Abstract
Purpose
An understanding of how hematopoietic cells respond to therapy that causes myelosuppression will help develop approaches to prevent this potentially life-threatening toxicity. The goal of this study was to determine how human myeloid precursor cells (MP) respond to temozolomide (TMZ)-induced DNA damage.
Experimental Design
We developed an ex vivo primary human MP cells model system to investigate the involvement of cell-death pathways using a known myelosuppressive regimen of O6-benzylguanine (6BG) and TMZ.
Results
Exposure to 6BG/TMZ led to increases in p53, p21, γ-H2AX, and mitochondrial DNA damage. Increases in mitochondrial membrane depolarization correlated with increased caspase-9 and caspase-3 activities following 6BG/TMZ treatment. These events correlated with decreases in activated AKT, downregulation of the DNA repair protein O6methylguanine-DNA methyltransferase (MGMT), and increased cell death. During MP cell expansion, FAS/CD95/APO1(FAS) expression increased over time and was present on ~100% of the cells following exposure to 6BG/TMZ. While c-flipshort, an endogenous inhibitor of FAS-mediated signaling, was decreased in 6BG/TMZ-treated versus control, 6BG-, or TMZ alone-treated cells, there were no changes in caspase-8 activity. Additionally, there were no changes in the extent of cell death in MP cells exposed to 6BG/TMZ in the presence of neutralizing or agonistic anti-FAS antibodies, indicating that FAS-mediated signaling was not operative.
Conclusions
In human MP cells, 6BG/TMZ-initiated apoptosis occurred by intrinsic, mitochondrial-mediated and not extrinsic, FAS-mediated apoptosis. Human MP cells represent a clinically relevant model system for gaining insight into how hematopoietic cells respond to chemotherapeutics and offer an approach for selecting effective chemotherapeutic regimens with limited hematopoietic toxicity.
INTRODUCTION
A major dose-limiting toxicity in anti-cancer chemotherapeutics is the induction of persistent DNA damage that leads to programmed cell death of hematopoietic cells in the blood, spleen, and bone marrow. (1) Additionally, the survival of rare hematopoietic-derived clonal populations with transforming DNA mutations due to chemotherapy exposure can lead to emergence of leukemic cells. (2) A primary contributing factor responsible for these deleterious outcomes is that hematopoietic cells typically express low levels of DNA repair proteins and therefore are highly susceptible to DNA damage caused by therapeutics targeting cancer cells. (3, 4) Understanding molecular processes that regulate how primary human hematopoietic cells respond to DNA damage could provide key information towards development of cancer treatments that specifically target cancer cells with minimal effects to normal hematopoietic cells.
Myeloid cells represent a diverse population of hematopoietic cells consisting of granulocyte and monocyte/macrophage lineages derived from pluripotent hematopoietic stem cells. (1, 2) Upon maturation, myeloid cells play critical roles in regulating immune responses, bone remodeling, and inflammation. Therefore, if left unrepaired, chemotherapy-mediated DNA damage can be highly detrimental to myeloid cell function. In this study, we examine the response of human myeloid precursor (MP) cells to temozolomide (TMZ) since it is routinely used as a front-line chemotherapeutic agent for the treatment of glioblastoma multiforme. (5) In particular, the molecular effects of TMZ-mediated myelosuppression in the presence of the O6methylguanine-DNA methyltransferase (MGMT) inhibitor, O6benzylguanine (6BG), were studied since 1) myelosuppression is observed in the clinic with this regimen; and 2) dependence of DNA repair and cell survival on MGMT expression could be assessed pharmacologically. (6, 7) TMZ is a pro-drug that hydrolyzes to its active metabolite (3-methyl-(triazen-1-yl)imidazole-4-carboxamide (MTIC) at physiological pH. (8) The main mechanism of TMZ-mediated cytotoxicity is the generation of a variety of DNA adducts including N7-methylguanine, N3-methyladenine, and O6-methylguanine (O6MeG). However, how the presence of methylated adducts leads to cell death is complex and not completely understood. (9) While the base-excision repair system is responsible for repairing N7-methylguanine and N3-methyladenine adducts, the direct repair protein, MGMT, repairs O6MeG adducts. If left unrepaired, the O6MeG adduct can be highly cytotoxic and is the most critical DNA lesion contributing to cell death when cells are exposed to alkylating reagents such as TMZ. This adduct can mispair with a thymine instead of the cytosine residue during DNA replication which leads to the formation of O6MeG:thymine mismatches. While the mismatches are recognized by the mismatch repair (MMR) system,(10) a futile cycle of repair ensues in which thymine is excised only to have another thymine reinserted opposite of the O6MeG adduct. This continues as long as O6MeG adducts are present and eventually leads to increased double-strand DNA breaks and ultimately cell death. O6MeG adducts can be directly repaired by MGMT by transfer of the methyl group from the oxygen in guanine to cysteine residue-145 in the active site of MGMT. (9) When cells with non-repaired O6MeG adducts enter DNA replication in the absence of adequate DNA repair, replication stalls at the O6MeG adducts resulting in an increase in double-strand DNA breaks and ultimately apoptosis.
