Abstract
Patients with cystic fibrosis, caused by mutations in CFTR, exhibit specific and consistent alterations in the levels of particular unsaturated fatty acids compared with healthy controls. Evidence suggests that these changes may play a role in the pathogenesis of this disease. Among these abnormalities are increases in the levels of n-7 and n-9 fatty acids, particularly palmitoleate (16:1n-7), oleate (18:1n-9), and eicosatrienoate or mead acid (20:3n-9). The underlying mechanisms of these particular changes are unknown, but similar changes in the n-3 and n-6 fatty acid families have been correlated with increased expression of fatty acid metabolic enzymes. This study demonstrated that cystic fibrosis cells in culture exhibit increased metabolism along the metabolic pathways leading to 16:1n-7, 18:1n-9, and 20:3n-9 compared with wild-type cells. Furthermore, these changes are accompanied by increased expression of the enzymes that produce these fatty acids, namely Δ5, Δ6, and Δ9 desaturases and elongases 5 and 6. Taken together, these findings suggest that fatty acid abnormalities of the n-7 and n-9 series in cystic fibrosis are as a result, at least in part, of increased expression and activity of these metabolic enzymes in CFTR-mutated cells.
Keywords: Fatty acid metabolism, Gene expression, Elongases, Desaturases, Monounsaturated fatty acids, Cystic fibrosis
Introduction
Cystic fibrosis (CF) is the most common life-threatening genetic disease in the Caucasian population, affecting 1 in every 3,000 newborns [1]. CF is caused by mutations in the cystic fibrosis transmembrane regulator (CFTR) gene [2]. The protein product of this gene is a cAMP-regulated chloride channel that belongs to the ATP binding cassette family. This protein also transports bicarbonate and indirectly regulates sodium flux. The mutations in CFTR cause accumulation of viscous secretions in the body, ultimately leading to pancreatic insufficiency, intestinal malabsorption, chronic airway infection, inflammation, and progressive lung disease [1].
In addition to the classic clinical findings in CF, specific fatty acid abnormalities have been consistently identified in the plasma and tissues of CF animal models, human patients, and cultured cell models compared with wild-type controls (reviewed in Refs. [3, 4]). The most noted of these are decreases in linoleate (18:2n-6) and docosahexaenoate (22:6n-3), and, in most cases, elevation of arachidonate (20:4n-6). Studies in mice suggest that these abnormalities are involved in the pathophysiology of CF [5].
Less attention has been paid to important fatty acid changes outside of the n-6 and n-3 pathway. These include increased concentrations of palmitoleate (16:1n-7), oleate (18:1n-9), and eicosatrienoate or mead acid (20:3n-9) in the cells and plasma of CF patients. In some studies, these changes were present along with pancreatic insufficiency and intestinal malabsorption [6, 7], and elevations of these fatty acids are a known marker of essential fatty acid deficiency [8, 9]. However, similar observations have been made in well-nourished CF patients [10-12] and some cultured CF cells [13], suggesting additional underlying causes exist to explain these alterations. Importantly, DHA therapy, which is known to reverse the n-3 and n-6 fatty acid abnormalities in CF patients and correct CF pathology in mice, also reduces 20:3n-9 levels [14].
These fatty acids are of particular interest in CF pathophysiology because of their functional properties outside cell membranes. A recent study revealed that 16:1n-7 acts as a lipid hormone, a so-called lipokine, providing communication between adipose tissue and distant organs to regulate systemic metabolic homeostasis [15]. Furthermore, there is evidence suggesting that products of Δ9-desaturase, which include 16:1n-7 and 18:1n-9, are involved in modulating inflammation [16-18], as are oxygenated products of 20:3n-9 [19].
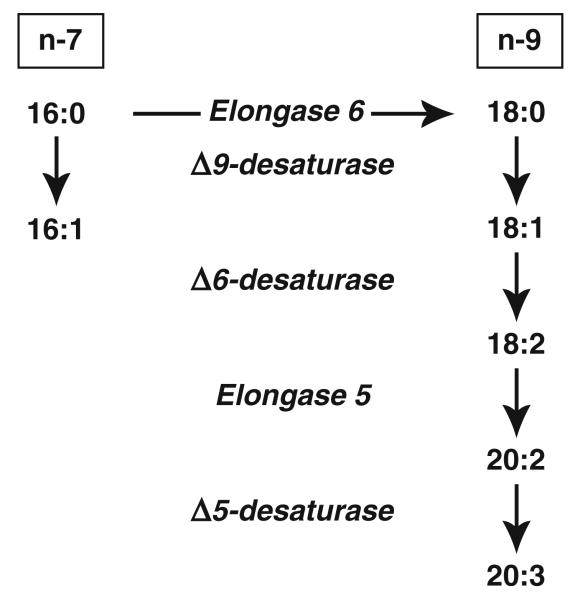
Unlike the n-3 and n-6 series fatty acids, 16:1n-7, 18:1n-9, and 20:3n-9 can be synthesized de novo. Palmitate (16:0; PAM), synthesized from acetyl CoA, can be converted to these fatty acids via the pathways indicated in Fig. 1. Our group has shown that alterations in n-3 and n-6 fatty acids in CF cells are associated with increased expression and activity of Δ6 and Δ5 desaturases compared with wild-type cells [20]. The objective if this study was to test whether the above-described changes in n-7 and n-9 fatty acids are also a result of altered activity of fatty acid metabolizing enzymes.
Fig. 1.
