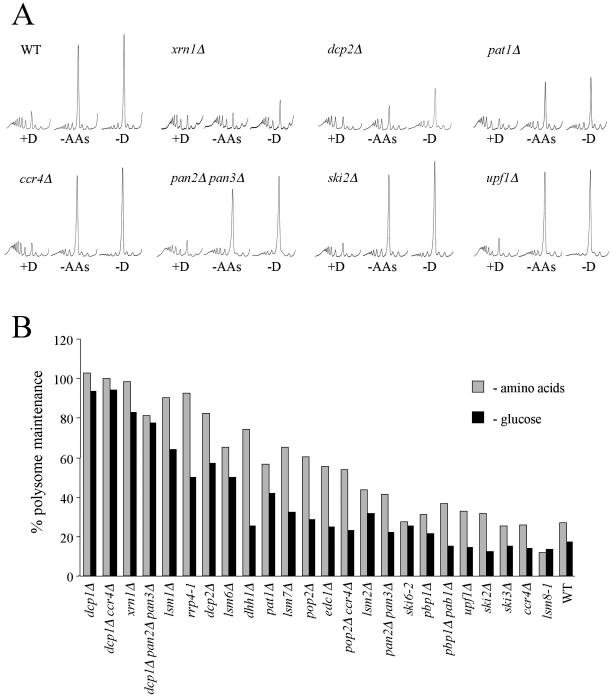
To further investigate the connection between mRNA decay and the regulation of translation initiation in response to nutritional stress, we analyzed the translational phenotypes of a series of mRNA decay mutants. The mutants used harbored deficiencies in the nonsense-mediated, exosome-mediated, or major 5′ to 3′ pathway of mRNA decay. Mutants were resuspended in rich medium and medium lacking either glucose or amino acids and were analyzed by polysomal profiling (Fig. 3A). Figure 3B depicts a graph summarizing these data in which polysome maintenance is a measure of the level of polysomes remaining after each stress relative to the level of polysomes present in unstressed cells.
FIG. 3.
Major mRNA decay pathway mutants affecting stages downstream of deadenylation are particularly resistant to translational stress. (A) Polysome analyses for a wild-type (WT) strain (yMK116) and various mutant strains (yMK380 [xrn1Δ], yMK704 [dcp2Δ], yMK703 [pat1Δ], yMK711 [ccr4Δ], yMK793 [pan2Δ pan3Δ], yMK705 [ski2Δ], and yMK710 [upf1Δ]). Each strain was grown in SCD medium, harvested, and then incubated for 10 min in SCD medium (+D), for 10 min in medium without amino acids (−AAs), or for 5 min in medium without glucose (−D). (B) A bar chart summarizing polysome analyses for a large number of mutant strains (see Table 1 for strain data). The percentage of polysomes remaining after either amino acid starvation for 10 min or glucose starvation for 5 min was calculated by dividing the area under the polysomal traces following stress (established using National Institutes of Health image J software) by the area found in the absence of stress.
A comparison of the effects of amino acid and glucose removal across this set of strains shows that each individual strain responds similarly to both nutritional stresses (Fig. 3B). Where inhibition of translation is observed, it is often more severe after glucose removal than after amino acid removal.
For mutants specifically deficient in the exosome (e.g., ski2Δ, ski3Δ, and ski6-2) or nonsense-mediated mRNA decay pathways (e.g., upf1Δ), translation was inhibited following either glucose or amino acid withdrawal in a manner similar to that seen with wild-type strains (Fig. 3). However, one exception to this was noted: that of the rrp4-1 mutant. This strain has multiple phenotypes (26) and is analyzed further below.
The lsm1Δ and dcp1Δ mutants thus far identified are deficient in the decapping step of the major 5′ to 3′ pathway of mRNA decay. As deadenylation is a prerequisite to decapping in this decay pathway, deadenylation mutants were also analyzed. For mutants such as the poly(A) nuclease pan2Δ pan3Δ mutant or ccr4Δ, the deadenylation mutant, translation was largely inhibited following either glucose or amino acid removal (Fig. 3). Pop2p has also been found as part of the deadenylase complex. In this mutant, partial resistance to the nutritional stresses was observed. This agrees with previously published observations and probably relates more to another function of Pop2p in glucose repression (42). Indeed, previously we have found that mutants which are constitutively derepressed for glucose repression genes are resistant to these stress-induced translational controls (3). Other mRNA decay mutants, affecting steps downstream of deadenylation (such as lsm mutants, pat1Δ, edc1Δ, and dhh1Δ), are at least partially resistant to the translational effects of glucose or amino acid removal. Mutants which directly affect either decapping (e.g., dcp1Δ and dcp2Δ) or 5′ to 3′ exonucleolytic digestion of the RNA (e.g., xrn1Δ) are almost completely resistant to these translational stresses. However, mutation of the nucleus-specific LSM8 gene (lsm8-1) showed wild-type levels of translational inhibition. The majority of the mutants that have been identified as resistant or partially resistant to translational stress influence the decapping step of the major mRNA decay pathway. As Xrn1p acts downstream of this step by degrading the body of the mRNA, it is intriguing that an xrn1Δ mutant is resistant to stress-induced translational regulation. Recently, it has been shown that Xrn1p is found associated with both the Lsm complex and the decapping complex (13). This correlates with recent evidence suggesting that many of the mRNA decay factors colocalize to large complexes called cytoplasmic processing bodies (35). Therefore, mutation of XRN1 is likely to have more complex effects than previously anticipated and may influence upstream stages of the mRNA decay process.
