Abstract
The repair of DNA double strand breaks (DSBs) is critical for the maintenance of genome integrity. The first step in DSB repair by homologous recombination is processing of the ends by one of two resection pathways, exemplified by Saccharomyces cerevisiae Exo1 and Sgs1–Dna2. Here we report in vitro and in vivo studies that characterize the impact of chromatin on each resection pathway. We find that efficient resection by the Sgs1-Dna2 -dependent machinery requires a nucleosome-free gap adjacent to the DSB. Resection by Exo1 is blocked by nucleosomes, and processing activity can be partially restored by removal of the H2A-H2B dimers. Our study also supports a role for the dynamic incorporation of the H2A.Z histone variant in Exo1 processing, and it further suggests that the two resection pathways require distinct chromatin remodeling events in order to navigate chromatin structure.
Keywords: DNA repair, Sgs1, Exo1, H2A.Z, homologous recombination, chromatin
DSBs, if not repaired properly, pose a serious threat to genome integrity. Improperly repaired DSBs can lead to loss of genetic material, chromosomal duplications or translocations, and carcinogenesis1. The yeast Mre11–Rad50–Xrs2 (MRX) complex facilitates the recognition of DNA ends and commitment to repair by homologous recombination (HR). Subsequently, the nucleolytic processing of the ends results in a 3′-single stranded DNA (ssDNA) intermediate that is bound by replication protein A (RPA) to provide the signal for DNA damage checkpoint activation2. The Rad52 protein helps displace RPA from ssDNA to promote assembly of a polymer of the Rad51 recombinase protein. The Rad51–ssDNA nucleoprotein filament then performs a search for a homologous DNA sequence to initiate error-free repair3.
Recent genetic studies have identified two redundant pathways for DNA end resection during HR, characterized by the yeast Sgs1–Dna2 and Exo1 enzymes4-6. In addition to DSB processing, Dna2 plays an essential role during DNA replication, and Exo1 is involved in DNA mismatch repair (MMR), meiotic crossovers, and the processing of stalled replication forks and improperly capped telomeres7-12. Recently, in vitro studies have demonstrated that efficient resection of DNA by the yeast Sgs1–Dna2 pathway requires a large contingent of proteins, including the MRX complex, RPA, and the Top3–Rmi1 complex13. In contrast, Exo1 is sufficient to resect dsDNA ends in vitro14,15. The components of both S. cerevisiae resection pathways are conserved among eukaryotes, and defects in the human homologs of Sgs1 (BLM, WRN, and RECQ4) have been linked with disease pathologies resulting in cancer predisposition and premature aging16.
ATP-dependent chromatin remodeling enzymes use the energy from ATP hydrolysis to disrupt histone-DNA contacts, resulting in nucleosome sliding, eviction, and/or histone exchange. In Saccharomyces cerevisiae, a large number of remodeling enzymes, including RSC, SWI/SNF, INO80, SWR-C, and Fun30 are recruited to chromatin regions adjacent to an HO endonuclease induced DSB17-23. RSC appears to catalyze the eviction or mobilization of nucleosomes directly adjacent to the DSB, promoting the recruitment of the MRX complex and subsequent DNA processing18. The Ino80 complex is also required for efficient DNA resection, though the Fun30 enzyme plays a more dominant role in these events22,23. The Swr1 and Ino80 complexes regulate the dynamic incorporation of the histone variant H2A.Z within DSB chromatin, and H2A.Z has been reported to also impact DNA processing efficiency24,25. While these ATP-dependent chromatin remodeling enzymes have been linked to DSB processing, it is not yet clear how they might facilitate this process.
Here we test how chromatin structure impacts DNA processing pathways by a combination of in vitro reconstituted chromatin assays and yeast gene deletion studies. We find that the helicase activity of yeast Sgs1 and its human homolog, BLM, is reduced on nucleosomal substrates, and that efficient resection by the Sgs1-Dna2 -dependent machinery requires a nucleosome-free gap adjacent to the DSB. We also report that resection by Exo1 is blocked by nucleosomes, and that processing activity can be partially restored by removal of the H2A/H2B dimers or incorporation of the histone variant H2A.Z. The SWR1-dependent incorporation of H2A.Z is found to also play a role in Exo1-dependent resection in vivo. Our study suggests that these two DNA processing pathways require distinct chromatin remodeling events in order to navigate chromatin structure, indicating complex interactions between chromatin dynamics and DNA repair.
Results
DNA resection is impaired on chromatin substrates
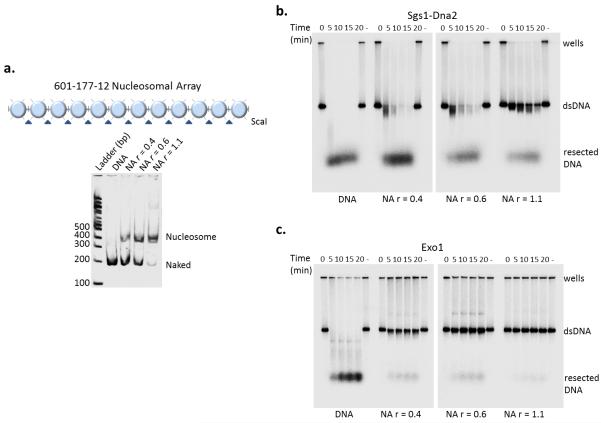
To investigate how chromatin might impact DSB processing, we assessed DNA resection by the Sgs1–Dna2 machinery and Exo1 on nucleosomal substrates. Chromatin fibers that harbor a varying number of positioned nucleosomes on a 32P 3′ end-labeled DNA template with 12 copies of the 601 nucleosome positioning sequence were reconstituted by salt dialysis (Fig. 1a). In the absence of nucleosomes, the Sgs1–Dna2 machinery and Exo1 rapidly processed dsDNA, consistent with previous biochemical studies (Fig. 1b, c, left lanes)13-15. However, addition of only a few nucleosomes (r=0.4 and 0.6) efficiently blocked resection catalyzed by Exo1, whereas the Sgs1–Dna2 processing machinery was relatively unimpeded (Fig. 1b, c). Assembly of a nucleosomal array fully loaded with nucleosomes (r=1.1) inhibited the Sgs1–Dna2-dependent resection. Thus, both resection pathways are inhibited by chromatin, with the Exo1 pathway being the more sensitive.
Figure 1.