Fatty acid metabolism through the n-7 (left) and n-9 (right) pathways
This hypothesis was tested in two cell culture models of CF with similar fatty acid abnormalities to those present in humans [13, 21]. To investigate the relative activities of fatty acid-metabolizing enzymes, the conversion of radiolabeled palmitate (16:0), stearate (18:0), and 18:1n-9 to downstream fatty acid products was measured and correlated with the expression of desaturase and elongase enzymes in CF cells. The results show increased metabolism of these fatty acids to 16:1n-7, 18:1n-9, and 20:3n-9 in CF compared with wild-type cells. There was also increased expression of Δ9, Δ6, and Δ5 desaturases and elongase 6. These findings suggest that increases in the expression and activity of these fatty acid metabolic enzymes are at least in part responsible for changes in the levels of these fatty acids observed in CF.
Materials and Methods
Materials
Radiolabeled fatty acids, [1-14C]16:0 (55 mCi/mmol), [1-14C]18:0 (55 mCi/mmol), and [1-14C]18:1n-9 (55 mCi/mmol) were obtained from American Radiolabeled Chemicals (St Louis, MO, USA). Fatty acid methyl ester (FAME) standards were purchased from NuChek Prep (Elysian, MN, USA). HPLC-grade solvents were purchased from Fisher Scientific (Pittsburgh, PA, USA) and liquid scintillation cocktail (IN-flow 2:1) was purchased from IN/US Systems (Tampa, FL, USA).
Cell Culture
16HBEo− sense and antisense cells were a gift from Dr Pamela Davis (Case Western Reserve University School of Medicine, Cleveland, OH, USA). These are human bronchial epithelial cells stably transfected with plasmids expressing the first 131 nucleotides of human CFTR in the sense or antisense orientation such that sense cells express wild-type CFTR and antisense cells lack CFTR expression [22]. IB3 and C38 cells were obtained from ATCC (Manassas, VA, USA). IB3 cells are compound heterozygotes which contain one DF508 allele and one nonsense mutation, W1282X, with a premature termination signal [23]. The CF phenotype present in the IB3-1 cells has been corrected in the C38 cell line by transfection with wild-type CFTR [24]. Cell culture techniques were performed as previously described [13, 21]. Cells were grown in tissue-culture flasks coated with LHC basal media (Invitrosgen, Carlsbad, CA, USA) containing 0.1 mg/mL BSA (Sigma–Aldrich, St Louis, MO, USA), 10 μg/mL human fibronectin (Sigma–Aldrich), and 3 μg/mL vitrogen (Angiotech Biomaterials, Palo Alto, CA, USA). Complete culture medium comprised minimum essential medium? glutamax (Invitrogen) supplemented with 100 μg/mL streptomycin, 100 U/mL penicillin, and 10% horse serum (Atlanta Biologicals, Lawrenceville, GA, USA). Cells were grown at 37 °C in a 5% CO2 humidified incubator. Medium was changed three times weekly.
For experiments, cells were seeded in six-well experimental plates. Sense cells were seeded at 3 × 105 cells/well and antisense cells were seeded at 1 × 105 cells/well. This enables the sense cells, which are smaller in size, to reach confluence at approximately the same time as antisense cells. C38 and IB3 cells were seeded at 1 × 105 cells/well. All cells were allowed to grow until two days post-confluence (7–8 days) before being used for experiments. This time point was chosen because it is the point at which the fatty acid abnormalities are most distinct [13].
Fatty Acid Composition Analysis
Sense and antisense cells were cultured as above until two days post-confluence, after which they were washed twice in ice-cold PBS, scraped, and transferred to a glass tube. The saturated fatty acid 17:0 (10 μg) was added as an internal standard. Lipids were extracted using a modification of the method of Folch et al. [25]. Cells were harvested by rinsing twice with ice-cold phosphate-buffered saline (Invitrogen), then scraping on ice with a rubber policeman. The cells were pelleted by centrifugation (100×g for 8 min). The cell pellet was resuspended in 0.5 mL cold phosphate-buffered saline, and lipids were extracted by adding six volumes of chloroform–methanol (2:1, v/v). These samples were incubated on ice for 10 min, vortex mixed, and centrifuged (1,100×g for 10 min). The lower, organic, phase was transferred to a new glass tube and dried down completely under a stream of nitrogen. Fatty acids were methylated using boron trifluoride (BF3; 14% in methanol; Sigma–Aldrich) and a methanolic-base reagent [26] as follows: 0.5 mL 0.5 M methanolic NaOH (Acros Organics, Geel, Belgium) was added to the sample, vortex mixed, and heated at 100 °C for 3 min, followed by addition of 0.5 mL BF3 at 100 °C for 1 min. To extract the fatty-acid methyl esters (FAMEs), 1 mL hexane was added to the mixture which was then incubated at 100 °C for 1 min, followed by addition of 6.5 mL saturated NaCl solution. The sample was then vortex mixed and centrifuged (500×g for 4 min) to separate the liquid phases. The upper, hexane, layer was used for quantification of FAMEs by gas chromatography (GC) using an Agilent 7980A GC system (Agilent Technologies, Santa Clara, CA, USA) equipped with a Supelcowax SP-10 capillary column (Supelco, Bellefonte, PA, USA) coupled to a mass spectrometer (model 5975c, Agilent Technologies). FAME mass was determined by comparing areas of unknown FAMEs with that of a fixed concentration of the 17:0 internal standard. Results were expressed as the molar percentage (mol%) of each FAME relative to the total FAME mass of the sample.
