During the course of a research project on full viral genome sequencing of porcine reproductive and respiratory syndrome (PRRS) virus, 62 samples of swine sera were collected from piglets from various Canadian farms that were experiencing PRRS outbreaks. In order to lower the viral genome contamination of serum samples by the host genome (i.e., swine genome) for the high-throughput sequencing (HTS) procedure, viral particles were purified by ultracentrifugation (113 000 × g on a 20% sucrose cushion) followed by a nuclease treatment (1 h at 37°C with 500 μg/mL DNase and 250 μg/mL RNase A). The viral genome was then extracted with Trizol reagent (Invitrogen, Burlington, Ontario) according to the manufacturer’s recommended procedure. Genomic material was subjected to random polymerase chain reaction (PCR) amplification as previously described (1). Qualitative and quantitative evaluations of the random amplification products were respectively assessed by agarose gel electrophoresis and by real-time PCR (EZ-PRRSVTM MPX 4.0 Real Time RT-PCR Target-Specific Reagents for the Rapid Identification and Differentiation of North American and European PRRS Viral RNA, Tetracore, Rockville, Maryland, USA). Based on those results, a total of 11 samples, comprised of 6 samples from Quebec, 4 from Ontario, and 1 from Alberta, were selected and sent to the “Institut de biologie integrative et des systèmes” (IBIS) for HTS (Table 1). For each sample, a GS-FLX rapid library was produced from amplified genomic material using the manufacturer’s instructions provided with the kit (Roche/454 Sequencing, Brandford, Connecticut, USA). Emulsion PCR and GS-FLX titanium sequencing was performed according to the manufacturer’s instructions (Roche/454 Sequencing) at the Plateforme d’Analyses Génomiques of the IBIS (Laval University, Quebec City, Quebec). Raw sequencing reads were assembled using the gsAssembler module of Newbler v.2.5.3.
Table 1.
Description of PRRS virus positive swine sera tested by high-throughput sequencing (HTS) and their porcine partetravirus status
| Province of origin | Years | HTS tested sera | Porcine partetravirus | |
|---|---|---|---|---|
|
| ||||
| Positive sera | Prevalence | |||
| Quebec | 2012–2013 | 6 | 0 | |
| Ontario | 2009–2010 | 4 | 1 | |
| Alberta | 2013 | 1 | 0 | |
| Total | 11 | 1 | 9.09% | |
In addition to PRRS virus genome sequences, HTS results revealed the presence of a swine parvovirus highly similar to the porcine partetravirus (a parvovirus also known as porcine hokovirus, porcine PARV4-like virus and porcine parvovirus 3) in one of the sera collected in Ontario (Table 1). Parvoviruses are small non-enveloped icosahedral viruses with a linear single-stranded DNA genome of about 4 to 6.3 kb (2,3). Based on this result, porcine partetravirus prevalence in Canadian swine herds was established to be 9.09% (Table 1). However, this estimate of the prevalence of porcine partetravirus in Canada may be biased because: i) the sera were selected in regards to their PRRS virus positive status, ii) the number of samples tested was low, and iii) the geographical origins of the tested sera did not accurately represent the overall Canadian swine industry. To our knowledge, the porcine partetravirus has never been isolated in cell culture, although virus isolation has been attempted by Lau et al (2008) and Streck et al (2013)(4,5). Currently, 5 porcine parvoviruses (PPV) have been identified in swine: porcine par-vovirus 1 (PPV1), PPV2, porcine partetravirus (PPV3), PPV4, and PPV5, which has been recently reported in the United States (2). When compared with one another, PPV1 through PPV5 have substantial genetic divergence. The porcine partetravirus is genomically related to human parvovirus 4 (60% to 65% nucleotide homology). Human parvovirus 4 was identified in 2005 from a plasma sample of a homeless drug user who had an acute viral infection (6). The porcine partetravirus was reported for the first time in China in 2008 (4). Since then, the porcine partetravirus has been reported in other countries, including United States, Great Britain, Romania, Hungary, and Germany (4,7–10). To our knowledge, the porcine partetravirus has not previously been reported in Canada.
