Abstract
The communities constituting our microbiotas are emerging as mediators of the health-disease continuum. However, deciphering the functional impact of microbial communities on host pathophysiology represents a formidable challenge, due to the heterogeneous distribution of chemical and microbial species within the gastrointestinal (GI) tract. Herein, we apply imaging mass spectrometry (IMS) to localize metabolites from the interaction between the host and colonizing microbiota. This approach complements other molecular imaging methodologies in that analytes need not be known a priori, offering the possibility of untargeted analysis. Localized molecules within the GI tract were then identified in situ by surface sampling with nano-desorption electrospray ionization (nanoDESI) FT-MS. Products from diverse structural classes were identified including cholesterol-derived lipids, glycans, and polar metabolites. Specific chemical transformations performed by the microbiota were validated with bacteria in culture. This study illustrates how untargeted spatial characterization of metabolites can be applied to the molecular dissection of complex biology in situ.
INTRODUCTION
The mammalian cells within our bodies are outnumbered 10–100 fold by the bacteria that constitute our gut microbiotas.1 Those microbial communities are emerging as a key players in the pathogenesis of multiple diseases including ulcerative colitis, Crohn’s disease, food allergies, GI cancers, infections and obesity.2 Acquisition and development of the microbiota is poorly understood, as microbes are acquired from the environment and thrive or perish in gut ecosystem under the review of the host immune system and a variety of selective pressures. One key approach to understand this complexity has been exemplified in the Human Microbiome Project—where tremendous effort has been applied to characterize these communities at the nucleic acid level.3 However, developing an understanding of the role of a given community member and its contribution to chemical transformations is challenging due to complexity of wild microbiotas with 500–1,000 species present.
Our study represents three firsts: the first application of MALDI-TOF IMS to study the gut microbiota, the first untargeted MALDI-TOF IMS study of metabolites, and novel nanoDESI methodology to identify molecules detected by IMS. Our investigation utilizes three established mouse models of the developing infant microbiota:4 germ-free, mono-associated with Bacteroides thetaiotaomicron (Bt), and bi-associated Bt and Bifidobacterium longum (Bl) (Figure 1A).5 Gram negative Bacteroides are members of the Bacteroidetes, one of the two dominant phyla in the typical healthy adult microbiota. Bt is also an early colonizer of the infant gut, and contributes to host caloric needs by processing complex glycans found in the diet, host mucus, or derived from breast milk into usable simple sugars.6, 7 Gram-positive Bifidobacteria such as Bl are members of the Actinobacteria, which are often abundant in the infant gut microbiota and are a subdominant phylum of the adult microbiota. Similar to the Bacteroides spp., many Bifidobacterium spp. are accomplished degraders of complex carbohydrates.5
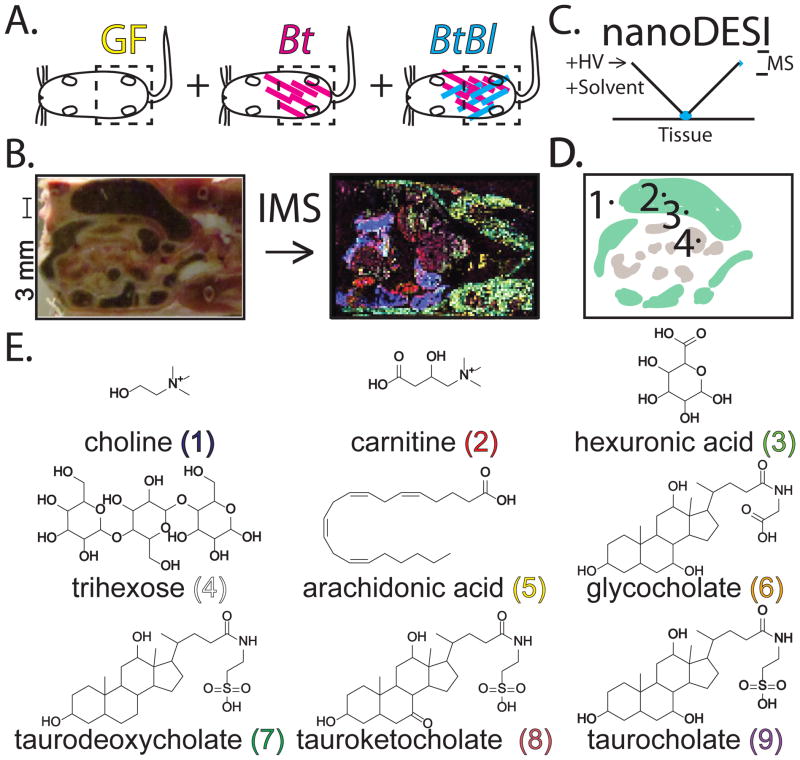
Figure 1.
Experimental design. (A) GF: Germ-free, Bt: mono-associated Bacteroides thetaiotaomicron, and BtBl: bi-associated associated Bacteroides thetaiotaomicron plus Bifidobacterium longum mice. Color-coding is noted and conserved in Figures 2–3. (B) Gut tissue sections were imaged via mass spectrometry to reveal metabolite distributions in false color. A scale bar is noted for all images. (C) NanoDESI FT-MS was applied for in situ compound identification. The capillaries are drawn to scale with the tissue section, however the actual droplet contact area is substantially smaller. HV: high voltage, MS: mass spectrometer. (D) Regions of interest from a mouse gut tissue section: 1. host connective tissue, 2. large intestine lumen, 3. large intestine/epithelial border, and 4. small intestine. (E) Selective compounds (1–9) identified by IMS and nanoDESI FT-MS. Stereochemistry is not illustrated as MS-based methods cannot readily distinguish contributions from different isomers. The exact contributions from specific regioisomers (C3, C7, C12) for (7–8) are not known—only the C7 modified isomer is illustrated. Compounds are with conserved color-coding as Figure 3. Observed m/z values are provided in Figure 3.