Fatty Acid Metabolism Experiments
For fatty acid metabolism experiments, medium containing radiolabeled fatty acids was prepared by drying the fatty acids dissolved in ethanol under a continuous stream of nitrogen gas. Reduced-lipid medium was added to the tube and sonicated three times for 5 s each. At two days post-confluence, each well was incubated for 4 h with media supplemented with reduced-lipid fetal bovine serum (Hyclone, Logan, UT, USA) and containing 0.5 μCi/well radiolabeled 16:0, 18:0, or 18:1n-9 and either harvested (4 h samples) or washed with PBS and incubated in complete medium for an additional 20 h before harvest (24 h samples). Lipids were then extracted and methylated as above. After methylation, the samples were dried under nitrogen, reconstituted in 200 μL methanol, then dried to a volume of approximately 50 μL. The mixture was vortex mixed, and 20 μL was injected for HPLC analysis (Agilent Technologies 1200 series instrument) on a 4.6 × 250 mm, 5 μm, Agilent Zorbax Eclipse XDB-C18 column. A 4.6 × 12.5 mm, 5 μm, guard column was used in conjunction with the analytical column. The fatty acids were separated using a binary mobile phase with a constant flow rate of 1 mL per minute. The solvent program began with 90% solvent A (100% HPLC-grade methanol) and 10% solvent B (HPLC-grade H2O) for 40 min, followed by 100% solvent A for 20 min. Peaks were detected using ultraviolet detection at 205 nm and identified by comparing retention times with those of unlabeled FAME standards. Quantification of the radiolabeled peaks was performed by use of a scintillation detector (β-RAM Model 4, IN/US Systems) coupled to the HPLC. Data are reported as percentage of total counts.
Quantitative Real-Time PCR
Specific primers for each gene of interest were designed using Beacon Designer software (Premier Biosoft International, Palo Alto, CA, USA), the sequences of which are listed in Table 1. To prevent amplification of genomic DNA, forward and reverse primers were designed from adjacent exons. Primer fidelity and efficiency were tested using an iCycler iQ system (Bio-Rad Laboratories, Hercules, CA, USA) using iQ5 software (Bio-Rad). Each primer pair produced a single product on melt curve analysis and had amplification efficiencies >94%.
Table 1.
Primer sequences used for quantitative real-time PCR
| Gene name |
Product | Sequence of forward and reverse primers (5′–3′) |
Genbank accession no. |
|---|---|---|---|
| RPLP0 | Ribosomal protein, large, P0 | ATGGCAGCATCTACAACCC GACAGACACTGGCAACATTG |
NM_001002 |
| SCD | Δ9- (stearoyl-CoA) desaturase | CCCAAGCCCCAAGGTTGAATATG CCCCAAAGCCAGGTGTAGAAC |
NM_005063 |
| ELOVL6 | Fatty acid elongase 6 | CAACGAGAATGAAGCCATC GCAGCATACAGAGCAGAA |
NM_024090 |
At two days post-confluence, total RNA was prepared from cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Residual DNA was removed by treatment with DNase I (DNA-free kit; Ambion, Austin, TX, USA). First-strand cDNA was synthesized from 2 μg total RNA using TaqMan reverse transcription reagents with random hexamer primers (Applied Biosystems, Foster City, CA, USA).
Quantitative real-time PCR was performed in a 20 μL reaction containing 50 ng reverse-transcribed total RNA, 156 nM forward and reverse primers, and 10 μL 2× SYBR green PCR Master Mix (Applied Biosystems). PCR reactions were performed in triplicate in 96-well plates using the iCycler iQ or CFX96 system (Bio-Rad). Results were analyzed using iQ5 or CFX Manager software (Bio-Rad). The relative amount of mRNAs was calculated using the comparative CT method [27]. RPLP0 mRNA was used as an invariant control.
Statistical Analysis
Data are presented as mean ± standard error of the mean (SEM). Statistically significant differences between groups were evaluated by use of Student’s t test to compare means in the sense and antisense groups using Excel (Microsoft, Redmond, WA, USA). P < 0.05 was interpreted as statistically significant.
Results
The metabolic pathways leading to the formation of 16:1n-7, 18:1n-9, and 20:3n-9 are shown in Fig. 1. These pathways were evaluated first in 16HBEo− human bronchial epithelial cells stably transfected with plasmids expressing the first 131 nucleotides of the cftr gene in either the sense or antisense orientation [22]. Consequently, the sense cells express CFTR protein whereas the antisense cells show nearly complete loss of both CFTR expression and activity [13, 22].
The relative fatty acid composition of sense and antisense cells is shown in Table 2. A small, but statistically significant increase in the saturated fatty acids 16:0 and 18:0 was observed for antisense cells (3.2 and 4.1% increase, respectively, compared with sense cells). Larger increases are seen in the monounsaturated derivatives of these fatty acids, 16:1n-7 and 18:1n-9 (13 and 15% increase, respectively). The largest relative increase in antisense cells was for 20:3n-9, which was 80% higher in antisense than sense cells. Of note, the 20:3n-9/20:4n-6 ratio, a marker of EFA deficiency [28], is significantly increased in antisense versus sense cells (0.09 vs. 0.05; P < 0.001).
Table 2.