All of the mRNA decay mutants described above that show resistance to translational stress actively deadenylate mRNAs. Therefore, these mutants accumulate mRNAs with reduced lengths of 3′ poly(A) tail (19, 38). The poly(A) tail impacts upon translation initiation via Pab1p in the closed loop model of translation initiation (31), possibly by facilitating the recruitment of the 60S large ribosomal subunit (32, 34). Therefore, it is plausible that the deadenylated mRNAs that accumulate in mRNA decay mutants might influence the control of translation initiation. We tested this model in a number of ways. We made use of the poly(A) polymerase mutant pap1-1, which at the nonpermissive temperature should accumulate mRNAs, without poly(A) tails, that get incorporated into polysomes in the cytoplasm (29). However, the translation rate for this mutant following a switch to nonpermissive temperature was already too low to allow assessment of whether this mutant is capable of responding to stress in terms of translation (data not shown). Another approach was to attempt to rescue the control of translation in the mRNA decay mutants by inactivating the deadenylation process. Therefore, we generated deadenylation mutants in the dcp1Δ background. Both the dcp1Δ ccr4Δ mutant strains and the dcp1Δ pan2Δ pan3Δ mutant strains were as resistant to the stress-induced translational inhibition as the dcp1Δ strain (Fig. 3B). However, it is unclear whether disruption of these nucleases is sufficient to reverse the poly(A) tail deficiency in a dcp1Δ mutant.
A further argument against the poly(A) deficiency model comes from studies of Pab1p. It seems likely that if there is a function for the poly(A) tail in maintaining the capacity for translational control, then this function would be mediated by Pab1p. Deletion of PAB1 is lethal for S. cerevisiae, but viability can be maintained by a number of suppressor mutations: for example, deletion of pbp1 (spb9) (25). Translation in both the pbp1Δ strains and pbp1Δ pab1Δ strains is regulated normally following glucose and amino acid removal (Fig. 3B). This shows that Pab1p is not required for translational regulation and suggests that the resistance of mRNA decay mutants does not result from an accumulation of deadenylated mRNA transcripts.
Therefore, this comprehensive analysis of many mRNA decay mutants shows that mutants affecting the major pathway of mRNA decay at stages downstream of deadenylation are resistant to certain translational stresses.
Mutation of DCP1 generates resistance to a host of translational controls.
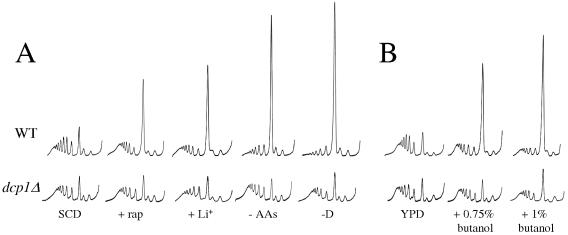
A number of stress conditions have been shown to elicit effects on translational initiation. To further examine how compromised mRNA decay pathways might influence these processes, the dcp1Δ mutant was subjected to a host of these translational inhibitory stresses. Figure 4A shows that following the addition of rapamycin or lithium (in galactose medium) or the removal of amino acids or glucose, a redistribution of polysomes into the monosome peak is observed for the wild-type strain. In contrast, when the dcp1Δ mutant was subjected to these stresses no change in the polysome profile was observed.
FIG. 4.
dcp1Δ is resistant to many forms of translational stress. (A) Polysome analyses for the wild-type (WT) strain (yMK329) and the dcp1Δ strain (yMK330). Strains were grown in SCD medium and then subjected to a number of stresses. Rapamycin (+ rap) was added to achieve a final concentration of 200 ng/ml for 1 h. A drug vehicle control gave a polysome trace identical to that of unstressed cells (data not shown). Lithium (+Li+) was added to achieve a final concentration of 30 mM for 15 min to yeast grown with galactose as the sole carbon source (the nontreated galactose grown control is not shown for simplicity, as the polysome profiles are indistinguishable from those of glucose-grown strains). The removal of amino acids for 10 min (−AAs) or glucose for 5 min (−D) was performed as described in previous figures. (B) Polysome analyses for the BUTS strain background, yMK36 (WT), and yMK863 (dcp1Δ). Strains were grown in rich medium (YPD), and 1-butanol was added to achieve a final concentration of 0.75% (vol/vol) (+0.75% butanol) or 1% (vol/vol) (+1% butanol) for 10 min.