Increasing nucleosome density inhibits resection. (a) Top: schematic of the 601-177-12 nucleosomal array (NA). Bottom: Native 4% PAGE of nucleosomal array ScaI digests after reconstitution by salt step dialysis. DNA template utilized for chromatin reconstitution comprised of 12 repeats of 177 bp (each flanked by ScaI restriction site) containing the “601” nucleosome positioning sequence. (b, c). Resection assays with 3′-radiolabelled naked DNA and chromatin at increasing ratios (r) of histone octamer to repeat sequence (0.4, 0.6, and 1.1). Time course of resection for both pathways, Sgs1-Dna2 (10 nM Mre11-Rad50-Xrs2 complex, 10 nM Sgs1, 10 nM Top3-Rmi1 complex, 20 nM Dna2 and 100 nM RPA) (b) and Exo1 (6 nM) (c), resulting in intact (dsDNA) or digested (resected) substrates.
Sgs1-Dna2 activity requires nucleosome free regions
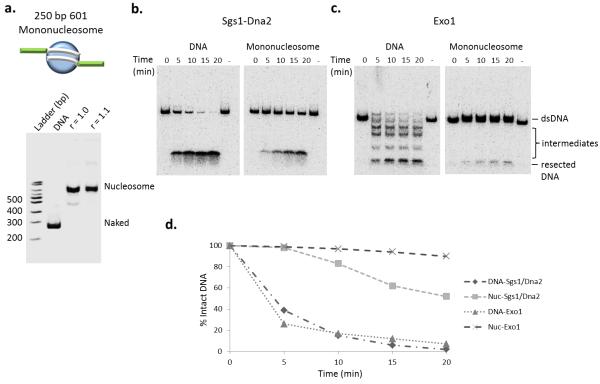
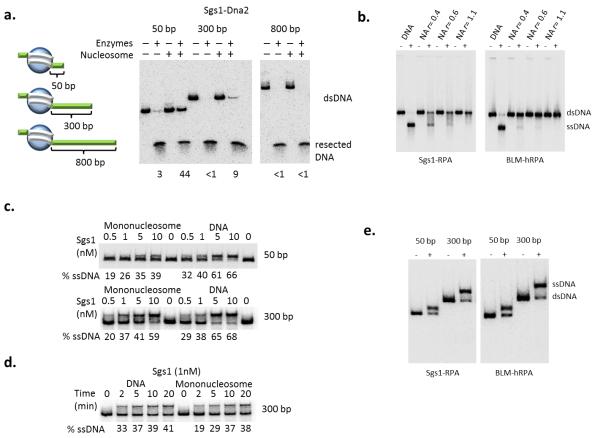
To further detail the role that nucleosomes play during chromatin fiber resection, center-positioned mononucleosomes were reconstituted on a 250 bp nucleosome positioning sequence (Fig. 2a). While the Sgs1–Dna2 ensemble rapidly degraded the free, 250 bp DNA fragment, much less digestion occurred on the mononucleosome substrate, even following extended incubation (Fig. 2). Notably, inhibition was not due to decreased substrate binding, as the Sgs1– Dna2 machinery bound equally well to free DNA and the 250 bp mononucleosome, as revealed in a streptavidin bead binding assay (Supplemental Fig. 1 online). Similar to the case for nucleosomal arrays, Exo1 was unable to process the mononucleosome substrate (Fig. 2). Strikingly, the inhibition of Sgs1–Dna2 activity was relieved when nucleosomes were reconstituted on a DNA fragment in which a 300 or 800 bp DNA segment was positioned adjacent to the nucleosome (Fig. 3a, b, c, Supplemental Fig. 2 online). Together, these results indicate that the Sgs1–Dna2 resection machinery can interact with a DNA end within chromatin, and with enough adjacent free DNA, this machinery can traverse a nucleosome.
Figure 2.
Mononucleosomes inhibit resection pathways differentially (a) Top: schematic of the 601-250 mononucleosome. Bottom: Native PAGE analysis of chromatin reconstitutions with Xenopus histone octamers. Template DNA consisted of one 601 positioning sequence in the middle of a 250 bp DNA fragment.(b) Resection assay using 601-250 naked DNA and mononucleosomes (3′ radiolabel on one end). Resection reaction conditions on mononucleosome are identical to those in Fig. 1b, c. (c) Quantification of signal remaining was calculated and graphed as percent of intact radiolabel DNA at indicated times compared to the 0 time point of each assay.
Figure 3.
Increasing free DNA adjacent to a nucleosome enhances the helicase activity of Sgs1. (a) Left: schematic of mononucleosome substrates depicting varying amounts of nucleosome-adjacent free DNA (50 bp, 300 bp, and 800 bp). Right: resection assay for the Sgs1-Dna2 pathway using the 50-, 300-, and 800-bp free DNA mononucleosomal substrates after 20 minutes at 30° C. Percent remaining after resection is indicated for each reaction. (b) Sgs1 (10 nM) and BLM (20 nM) helicase activity in the presence of RPA on identical NAs to Fig. 1a after 20 min at 30° C. (c, d) Helicase assay of Sgs1 on DNA and mononucleosomes of indicated sizes. Substrates were incubated with 100 nM RPA and the indicated concentrations of Sgs1 at 30° C for 20 minutes (c) or indicated times (d). (e) The helicase activity of Sgs1 (left panel) and human homolog BLM (right panel) in the presence of RPA on chromatin substrates described in Fig. 2a.
To further define how nucleosome assembly inhibits the Sgs1–Dna2 reaction, we assessed the helicase activity of Sgs1 by omitting the Dna2 nuclease from the reaction (Fig. 3d, Supplemental Fig. 3a online). First, we found that Sgs1, together with RPA, efficiently unwound the DNA of sub-saturated nucleosomal arrays (r=0.4). Furthermore, and similar to the complete resection reaction, Sgs1 helicase activity was inhibited on the fully saturated array (Fig. 3d). Sgs1 helicase activity was also inhibited on the 250 bp mononucleosome that contains only 50 bp of adjacent free DNA (Fig. 3b, top panel), but activity was restored by an adjacent 300 bp nucleosome-free region (Fig. 3b, c). Importantly, the requirement for a nucleosome-free region adjacent to the DSB is shared by human BLM, the orthologue of Sgs1, although BLM was more sensitive to nucleosomes on sub-saturated arrays (Fig. 3d, e). These data are consistent with Dna2 functioning as a nuclease in these resection reactions; indeed, the ATPase- and helicase-defective variant, (dna2-K1080E) that has previously been shown to resect DNA with Sgs1, also efficiently substituted for Dna2 in the chromatin resection reactions (Supplemental Fig. 3b online)7,13. These results also indicate that the helicase activity of Sgs1 is inhibited when nucleosomes are located adjacent to a DSB, and they suggest that this reaction requires chromatin remodeling events that generate a short nucleosome-free region.