Imaging mass spectrometry (IMS) was our key tool to localize metabolites of interest in an untargeted fashion (Figure 1B). In IMS, a raster of mass spectra is taken across a prepared tissue section.8 After data processing, spectra intensities at a particular mass-to-charge value (m/z) and X-Y coordinates can be viewed as a false-color image, where high color brightness of pixels represents high intensities of spectra at these pixels. Results are analogous to fluorescence microscopy, except this method (i) does not require staining or labeling prior to analysis and (ii) provides hundreds of simultaneous channels in parallel separated by molecular mass. We applied an untargeted metabolomics approach to generate biological hypotheses for downstream investigation.9 Metabolomics (e.g. GC-MS, LC-MS and NMR) have helped elucidate and quantify chemical changes induced by the microbiota in homogenized samples.10, 11 MALDI-TOF IMS is a complimentary technique allowing for the localization and identification of analytes such as: drugs, lipids, peptides, and proteins.12 IMS has been applied to examine changes in protein expression upon S. aureus infection.13 Our untargeted MALDI-TOF IMS metabolomics approach offers the exciting possibility of localizing key biology in a complex system.
EXPERIMENTAL SECTION
General
All solvents used in this study were LC-MS grade. Authentic standards were purchased from Sigma or Fisher.
Gnotobiotic mice
Swiss-Webster germ-free mice were maintained in gnotobiotic isolators as previously described14 and fed an autoclaved standard diet (Purina LabDiet 5K67) ad libitum. GF mice were colonized using oral gavage of 200 μL of a saturated overnight culture of Bt or 100 μL each Bt and Bl. Mice were sacrificed by CO2 asphyxiation 10 days after colonization, frozen in liquid nitrogen, and stored at −80 °C. All animal protocols were in accordance with A-PLAC, the Stanford IACUC.
Tissue section IMS sample preparation
Whole frozen mice (−80 °C) were skinned and then cut in half. The posterior portion of the carcass was affixed to the chuck in a Leica CM1850 cryostat with a minimal amount of OCT. To avoid OCT signals, OCT was used as glue without touching the regions that were being sectioned. No embedding material was used and the samples were allowed to equilibrate to the chamber temperature for several hours. 25 μm thick sections were cut between −22 °C to −25 °C along the dorsal-ventral axis inwards from the chest towards the spine. Sections were transferred to cold ITO-coated conductive glass slides (Bruker Daltonics) and then briefly warmed with the operator’s thumb (2–3 s) to affix the tissue to the slide. Samples were promptly refrozen on the cold plate in the cryostat to minimize molecular diffusion. Sample slides were then placed in a vacuum desiccator above a bed of fresh dry-rite. The desiccator was kept cold in a cooler of dry ice, which was allowed to evaporate over 12 hours. The samples were dried for an additional 12 hours after the dry ice had evaporated. Sample slides were then coated with either DHB matrix for positive mode analysis, or 9-aminoacridine for negative mode analysis. DHB was applied in a sublimation chamber at 50 Torr for 8.5 min at 115 °C.15 9AA was applied with an airbrush at 10 mg/mL in 70% EtOH/30% DI/1% TFA. Coating was applied in 20 passes of 10 seconds, and dried under N2 between passes. Samples were rehydrated in a humidity chamber at 50 °C for 2 minutes. Two mice were examined from each sample condition and only results common across both animals are reported.
Agar plate IMS sample preparation
Overnight cultures of Bt and Bl were diluted to 0.1 OD600, and then 5 μL of each culture was spotted on thin TYG agar plates (11 mL) with or without taurocholate or taurodeoxycholate (1 mg/mL). Cultures were incubated for 48 hours at 37 °C at which point colonies or blank agar controls were excised from the plate with a razor blade and transferred to a MALDI target with a spatula. Universal matrix (positive ion mode) or 9-aminoacridine (negative ion mode) were then applied through 50 μm sieves to thoroughly coat the plates. Samples were dried for 4 hours at 50 °C and then subjected to IMS analysis as described below.
IMS analysis
Imaging mass spectrometry experiments were performed on a Bruker Autoflex Speed TOF/TOF instrument. Mass calibration and tuning was performed with a PepMix II standard solution (Bruker Daltonics) in the quadratic mode. For reflectron positive mode analysis the following settings were used: laser size medium, laser offset 45%, laser range 40%, laser power 70%, laser frequency 1000 Hz (tissue sections) or 200 Hz (agar plates), laser summed shots 128 total at 32 shots in 4 groups on random walk, mass range m/z 0 to m/z 4,000, deflectron off, gain enhanced 14.3x 4GS/s, ionsource 1 19 kV, ionsource 2 16.7 kV, lens 8.2 kV, reflectron 1 21 kV, reflectron 2 9.65 kV, delay 150 nS. For linear negative mode analysis the following settings were used: laser size medium, laser offset 45%, laser range 40%, laser power 70%, laser frequency 1000 Hz (tissue sections) or 200 Hz (agar plates), laser shots 128 at 32 shots in 5 groups on random walk, mass range 0 m/z 0 to m/z 1,500, deflectron off, gain enhanced 40x 2GS/s, ionsource 1 19.5 kV, ionsource 2 18.3 kV, lens 6 kV, delay 130 ns.