Recently we have found that the addition of fusel alcohols such as 1-butanol to yeast can inhibit translation initiation, leading to a redistribution of polysomes (4). This regulation relies on specific allelic variation in the GCD1 gene, which encodes the γ subunit of eIF2B. Therefore, we disrupted the DCP1 gene in a butanol-sensitive (BUTS) strain background. Addition of 1-butanol to the parental strain for as little as 10 min leads to a dramatic inhibition of translation initiation, as shown by the change in polysome profile (Fig. 4B). However, in the dcp1Δ mutant the same stress had little effect on the polysome profile, suggesting that translation is maintained in this mutant (Fig. 4B). Indeed, we have now shown that DCP1 deletion in three different strain backgrounds leads to resistance to translational inhibition. In terms of growth this disruption has background-dependent effects, resulting in quite different growth rates for individual dcp1Δ strains. However, the translational regulation characteristics and shapes of the polysome profiles across these different backgrounds were indistinguishable (data not shown). Therefore, mutation of the DCP1 gene of the major mRNA decay pathway causes resistance to many different forms of translational stress.
Phosphorylation of eIF2α and decreased ternary complex levels do not inhibit translation initiation in the dcp1Δ mutant.
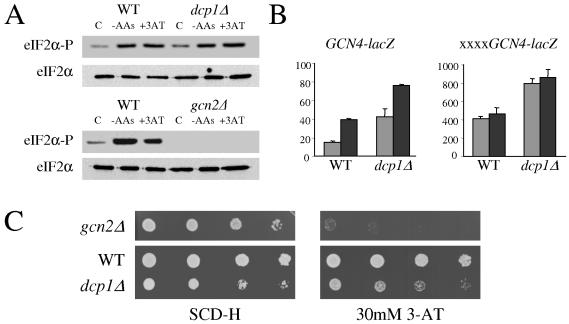
Amino acid starvation activates the Gcn2p protein kinase and eIF2α phosphorylation, inhibiting general translation initiation and yet activating the translation of Gcn4p via four upstream ORFs (17). The translational resistance of the dcp1Δ mutant to amino acid starvation is similar to that of mutants in this pathway with Gcn− phenotypes (17). For example, the gcn2Δ mutant cannot phosphorylate eIF2α and maintains polysomes following severe amino acid starvation (12) (Fig. 2C). Surprisingly, the dcp1Δ mutant was capable of phosphorylating eIF2α to an extent similar to that seen with the wild-type strain in response to either severe amino acid starvation or addition of the competitive inhibitor of histidine biosynthesis 3-AT for 15 min (Fig. 5A). The rapid phosphorylation of eIF2α by Gcn2p following amino acid starvation inhibits eIF2B, leading to lower levels of eIF2 GTP 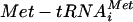 , the ternary complex. Translation of the GCN4 mRNA is exquisitely sensitive to changes in ternary complex abundance. Over a period of 6 h following amino acid starvation, there is a gradual increase in Gcn4p protein levels which relies upon this decrease in ternary complex abundance (1). As the level of ternary complex has never been directly assayed, the GCN4-lacZ reporter (containing the GCN4 5′ leader that includes the four uORFs) has therefore become an accepted indirect in vivo predictor for ternary complex levels (17). A dcp1Δ mutant maintained polysomes and protein synthesis for more than 1 h following amino acid starvation (data not shown); therefore, we used a 1-h time point to evaluate GCN4-lacZ levels. In keeping with the results seen with previous analyses, it is clear that amino acid starvation induces GCN4-lacZ approximately two- to threefold in the wild-type strain over this time frame (Fig. 5B). This induction was not observed when a gcn2Δ mutant in the same strain background was used (data not shown). However, even though in terms of general translation the dcp1Δ mutant behaved like a gcn mutant, GCN4-lacZ was still induced upon amino acid starvation (Fig. 5B). This induction relies upon the four upstream ORFs, as no induction was observed in a mutant lacking these (Fig. 5B).
, the ternary complex. Translation of the GCN4 mRNA is exquisitely sensitive to changes in ternary complex abundance. Over a period of 6 h following amino acid starvation, there is a gradual increase in Gcn4p protein levels which relies upon this decrease in ternary complex abundance (1). As the level of ternary complex has never been directly assayed, the GCN4-lacZ reporter (containing the GCN4 5′ leader that includes the four uORFs) has therefore become an accepted indirect in vivo predictor for ternary complex levels (17). A dcp1Δ mutant maintained polysomes and protein synthesis for more than 1 h following amino acid starvation (data not shown); therefore, we used a 1-h time point to evaluate GCN4-lacZ levels. In keeping with the results seen with previous analyses, it is clear that amino acid starvation induces GCN4-lacZ approximately two- to threefold in the wild-type strain over this time frame (Fig. 5B). This induction was not observed when a gcn2Δ mutant in the same strain background was used (data not shown). However, even though in terms of general translation the dcp1Δ mutant behaved like a gcn mutant, GCN4-lacZ was still induced upon amino acid starvation (Fig. 5B). This induction relies upon the four upstream ORFs, as no induction was observed in a mutant lacking these (Fig. 5B).