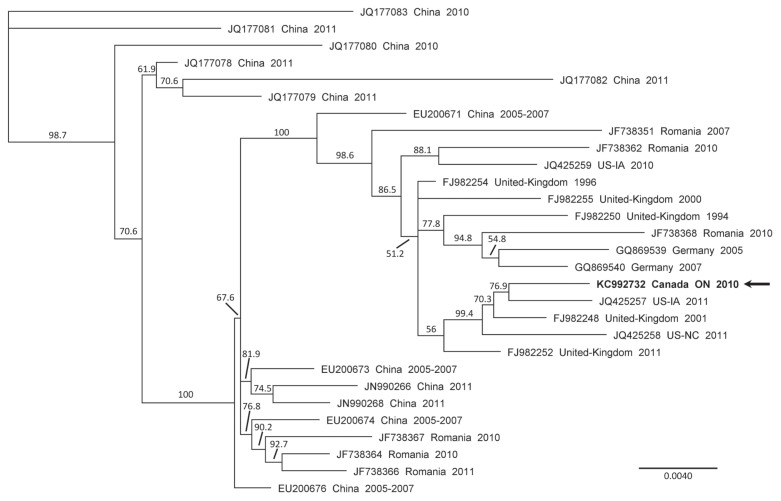
The identified Canadian porcine partetravirus strain, named FMV10-1437266, possesses a genome length of 5402 nucleotides (nt) (GenBank accession number KC992732) and has a GC content of 51.29%. After HTS procedure, the FMV10-1437266 genomic sequence was assembled from 938 reads, with a mean coverage of 48 reads per nt (minimum coverage of 8 reads and maximum coverage of 85 reads). The FMV10-1437266 partetravirus strain has a genome nearly 300 nt larger than that of the longest reported porcine partetravirus that was identified in China in 2009 (GenBank accession number EU200677). This increased nt genomic length of the FMV10-1437266 gives a better insight into the inverted terminal repeats (ITR) found at both 5′ and 3′ ends of the porcine partetravirus genome. The ITR are involved in parvovirus genome replication (3). Genomic sequence comparison of the porcine partetravirus FMV10-1437266 with previously reported strain sequences gathered from the GenBank database revealed sequence identities varying between 95.4% and 99.3% (data not shown), which is in accordance with previous reports (5,8,10,11). A phylogenetic tree (Figure 1) was constructed by the neighbor-joining method with 1000 bootstrap replicates with the Geneious Pro (version 5.6.6) software. The genomic comparison was done using the longest common nt sequences between all compared partetravirus genomic sequences (which correspond to nt positions 292 to 5074 of FMV10-1437266). Phylogenetic analysis revealed that the porcine partetravirus FMV10-1437266 is highly similar to a USA strain detected in Iowa in 2011 (GenBank accession number JQ425257). In the USA, porcine partetravirus has also been reported in North Carolina (7).
Figure 1.
Phylogenetic tree of the genomic nucleotide sequences of porcine partetravirus strains. The phylogenetic tree was generated by the neighbor-joining method using the Geneious V5.6.6 software with a bootstrap resampling method (1000 replications) using the longest common nucleotide sequences between all compared partetravirus genomic sequences. Bootstrap confidence levels are indicated at the nodes of the phylogenetic tree. GenBank accession numbers for the sequences of all viruses are provided as well as their countries of origin and their date of collection (year). The horizontal scale indicates the distances between strains; a distance of 0.0040 means that the strains possess 99.6% nucleotide identity. The arrow indicates the porcine partetravirus detected in the Ontario swine serum.
The classical porcine parvovirus (PPV1) is an important pathogen associated with reproductive disorders of sows and is widely distributed around the world, including Canada (10). The identification of novel porcine parvoviruses (such as PPV2, porcine partetravirus, PPV4, and PPV5) raises concerns about their potential involvement in swine health and manifestations of clinical signs or disease. At present, the data are sparse and involvement is unknown (12). Porcine partetravirus has not been found to be an etiological agent of swine disease and is currently considered to be non-pathogenic. Nonetheless, Dr. Opriessnig’s research team has revealed that the prevalence of porcine partetravirus in pigs with respiratory diseases, systemic/central nervous system diseases, and enteric diseases was 14.4%, 11.6%, and 2.7%, respectively (7). These results suggest that the porcine partetravirus may be associated with diseases of these systems but this still needs to be proven. Porcine partetravirus has been detected in porcine blood pharmaceutical by-products such as plasma and Factor VIII (FVIII). Porcine-derived FVIII is being used to treat a human autoimmune hemophilia in which patients possess antibodies against FVIII. Thus, the presence of porcine partetravirus in swine blood and subsequently in pharmaceutical by-products may represent a potential threat for human health (11). In swine, the porcine partetravirus was detected in adult pigs and nursery pigs, and was not detected in fetuses or suckling pigs (7). In the near future, experiments will be conducted to establish with precision the prevalence of porcine partetravirus in the Canadian swine population. Moreover, further experiments are needed to ascertain the involvement of porcine partetravirus in swine diseases.