Exo1 is stimulated by removal of H2A–H2B dimers
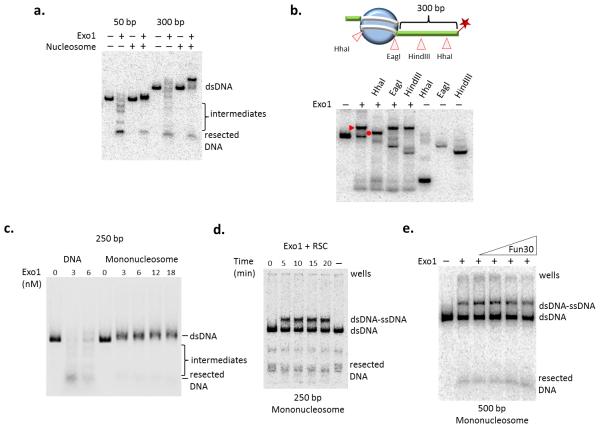
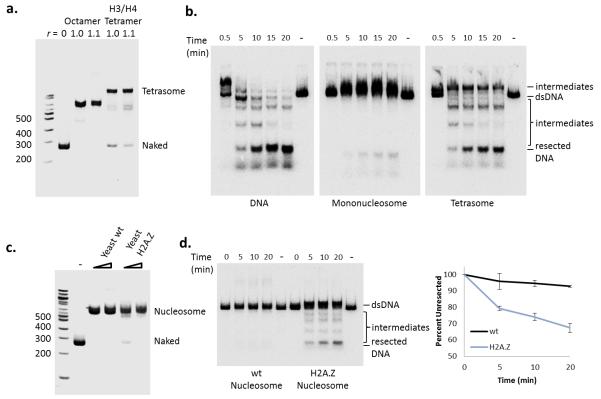
Next we further characterized how nucleosome assembly blocks Exo1 activity. As shown above, Exo1 activity was blocked when only a few nucleosomes were present on a long DNA fragment (Fig. 1c). Consistent with this, resection by Exo1 was also blocked on a mononucleosome regardless of the length of adjacent free DNA (Fig. 4a). Interestingly, on the longer mononucleosome template, the Exo1 reaction produced a slowly migrating DNA species. Digestion with several restriction enzymes demonstrated that this product is a hybrid ssDNA-dsDNA molecule resulting from Exo1 processing of the free DNA end, with the resection reaction terminating at the edge of the nucleosome (Fig. 4b). The nuclease activity of Exo1 could not substitute for Dna2 in the Sgs1 chromatin resection reaction, indicating a separate means of navigating chromatin barriers for Exo1 (Supplemental Fig. 4a online). Addition of the MRX complex, Sae2, and/or RPA to the Exo1 reaction did not stimulate nucleosomal resection (Supplemental Fig. 4b online), nor did increased Exo1 concentrations (Fig. 4c). Likewise, addition of either the RSC or Fun30 chromatin remodeling enzyme was unable to relieve the nucleosomal block (Fig. 4d, e). RSC was also unable to stimulate the activity of the Sgs1–Dna2-dependent reaction (Supplemental Fig. 4c online). In reactions with a 250 bp mononucleosome, RSC appeared to catalyze sliding of the nucleosome to one or both DNA ends. Nucleosome sliding allowed Exo1 to process the resulting free DNA end, but it remained blocked by the nucleosome, generating a dsDNA/ssDNA hybrid product (Fig. 4d). In contrast, Exo1 activity was dramatically enhanced on a substrate reconstituted with only an H3–H4 tetramer, indicating that the H2A–H2B dimers are largely responsible for nucleosomal inhibition of Exo1 activity (Fig. 5a, b).
Figure 4.
Nucleosomes act as a barrier to Exo1 DNA resection. (a) Resection assay with increasing concentration of Exo1 on 250 bp DNA or mononucleosome (50 bp end) after 20 minutes at 30° C. (b) Exo1 resection assay using naked DNA or mononucleosome containing either 50- or 300-bp of nucleosome-adjacent free DNA after 20 minutes at 30°C. (c) Top: schematic of restriction enzyme site locations on the 500 bp mononucleosome in relation to nucleosome positioning. Star indicates location of the 3′-radiolabel. Bottom: restriction enzyme (RE) digests following Exo1 resection and Proteinase K treatment to map ssDNA formation on the 601-500 mononucleosome. Exo1 resection intermediate and the RE cleaved resection intermediate are indicated by a closed triangle and closed circle respectively. (d) 250 bp mononucleosome incubated with Exo1 (6 nM) and RSC (1 nM). The lane labeled (-) received no Exo1 or RSC. Note that the appearance of the slowly migrating species following RSC treatment is likely due to sliding of the additional free DNA to form a ssDNA-dsDNA intermediate similar to Exo1 product in (b). (e) 500 bp mononucleosome incubated with Exo1 (6 nM) with increasing amounts of Fun30 (0.025 nM to 0.5 nM) after 20 minutes at 30°C.
Figure 5.
Loss of H2A-H2B dimers and incorporation of H2A.Z dimers promotes chromatin resection by Exo1. (a) Native PAGE analysis of chromatin reconstitutions with Xenopus histone octamers and H3/H4 tetramers on a 250 bp DNA template. (b) Time-course of Exo1 resection analysis on naked DNA, mononucleosomes, and tetrasomes. (c) Reconstitution of yeast mononucleosomes with either wild type (wt) H2A or H2A.Z-containing octamers. (d) Left: time-course of Exo1 resection analysis using either wild type (H2A) or H2A.Z-containing mononucleosomes. Right: Quantification of dsDNA signal after Exo1 resection provided as mean values +/− SEM (n=3).