The overall hypothesis of the present study is that persistence of TMZ-mediated DNA damage in human MP cells results in the activation of a predominant cell-death pathway. To address this hypothesis, we first developed a primary hematopoietic culture system of human origin that could be used to investigate regulation of signaling pathways following exposure to DNA-damaging agents. Human MP cells were chosen since these cells represent a population of bone-marrow precursor cells responsible for development of all myeloid lineages. Large numbers of MP cells were efficiently generated by incubating human CD34+ cells with G-CSF and SCF. Under these conditions, CD34+ cells underwent robust proliferation starting at day 3 and partial differentiation down the myeloid lineage differentiation pathway by days 7-10. We chose to focus on DNA-damage responses elicited via a myelosupressive TMZ-based regimen previously used in the clinic. (6) In this system, we were able to examine a variety of DNA-damage and cell-death response pathways. As a result of 6BG/TMZ treatment, the p53 pathway was activated and DNA damage in both the mitochondrial and nuclear genomes increased. Increased genome damage correlated with MGMT downregulation and increased apoptosis of 6BG/TMZ-treated MP cells. In contrast to MP cells treated with vehicle control, 6BG, or TMZ, FAS expression increased and expression of the endogenous caspase-8 inhibitor, c-FLIPshort, decreased in 6BG/TMZ-treated MP cells. However, caspase-8 activity remained unchanged and anti-FAS antibodies (agonistic and neutralizing) had no effect on 6BG/TMZ-mediated MP cell death indicating that death-inducing signals are not delivered through FAS. In contrast, the intrinsic, mitochondrial-mediated cell-death pathway was activated in MP cells since mitochondrial membrane depolarization and caspase-9 activity increased following 6BG/TMZ exposure.
Material and Methods
Isolation of umbilical cord blood (UCB) CD34+ cells and expansion of primary human MP cells
All protocols were approved by Indiana University School of Medicine’s Institutional Review Board (IRB) and St. Vincent Hospital’s IRB (Indianapolis, IN). Samples of UCB were collected from normal, full-term infants delivered by cesarean section and the CD34+ cells isolated using the CD34 MicroBead kit and VarioMACS Separator™ (Miltenyi Biotech Inc., Auburn, CA) according to the manufacturer’s instructions. Isolated CD34+ cells were plated at 0.5 × 106 cells per ml and expanded in the presence of 100 ng/ml G-CSF and SCF (PeproTech, Rocky Hill, NJ) for 8-12 days in BioWhittaker X-vivo 10 serum-free medium (Lonza Walkersville, MD) containing 1% human serum albumin.
Formulation of O6-benzylguanine (6BG) and temozolomide (TMZ)
6BG (Sigma, St. Louis, MO, USA) was dissolved in 40% polyethylene glycol-400 (PEG) (v/v) and 60% PBS (v/v) and sonicated; final stock concentration was 8.3 mM. The 40% PEG formulation served as the vehicle control (control) in all experiments. TMZ (LKT Laboratories,Inc) was dissolved in PBS and sonicated at 37°C for 5min; final stock was concentration was 30.9 mM.
Colony forming-Unit (CFU) assays
CFU assays were performed using isolated human CD34+ cells and expanded CB cells (Methocult GF, H4434, Stem Cell Technologies, Inc.) as previously described.(11) The cells were seeded in triplicate in 35 mm dishes at concentrations of 2 × 103. After 10 - 14 days of incubation at 37°C in 5% CO2, colony forming units-granulocyte-macrophage (CFU-GM) and burst-forming units-erythroid (BFU-E) and colony forming unit-granulocyte/erythrocyte/monocyte/megakaryocyte (CFU-GEMM) were enumerated using the Axiovert 25 inverted-light microscope.