NanoDESI FTICR-MS/MS
nanoDESI was performed on a Thermo LTQ-FT 7T instrument using a modified Prosolia Omnispray DESI source with 2D automated stage.16 150 μm ID, 360 μm OD fused silica capillary tubing was used for the primary capillary and 75 μm ID, 150 μm OD fused silica capillary tubing secondary capillaries and solvent (60% MeCN/40% Di/0.1% Formic Acid) was infused at 1.5 μL/min with an applied voltage of 2 kV. Gas settings were all set to 0 with an inlet temperature of 250 °C. The instrument scan cycle consisted of two segments. The first segment had a duration of 3 min during which three profile mode MS scans were cycled: m/z 100–350 at 25,000 resolution in the FT cell with three microscans and a max inject time of 2 s, m/z 300–1,000 at 50,000 resolution in the FT cell with three microscans and a max inject time of 2 s, and one full scan in the IT with 5 microscans from m/z 100–2,000 with 1 s max fill time. The second MS/MS segment was allowed to run for 397 scans (10–20 minutes), with a m/z 3 isolation window swept through the mass range by m/z 2 units (e.g 101, 103, 105, … 889, 891). These scans consisted of 1 microscan with a maximum injection time of 2,000 ms, 25% normalized collision energy, 0.25 activation Q, and 0.05 s activation time. For nanoDESI analysis, we considered sections consecutive to those analyzed by IMS, which were mounted on insulating plastic microscope slides.
LC-MS
LC-MS was performed on a Thermo LTQ-FT 7T instrument using the IonMax source with a solvent flow rate of 200 μL/min and 4 kV of ESI voltage. Gas settings were 30 units for sheath, 18 units for aux, and 3 units for sweep at 300 °C. The instrument scan cycle consisted of one full scan in the FT-cell at 25,000 resolution with 2 microscans from m/z 100–2,000 with 1 s max fill time, followed by one full scan in the IT with 3 microscans from m/z 100–2,000 with 0.5 s max fill time—scans were performed in the profile mode. Five data-independent MS/MS scans were then performed in the IT in centroid mode using an inclusion list generated for masses with prominent localizations discovered from the IMS dataset with a 0.5 s max fill time and 25% normalized collision energy, 0.25 activation Q, and 0.05 s activation time. For the compound identity validation study, Figure S3–4, four data-independent scans were run instead in positive ion mode (m/z +104, m/z +162, m/z +217, m/z +527) and negative ion mode (m/z −303, m/z −464, m/z −498, m/z −514). Negative mode solvent consisted of A: 99% DI/1% MeOH and B: 99% MeOH/1% DI whereas in positive mode solvents consisted of A: 99% DI/1% MeCN/0.1% FA and B: 99% MeCN/1% DI/0.1% FA. A Thermo Surveyor LC system was utilized with a Synergi 4 μm Hydro RP 80Å 2 × 250 mm column. 20 μL of sample was injected and analyzed using the following gradient: 1% B (0–4 min), 1% → 99% B (4–29 min), 99% B (29–30 min), 99% → 1% (30–30.5 min), 1% (30.5–35 min). A divert valve was utilized to send the first 4 min to waste instead of the detector in some cases. Samples consisted of three pooled microscope slides matching those analyzed by IMS (above) that were scraped from the slide surface instead of coated with matrix. Samples were vortexed at maximum speed 3 × 1 min, and then sonicated for 5 min in 70% MeOH. Samples were then left at −20 °C for 1 hour, centrifuged at 4 °C and 14,000 RCF x G for 30 min, and then transferred to fresh tubes. Tissue sample concentrations were normalized at 60 μg/mL. Authentic standards were injected at 10 μg/mL under identical conditions.
Lipidomics assay
Reagents and materials: 132 eicosanoids and 28 isotope-labeled internal standards were purchased from Cayman Chemical. Sample preparation: Tissue sections were thoroughly homogenized with steel beads in 70% MeOH. 1 ng of isotope-labeled internal standards were added to the supernatant immediately. The mixtures were diluted with 10% MeOH in HO to 1 mL followed by the solid phase extraction (SPE). Strata-X 33 μm polymeric reversed phase 96-well plate (60 mg/well, Phenomenex) was pretreated with MeOH, and equilibrated with H2O. After the samples were extracted, the plate was washed with 10% MeOH. The lipids were eluted from the plate with 1 mL MeOH and dried in a SpeedVac. The dried samples were stored at −80 °C freezers before analysis. Preparation of standards for calibration curves: The combined stock standard solutions of 132 lipids were prepared by adding 10 μg of each lipid to a glass tube and made up to 10 mL with MeOH to obtain the final concentration of 1 μg/mL for each component. The stock solution was then serially diluted with MeOH to 8 different concentrations at 300, 100, 30, 10, 3, 1, 0.3, 0.1 ng/mL. The combined stock solution of internal standards (IS) was prepared by adding of 5 μg of each lipid to a glass tube and made up to 100 mL to obtain the final concentration of 50 ng/mL for each standard. An aliquot of 20 μL of the IS stock solution was spiked to samples, as well as to 40 μL of the standard stock solutions at different concentrations. The standards mix were extracted by SPE and dried by SpeedVac in the same 96-well plate as the samples. The stock solutions were stored at −80°C until the analysis. Chromatographic conditions: Both samples and standards were resuspended in MeOH-mobile phase A (50:50, v/v). An aliquot of 5 μL of sample was injected on the Acquity ultra performance liquid chromatography (Waters). Lipids were separated on a Kinetex C18 column (150 mm × 2.1 mm, 1.7 μm, Phenomenex) using a flow rate of 400 μL/min at 50 °C. The lipids were eluted using a stepwise gradient with mobile phase A consisting of H2O-ACN-acetic acid (70:30:0.1, v/v/v) and mobile phase B consisting of ACN-IPA-acetic acid (50:50:0.02, v/v/v). The gradient program was employed as follows: 0–0.24 min 100% A, 0.6 min 70% A, 2.75 min 55% A, 3.25 min 40% A, 4.50 min 25% A, 4.62 min 5% A, 5.2 min 5% A, 5.3 min 100% A, from 5.3 to 6.5 min, 100% A. The gradient was linear between each step. Mass spectrometry conditions: The mass spectrometric detection was operated on an AB SCIEX API 4000 system equipped with a Turbo Ionspray source. The data acquisition software used to operate the mass spectrometry was Analyst version 1.5. Quantitation was performed by scheduled multiple reaction monitoring (sMRM) in negative mode with parameters as follows: −4500 V ion spray voltage; 20 psi curtain gas; 30 psi ion source gas 1 (GS1); 30 psi ion source gas 2 (GS2); 6 psi collision gas (CAD); −10 V entrance potential (EP); −10 V collision cell exit potential (CXP); 600 °C and 20 s MRM detection window. The optimized MRM fragmentation transitions and parameters for declustering potential (DP) and collision energy (CE) were tuned for each lipid.