FIG. 5.
dcp1Δ responds normally to amino acid starvation in terms of eIF2α phosphorylation and GCN4 induction. (A) Protein extracts from strains yMK329 (WT) and yMK330 (dcp1Δ) (top panels) and strains yMK429 (WT) and yMK513 (gcn2Δ) (bottom panels) were blotted and probed with antibodies to eIF2α and phosphospecific antibodies to phosphoserine 51 on eIF2α. C, control. (B) β-Galactosidase assays measured in Miller units from extracts prepared from strains yMK926 and yMK936 (GCN4-lacZ reporter) or strains yMK927 and yMK937 (xxxxGCN4-lacZ reporter). The light-gray bars represent cells which were transferred into complete medium for 1 h, whereas the dark-gray bars represent cells transferred into medium lacking amino acids for 1 h. Error bars represent the standard deviations from three independent experiments. (C) Serial dilution plate assays for yMK513 (gcn2Δ), yMK431 (WT), and yMK861 (dcp1Δ). The SCD medium lacking histidine (SCD-H) plates were incubated for 2 days, whereas the SCD-H plates with 30 mM 3-amino triazole added were incubated for 3 days.
Interestingly, the basal level of activity from both the GCN4-lacZ reporter and a reporter bearing mutant uORFs was higher in the dcp1Δ strain than in the wild type (Fig. 5B). The results of Northern analysis of GCN4 mRNA levels suggest that this difference is due to higher levels of the GCN4 mRNA in this mutant, probably resulting from deficient mRNA decay (data not shown).
The induction of GCN4 reporter activity following amino acid starvation in the dcp1Δ mutant correlates with the fact that unlike the gcn2Δ mutant, the dcp1Δ strain is not sensitive to 3-AT, the histidine starvation inducer, in terms of growth (Fig. 5C). Under conditions of severe starvation for amino acids, therefore, a dcp1Δ mutant strain responds by increasing the phosphorylation of eIF2α and the subsequent levels of Gcn4p. As eIF2α phosphorylation is known to decrease ternary complex levels (leading to an induction of GCN4 translation), these results suggest that the ternary complex levels decrease in a dcp1Δ mutant strain following amino acid starvation and yet that global protein synthesis is sustained.
Both 60S ribosomal subunit biogenesis mutant and eIF4G overexpression strains are translationally resistant to nutritional stress.
Given the apparent normal response to amino acid starvation in the dcp1Δ strain (as measured using the Gcn4p regulatory system), the obvious question is why do global rates of translation initiation not decrease following translational stress? It seems that this mutant strain background is somehow capable of maintaining translation even though the levels of ternary complex decrease. This further indicates that in this mutant strain background, the rate of translation initiation is less dependent upon the rate of ternary complex recruitment to the 40S ribosomal subunit.
One possible explanation is that increased levels of other components in the translation initiation process (such as the 40S ribosomal subunit or the eIF4F-bound mRNA) might overcome the decreased ternary complex and thus the translational inhibition by a mass action mechanism. To test this idea we have taken two broad approaches.
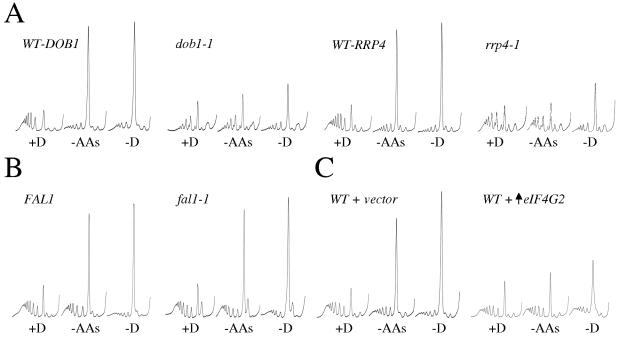
First, we have used mutants in the 60S ribosomal biogenesis pathway, reasoning that the excess of 40S ribosomal subunits in these strains could facilitate a level of translational resistance similar to that observed in the mRNA decay mutants. More specifically, Dob1p is a putative ATP-dependent RNA helicase that is involved in biogenesis of the 5.8S rRNA. Mutation of this gene leads to a deficiency in 60S ribosomal subunits and an increase in both the free 40S ribosomal subunits and “half-mers” (10). Half-mers are characteristic peaks which sediment just after each ribosomal peak and represent mRNAs with a 40S subunit that has not yet complexed with the 60S subunit. In contrast to the results seen with a wild-type strain, polysomes are maintained or partially maintained in the dob1-1 mutant following amino acid starvation or glucose starvation, respectively (Fig. 6A). Interestingly, Dob1p interacts with components of the nuclear exosome (including Rrp4p) and is thought to facilitate 5.8S rRNA biogenesis without affecting cytoplasmic 3′ to 5′ mRNA decay (2, 10). This suggests that the explanation for the translational resistance of the rrp4-1 mutant (Fig. 3B and 6A) lies more in the 60S ribosomal subunit biogenesis defect for this strain rather than in a 3′ to 5′ mRNA decay defect.