Fatty acid composition (mol %a) of sense and antisense cells
| Acid | Sense | Antisense | Acid | Sense | Antisense |
|---|---|---|---|---|---|
| 14:0 | 2.04 ± 0.04 | 1.60 ± 0.01d | 18:3n-3 | 0.20 ± 0.09 | 0.09 ± 0.01 |
| 15:0 | 1.11 ± 0.02 | 0.51 ± 0.01d | 20:3n-3 | 0.12 ± 0.01 | 0.04 ± 0.00b |
| 16:0 | 16.45 ± 0.10 | 16.97 ± 0.07 b | 20:5n-3 | 0.34 ± 0.01 | 3.20 ± 0.02d |
| 18:0 | 16.06 ± 0.11 | 16.72 ± 0.04 c | 22:5n-3 | 4.40 ± 0.05 | 5.15 ± 0.03d |
| 20:0 | 0.21 ± 0.07 | 0.30 ± 0.01 | 22:6n-3 | 1.00 ± 0.02 | 0.58 ± 0.01d |
| 22:0 | 0.18 ± 0.01 | 0.26 ± 0.01c | 18:2n-6 | 14.37 ± 0.24 | 11.88 ± 0.08d |
| 24:0 | 0.19 ± 0.01 | 0.26 ± 0.02 | 20:2n-6 | 1.17 ± 0.06 | 0.40 ± 0.07c |
| 16:1n-7 | 2.47 ± 0.05 | 2.79 ± 0.05 c | 18:3n-6 | 0.38 ± 0.00 | 0.69 ± 0.01d |
| 18:1n-7 | 3.65 ± 0.07 | 2.79 ± 0.04d | 20:3n-6 | 1.50 ± 0.05 | 2.27 ± 0.02d |
| 18:1n-9 | 17.83 ± 0.07 | 20.43 ± 0.12 d | 20:4n-6 | 10.73 ± 0.24 | 10.71 ± 0.06 |
| 20:1n-9 | 0.59 ± 0.13 | 0.32 ± 0.01 | 22:4n-6 | 2.61 ± 0.05 | 0.48 ± 0.01d |
| 20:3n-9 | 0.53 ± 0.02 | 0.96 ± 0.02 d | 22:5n-6 | 1.68 ± 0.08 | 0.11 ± 0.01d |
| 22:1n-9 | 0.12 ± 0.00 | 0.29 ± 0.01d | |||
| 24:1n-9 | 0.06 ± 0.01 | 0.19 ± 0.00d |
Data are given as mean ± SEM of three replicates. Fatty acids of interest to this study are in bold
Molar percentage of total fatty acids
P<0.05
P<0.01
P<0.001
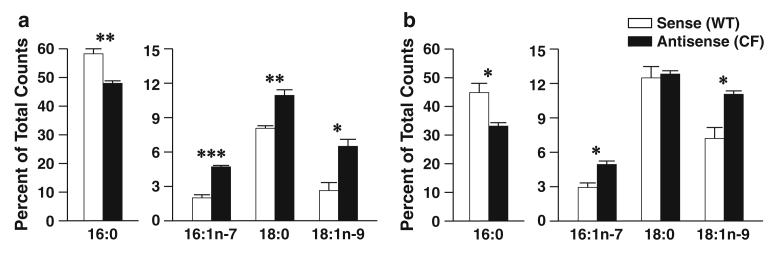
To determine if these fatty acid changes in antisense cells were a result of alterations in enzymatic metabolism, sense and antisense cells were incubated with radiolabeled 16:0, 18:0, and 18:1n-9, and conversion to downstream metabolites was measured. The results of labeling experiments with [1-14C]16:0 are shown in Fig. 2. After 4 h incubation, there was significant diminution of 16:0 with increased conversion to 16:1n-7, 18:0, and 18:1n-9 in antisense cells compared with sense cells (Fig. 2a). This effect persisted for all but 18:0 at 24 h (Fig. 2b), perhaps because of subsequent conversion to 18:1n-9.
Fig. 2.
Palmitate (16:0) metabolism through the n-7 and n-9 pathways in 16HBE cells. Sense (wildtype, WT) and antisense (cystic fibrosis, CF) cells were cultured in complete medium for seven days and then incubated with 4.1 μM [1-14C]16:0 in reduced-lipid cell culture medium for 4 h and harvested (a) or washed twice in PBS and incubated for an additional 20 h in serum-containing medium (b). Levels of labeled 16:0, 16:1n-7, 18:0, and 18:1n-9 were determined by HPLC as described in “Materials and Methods”. Data are expressed as percentages of total counts and bars represent mean ± SEM (n = 3). The findings are representative of at least two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 for sense versus antisense cells
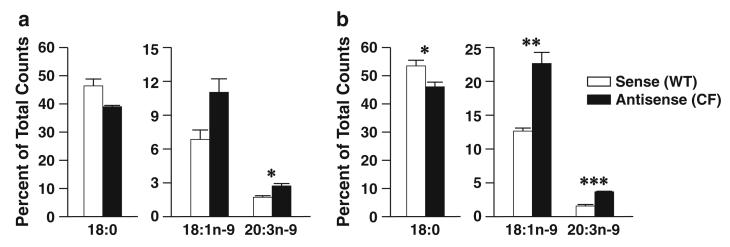
Similar results were observed when cells were incubated with [1-14C]18:0. Metabolism of 18:0 to 18:1n-9 was increased in antisense versus sense cells at both time points (Fig. 3a, b), although these differences were statistically significant at 24 h only. Further metabolism to 20:3n-9 was significantly increased in antisense cells at both time points.
Fig. 3.
Stearate (18:0) metabolism through the n-9 pathway in 16HBE cells. Sense (WT) and antisense (CF) cells were cultured in complete medium for seven days and then incubated with 4.1 μM [1-14C]18:0 in reduced-lipid cell culture medium for 4 h and harvested (a) or washed twice in PBS and incubated for an additional 20 h in serum-containing medium (b). Levels of labeled 18:0, 18:1n-9, and 20:3n-9 were determined by HPLC as described in “Materials and Methods”. Data are expressed as percentages of total counts and bars represent mean ± SEM (n = 3). The findings are representative of at least two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 for sense versus antisense cells
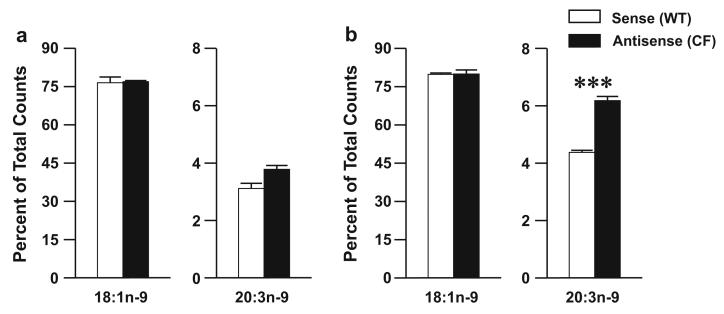
Increased production of 20:3n-9 is, in part, because of increased metabolism of 18:1n-9. When incubated with [1-14C]18:1n-9, antisense cells showed increased conversion of this substrate to 20:3n-9 at both time points, with difference reaching statistical significance at 24 h (Fig. 4a, b).