H2A.Z incorporation enhances Exo1 chromatin resection
Previous studies have demonstrated that the histone variant, H2A.Z, is incorporated into chromatin adjacent to a DSB, and that the level of H2A.Z is regulated by both the Swr1 and INO80 chromatin remodeling enzymes21,24. DSB resection is also slower when the gene (HTZ1) encoding H2A.Z is deleted21, though the results of several genetic studies raise the question of whether the phenotypes of an htz1 mutant faithfully report on direct roles for H2A.Z 22,25,26. To determine if H2A.Z impacts resection in vitro, we reconstituted yeast mononucleosomes that contain either H2A or H2A.Z (Fig. 5c). Whereas Exo1 activity was efficiently blocked by the canonical yeast nucleosome, incorporation of H2A.Z led to a stimulation of Exo1 activity, with nearly 30% resection in the 20 minute time course (Fig. 5d). In contrast, H2A.Z incorporation did not markedly stimulate the Sgs1-Dna2 resection or the Sgs1 helicase reactions using a 250 bp mononucleosome substrate (Supplemental Fig. 4d online). Thus, these results suggest that H2A.Z specifically stimulates the Exo1 resection pathway. Yeast nucleosomes that harbor H2A.Z are more salt labile in vivo27 and in vitro28, suggesting a model in which the lower stability of the H2A.Z/H2B-H3/H4 interface allows Exo1 to specifically invade an H2A.Z nucleosomal substrate.
Swr1 facilitates Exo1 processing in vivo
In vivo studies have demonstrated that inactivation of either the Sgs1 helicase or the Exo1 nuclease has only minor effects on DSB resection kinetics, but removal of both enzymes eliminates long-range DSB processing4-6. To test whether H2A.Z specifically stimulates the Exo1 resection pathway in vivo, we created an isogenic set of yeast strains that harbor a galactose-inducible HO endonuclease and EXO1, SGS1, HTZ1, or SWR1 gene deletions. SWR1 encodes the catalytic subunit of the SWR-C ATP-dependent chromatin remodeling complex that is responsible for the deposition of H2A.Z into chromatin, and inactivation of Swr1 eliminates H2A.Z incorporation in vivo21,24,27,29. Note that use of a swr1 deletion eliminates the complex genetic interactions that appear to result from deletion of HTZ125,26.
As an initial test for whether Swr1 might function together with Exo1 in a DNA damage pathway, we monitored sensitivity to exposure to zeocin, which induces DNA double strand breaks, and to UV damage. Isogenic strains were grown to mid-log phase in rich media, and serial dilutions were spotted on solid media containing zeocin or exposed to UV. As shown in Supplemental Fig. 5a, the sgs1, exo1, and swr1 single mutants show mild sensitivity to UV, and the exo1 sgs1 double mutant shows the expected enhanced sensitivity. Interestingly, the swr1 sgs1 double mutant also showed increased sensitivity to UV compared to either single mutant, whereas the swr1 exo1 double mutant had UV sensitivity similar to the exo1 single mutant. Likewise, the zeocin sensitivity of the sgs1 swr1 double mutant was quite similar to that of the sgs1 exo1 double mutant, whereas the exo1 swr1 double mutant was only slightly more sensitive than each single mutant (Supplemental Fig. 5b online). These data are consistent with SWR1 and SGS1 functioning in different pathways that support UV and zeocin resistance, and they are consistent with our in vitro data indicating that Swr1 facilitates Exo1 function.
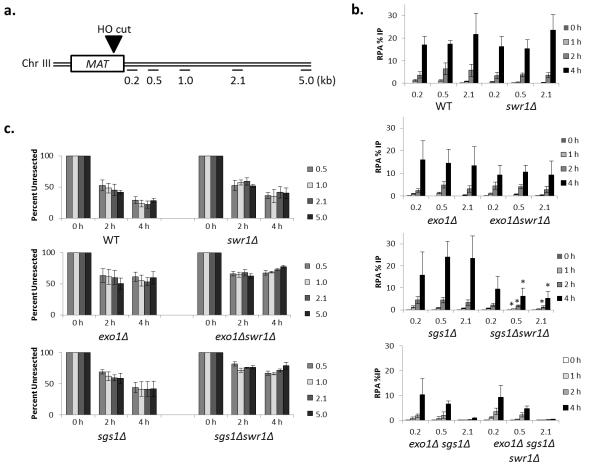
To more directly assess the role of H2A.Z in DSB resection, an unrepairable DSB was induced by galactose induction of HO (Fig. 6a, Supplemental Fig. 6 online), and the kinetics of DSB resection were monitored with two independent assays: (1) by following recruitment of the single-stranded DNA binding protein RPA by chromatin immunoprecipitation (ChIP); and (2) by monitoring genomic DNA levels adjacent to the HO cut site by qPCR. Consistent with previous studies4-6, DSB resection rates monitored by either assay were similar to wildtype levels in exo1 or sgs1 single mutants, whereas long-range resection was abolished in the sgs1 exo1 double mutant (Fig. 6b, c). Also consistent with previous studies 21, DSB resection in an htz1Δ strain was less effective, as measured by the qPCR assay (Supplemental Fig. 7a online). Interestingly, the sgs1Δ htz1Δ double mutant exhibited a more severe resection defect than either single mutant, and this additive effect was less apparent with the exo1Δ htz1Δ double mutant (Supplemental Fig. 7a online). We extended these findings with a more extensive analysis of a swr1Δ mutant which not only eliminates H2A.Z deposition in vivo, but it also disrupts the SWR-C complex which appears to cause aberrant genetic phenotypes in the absence of H2A.Z22,25,26. Consistent with a recent report, the swr1Δ mutant did not display a significant defect in resection22. Likewise, the swr1 exo1 double mutant had resection rates similar to the exo1 single mutant (Fig. 6b, c). These results are consistent with our in vitro data showing that H2A.Z incorporation does not dramatically impact Sgs1/Dna2-dependent resection. In contrast, the swr1 sgs1 double mutant exhibited a resection defect that was more severe than the sgs1 single mutant, indicative of a synergistic resection defect (Fig. 6b, c). Furthermore, loss of Swr1 did not further diminish resection in the sgs1Δ exo1Δ double mutant (Fig. 6b). Notable, all strains showed similar cell cycle profiles by FACS analysis prior to DSB formation, indicating differences in resection rates are not due to DNA repair choice being influenced by cell cycle (Supplemental Fig. 6b online). To further exclude this possibility, we also monitored DSB resection in cells synchronized in G2/M with nocodazole (Supplemental Fig. 7b online). Similar to data obtained with asynchronous cell populations, the sgs1 swr1 double mutant showed a larger defect in RPA recruitment in synchronized cells compared to the sgs1 single mutant, whereas the exo1 swr1 double mutant was equivalent to the exo1 mutant. These data are consistent with SWR1 and EXO1 functioning in the same genetic pathway for DSB resection.