FIG. 6.
Strains that accumulate eIF4G or 40S ribosomal subunits are resistant to translational stress. All strains were grown in SCD medium, selecting for plasmids where appropriate. They were harvested and then resuspended in the same medium (+D), in medium without amino acids (−AAs), or in medium without glucose (−D). (A) Polysome traces from the 60S biogenesis mutants yMK899 (dob1-1) and yMK709 (rrp4-1) as well as from their respective isogenic wild-type strains yMK898 (WT-DOB1) and yMK116 (WT-RRP4). (B) Polysome traces from the 40S biogenesis mutant yMK1046 (fal1-1) and the isogenic wild-type yMK1045 (FAL1). (C) Polysome traces from strains yMK158 and yMK157, which are wild-type (WT) strains bearing either an eIF4G2 overexpression plasmid (↑eIF4G2) or an empty vector (+vector).
This translational resistance observed for the 60S biogenesis mutants might be due to a rate-limiting step at the 60S joining step of translation initiation rather than to an excess of free 40S subunits. However, a fun12Δ mutant (which has a 60S subunit joining deficiency) was completely inhibited for translation initiation following glucose removal (data not shown). Therefore it seems more likely that an accumulation of 40S ribosomal subunits can overcome translational inhibition.
It is also possible that general alterations to the relative levels of ribosomal subunits could allow resistance to translational stress. To investigate this possibility we analyzed a 40S subunit biogenesis mutant, fal1-1, which accumulates free 60S subunits (21). In contrast to the results seen with 60S biogenesis mutants, fal1-1 was largely inhibited following either amino acid or glucose withdrawal (Fig. 6B). Therefore, the accumulation of 60S subunits (in 40S biogenesis mutants) did not give the same resistance to translational stress as the accumulation of 40S subunits (in 60S biogenesis mutants).
In a second approach designed to test whether excess downstream components can increase the level of preinitiation complex formation (especially under conditions of limiting ternary complex), we overexpressed TIF4632. This encodes eIF4G, the least abundant translation initiation factor in the eIF4F cap binding complex (46). We reasoned that accumulation of the eIF4F complex in a wild-type strain might alter the existing cap competition between Dcp1p and eIF4F, thereby allowing ultimately greater concentrations of mRNA/eIF4F complexes. Therefore, this scenario may mimic the results seen with a dcp1Δ mutant. As shown in Fig. 6C and in contrast to the results seen with a strain containing plasmid vector alone, overexpression of eIF4G from a plasmid vector allows continued translation following both amino acid and glucose starvation. Clearly this effect is less substantial than that seen with the dcp1Δ mutant, but it does suggest a plausible explanation for the continued synthesis of proteins in mRNA decay mutants under stress conditions. That is, an excess of downstream components of the translation pathway can counter the stress-dependent inhibition of translation initiation. Ultimately, these results suggest that there is a delicate stoichiometrical balance of translational, ribosomal, and mRNA decay complexes which gives cells the capacity to rapidly control translation initiation.
DISCUSSION
Many studies over the years have highlighted the connection between translation and mRNA decay (33). The most obvious example is nonsense-mediated mRNA decay, in which a prerequisite to this decay pathway is the translational recognition of a termination codon (20). Studies on the connection between the general pathways of mRNA decay and translation have usually assessed the effects of alterations in translation on mRNA stability. For instance, the inhibition (using cycloheximide or via conditional mutations) of translation elongation results in mRNA stabilization. In addition, it has been shown that in a variety of translation initiation mutants, the rate of decay for specific mRNAs increases (reviewed in reference 33). This has led to the hypothesis that mRNA decay and translation initiation in some way compete for the capped end of a mRNA species (33).
In this paper, we describe studies that approach the problem of translation and mRNA decay coordination from a different angle. Our goal was to determine how the various pathways of mRNA decay might impact upon the control of translation initiation. We show that specific mutants in the major 5′ to 3′ pathway of mRNA decay (most notably a dcp1Δ mutant strain) are almost completely resistant to stress-induced translational inhibitory pathways. To further evaluate the mechanism involved in this connection between translational control and mRNA decay, we studied the well-characterized amino acid starvation pathway of global translational inhibition and Gcn4p induction. We show that even though the dcp1Δ strain is translationally resistant to severe amino acid starvation, this mutant does not bear other hallmarks of a gcn mutant. For instance, this mutant is capable of phosphorylating eIF2α and activating GCN4 translation. As eIF2α phosphorylation is known to decrease ternary complex levels and consequently activate GCN4 translation, these results suggest that (in terms of global translation initiation) dcp1Δ mutants are resistant to decreases in ternary complex abundance.