Bioinformatics
Manual IMS data analysis was performed in Bruker Daltonics FlexImaging 2.0 and nanoDESI and ESI analysis was performed in Thermo QualBrowser 2.0. Database searching for manual metabolite identification was performed with Human Metabolome Database, NIST, Metlin, Massbank, and Lipidmaps. Spatial segmentation of IMS data and the search for co-localized m/z-values for the segments of interest was performed using the algorithm proposed17 and implemented as custom made scripts in the Matlab software (The Mathworks Inc.). In short, the algorithm selects dataset-relevant peaks, applies edge-preserving image denoising to the m/z-images of selected peaks, and clusters the processed spectra by their similarity, representing the results as a segmentation map pseudo-coloring pixels of the same cluster. Then, the co-localized m/z-values are found by using correlation of their m/z-images with the spatial regions of the segmentation map. The nanoDESI MS/MS plots were generated using custom made scripts in Matlab by creating a table of MS/MS spectra, where each column corresponds to a precursor ion m/z and contains an MS/MS spectrum; columns are sorted by the precursor m/z-values. Spectral networking was performed as described.16
RESULTS AND DISCUSSION
Exploring the metabolic output of the gut microbiota
Mouse tissue sections were subjected to MALDI-TOF IMS in the positive and negative ion modes (Figure 1A–B). The posterior half was sectioned along the dorsal-ventral axis from the ventral side inwards to reveal the small and large intestines. Care was taken to match sections between different mice, although this was challenging due to individual variation and the differential effects of mouse colonization state on gut physiology—such as the enlarged cecums of germ-free animals. 25 μm-thick frozen tissue sections were briefly thawed (1–3 seconds) then frozen to fix them to the conductive indium tin oxide (ITO) slide surface while minimizing molecular diffusion. Samples were desiccated under vacuum while frozen, and coated with matrix. Matched tissue sections from all three conditions were then analyzed simultaneously in the reflectron positive and linear negative ion modes in a MALDI-TOF MS at 300 μm spatial resolution. IMS resulted in hundreds of observed molecular signals—detected through automated data processing and subsequent manual analysis.17 Accurate mass m/z values were identified through nanoDESI FT-MS (Figure 1C). This ambient ionization technique allowed for in situ accurate mass and MS/MS analysis at regions of interest (Figure 1D).16, 18 Regions were identified based on IMS data as having distinct chemical localization patterns.
Identifying molecules detected from IMS datasets
Identifying metabolites detected from IMS datasets (Figure 2A, Figure 3 (1–9)) remains a challenge. Molecules at m/z values exhibiting interesting localization patterns were first fragmented by MALDI-TOF/TOF MS/MS in situ, however this low mass-accuracy, low-resolution data is not always able to provide a unique identifier of metabolites (as opposed to peptides). We complemented this analysis with nanoDESI FT-MS and nanoDESI iontrap (IT)-MS/MS (Figure 1C–D, Figure 2B–D). NanoDESI can be coupled to high-mass accuracy/resolution FT-MS detector (Figure 2B, 2D). The high-resolution mass-accuracy data proved to be quite useful for identifying compounds of interest, and exhibited similar peaks as compared to MALDI (with the exception of matrix ions). The experimental FT-MS parent mass of compounds identified by MS/MS were within 12 ppm of theoretical molecular mass and presence/absence of peaks correlated with IMS results in the large intestine/epithelial border region are displayed (Figure 1D–3, Figure 3). The lack of sample preparation in this ambient ionization experiment is critical for our studies, as we can directly target regions identified by IMS for analysis without the need for any biased extraction technique. Indeed, no extraction would likely provide (1–9) in a single fraction. Similar analytical limitations are also present in both MALDI and nanoDESI which may help support the high-degree of similarity between our MS spectra—notably the bias towards signals from abundant ions with high ionization efficiencies. Indeed, all identified ions (1–9) fit these criteria.
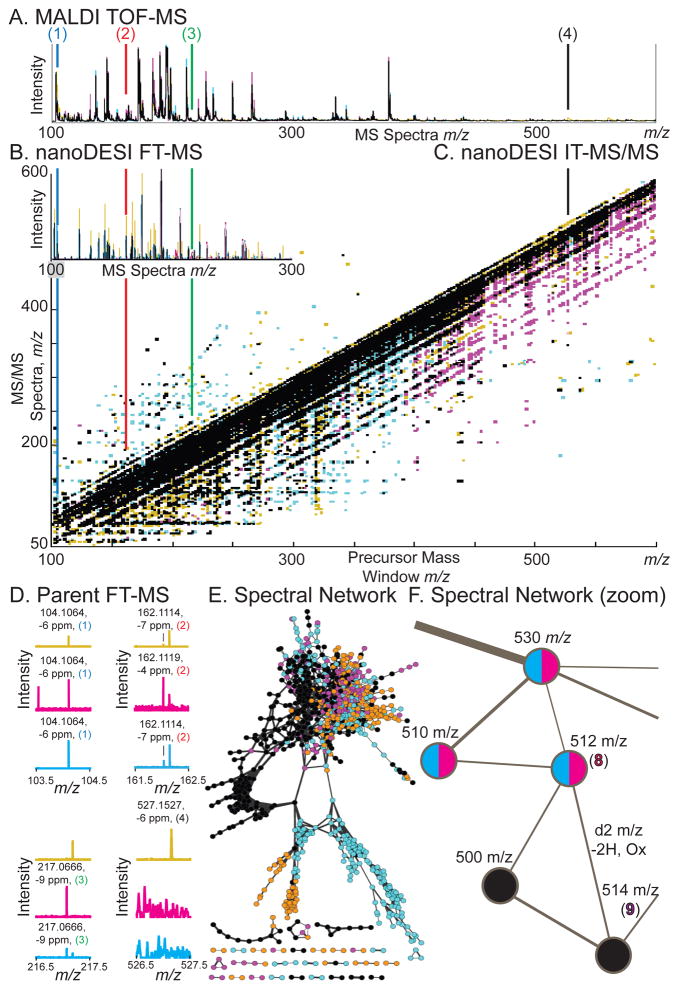
Figure 2.