Figure 6.
Swr1 stimulates DSB repair through the Exo1 pathway. (a) Schematic of MAT locus with sites of HO cut and primer location indicated. (b) ChIP analysis of RPA occupancy at indicated distances and times in indicated yeast strains. RPA levels were measured at increasing distances from the DSB (0.2, 0.5, and 2.1 kb) and normalized to the percentage of DSB in each strain (WT = wild type, JKM139). Graph values reflect the percent of precipitated DNA relative to the input at each region +/− SEM (n=3). Bars with * indicate P value < 0.02 compared to sgs1 single mutant.(c) DNA levels at indicated distances to the right of a galactose-inducible DSB were monitored by quantitative real-time PCR. DNA levels at indicated times were normalized to actin and the 0 hour time point signals were set at 100% with subsequent hours signal indicated as percent with +/− SEM (n=3).
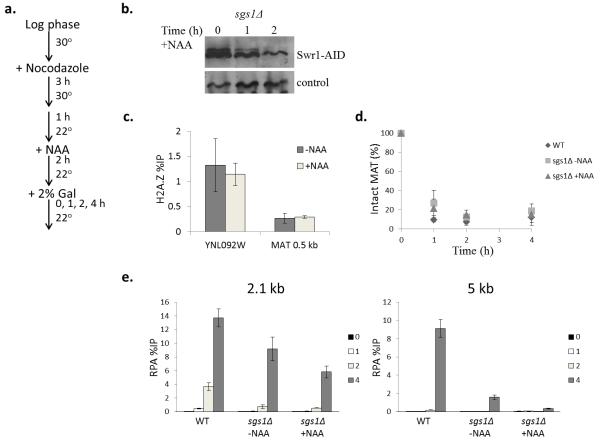
In addition to the dynamic incorporation of H2A.Z at DSBs, the SWR-C remodeling enzyme also deposits H2A.Z within nucleosomes that flank promoters of genes transcribed by RNA polymerase II, as well as nucleosomes that flank chromatin boundary elements, centromeres, and replication origins. To investigate whether the dynamic incorporation of H2A.Z is required for the Exo1-dependent resection pathway, we employed an auxin-based degron system to induce the degradation of Swr1 in synchronized sgs1Δ cells just prior to DSB formation30. A yeast strain was constructed in which the Arabidopsis thaliana TIR1 gene is expressed from the constitutive ADH1 promoter and an auxin-inducible degron (AID) cassette is fused in frame to the C-terminus of Swr1. AtTir1 can form a complex with yeast Skp1, and the resulting SCF-TIR1 complex targets proteins harboring an AID domain for ubiquitin-dependent degradation. WT and sgs1Δ SWR1-AID cells were arrested with nocodazole, incubated for two hours at 22°C with 1% ethanol carrier or the synthetic auxin, NAA, and then galactose was added to induce a DSB at the MAT locus (Fig. 7). Importantly, cells remained efficiently arrested in G2/M, as monitored by the persistence of large budded cells. Samples were processed for ChIP to monitor RPA levels at the DSB, as a measure of resection, and for western blot to monitor Swr1 levels. In this strain background, two hours of auxin treatment reduced Swr1 levels to ~15% of normal levels (Fig. 7b). Notably, depletion of Swr1 in synchronized cells did not decrease the levels of H2A.Z at a promoter proximal region or at the MAT locus (Fig. 7c), indicating that the H2A.Z that was deposited prior to DSB formation is not depleted by this experimental regimen. Strikingly, Swr1 depletion caused an additional resection defect in the sgs1Δ mutant, and this defect was quite pronounced at 5 kb distal from the DSB (+NAA columns, Fig. 7d). These data are fully consistent with a role for Swr1 (and H2A.Z) in facilitating the Exo1-dependent resection pathway, and they indicate that dynamic incorporation of H2A.Z plays a key role.
Figure 7.
Depletion of Swr1 inhibits Exo1 resection. (a) Schematic of yeast culture growth and treatments. (b) Western blotting of Swr1 depletion during addition of 1 mM 1-naphthaleneacetic acid (NAA), a synthetic auxin hormone. (c) ChIP analysis of H2A.Z occupancy after 2 hrs. of auxin treatment at the YNL092W promoter and +0.5 kb at MAT HO cut site before Gal induction. Error bars represent +/− SEM (n=3). (d) DSB induction kinetics after auxin treatment. Error bars represent +/− SEM (n=3). (e) ChIP analysis of RPA occupancy in the indicated yeast strains (WT = wild type, CY2049) arrested in G2/M with nocodazole. RPA levels were measured at increasing distances from the DSB (2.1 and 5 kb) and normalized to the percentage of DSB in each strain. Graph values reflect the percent of precipitated DNA relative to the input at each region +/− SEM (n=3).
Discussion
Our studies have demonstrated that nucleosomes present a block to DNA processing enzymes, though the inhibition is more severe for Exo1. Resection by the Sgs1–Dna2 machinery remains efficient when chromatin fibers are sub-saturated with nucleosomes, and our data indicate that initiation of resection by this pathway may simply require a single nucleosome-free gap next to the DSB. This further suggests that once resection has been initiated, extensive processing by the Sgs1–Dna2 machinery may not require additional chromatin remodeling events. This conclusion is consistent with previous in vivo studies that demonstrate a key role for the RSC remodeling enzyme in the removal or sliding of a single nucleosome next to an HO-induced DSB31. Although we do not see stimulation by RSC in vitro, ATP-dependent sliding of nucleosomes, not eviction, predominates in these in vitro reactions. How does this nucleosome-free gap stimulate Sgs1–Dna2? This requirement does not appear to reflect a need to load multiple helicase molecules, as the concentration of Sgs1 required for unwinding the 500 bp nucleosome is quite similar to that required for naked DNA (Fig. 3c, d). We favor a model in which the Sgs1-dependent unwinding of free DNA leads to superhelical torsion that disrupts the adjacent nucleosome. Interestingly, the potent activity of Sgs1–Dna2 on sub-saturated chromatin fibers is similar to the activity of the bacterial recombination enzyme, RecBCD, which contains a Sgs1-related helicase that is also able to induce histone sliding and eviction on sub-saturated chromatin templates in vitro32,33.