The fact that we have identified mutants such as dcp1Δ that are resistant to almost all forms of translational stress in yeast might mean that these translational regulatory responses have a common mechanism. For amino acid starvation, the pathway of translational regulation is well characterized and ultimately impacts upon eIF2B activity and ternary complex levels (17). It seems likely that fusel alcohols affect translation initiation via a similar mechanism except that this regulation does not rely on eIF2α kinases but instead probably affects the activity of eIF2B more directly (4). Rapamycin has recently been shown to inhibit translation initiation by activating the yeast eIF2α kinase, Gcn2p, and therefore also inhibits eIF2B activity, leading to lower ternary complex levels (8). All three of these translational stresses lead to increased translation of GCN4, which demonstrates the reduction in ternary complex levels (4, 8, 17, 22, 44). Glucose starvation also leads to increased translation of GCN4 (48). However, the kinetics of this increased GCN4 translation are relatively slow compared with the results seen with other stresses such as amino acid starvation and fusel alcohol addition (data not shown). This suggests that the primary cause of the translational inhibition following glucose removal may not be the decreased ternary complex levels. The other stress that we have used in this study was the addition of lithium when yeast was grown in galactose medium. For this stress it has been found that overexpression of the gene for eIF4A suppresses the translational inhibitory effects (28). In addition, we have measured GCN4-lacZ reporter activity; according to the results seen with this indirect assay, lithium addition does not decrease ternary complex levels (data not shown). Overall it seems that even though mRNA decay mutants are resistant to many forms of translational stress, it is likely that these stresses affect translation via distinct mechanisms.
Translational control via eIF2α kinases is an evolutionarily conserved response to stress. For example, four eIF2α kinases have been identified in mammalian cells: HRI (which responds to the depletion of heme), PKR (which is activated during viral infection), PEK/PERK (which responds to misfolded proteins within the lumen of the endoplasmic reticulum), and mGCN2 (which responds to nutrient availability). All of these kinases target the guanine nucleotide exchange function of eIF2B, highlighting the role of eIF2B and the subsequent regulation of ternary complex levels as a major control point of protein synthesis (reviewed in reference 11). Furthermore, inherited mutations in eIF2B have recently been shown to cause the fatal human brain disease childhood ataxia with central nervous system hypomyelination (45). Therefore, the ternary complex level within eukaryotic cells is considered a key rate-limiting determinant for translation initiation. In this study we have identified specific yeast mutants where cells can maintain protein synthesis even though ternary complex levels are decreased. In addition, these mutants are resistant to translational stresses that have no obvious impact upon ternary complex levels (as described above). One possible explanation for these observations is that a specific mRNA(s) might be upregulated in mRNA decay mutants and have direct effects upon translational regulation. However, it seems unlikely that changes in expression of specific mRNAs can generate resistance to so many divergent translational stresses that are thought to act via distinct mechanisms. Therefore, we favor a model in which the mRNA decay mutants have global effects on the gene expression profile of cells. Indeed, these mutants have globally relevant characteristics which may affect translation initiation and prevent translation regulation. These include an increased abundance of mRNA and accumulation of mRNA decay intermediates (19).
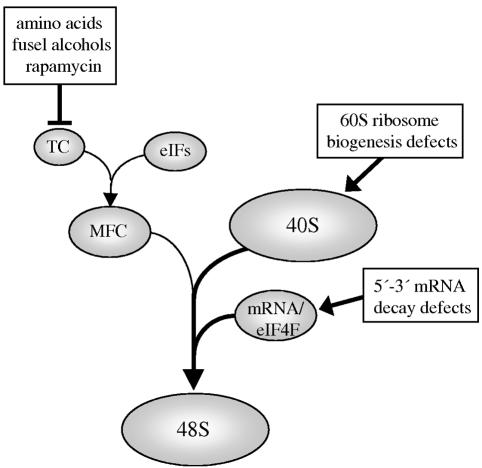
A model that has been proposed to explain the connection between translation initiation and mRNA decay is the cap competition model. In this scenario, the lack of decapping in a dcp1Δ mutant would allow an accumulation of translationally competent or eIF4F-bound mRNA (33). Here we have shown that stabilization of the eIF4F complex in a wild-type strain reduces the extent of translational regulation following stress. It seems that decreasing either the activity of Dcp1p (Fig. 2) or its access to the 5′ cap (Fig. 6C) allows resistance to reduced levels of ternary complex and to translational stresses considered to act via alternate mechanisms. We favor a model whereby an accumulation of eIF4F bound mRNA might globally increase the recruitment rate of 43S complexes by mass action, therefore negating the effect of changes in ternary complex level (Fig. 7).
FIG. 7.
A model depicting how 5′ to 3′ mRNA decay or 60S biogenesis defects might overcome decreases in ternary complex. Mutants that accumulate either 40S subunits or mRNA/eIF4F complexes drive the assembly of the preinitiation machinery by mass action. TC and MFC represent the ternary complex (eIF2-GTP 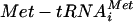 ) and the multifactor complex (containing ternary complex, eIF3, eIF1, and eIF5), respectively. Arrows represent the effects of the presence of excess 40S ribosomal subunits and eIF4F-bound mRNA on the rate of translation initiation.
) and the multifactor complex (containing ternary complex, eIF3, eIF1, and eIF5), respectively. Arrows represent the effects of the presence of excess 40S ribosomal subunits and eIF4F-bound mRNA on the rate of translation initiation.