Identification of metabolites detected from IMS experiments. Germ-free, mono-associated Bacteroides thetaiotaomicron, and BtBl: bi-associated associated Bacteroides thetaiotaomicron plus Bifidobacterium longum mice are noted by color, with overlap in black. Compounds (1–4, 8–9) are displayed with conserved color coding as in Figures 1, 3. (A) MALDI-TOF MS average spectra displayed as intensity versus m/z. (B) NanoDESI-FTICR MS displayed as intensity versus m/z. (C) Data-independent iontrap (IT)-MS/MS data from the large intestine/epithelial border (Figure 1D–3.) displayed as intensity in Z, MS/MS spectra in m/z in Y, versus precursor mass window in X in m/z. (D) Zoomed nanoDESI FT-MS spectra from (B) for (1–4) displayed as intensity versus MS spectra in m/z. Accurate mass assignments are noted, correlating with IMS data. See Figure S1 for zoomed nanoDESI-MS/MS spectra from (C). (E) Spectral network clustering of data-independent MS/MS data. Nodes are linked based on MS/MS spectral similarity as a proxy for chemical similarity. This figure is an alternative view of (C), illustrating some of the complexity of the dataset. (F) Spectral network zoomed, illustrating a node at m/z −512 (8) connected to a node at m/z −514 (9). This data illustrates how spectral networking can be used to link known compounds to unknowns—forming visual data hypotheses for manual validation.
Figure 3.
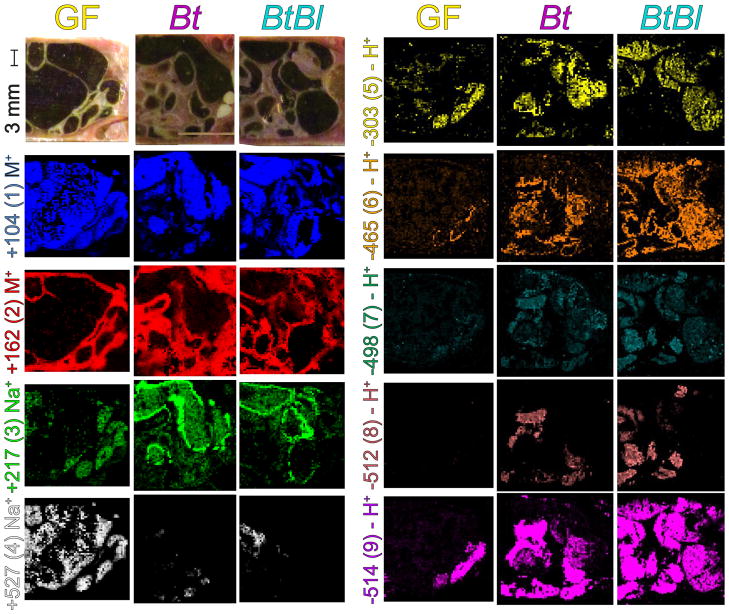
Imaging mass spectrometry of the gut of three mouse microbiota models. A series of panels are provided for GF, Bt, and BtBl associated mice. The panels represent the ventral view of the mouse with the anterior facing left and the posterior facing right (Figure 1A). Selected anatomical features are noted (Figure 1D).Orientation and color of IMS data match (Figure 1E). Optical images were taken at the block face with a universal scale provided. Each false-colored IMS image represents the normalized signal intensity for the m/z value illustrated corresponding to (1–9) as a particular ion or charged adduct. Identities of m/z values were assigned by nanoDESI MS/MS and LC-MS (Figures 2 and S1–4). This figure thus depicts differential signal and localization patterns of (1–9) across three sample conditions.
Data-independent IT-MS/MS spectra were obtained by sweeping a m/z 3 window in m/z 2 increments (Figure 2C). This prolonged analysis (15–30 min) on a single region was possible due to the low rate of sample consumption and high sensitivity of nanoDESI.16 Data-independent acquisition methods with a defined series of MS/MS events utilized in every run may offer advantages over typical data-dependent techniques and are worth further investigation. One key advantage is non-stochastic acquisition, allowing for systematic comparisons of identical data sets, as illustrated with the overlaid spectra (Figure 2C).19 A further advantage may be greater sampling of low-abundance ions, however, direct comparisons with data-dependent methods will be required to evaluate this. This approach facilitates comparisons of entire MS/MS datasets as single multi-dimensional objects, an under-utilized technique of analysis.
For organization and identification a spectral network approach was exploited (Figure 2E).16 A spectral network is generated from an MS/MS data-independent run representing similarities between individual MS/MS spectra, where spectral similarity is used as a proxy for chemical similarity. These networks can be interactively explored and annotated in the program Cytoscape, allowing for direct intuitive interaction with the data. This enables for the direct generation of chemical hypotheses from data as illustrated in a zoomed view (Figure 2F). Here an MS/MS spectrum of the node at m/z -514 was found to match a reference library MS/MS spectra in the MassBank20 metabolomics spectral repository. This node, taurocholate (9), could then be checked against accurate mass nanoDESI FT-MS data (Figure 2D). The node can then be revisited in the spectral network view, where it was linked to two nodes, once at m/z −500 present in all samples and one at m/z −512 present only with Bt or BtBl (8). Given the mass shift of 2 m/z units one possible hypothesis is oxidation, suggesting that the compound is tauroketocholate (8). This illustrates the ability of spectral networks to link “known-unknowns” (compounds present in spectral databases) to “unknown-unknowns” (compounds not present in spectral databases) in raw experimental data.21 Furthermore, this identification can likely be spread out even further into the local network cluster of tightly connected nodes, for example, the compound at m/z 500 may be a desmethyl bile acid analog. Thus the combination of nanoDESI, providing prolonged analysis at a specific location targeted by IMS, comprehensive data-independent MS/MS datasets, and spectral networking allows us to uncover molecular transformations that are observed upon microbial colonization. To provide additional support for the molecular identities or structural similarity, homogenized tissue slices were subjected to LC-MS for co-elution with commercial authentic standards for compound validation (Figure S3–4).22 Thus, our novel workflow allows us to rapidly identify molecules from tissue, so as to rationalize and interpret our IMS results.