Fig. 4.
Oleate (18:1n-9) metabolism through the n-9 pathway in 16HBE cells. Sense (WT) and antisense (CF) cells were cultured in complete medium for seven days and then incubated with 4.1 μM [1-14C]18:1n-9 in reduced-lipid cell culture medium for 4 h and harvested (a) or washed twice in PBS and incubated for an additional 20 h in serum-containing medium (b). Levels of labeled 18:1n-9 and 20:3n-9 were determined by HPLC as described in “Materials and Methods”. Data are expressed as percentages of total counts and bars represent mean ± SEM (n = 3). The findings are representative of at least two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 for sense versus antisense cells
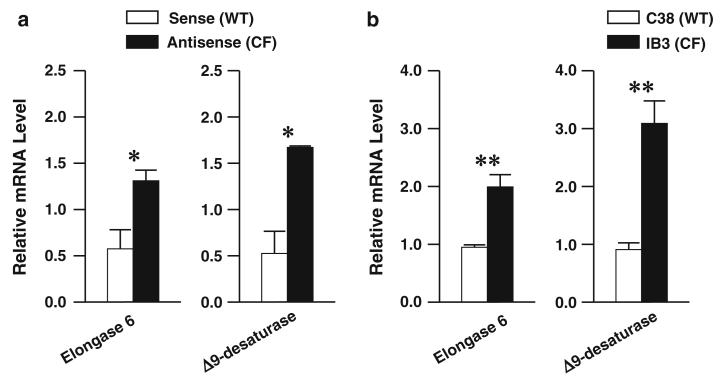
Taken together, these fatty acid metabolism results indicate that antisense cells exhibit increased enzymatic activity catalyzing the conversion of 16:0 to 16:1n-7 and from 16:0 through 18:0 to 18:1n-9. The enzymes responsible for these metabolic conversions are Δ9-desaturase and elongase 6, which are regulated almost entirely at the transcriptional level [29, 30]. To test potential changes in the expression of these enzymes, quantitative RT-PCR was performed on total RNA extracted from sense and antisense cells using primers specific to these two enzymes. As predicted, there was a significant increase in the relative mRNA levels corresponding to both elongase 6 and Δ9-desaturase (Fig. 5a).
Fig. 5.
Relative mRNA expression of metabolic enzymes in the n-7 and n-9 pathways. Sense (WT) and antisense (CF) cells (a) or C38 (WT) and IB3 (CF) cells (b) were incubated in complete medium for seven days, after which RNA was extracted and cDNA synthesized as described in “Materials and Methods”. qRT-PCR was performed using primers for the mRNA sequences of elongase 6 (ELOVL6) and Δ9-desaturase (SCD). Relative expression was determined by the ΔΔCT method using ribosomal protein RPLP0 as a control. Bars represent mean ± SEM (n = 3). The findings are representative of at least two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 for sense versus antisense or IB3 versus C38 cells
To confirm these findings, expression of these enzymes was measured in a second cell-culture system. The IB3 cell line consists of immortalized bronchial epithelial cells from an actual CF patient that carried a compound heterozygous genotype (ΔF508/W1282X) [23]. C38 cells are IB3 cells stably transfected with a wild-type cftr gene, restoring normal CFTR function [24]. They were used as controls. Similar to the sense and antisense cells, the IB3 cells exhibit increased expression of both elongase 6 and Δ9-desaturase compared with C38 cells (Fig. 5b).
Further metabolism of 18:1n-9 is catalyzed by Δ6-desaturase, elongase 5, and Δ5-desaturase (Fig. 1). Previous studies [20] in these cell culture systems demonstrated that expression of the two desaturase enzymes is significantly increased in antisense versus sense cells. All three enzymes show increased expression in IB3 versus C38 cells.
Discussion
Changes in fatty acid levels are a consistent feature of CF and there is increasing evidence that they are involved in the pathogenesis of the disease. For example, studies have shown that fatty acid alterations are more pronounced in patients carrying genotypes associated with more severe disease [12, 31]. Accordingly, the degree of fatty acid alteration correlates with the severity of CF-associated clinical data [32-34]. Furthermore, correction of fatty acid alterations by docosahexaenoate (22:6n-3) in a mouse model of CF also corrects CF-associated respiratory and gastrointestinal tract pathology [5].
If fatty acid imbalances play a role in CF pathogenesis, it is important to understand their underlying molecular mechanisms. A recent study demonstrated that changes in n-3 and n-6 polyunsaturated fatty acids in CF cells are a result of increased expression of fatty acid metabolic enzymes, specifically the Δ5 and Δ6 desaturases [20]. Our study extends this analysis to alterations observed in n-7 and n-9 fatty acids. It demonstrates increased metabolism of 16:0 to 16:1n-7 and of 16:0 through 18:0 to 18:1n-9 and 20:3n-9 in antisense cells. These changes are associated with increased expression of fatty acid metabolic enzymes, including Δ5, Δ6, and Δ9 desaturases and elongase 6 both in antisense cells and in IB3 cells. Combined, the findings of these two studies strongly support the notion that induction of metabolic enzymes underlies alterations of fatty acid composition in CF. Furthermore, the detection of these changes in an isolated, homogenous cell culture system confirms that the alterations are a result of intrinsic alterations in the CFTR-mutated cells, and not caused by malabsorption or other physiologic abnormalities of CF patients.