The hypothesis that other components of the translational initiation process can act by mass action to overcome changes in ternary complex level is further supported by an analysis of mutants in the biogenesis of the large ribosomal subunit. These mutants accumulate 40S ribosomal subunits and (in similarity to mRNA decay mutants) are resistant to translational stress (Fig. 6A). The excess 40S subunits may drive the various equilibria within the translation initiation process towards the preinitiation complex, thus overcoming effects on ternary complex levels (Fig. 7).
The results presented in this study graphically demonstrate the intimate relationship that exists between protein synthesis, mRNA decay, and ribosomal biogenesis. Although these connections have been implicated previously in a number of studies, this study represents the first analysis in which the global control of translation in response to stress has been added to the equation. Surprisingly, mRNA decay mutants can maintain translation under stress even though ternary complex levels appear to decrease normally. As regulation of ternary complex is a highly conserved mechanism of translational control, this may have wide ranging implications. Our interpretation of these results is that there is a delicate balance in the stoichiometry of different translation initiation components (such as MFC, 40S ribosomes, and mRNA) and that the maintenance of this equilibrium is critical in preserving the cell's capacity to globally control protein synthesis.
Acknowledgments
This work was supported largely by Wellcome Trust project grant 061867/Z/00/Z to M.P.A. L.E.A.H. is supported by an MRC studentship. S.G.C. is supported by Wellcome Trust project grant 067328/Z/02/Z to M.P.A. In addition, the work was supported by NIH grant GM50308 to A.B.S. and by an EMBO long-term fellowship to M.P.A. while at University of California at Berkeley.
We thank G. Pavitt, C. Grant, A. Johnson, C. Wilutz, S. Peltz, S. Butler, and P. Linder for reagents and advice. We especially thank R. Parker for his generous donation of yeast strains and plasmids, without which this work would not have been possible.
REFERENCES
- 1.Albrecht, G., H. U. Mosch, B. Hoffmann, U. Reusser, and G. H. Braus. 1998. Monitoring the Gcn4 protein-mediated response in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 273:12696-12702. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, J. S. J., and R. P. Parker. 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17:1497-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashe, M. P., S. K. De Long, and A. B. Sachs. 2000. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell 11:833-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashe, M. P., J. W. Slaven, S. K. De Long, S. Ibrahimo, and A. B. Sachs. 2001. A novel eIF2B-dependent mechanism of translational control in yeast as a response to fusel alcohols. EMBO J. 20:6464-6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbet, N. C., U. Schneider, S. B. Helliwell, I. Stansfield, M. F. Tuite, and M. N. Hall. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7:25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beelman, C. A., A. Stevens, G. Caponigro, T. E. LaGrandeur, L. Hatfield, D. M. Fortner, and R. Parker. 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382:642-646. [DOI] [PubMed] [Google Scholar]
- 7.Boeck, R., B. Lapeyre, C. E. Brown, and A. B. Sachs. 1998. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell. Biol. 18:5062-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherkasova, V. A., and A. G. Hinnebusch. 2003. Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2. Genes Dev. 17:859-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, S. K., J. H. Lee, W. L. Zoll, W. C. Merrick, and T. E. Dever. 1998. Promotion of met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science 280:1757-1760. [DOI] [PubMed] [Google Scholar]
- 10.de la Cruz, J., D. Kressler, D. Tollervey, and P. Linder. 1998. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 17:1128-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dever, T. E. 1999. Translation initiation: adept at adapting. Trends Biochem. Sci. 24:398-403. [DOI] [PubMed] [Google Scholar]
- 12.Dever, T. E., L. Feng, R. C. Wek, A. M. Cigan, T. F. Donahue, and A. G. Hinnebusch. 1992. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68:585-596. [DOI] [PubMed] [Google Scholar]
- 13.Fromont-Racine, M., A. E. Mayes, A. Brunet-Simon, J. C. Rain, A. Colley, I. Dix, L. Decourty, N. Joly, F. Ricard, J. D. Beggs, and P. Legrain. 2000. Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast 17:95-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González, C. I., A. Bhattacharya, W. Wang, and S. W. Peltz. 2001. Nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Gene 274:15-25. [DOI] [PubMed] [Google Scholar]
- 15.Goossens, A., T. E. Dever, A. Pascual-Ahuir, and R. Serrano. 2001. The protein kinase Gcn2p mediates sodium toxicity in yeast. J. Biol. Chem. 276:30753-30760. [DOI] [PubMed] [Google Scholar]
- 16.Guthrie, C., and G. R. Fink (ed.). 1991. Guide to yeast genetics and molecular biology. Academic Press, San Diego, Calif.