Metabolites identified as tissue markers
The three predominant tissue types observed in the analysis include large intestine, small intestine, and host tissue (Figure 3 optical). Ions such as choline (Figure 3, (1)) can be localized to the luminal contents within the small and large intestine across GF, Bt, and BtBl samples, serving as a fecal marker. Choline (1) is a key dietary metabolite rapidly transformed to compounds such as acetylcholine, betaine, and phosphatidylcholines by the host, and is present at 2,000 ppm in the mouse chow used in this study. Phosphocholine conversion by gut bacteria can lead to downstream metabolites that are indicative of cardiovascular disease.23 In contrast, the metabolite carnitine (Figure 3, (2)) was selected as a marker for host tissue since 99% of it is intercellular.24 Carnitine (2) is a key metabolite in oxidative metabolism where it shuttles acetate and other acyl metabolites into the mitochondria.
Glycans displaying altered localization patterns dependent upon microbiota status
Two glycans with prominent localization patterns were detected by IMS at m/z +217, and m/z +527. Hexuronic acid (Figure 3, (3)) is predominant only in the Bt and BtBl samples, localized to the inner epithelial wall of the intestine. This oxidized hexose likely consists of a mixture of the isobaric, naturally-occurring hexuronic acids: galacturonic acid, glucuronic acid, and iduronic acid as all three of these sugars are components of the capsule of Bt.25 Galacturonic acid is also produced by the enzymatic degradation of complex plant dietary fibers such as pectin—a known dietary substrate of Bt and Bl.26 Glucuronic acid may also be contributed through mucosal secretion since glucuronidation is a key pathway for the body to eliminate xenometabolites and gut microbiota often contain enzymes to cleave these sugars.27 Iduronic acid is part of glycosaminoglycans (GAGs) component of host mucus, which can be scavenged by Bt.25 Mannuronic acid is primarily derived from alginates. Glucuronic acid has been reported to be upregulated in conventional versus GF mice in serum by GC-MS.28 Numerous enzymes for processing pectin, galacturonic acid, and glucuronic acid are upregulated when Bt mice are fed a diet of complex plant sugars instead of simple sugars.7
The second carbohydrate, a trihexose (Figure 3, (4)), which is ubiquitous within the gut lumen of the germ-free sample. This ion represents a pool of different trisaccharides, highlighting the buildup of unprocessed dietary glycans in the absence of carbohydrate consumers such as Bt and Bl. Indeed, a germ-free mouse is essentially a knock-out of glycan processing capabilities in the host-microbe assemblage since these glycolytic enzymes constitute >5% and >9% of the genomes of Bt and Bl. This finding is supported by GC-MS analysis where it was found that after degradation of cecum polysaccharides to monosaccharides, germ-free mice had substantially more sugars present as compared to Bt associated mice.7
Arachidonic acid IMS and follow-up of related metabolites with targeted LC-MS
The next identified metabolite is key dietary fatty acid arachidonic acid (Figure 3, (5)) which sits at the head of signaling cascades such as the leukotrienes and prostaglandins.29 While there were relatively small gross differences in metabolite levels between samples, the abundant presence of this molecule in the gut and tissue led us to investigate downstream signaling with a targeted assay. We applied an isotopic dilution LC-MS scheduled multiple reaction monitoring (sMRM) assay for simultaneous quantitation of 132 arachidonic acid derived metabolites (Figure S5, Table S1). Some of the changes we saw included arachidonic acid levels, which were 3 times lower in GF as compared to Bt or BtBl. Interestingly, one of the most biologically active prostaglandins, PGE2, was more than 100 times lower in GF as compared to Bt or BtBl. Gut microbes have been shown to stimulate PGE2 secretion which can mediate intestinal permeability.30 The relative difference in arachidonic acid versus PGE2 levels between samples illustrates that precursor levels do not necessary correlate with downstream metabolites—suggesting differences in regulation specific to inflammatory responses.
Microbiota transformations of host-derived bile acids
A series of bile acids were also identified (Figure 3, (6–9)). Bile acids are derived from cholesterol and play a key role in the digestive process by solubilizing dietary fats for uptake. They also act as key molecules in signaling pathways under control of nuclear hormone receptors, such as the FXR receptor in the gut, and TGR5, a dedicated bile acid receptor.31 Conjugated glycocholate (6) and taurocholate (9) are primary bile acids released from the host. Primary bile acids are deconjugated, dehydroxylated, or oxidized by gut microbes to form secondary bile acids. (7) represents contributions from different dehydroxylation pattern at C3, C7 (shown), or C12 which can include secondary bile acids such as taurodeoxycholate, tertiary bile acid or primary bile acids such as taurochenodeoxycholate. Signal intensities for the primary bile acids (6) and (9) are lower in the germ-free sample but still present. However, (7) is lacking in the absence of gut microbiota. This suggests that the major isomeric contributor to (7) may be the bacterial-derived secondary bile acid taurodeoxycholate.32 The secondary bile acid, ketotaurocholate (8), is the product of bacterial oxidation and is not detected in the germ-free sample. Reportedly, bile acids are abundant in the intestines of microbiota-associated mice as compared to germ-free models.10 Bacterial production and regiochemistry of tauroketocholate was investigated in agar culture (Figure 4).