Acknowledgments
The authors are grateful to the Canadian researchers, diagnosticians, and veterinarians (in particular: Markus Czub, Sylvain Messier, Lori Moser, Pete Pawluk, Leigh Rosengren) who provided the PRRSV positive serum samples for the Canadian PRRS virus HTS research project. Dr. Gagnon was financially supported by the Canadian Swine Health Board (CSHB).
Footnotes
Use of this article is limited to a single copy for personal study. Anyone interested in obtaining reprints should contact the CVMA office (hbroughton@cvma-acmv.org) for additional copies or permission to use this material elsewhere.
References
- 1.Chen EC, Miller SA, DeRisi JL, Chiu CY. Using a pan-viral microarray assay (Virochip) to screen clinical samples for viral pathogens. J Vis Exp. 2011 Apr 27;(50):2536. doi: 10.3791/2536. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao CT, Halbur PG, Opriessnig T. Complete genome sequence of a novel porcine parvovirus (PPV) provisionally designated PPV5. Genome announce. 2013;1(1):e00021–12. doi: 10.1128/genomeA.00021-12. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tijssen P, Agbandje-McKenna M, Almendral J, et al. Family Parvoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy; 9th Report of the International Committee on Taxonomy of Viruses; London. Waltham, Massachussetts: Academic Press; 2011. pp. 405–425. [Google Scholar]
- 4.Lau SK, Woo PC, Tse H, et al. Identification of novel porcine and bovine parvoviruses closely related to human parvovirus 4. J Gen Virol. 2008;89:1840–1848. doi: 10.1099/vir.0.2008/000380-0. [DOI] [PubMed] [Google Scholar]
- 5.Streck AF, Homeier T, Foerster T, Fischer S, Truyen U. Analysis of porcine parvoviruses in tonsils and hearts from healthy pigs reveals high prevalence and genetic diversity in Germany. Arch Virol. 2013;158:1173–1180. doi: 10.1007/s00705-013-1603-0. [DOI] [PubMed] [Google Scholar]
- 6.Jones MS, Kapoor A, Lukashov VV, Simmonds P, Hecht F, Delwart E. New DNA viruses identified in patients with acute viral infection syndrome. J Virol. 2005;79:8230–8236. doi: 10.1128/JVI.79.13.8230-8236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao CT, Gimenez-Lirola LG, Halbur PG, Opriessnig T. Increasing porcine PARV4 prevalence with pig age in the US pig population. Vet Microbiol. 2012;160:290–296. doi: 10.1016/j.vetmic.2012.05.038. [DOI] [PubMed] [Google Scholar]
- 8.Cadar D, Csagola A, Lorincz M, Tombacz K, Spinu M, Tuboly T. Distribution and genetic diversity of porcine hokovirus in wild boars. Arch Virol. 2011;156:2233–2239. doi: 10.1007/s00705-011-1125-6. [DOI] [PubMed] [Google Scholar]
- 9.Adlhoch C, Kaiser M, Ellerbrok H, Pauli G. High prevalence of porcine Hokovirus in German wild boar populations. Virol J. 2010;7:171. doi: 10.1186/1743-422X-7-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Csagola A, Lorincz M, Cadar D, Tombacz K, Biksi I, Tuboly T. Detection, prevalence and analysis of emerging porcine parvovirus infections. Arch Virol. 2012;157:1003–1010. doi: 10.1007/s00705-012-1257-3. [DOI] [PubMed] [Google Scholar]
- 11.Szelei J, Liu K, Li Y, Fernandes S, Tijssen P. Parvovirus 4-like virus in blood products. Emerg Infect Dis. 2010;16:561–564. doi: 10.3201/eid1603.090746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao CT, Halbur PG, Opriessnig T. Molecular evolutionary genetic analysis of emerging parvoviruses identified in pigs. Infect Genet Evol. 2013;16C:369–376. doi: 10.1016/j.meegid.2013.03.017. [DOI] [PubMed] [Google Scholar]