In contrast to the Sgs1–Dna2 machinery, the Exo1 nuclease cannot overcome nucleosomal barriers, even when a nucleosome is bounded by large tracts of free DNA. Previous reports indicate that the BLM helicase can enhance the DNA resection activity of human Exo134, but we find that Sgs1 helicase does not aid chromatin resection by yeast Exo1 (Supplemental Fig. 4a online). We find that removal of H2A–H2B dimers dramatically enhances Exo1 activity, suggesting that an ATP-dependent chromatin remodeling enzyme that can remove histone dimers may regulate Exo1-mediated resection in vivo. We postulate that ATP-dependent dimer eviction may also aid the initiation of processing by the Sgs1–Dna2 pathway, as loss of dimers will release additional free DNA, thus increasing Sgs1 helicase activity.
Interestingly, Exo1 also plays a key role in the excision step of DNA mismatch repair35,36, and components of the MMR machinery have been associated with replication centers in vivo37,38. Our results suggest that H3–H4 deposition on newly synthesized DNA at a replication fork would not preclude Exo1 activity during DNA MMR. Furthermore, these data suggest that there may be a “window of opportunity” for efficient completion of MMR within nascent chromatin before nucleosomes have been fully matured by addition of H2A–H2B dimers.
A recent study evaluated the role of several yeast ATP-dependent chromatin remodeling enzymes in DSB resection22. This work identified the Fun30 remodeling enzyme as a positive regulator of both the Exo1- and Sgs1–Dna2-dependent processing pathways. Interestingly, the requirement for Fun30 was completely alleviated by removal of the Rad9 checkpoint factor, also known as an inhibitor of resection at DSBs and telomeres. Thus, it appears that the key role for Fun30 during DSB resection is not to disrupt nucleosomes per se, but to antagonize the resection inhibitor, Rad9. These data are consistent with our biochemical results where we find that Fun30 does not stimulate Exo1-dependent chromatin resection in vitro.
Previous studies have suggested dynamic incorporation of the H2A.Z variant within DSB chromatin21,24, and our study implicates such dynamics as a key regulator of Exo1 activity. Our finding that DSB resection is reduced in a swr1 sgs1 strain or when Swr1 is conditionally depleted in a sgs1 mutant is fully consistent with our biochemical studies that link H2A.Z and Exo1, and they implicate a key role for Swr1-dependent H2A.Z incorporation in enhancing Exo1-dependent DSB processing during recombinational DSB repair. We note that recent work in mammalian cells has also implicated H2A.Z in the DSB processing steps of HR39. Intriguingly, H2A.Z appears to inhibit resection at DSBs in mammalian cells, apparently by promoting the recruitment of the nonhomologous endjoining (NHEJ) machinery. Whether H2A.Z also promotes resection in the absence of NHEJ in mammalian cells is not yet clear. Our results may explain in part why dysregulation of human H2A.Z is linked to cancer, and depletion of H2A.Z compromises the stability of the human genome40.
Online Materials and Methods
Protein Purification
Resection proteins (Mre11-Rad50-Xrs2, Dna2, yeast and human RPA, Sgs1, Top3-Rmi1, BLM) were expressed in insect, yeast, or E. coli cells and purified as previously described13,41,42. For Exo1 purification, a DNA fragment encoding Exo1 with a C-terminal FLAG tag (plasmid kindly provided by Michael Liskay) was inserted into pFast-Bac1 vector (Invitrogen). A bacmid was generated in the E. coli strain DH10Bac (Invitrogen) and a recombinant baculovirus was made for expressing the tagged Exo1 in insect cells. All the purification steps were carried out at 0-4°C. The insect cell pellet (~15 g from 1 L culture) was resuspended in 100 ml of K buffer (20 mM KH2PO4, pH 7.4, 10% glycerol, 0.5 mM EDTA, 0.01% Igepal, 1 mM DTT) ) containing a cocktail of protease inhibitors (aprotinin, chymostatin, leupeptin, and pepstatin A at 5 μg/ml each, and also 1 mM phenyl-methylsulfonyl fluoride) and 150 mM KCl. Cells were disrupted by sonication for 30 seconds, and the lysate was clarified by ultracentrifugation (100,000Xg for 45 min) and then loaded onto a 10 ml SP Sepharose column. After washing the column with the same buffer, it was developed with 50 ml gradient from 150 to 500 mM KCl. The peak fractions were pooled and then incubated with 0.5 ml of anti-FLAG-M2 resin for 2 h. The matrix was washed once with 20 ml K buffer containing 500 mM KCl and 2 mM each of ATP and MgCl2, then washed three additional times with 10 ml of K buffer containing 500 mM KCl. Exo1 was eluted by incubating the matrix with 1 ml of K buffer containing 500 mM KCl and 200 μg/ml FLAG peptide (Sigma) for 1 h. The purified Exo1 protein (~ 30 μg) was frozen in liquid nitrogen and stored at −80°C in small portions. Xenopus and yeast histones were expressed in bacteria and purified according to standard protocols43. Briefly, histones are induced individually in BL21 (DE3) cells, isolated as inclusion bodies, purified by SP-HR ion exchange chromatography, and lyophilized to give pure recombinant histones. The ATP-dependent remodeling complexes, RSC and Fun30, were purified as previously described44. The complexes were quantified by ATPase activity and analyzed by SDS-PAGE and silver staining.
DNA Substrate Generation
DNA substrates were generated by plasmid digestion followed by size-exclusion chromatography (601-177-12) or by PCR (601-1; 250 bp, 500 bp, 1000 bp). The 601-177-12 positioning array was purified from plasmid (CP1088) by AlwNI, AhdI, Bsa I, Ssp I, NaeI and BamHI restriction enzyme digestion followed by size-exclusion chromatography (Sephacryl S-500 (GE)). The mononucleosome DNA templates were generated by PCR using the 601 pGem-3Z (CP1024) as the template. Biotinylation of the 5′ end was incorporated by using biotinylated forward primers. PCR products were treated with EcoRI or BsaI to yield one ssDNA overhang. DNA templates were 3′-labeled on one end with [alph-32P] dATP by Klenow fill-in at room temperature and purified through Sephadex G-25 columns after phenol-chloroform extraction.
Chromatin Reconstitution
Histone octamers were prepared by denaturing each histone followed by dialysis into 2.0 M NaCl TE buffer. Histone octamers were subsequently purified by size-exclusion chromatography (GE Superdex 200) and quantified as previously described43. To assemble nucleosomal arrays, nucleosomes were reconstituted at increasing ratios (r) of Xenopus histone octamer per 177 bp donor DNA by salt step dialysis. Mononucleosomes were reconstituted to saturation with both Xenopus octamers by salt step dialysis and S. cerevisiae octamers by salt gradient dialysis on indicated DNA substrates. Nucleosome saturation levels were monitored by ScaI digestion and analysis on 4% native polyacrylamide gel electrophoresis in 0.5X TBE with ethidium bromide staining.