Figure 4.
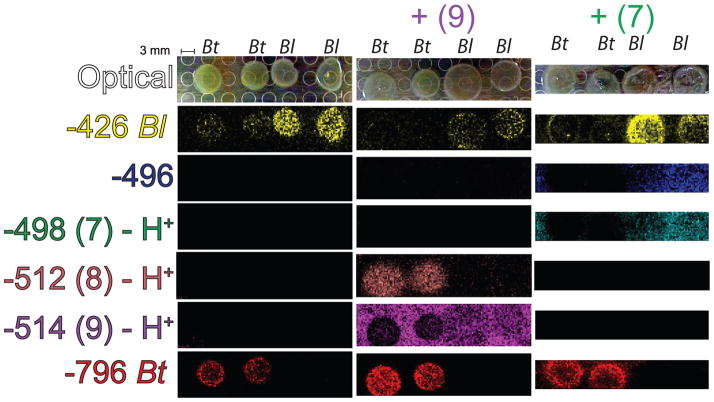
Imaging mass spectrometry to explore microbial bile acid transformations. A series of panels are provided for two colonies of Bt and two colonies of Bl on TYG agar, TYG agar + taurocholate (9), and TYG agar + deoxytaurocholate (7). The panels represent the portion of the Petri dish agar relocated to a MALDI target plate. Each false-colored image represents the normalized signal intensity for the m/z value illustrated. Identities of m/z values were assigned by nanoDESI MS/MS and LC-MS (Figures 2 and S1–4). For compounds (7–9) present in Figures 1–3, identical color coding is utilized.
Dissecting microbiota bile acid transformations in pure culture
To help further explore the direct role of our model microbiota in the hypothesized processing of host bile acids (Figure 3) we cultured Bt and Bl on TYG agar with bile acids33 and then analyzed the samples by microbial IMS.34 Two unidentified ions at m/z -426 and m/z −796 were used to indicate the Bl and Bt colonies respectively. On taurocholate (9) plates, the m/z −514 appears to be depreciated only under the Bt colonies in the resulting ion image. A new signal is observed under the Bt colony at m/z −512, consistent with the ability of Bt (but not Bl) to transform taurocholate (9) to tauroketocholate (8) in high yield. However, on taurodeoxycholate plates, (7) does not appear to be transformed to the analogous oxo- compound at m/z −496. This indicates that the transformation mediated by Bt may be stereospecific to the C7 ring location, as depicted (Figure 1, (7–8)).
This transformative capability of Bt is in agreement with previous reports of Bt capable of transforming cholate to 7-keto cholic acid in 83% yield.35 Various gut microbes including Bacteriodes have been noted to oxidize bile acids at the C3, C7, or C12 positions.36 The ability of Bl to dehydroxylate or oxidize bile acids has been noted, but was substantially less compared to Bt.37 Keto bile acids are also formed in low abundance <3% in mice carrying human fecal strains—however the primary activities reported are deconjugation and dehydroxylation.38 Bile hydrolase acid activity has been detected and characterized in Bl.39 Production of deconjugated bile acids is also likely occurring in our experiment (Figure 4). However, based upon studies of authentic standards, free cholate and deoxycholates could not be detected even at concentrations of 1 mg/mL in agar plates (data not shown).
CONCLUSIONS
IMS allowed us to explore the complex metabolic interactions in the gut in an untargeted manner while retaining spatial information. This is the first IMS study to localize bile acid, glycans, lipids, and polar metabolites in the gut. Our findings are consistent with NMR or MS studies on homogenates with the additional spatial information afforded by IMS. IMS and associated techniques in this integrated workflow allowed us to investigate the role of low-molecular-weight metabolites of varied structural classes across tissue types in a single experiment. Molecules detected by IMS in complex systems such as our mouse models can be used for targeted downstream investigations (arachidonic acids) or be studied in detail in microbial culture (bile acids). These workflows were enabled by the data-independent nanoDESI workflow using spectral networking which allowed rapid organization and identification of complex datasets. This study highlights the capabilities of IMS, as a hypothesis generating tool for exploring the biological complexity of gut microbiota/host interactions. For example, do identified compounds such as dietary fibers, lipids, and bile acids interact with each other? Are the transformative capabilities and metabolic response in the background of Bt and Bl general or specific? Future studies will focus on extending this work by obtaining higher spatial resolution within specific tissue. A greater diversity of microbial models will also be explored. Targeted methods will be used to identify low abundance metabolites of interest, or chemical markers for the localization of specific bacteria. Fermentation of sugars to short-chain fatty acids play an important role in maintaining gut homeostasis by inhibiting growth of harmful bacteria—what other metabolites change in concert with this biology?40, 41 Applying this methodology to extend our understanding of the fundamental nature of the gut microbiota may well help us recontextualize the important factors in problematic medical indications.
Supplementary Material
Acknowledgments
This work was supported by the Keck Foundation. Instrumentation in PCD’s laboratory is supported by Bruker Therapeutic Discovery Mass Spectrometry Center and NIH GM S10RR029121. Professor James Coleman and Professor Daniela Monti are graciously thanked for additional assistance with bile acid standards.