- 17.Hinnebusch, A. G. 2000. Mechanism and regulation of initiator methionyl-tRNA binding to ribosomes, p. 185-244. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Hinnebusch, A. G., and K. Natarajan. 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 1:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu, C. L., and A. Stevens. 1993. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13:4826-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobson, A., and S. W. Peltz. 2000. Destabilization of nonsense-containing transcripts in Saccharomyces cerevisiae, p. 827-847. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Kressler, D., J. de la Cruz, M. Rojo, and P. Linder. 1997. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:7283-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubota, H., T. Obata, K. Ota, T. Sasaki, and T. Ito. 2003. Rapamycin-induced translational derepression of GCN4 mRNA involves a novel mechanism for activation of the eIF2α kinase GCN2. J. Biol. Chem. 278:20457-20460. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn, K. M., J. L. DeRisi, P. O. Brown, and P. Sarnow. 2001. Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Mol. Cell. Biol. 21:916-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lucchini, G., A. G. Hinnebusch, C. Chen, and G. R. Fink. 1984. Positive regulatory interactions of the HIS4 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 4:1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mangus, D. A., N. Amrani, and A. Jacobson. 1998. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol. 18:7383-7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell, P., E. Petfalski, A. Shevchenko, M. Mann, and D. Tollervey. 1997. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91:457-466. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell, P., and D. Tollervey. 2003. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′→5′ degradation. Mol. Cell 11:1405-1413. [DOI] [PubMed] [Google Scholar]
- 28.Montero-Lomeli, M., B. L. Morais, D. L. Figueiredo, D. C. Neto, J. R. Martins, and C. A. Masuda. 2002. The initiation factor eIF4A is involved in the response to lithium stress in Saccharomyces cerevisiae. J. Biol. Chem. 277:21542-21548. [DOI] [PubMed] [Google Scholar]
- 29.Proweller, A., and S. Butler. 1994. Efficient translation of poly(A)-deficient mRNAs in Saccharomyces cerevisiae. Genes Dev. 8:2629-2640. [DOI] [PubMed] [Google Scholar]
- 30.Rolfes, R. J., and A. G. Hinnebusch. 1993. Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: implications for activation of the protein kinase GCN2. Mol. Cell. Biol. 13:5099-5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sachs, A. B. 2000. Physical and functional interactions between the mRNA cap structure and the poly(A) tail, p. 447-465. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Sachs, A. B., and R. W. Davis. 1989. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell 58:857-867. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz, D. C., and R. Parker. 2000. Interaction of mRNA translation and mRNA decay in Saccharomyces cerevisiae, p. 807-825. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Searfoss, A., T. E. Dever, and R. Wickner. 2001. Linking the 3′ poly(A) tail to the subunit joining step of translation initiation: relations of Pab1p, eukaryotic translation initiation factor 5b (Fun12p), and Ski2p-Slh1p. Mol. Cell. Biol. 21:4900-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steiger, M., A. Carr-Schmid, D. C. Schwartz, M. Kiledjian, and R. Parker. 2003. Analysis of recombinant yeast decapping enzyme. RNA 9:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tharun, S., and R. Parker. 1999. Analysis of mutations in the yeast mRNA decapping enzyme. Genetics 151:1273-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tharun, S., W. He, A. E. Mayes, P. Lennertz, J. D. Beggs, and R. Parker. 2000. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404:515-518. [DOI] [PubMed] [Google Scholar]
- 39.Tucker, M., and R. Parker. 2000. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu. Rev. Biochem. 69:571-595. [DOI] [PubMed] [Google Scholar]
- 40.Tucker, M., R. R. Staples, M. A. Valencia-Sanchez, D. Muhlrad, and R. Parker. 2002. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 21:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tzamarias, D., I. Roussou, and G. Thireos. 1989. Coupling of GCN4 mRNA translational activation with decreased rates of polypeptide chain initiation. Cell 57:947-954. [DOI] [PubMed] [Google Scholar]
- 42.Uesono, Y., and A. Toh-E. 2002. Transient inhibition of translation initiation by osmotic stress. J. Biol. Chem. 277:13848-13855. [DOI] [PubMed] [Google Scholar]
- 43.Valasek, L., A. A. Mathew, B. S. Shin, K. H. Nielsen, B. Szamecz, and A. G. Hinnebusch. 2003. The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev. 17:786-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valenzuela, L., C. Aranda, and A. Gonzalez. 2001. TOR modulates GCN4-dependent expression of genes turned on by nitrogen limitation. J. Bacteriol. 183:2331-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Knaap, M. S., P. A. Leegwater, A. A. Konst, A. Visser, S. Naidu, C. B. Oudejans, R. B. Schutgens, and J. C. Pronk. 2002. Mutations in each of the five subunits of translation initiation factor eIF2B can cause leukoencephalopathy with vanishing white matter. Ann. Neurol. 51:264-270. [DOI] [PubMed] [Google Scholar]
- 46.von der Haar, T., and J. E. McCarthy. 2002. Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol. Microbiol. 46:531-544. [DOI] [PubMed] [Google Scholar]
- 47.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 48.Yang, R., S. A. Wek, and R. C. Wek. 2000. Glucose limitation induces GCN4 translation by activation of Gcn2 protein kinase. Mol. Cell. Biol. 20:2706-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]