DNA Resection Assays
Assays were performed in either 10 μl or 25 μl reactions (40 mM Tris-HCl, pH 7.5, 2 mM ATP, 2 mM MgCl2, 50 mM KCl, 1 mM DTT, 100 μg/ml BSA, and ATP-regenerating system of 20 mM creatine phosphate and 20 μg/ml creatine kinase) with specified DNA or chromatin substrates (0.25 nM DNA) at 30°C for indicated times. For the Sgs1-Dna2 resection, each reaction contained 10 nM MRX complex, 10 nM Sgs1, 10 nM Top3-Rmi1 complex, 20 nM Dna2 and 100 nM RPA. For the Exo1 resection, reactions contained 6 nM Exo1 unless otherwise specified. The addition of 1 nM RSC was added to chromatin prior to resection enzymes and incubated 5 minutes at 30° C. Resection samples were deproteinized by SDS (1 %) and proteinase K (0.5 mg/ml) at 37°C for 10 min before analysis in a 1% agarose gel in 1X TAE or 4% polyacrylamide gel in 0.5X TBE. Gels were dried and analyzed by phosphorimaging (GE Storm 820) or x-ray film exposure.
DNA Helicase Assays
Sgs1 at indicated concentrations was incubated with yRPA (100 nM), and BLM (20 nM) with hRPA (100 nM) with the indicated DNA/chromatin substrates (0.25 nM) for 20 min at 30°C in 10 μl buffer (40 mM Tris-HCl, pH 7.5, 2 mM ATP, 2 mM MgCl2, 50 mM KCl, 1 mM DTT, 100 μg/ml BSA, and ATP-regenerating system of 20 mM creatine phosphate and 20 μg/ml creatine kinase). The reactions were deproteinized, and resolved in a 1% agarose gel in 1X TAE (NAs) or 0.5X TBE 4% native polyacrylamide gel (mononucleosomes) and exposed to phosphorimaging or x-ray film.
Resection Intermediate Mapping
To map the Exo1-histone block, Exo1-nucleosome resection products were restriction enzyme digested to determine Exo1 ssDNA production. A 500 bp mononucleosome (0.25 nM) was incubated with Exo1 as previously described. Reaction was deproteinized, phenol/chloroform extracted, ethanol precipitated, resuspended in TE, and divided into four separate samples. These divided samples were then incubated with the corresponding restriction enzyme and digestion buffer (New England BioLabs) at 37°C for 1 hour. Reactions were phenol-chloroform extracted, ethanol precipitated, resuspended in 10% glycerol, and resolved on a 4% native polyacrylamide electrophoresis. Gels were then dried and visualized by phosphorimaging.
ChIP Assay
Yeast cultures were grown in media containing 2% lactic acid to an optical density (OD)600 of 0.5-0.8. Cultures arrested in G2/M were grown to OD600 0.3-0.4 and then treated with nocodazole (3-5 hrs.) until >70% cells visually arrested (~ OD600 0.6-0.8). Expression of HO endonuclease was induced with the addition of 2% galactose with cells collected at indicated time points. DSB formation was monitored by QPCR with primers spanning the HO cut site and normalized to an internal control (actin). WT DSB was arbitrarily set to 100% at 0 time point (Supplemental Fig. 8 online). Formation of ssDNA was monitored by ChIP with a polyclonal RPA antibody (0.5 μl) (a gift from Valerie Borde) at sites 0.2 kb, 0.5 kb, 2.1, and 5.0 kb to the right of the HO-induced DSB. Cultures treated with auxin had H2A.Z occupancy monitored by ChIP (H2A.Z antibody, Active Motif). IP signals were normalized to percent DSB and calculated as percent of input chromatin24. Primer sequences are detailed in Supplemental Table 2.
Genomic DNA Purification
Yeast cultures were grown were grown and HO induced as described in the ChIP assay above. After HO induction, cells were collected at indicated times, pelleted and frozen at −80° C overnight. Genomic DNA was extracted by vortexing with glass beads and phenol. qPCR values were acquired from multiple sites adjacent to the induced DSB and normalized to Actin levels of each strain. DNA from 0 hour time point was set to 100% with subsequent times points measured relative to these samples signal.
Immunoblotting
Samples were collected from sgs1Δ Swr1-degron strains and extracted by TCA precipitation as previously described24. Extracted proteins were resolved on a 6% SDS-PAGE and transferred to nitrocellulose membrane (GE). Swr1-AID was detected with anti-AID (BioRois Co., Ltd.) and anti-RPA (Thermo Scientific) used for the loading control.
Yeast Strains
All strains used in this study were created in the JKM139 (hoΔ hml::ADE1 MATa, hmr::ADE1 ade1 leu2-3,112 lys5 trp1::hisG ura3-52 ade3::GAL10::HO) background. The wild type, and the sgs1, exo1, sgs1 exo1 deletion strains were provided by G. Ira’s lab4. The SWR1 gene was disrupted in these strains by replacing the coding region with NAT-MX6 (ref. 45). The Swr1 degron strain was created by inserting pMK76 containing the AtTIR1 gene at the URA3 locus and c-terminal tagging of Swr1 with IAA17 (AID) was as previously described30.
Affinity Pulldown Assays
To test the effect of short free DNA adjacent to a nucleosome on Sgs1 recruitment, MRX (200 ng), Sgs1 (80 ng), Dna2 (80 ng), and Top3-Rmi1 (40 ng) were incubated with streptavidin magnetic bead immobilized 250bp DNA and mononucleosomes (10 ng DNA) at room temperature for 20 min with agitation in 60 μl buffer (40 mM Tris-HCl, pH 7.5, 2 mM ATP, 2 mM MgCl2, 50 mM KCl, 1 mM DTT, 100 μg/ml Insulin). Beads were subsequently washed five times in 60 μl buffer. Proteins were eluted by 5 min. boiling in 20 μl SDS loading buffer, and proteins were resolved by 8% SDS polyacrylamide electrophoresis with visualization of bands by silver staining (Invitrogen).