Although this study begins to unravel the mechanisms of fatty acid changes in CF, the relationship between CFTR mutations and these alterations remains unclear. While CFTR, functions primarily as a chloride channel, it interacts with other proteins and is involved in several other physiologic processes [35]. Thus, changes in fatty acid metabolism could result from a number of different molecular and physiologic alterations caused by CFTR mutation. Consequently, it may be most sensible to begin with the fatty acid metabolic changes and work backwards, establishing this connection step by step. Our work contributes to early first steps in that process.
Others have described similar changes in fatty acid metabolism. Mailhot et al. [36], using a different cell type, a model of Caco-2/15 cells deficient in CFTR, showed altered regulation of lipid metabolism in the CFTR deficient cells. These cells had higher cellular fatty acid content and an elevated proportion of saturated and n-7 fatty acids. These changes were felt to be a result of increased de-novo lipogenesis, enhanced fatty acid uptake, and suppression of transcription factors PPAR-α, RXR-α, LXR-α, and LXR-β mRNA, which are involved in the regulation of lipid metabolism. Altered Δ9-desaturase expression in CF is also supported by the findings of Xu et al. [37], who examined the expression of selected lipid metabolizing enzymes in bone marrow-derived dendritic cells from wild-type and cftr−/− knockout mice. They did not detect a difference in Δ9-desaturase mRNA levels between wild-type and CF cells at baseline. However, when exposed to Pseudomonas infection, Δ9-desaturase expression was down-regulated in wild-type, but not CF cells. The net result was significantly increased expression of this enzyme in infected CF versus wild-type cells.
Some have speculated that abnormalities in n-3 and n-6 fatty acid levels contribute to CF pathogenesis by increasing production of oxygenated fatty acid species, notably the eicosanoids. The specific role that increased n-7 and n-9 fatty acids, for example 16:1n-7, 18:1n-9, and 20:3n-9, might play in the pathophysiology of CF is less obvious. However, there is a growing recognition that these lipids are important in mammalian physiology and disease. For example, several recent studies have demonstrated that 16:1n-7 increases the insulin sensitivity of peripheral tissues, particularly muscle and liver [15, 38]. In this “lipokine” role, it may also partially mediate the effect of insulin-sensitizing drugs, for example thiazolidinediones [39]. These findings may partially explain the described role of Δ9-desaturase in obesity and insulin resistance [40] and emphasize the role of 16:1n-7 as a signaling molecule.
The other major product of Δ9-desaturase metabolism, 18:1n-9, has been shown to be involved in inflammation, a process particularly germane to CF pathophysiology. Liu et al. [17] showed that decreased 18:1n-9 in Δ9-desaturase-deficient mice attenuated inflammation in adipocytes, macrophages, and endothelial cells. In contrast, Harvey et al. [41] showed that 18:1n-9 blocks 18:0-induced cell growth inhibition and inflammatory signaling. Other studies [16, 18] have found that decreased 18:1n-9 levels because of Δ9-desaturase inhibition or deletion exacerbate inflammation in mouse models of acute colitis and atherosclerosis. Thus, 18:1n-9 may have cell-specific effects in inflammation. In addition, Patel et al. [19] demonstrated that 20:3n-9 may mediate inflammation via oxygenated derivatives. This study found that the major product of 20:3n-9 metabolism was 5-hydroxy-20:3, a potent pro-inflammatory mediator and activator of neutrophils and eosinophils.
It should be noted that a high-fat diet is recommended for patients with CF, with no suggestion that the fat be of a specific type and therefore provide the patient with higher concentrations of specific fatty acids. Understanding the metabolic mechanisms responsible for the specific fatty acid alterations observed in CF may lead to better informed and more specific dietary and therapeutic recommendations that may be beneficial for patients.
Acknowledgments
The authors thank Jesse Gilliam for technical assistance and Eva Henderson and the Vanderbilt Molecular Cell Biology Core Laboratory for primer design and testing and qRT-PCR support. This work was funded in part by Clinical Translational Science Award 1UL-1RR024975 from the National Center for Research Resources and Training Grant in Gastroenterology 5T32DK007673-18 from the National Institutes of Health to K.F.T., the Edward and Nancy Fody Endowed Chair in Pathology (M.L.), and the Vanderbilt Physician Scientist Training Program (A.C.S.).
Abbreviations
- CF
Cystic fibrosis
- CFTR
Cystic fibrosis transmembrane regulator
- FAME
Fatty acid methyl ester
- HPLC
High-performance liquid chromatography
- GC
Gas chromatography
Contributor Information
Kelly F. Thomsen, Department of Pediatrics, Vanderbilt University Medical Center, Nashville, TN, USA
Michael Laposata, Department of Pathology, Vanderbilt University Medical Center, Nashville, TN, USA.
Sarah W. Njoroge, Department of Pathology, Vanderbilt University Medical Center, Nashville, TN, USA
Obi C. Umunakwe, Department of Pathology, Vanderbilt University Medical Center, Nashville, TN, USA
Waddah Katrangi, Department of Pathology, Vanderbilt University Medical Center, Nashville, TN, USA.