Footnotes
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Gill SR, Pop M, DeBoy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Science. 2006;312(5778):1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sekirov I, Russell SL, Antunes LCM, Finlay BB. Physiol Rev. 2010;90(3):859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 3.Human Microbiome Project Consortium. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.France de La Cochetiere M, Rouge C, Darmaun D, Christophe Roze J, Potel G, Gras Leguen C. Cur Ped Rev. 2007;3(1):21–34. [Google Scholar]
- 5.Sonnenburg JL, Chen CTL, Gordon JI. PLoS Biol. 2006;4(12):e413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JAB, Sonnenburg JL. Cell Host & Microbe. 2012;10(5):507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Science. 2005;307(5717):1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 8.Watrous JD, Alexandrov T, Dorrestein PC. J Mass Spec. 2012;46(2):209–222. doi: 10.1002/jms.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patti GJ, Yanes O, Siuzdak G. Nat Rev Mol Cell Biol. 2012;13(4):263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claus SP, Tsang TM, Wang Y, Cloarec O, Skordi E, Martin FP, Rezzi S, Ross A, Kochhar S, Holmes E, Nicholson JK. Mol Syst Biol. 2008;4:219–230. doi: 10.1038/msb.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Proc Nat Acad Sci USA. 2009;106(10):3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seeley EH, Caprioli RM. Trends Biotech. 2011;29(3):136–143. doi: 10.1016/j.tibtech.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Attia AS, Schroeder KA, Seeley EH, Wilson KJ, Hammer ND, Colvin DC, Manier ML, Nicklay JJ, Rose KL, Gore JC, Caprioli RM, Skaar EP. Cell Host & Microbe. 2012;11(6):664–673. doi: 10.1016/j.chom.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. Cell. 2012;141(7):1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hankin JA, Barkley RM, Murphy RC. J Am Soc Mass Spec. 2007;18(9):1646–1652. doi: 10.1016/j.jasms.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, Moore BS, Laskin J, Bandeira N, Dorrestein PC. Proc Nat Acad Sci USA. 2012;109(26m):E1743–E752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexandrov T, Becker M, Deininger Sr-O, Ernst Gn, Wehder L, Grasmair M, von Eggeling F, Thiele H, Maass P. J Proteome Res. 2010;9(12):6535–6546. doi: 10.1021/pr100734z. [DOI] [PubMed] [Google Scholar]
- 18.Roach PJ, Laskin J, Laskin A. Analyst. 2010;135(9):2233–2236. doi: 10.1039/c0an00312c. [DOI] [PubMed] [Google Scholar]
- 19.Gillet LC, Navarro P, Tate S, Roest H, Selevsek N, Reiter L, Bonner R, Aebersold R. Mol & Cell Proteomics. 2012 doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, Oda Y, Kakazu Y, Kusano M, Tohge T, Matsuda F, Sawada Y, Hirai MY, Nakanishi H, Ikeda K, Akimoto N, Maoka T, Takahashi H, Ara T, Sakurai N, Suzuki H, Shibata D, Neumann S, Iida T, Tanaka K, Funatsu K, Matsuura F, Soga T, Taguchi R, Saito K, Nishioka T. J Mass Spec. 2010;45(7):703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 21.Neumann S, Böcker S. Anal Bioanal Chem. 2010;398(7):2779–2788. doi: 10.1007/s00216-010-4142-5. [DOI] [PubMed] [Google Scholar]
- 22.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. Anal Chem. 2006;78(3):779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WHW, DiDonato JA, Lusis AJ, Hazen SL. Nature. 2011;472(7341):57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flanagan JL, Simmons PA, Vehige J, Willcox MD, Garrett Q. CORD Conf Prof. 2010;7:30–31. doi: 10.1186/1743-7075-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martens EC, Roth R, Heuser JE, Gordon JI. J Biol Chem. 2009;284(27):18445–18457. doi: 10.1074/jbc.M109.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dongowski G, Lorenz A, Anger H. Ap Environ Microbiol. 2000;66(4):1321–1327. doi: 10.1128/aem.66.4.1321-1327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritter JK. Chem-Biol Int. 2000;129:171–193. doi: 10.1016/s0009-2797(00)00198-8. [DOI] [PubMed] [Google Scholar]
- 28.Velagapudi VR, Hezaveh R, Reigstad CS, Gopalacharyulu P, Yetukuri L, Islam S, Felin J, Perkins R, Boren J, Oresic M, Backhed F. J Lipid Res. 2009;51(5):1101–1112. doi: 10.1194/jlr.M002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harizi H, Corcuff J-Bt, Gualde N. Trends Mol Med. 2008;14(10):461–469. doi: 10.1016/j.molmed.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Resta-Lenert S, Barrett KE. Gastroenterol. 2002;122(4):1070–1087. doi: 10.1053/gast.2002.32372. [DOI] [PubMed] [Google Scholar]
- 31.Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Nat Rev Drug Discov. 2008;7(8):678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 32.Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, Nicholson JK, Holmes E. Proc Nat Acad Sci USA. 2011;108(S1):4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones BV, Begley Mi, Hill C, Gahan CGM, Marchesi JR. Proc Nat Acad Sci USA. 2008;105(36):13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang YL, Xu Y, Straight P, Dorrestein PC. Nat Chem Biol. 2009;5(12):885–887. doi: 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edenharder R. J Steroid Biochem. 1984;21(4):413–420. doi: 10.1016/0022-4731(84)90304-2. [DOI] [PubMed] [Google Scholar]
- 36.Bortolini O, Medici A, Poli S. Steroids. 1997;62:564–577. doi: 10.1016/s0039-128x(97)00043-3. [DOI] [PubMed] [Google Scholar]
- 37.Chikai T, Nakao H, Uchida K. Lipids. 1987;22(9):669–671. doi: 10.1007/BF02533948. [DOI] [PubMed] [Google Scholar]
- 38.Narushima S, Itoh K, Miyamoto Y, Park SH, Nagata K, Kuruma K, Uchida K. Lipids. 2006;41(9):835–843. doi: 10.1007/s11745-006-5038-1. [DOI] [PubMed] [Google Scholar]
- 39.Grill J, Schneider F, Crociani J, Ballongue J. Ap Environ Microbiol. 1995;61(7):2577–82. doi: 10.1128/aem.61.7.2577-2582.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. Nature. 2011;469(7331):543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 41.Kinross J, von Roon A, Holmes E, Darzi A, Nicholson J. 2008;10(4):396–403. doi: 10.1007/s11894-008-0075-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.