UV and Zeocin Sensitivity Assays
Yeast cultures of indicated strains were plated in 10-fold serial dilutions on YPD plates. UV treated plates were subjected to 90 J/m2 ultraviolet in a UV Stratalinker 1800 (Stratagene). Zeocin treated plates contained 1 μg/ml zeocin (Invitrogen). Plates were then incubated at 30° C for 2-3 days.
Supplementary Material
Acknowledgements
This work was supported by grants from the US National Institutes of Health to N.L.A. (F32 GM096701), C.L.P. (RO1 GM54096), and P.S. (RO1 ES07061). We thank Y. Kwon (Yale University, New Haven, CT) for purified MRX, G Ira (Baylor University, Houston, TX) for yeast strains, V. Borde for RPA antibody (Institute Curie, Paris, France), C. Van (UMMS, Worcester, MA) for help with the degron experiments, and M. Liskay (The Ohio State University, Columbus, OH) for the Exo1 clone.
Footnotes
Author Contributions N.L.A. performed all experiments, P.S. and H.N. provided purified resection enzymes, and all authors were involved in data analysis and manuscript preparation.
References
- 1.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 2.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 3.Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 4.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budd ME, Choe WC, Campbell JL. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J Biol Chem. 1995;270:26766–26769. doi: 10.1074/jbc.270.45.26766. [DOI] [PubMed] [Google Scholar]
- 8.Szankazi P, Smith GR. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- 9.Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerecisiae. Mol Biol Cell. 2000;11:2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotta-Ramusino C, et al. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell. 2005;17:153–159. doi: 10.1016/j.molcel.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 11.Hackett JA, Greider CW. End resection initiates genomic instability in the absence of telomerase. Mol Cell Biol. 2003;23:8450–8461. doi: 10.1128/MCB.23.23.8450-8461.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertuch AA, Lundblad V. EXO1 contributes to telomere maintenance in both telomerase-proficient and telomerase-deficient Saccharomyces cerevisiae. Genetics. 2004;166:1651–1659. doi: 10.1534/genetics.166.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niu H, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szankasi P, Smith GR. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J Biol Chem. 1992;267:3014–3023. [PubMed] [Google Scholar]
- 15.Tran PT, Erdeniz N, Dudley S, Liskay RM. Characterization of nuclease-dependent functions of Exo1p in Saccharomyces cerevisiae. DNA Repair (Amst) 2002;1:895–912. doi: 10.1016/s1568-7864(02)00114-3. [DOI] [PubMed] [Google Scholar]
- 16.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat Rev Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 17.Chai B, Huang J, Cairns BR, Laurent BC. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005;19:1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shim EY, et al. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol Cell Biol. 2007;27:1602–1613. doi: 10.1128/MCB.01956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osley MA, Tsukuda T, Nickoloff JA. ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat Res. 2007;618:65–80. doi: 10.1016/j.mrfmmm.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsukuda T, et al. INO80-dependent chromatin remodeling regulates early and late stages of mitotic homologous recombination. DNA Repair (Amst) 2009;8:360–369. doi: 10.1016/j.dnarep.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 21.Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 22.Chen X, et al. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature. 2012;489:576–580. doi: 10.1038/nature11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costelloe T, et al. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature. 2012;489:581–584. doi: 10.1038/nature11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papamichos-Chronakis M, Krebs JE, Peterson CL. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 2006;20:2437–2449. doi: 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morillo-Huesca M, Clemente-Ruiz M, Andújar E, Prado F. The SWR1 histone replacement complex causes genetic instability and genome-wide transcription misregulation in the absence of H2A.Z. PLoS One. 2010;5:12143. doi: 10.1371/journal.pone.0012143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halley JE, Kaplan T, Wang AY, Kobor MS, Rine J. Roles for H2A.Z and its acetylation in GAL1 transcription and gene induction, but not GAL1-transcriptional memory. PLoS Biol. 2010;8:1000401. doi: 10.1371/journal.pbio.1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe S, Radman-Livaja M, Rando O, Peterson CL. A histone acetylation switch regulates H2A.Z deposition by the SWR-C remodeling enzyme. Science. 2013 Apr 12th; doi: 10.1126/science.1229758. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizuguchi G, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 30.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nature Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 31.Shim EY, et al. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol Cell Biol. 2007;27:1602–1613. doi: 10.1128/MCB.01956-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eggleston AK, O’Neill TE, Bradbury EM, Kowalczykowski SC. Unwinding of nucleosomal DNA by a DNA helicase. J Biol Chem. 1995;270:2024–2031. doi: 10.1074/jbc.270.5.2024. [DOI] [PubMed] [Google Scholar]
- 33.Finkelstein IJ, Visnapuu ML, Greene EC. Single-molecule imaging reveals mechanisms of protein disruption by a DNA translocase. Nature. 2010;468:983–987. doi: 10.1038/nature09561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nimonkar AV, et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei K, et al. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17:603–614. doi: 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giannattasio M, et al. Exo1 competes with repair synthesis, converts NER intermediates to long ssDNA gaps, and promotes checkpoint activation. Mol Cell. 2010;40:50–62. doi: 10.1016/j.molcel.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 37.Hombauer H, Campbell CS, Smith CE, Desai A, Kolodner RD. Visualization of eukaryotic DNA mismatch repair reveals distinct recognition and repair intermediates. Cell. 2011;147:1040–1053. doi: 10.1016/j.cell.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hombauer H, Srivatsan A, Putnam CD, Kolodner RD. Mismatch repair, but not heteroduplex rejection, is temporally coupled to DNA replication. Science. 2011;334:1713–1716. doi: 10.1126/science.1210770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Yiduo H, Price BD. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol Cell. 2012;48:723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rangasamy D. Histone variant H2A. Z can serve as a new target for breast cancer therapy. Curr Med Chem. 2010;17:3155–3161. doi: 10.2174/092986710792231941. [DOI] [PubMed] [Google Scholar]
Methods-only reference
- 41.Sigurdsson S, Trujillo K, Song B, Stratton S, Sung P. Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J Biol Chem. 2001;276:8798–8806. doi: 10.1074/jbc.M010011200. [DOI] [PubMed] [Google Scholar]
- 42.Raynard S, Bussen W, Sung PA. A double Holliday junction dissolvasome comprising BLM, topoisomerase III alpha, and BLAP75. J Biol Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- 43.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 44.Smith CL, Horowitz-Scherer R, Flanagan JF, Woodcock CL, Peterson CL. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat Struct Biol. 2003;10:141–145. doi: 10.1038/nsb888. [DOI] [PubMed] [Google Scholar]
- 45.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.