Adam C. Seegmiller, Department of Pathology, Vanderbilt University Medical Center, Nashville, TN, USA
References
- 1.O’Sullivan BP, Freedman SD. Cystic fibrosis. Lancet. 2009;373:1891–1904. doi: 10.1016/S0140-6736(09)60327-5. [DOI] [PubMed] [Google Scholar]
- 2.Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, et al. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 3.Al-Turkmani MR, Freedman SD, Laposata M. Fatty acid alterations and n-3 fatty acid supplementation in cystic fibrosis. Prostaglandins Leukot Essent Fatty Acids. 2007;77:309–318. doi: 10.1016/j.plefa.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Strandvik B. Fatty acid metabolism in cystic fibrosis. Prostaglandins Leukot Essent Fatty Acids. 2010;83:121–129. doi: 10.1016/j.plefa.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Freedman SD, Katz MH, Parker EM, Laposata M, Urman MY, et al. A membrane lipid imbalance plays a role in the phenotypic expression of cystic fibrosis in cftr(−/−) mice. Proc Natl Acad Sci USA. 1999;96:13995–14000. doi: 10.1073/pnas.96.24.13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubbard VS, Dunn GD. Fatty acid composition of erythrocyte phospholipids from patients with cystic fibrosis. Clin Chim Acta. 1980;102:115–118. doi: 10.1016/0009-8981(80)90441-6. [DOI] [PubMed] [Google Scholar]
- 7.Lepage G, Levy E, Ronco N, Smith L, Galeano N, et al. Direct transesterification of plasma fatty acids for the diagnosis of essential fatty acid deficiency in cystic fibrosis. J Lipid Res. 1989;30:1483–1490. [PubMed] [Google Scholar]
- 8.Fokkema MR, Smit EN, Martini IA, Woltil HA, Boersma ER, et al. Assessment of essential fatty acid and omega3-fatty acid status by measurement of erythrocyte 20:3omega9 (mead acid), 22:5omega6/20:4omega6 and 22:5omega6/22:6omega3. Prostaglandins Leukot Essent Fatty Acids. 2002;67:345–356. doi: 10.1054/plef.2002.0440. [DOI] [PubMed] [Google Scholar]
- 9.Siguel EN, Chee KM, Gong JX, Schaefer EJ. Criteria for essential fatty acid deficiency in plasma as assessed by capillary column gas-liquid chromatography. Clin Chem. 1987;33:1869–1873. [PubMed] [Google Scholar]
- 10.Aldamiz-Echevarria L, Prieto JA, Andrade F, Elorz J, Sojo A, et al. Persistence of essential fatty acid deficiency in cystic fibrosis despite nutritional therapy. Pediatr Res. 2009;66:585–589. doi: 10.1203/PDR.0b013e3181b4e8d3. [DOI] [PubMed] [Google Scholar]
- 11.Roulet M, Frascarolo P, Rappaz I, Pilet M. Essential fatty acid deficiency in well nourished young cystic fibrosis patients. Eur J Pediatr. 1997;156:952–956. doi: 10.1007/s004310050750. [DOI] [PubMed] [Google Scholar]
- 12.Van Biervliet S, Vanbillemont G, Van Biervliet JP, Declercq D, Robberecht E, et al. Relation between fatty acid composition and clinical status or genotype in cystic fibrosis patients. Ann Nutr Metab. 2007;51:541–549. doi: 10.1159/000114208. [DOI] [PubMed] [Google Scholar]
- 13.Andersson C, Al-Turkmani MR, Savaille JE, Alturkmani R, Katrangi W, et al. Cell culture models demonstrate that CFTR dysfunction leads to defective fatty acid composition and metabolism. J Lipid Res. 2008;49:1692–1700. doi: 10.1194/jlr.M700388-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Biervliet S, Devos M, Delhaye T, Van Biervliet JP, Robberecht E, et al. Oral DHA supplementation in DeltaF508 homozygous cystic fibrosis patients. Prostaglandins Leukot Essent Fatty Acids. 2008;78:109–115. doi: 10.1016/j.plefa.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Cao H, Gerhold K, Mayers JR, Wiest MM, Watkins SM, et al. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell. 2008;134:933–944. doi: 10.1016/j.cell.2008.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen C, Shah YM, Morimura K, Krausz KW, Miyazaki M, et al. Metabolomics reveals that hepatic stearoyl-CoA desaturase 1 downregulation exacerbates inflammation and acute colitis. Cell Metab. 2008;7:135–147. doi: 10.1016/j.cmet.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Miyazaki M, Flowers MT, Sampath H, Zhao M, et al. Loss of Stearoyl-CoA desaturase-1 attenuates adipocyte inflammation: effects of adipocyte-derived oleate. Arterioscler Thromb Vasc Biol. 2010;30:31–38. doi: 10.1161/ATVBAHA.109.195636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacDonald ML, van Eck M, Hildebrand RB, Wong BW, Bissada N, et al. Despite antiatherogenic metabolic characteristics, SCD1-deficient mice have increased inflammation and atherosclerosis. Arterioscler Thromb Vasc Biol. 2009;29:341–347. doi: 10.1161/ATVBAHA.108.181099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel P, Cossette C, Anumolu JR, Gravel S, Lesimple A, et al. Structural requirements for activation of the 5-oxo-6E, 8Z, 11Z, 14Z-eicosatetraenoic acid (5-oxo-ETE) receptor: identification of a mead acid metabolite with potent agonist activity. J Pharmacol Exp Ther. 2008;325:698–707. doi: 10.1124/jpet.107.134908. [DOI] [PubMed] [Google Scholar]
- 20.Njoroge SW, Seegmiller AC, Laposata M. Fatty acid changes in cultured cystic fibrosis cells result from increased expression of Δ5- and Δ6-desaturases and eicosanoid forming enzymes. Pediatr Pulmonol. 2010;45:279. [Google Scholar]
- 21.Al-Turkmani MR, Andersson C, Alturkmani R, Katrangi W, Cluette-Brown JE, et al. A mechanism accounting for the low cellular level of linoleic acid in cystic fibrosis and its reversal by DHA. J Lipid Res. 2008;49:1946–1954. doi: 10.1194/jlr.M800035-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajan S, Cacalano G, Bryan R, Ratner AJ, Sontich CU, et al. Pseudomonas aeruginosa induction of apoptosis in respiratory epithelial cells: analysis of the effects of cystic fibrosis transmembrane conductance regulator dysfunction and bacterial virulence factors. Am J Respir Cell Mol Biol. 2000;23:304–312. doi: 10.1165/ajrcmb.23.3.4098. [DOI] [PubMed] [Google Scholar]
- 23.Zeitlin PL, Lu L, Rhim J, Cutting G, Stetten G, et al. A cystic fibrosis bronchial epithelial cell line: immortalization by adeno-12-SV40 infection. Am J Respir Cell Mol Biol. 1991;4:313–319. doi: 10.1165/ajrcmb/4.4.313. [DOI] [PubMed] [Google Scholar]
- 24.Egan M, Flotte T, Afione S, Solow R, Zeitlin PL, et al. Defective regulation of outwardly rectifying Cl-channels by protein kinase A corrected by insertion of CFTR. Nature. 1992;358:581–584. doi: 10.1038/358581a0. [DOI] [PubMed] [Google Scholar]
- 25.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 26.Alvarez JG, Storey BT. Differential incorporation of fatty acids into and peroxidative loss of fatty acids from phospholipids of human spermatozoa. Mol Reprod Dev. 1995;42:334–346. doi: 10.1002/mrd.1080420311. [DOI] [PubMed] [Google Scholar]
- 27.Applied-Biosystems . User bulletin No. 2, Rev. B. Applied Biosystems; Forster City: 2001. [Google Scholar]
- 28.Holman RT. The ratio of trienoic: tetraenoic acids in tissue lipids as a measure of essential fatty acid requirement. J Nutr. 1960;70:405–410. doi: 10.1093/jn/70.3.405. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura MT, Nara TY. Gene regulation of mammalian desaturases. Biochem Soc Trans. 2002;30:1076–1079. doi: 10.1042/bst0301076. [DOI] [PubMed] [Google Scholar]
- 30.Jakobsson A, Westerberg R, Jacobsson A. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog Lipid Res. 2006;45:237–249. doi: 10.1016/j.plipres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Strandvik B, Gronowitz E, Enlund F, Martinsson T, Wahlstrom J. Essential fatty acid deficiency in relation to genotype in patients with cystic fibrosis. J Pediatr. 2001;139:650–655. doi: 10.1067/mpd.2001.118890. [DOI] [PubMed] [Google Scholar]
- 32.Guilbault C, Wojewodka G, Saeed Z, Hajduch M, Matouk E, et al. Cystic fibrosis fatty acid imbalance is linked to ceramide deficiency and corrected by fenretinide. Am J Respir Cell Mol Biol. 2009;41:100–106. doi: 10.1165/rcmb.2008-0279OC. [DOI] [PubMed] [Google Scholar]
- 33.Maqbool A, Schall JI, Garcia-Espana JF, Zemel BS, Strandvik B, et al. Serum linoleic acid status as a clinical indicator of essential fatty acid status in children with cystic fibrosis. J Pediatr Gastroenterol Nutr. 2008;47:635–644. doi: 10.1097/MPG.0b013e31817fb76b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olveira G, Dorado A, Olveira C, Padilla A, Rojo-Martinez G, et al. Serum phospholipid fatty acid profile and dietary intake in an adult Mediterranean population with cystic fibrosis. Br J Nutr. 2006;96:343–349. doi: 10.1079/bjn20051655. [DOI] [PubMed] [Google Scholar]
- 35.Vankeerberghen A, Cuppens H, Cassiman JJ. The cystic fibrosis transmembrane conductance regulator: an intriguing protein with pleiotropic functions. J Cyst Fibros. 2002;1:13–29. doi: 10.1016/s1569-1993(01)00003-0. [DOI] [PubMed] [Google Scholar]
- 36.Mailhot G, Rabasa-Lhoret R, Moreau A, Berthiaume Y, Levy E. CFTR depletion results in changes in fatty acid composition and promotes lipogenesis in intestinal Caco 2/15 cells. PLoS One. 2010;5:e10446. doi: 10.1371/journal.pone.0010446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Y, Tertilt C, Krause A, Quadri LE, Crystal RG, et al. Influence of the cystic fibrosis transmembrane conductance regulator on expression of lipid metabolism-related genes in dendritic cells. Respir Res. 2009;10:26. doi: 10.1186/1465-9921-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stefan N, Kantartzis K, Celebi N, Staiger H, Machann J, et al. Circulating palmitoleate strongly and independently predicts insulin sensitivity in humans. Diabetes Care. 2010;33:405–407. doi: 10.2337/dc09-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuda O, Stankova B, Tvrzicka E, Hensler M, Jelenik T, et al. Prominent role of liver in elevated plasma palmitoleate levels in response to rosiglitazone in mice fed high-fat diet. J Physiol Pharmacol. 2009;60:135–140. [PubMed] [Google Scholar]
- 40.Flowers MT, Ntambi JM. Role of stearoyl-coenzyme A desaturase in regulating lipid metabolism. Curr Opin Lipidol. 2008;19:248–256. doi: 10.1097/MOL.0b013e3282f9b54d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harvey KA, Walker CL, Xu Z, Whitley P, Pavlina TM, et al. Oleic acid inhibits stearic acid-induced inhibition of cell growth and pro-inflammatory responses in human aortic endothelial cells. J Lipid Res. 2010;51:3470–3480. doi: 10.1194/jlr.M010371. [DOI] [PMC free article] [PubMed] [Google Scholar]