Immunophenotyping by flow cytometry
Phenotype of expanded primary human myeloid cells was performed in the flow cytometry laboratory. Aliquots of 1–2 × 105 cells/tube were stained with various antibodies (2 μl per sample) for 25 min at 4°C in complete medium and washed one time in PBS containing 1% FBS. Samples were analyzed using four-color flow cytometry. Total cell count was determined and the viability of the sample was analyzed with 7-AAD staining. The antibodies were directly conjugated with fluorescein isothiocyanate (FITC), phycoerythrin (PE), PE-Cy5 (PC5) or PE-Texas Red (ECD). The following antibodies were analyzed: CD45, CD34, HLA-DR, CD117, CD38, CD71, CD33, CD13, CD15, myeloperoxidase, CD64, CD14, CD3, and CD19. All antibodies were purchased from BD Biosciences (San Jose, CA). Data acquisition was performed using COULTER FC-500 flow cytometers and CXP software (Beckman Coulter, Miami, FL, USA). The cellular debris was excluded before the analysis. The cells of interest were gated on right angle light scatter vs CD45 display.
Determination of cell death
After treatment, apoptotic cells were detected using APC Annexin V/Dead Cell Apoptosis Kit with APC Annexin V and SYTOX® Green (Invitrogen, Eugene, OR). MP cells were resuspended at a concentration of 1 × 106 cells/mL in 1X Annexin-binding buffer; 5 μL APC-Annexin V and 1 μL of the 1 μM SYTOX® Green stain working solution were added to each 100 μL cell suspension. The cells were incubated at 37°C in an atmosphere of 5% CO2 for 15 minutes, and then analyzed by flow cytometry. For antibody blocking studies of FAS-mediated signaling, the agonistic anti-FAS clone CH-11 (EMD Millipore) and the blocking anti-FAS antibody ZB4 (Kamiya Biomedical, Seattle, WA) were used. The Jurkat T-lymphoblastoid cell line was purchased from ATCC (Manassas, Va) and used at low passage.
Measurement of the mitochondrial membrane potential (MMP)
MMP was determined by the MitoProbe™ DiIC1(5) Assay Kit for Flow Cytometry (Molecular Probes products, Invitrogen) according to manufacturer’s instructions. MP cells were collected after treatment and incubated with 50 nM DiIC1(5) at 37°C, 5% CO2, for 30 min. Cells were washed twice with PBS and analyzed with flow cytometry with 633 nm excitation using emission filters appropriate for Alexa Fluor® 633 dye to detect the cyanine dye 1,1’,3,3,3’3’-hexamethylindodicarbocyanine iodide (DiIC1). (12) The membrane depolarization agent, carbonyl cyanide m-chlorophenylhydrazone (CCCP) served as a positive control for mitochondrial depolarization.
Cell cycle analysis
Following treatment, cell cycle analysis was performed with FITC BrdU Flow Kits (BD Pharmingen™ San Diego, CA) according to manufacturer’s instructions. Briefly, 10 μl of BrdU solution (1 mM BrdU in DPBS) was added to each ml of cell culture media; cell culture density was always maintained at < 2 × 106 cells/ml. The treated cells were incubated for 45 minutes and then fixed and permeabilized with BD Cytofix/Cytoperm Buffer. The cells were then incubated with DNase followed by 20-min incubation with fluorescent anti-BrdU antibodies to detect incorporation of BrdU. Samples were washed and resuspended in buffer. Following the addition of 20 μl 7-AAD solution, the cells were analyzed by flow cytometry.
Western Analyses
After treatment, the cells were harvested and protein lysates were prepared using 1X Cell Lysis Buffer from Cell Signaling Technology (Danvers, MA). Due to the large number of proteases in myeloid cells, the following protease inhibitors were included in the lysis buffer: Diisopropyl fluorophosphate, Phenylmethylsulfornyl fluoride, Chymostatin, 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride, Chymostatin, Sodium orthovanadate, Sodium fluoride (all purchased from Sigma, St.Louis, MO) and Leupeptin (Roche Applied Science, USA). Lysates were separated on 12% Tris-HCl polyacrylamide gels (Bio-Rad Laboratories, Inc., Hercules, CA), and proteins transferred to a nitrocellulose membrane with a 0.45 μm pore size (Bio-Rad). The membrane was then blocked with 5% milk containing 0.1% Tween 20 (Sigma-Aldrich Co., St. Louis, MO) and probed overnight at 4 °C with antibodies specific for the following human proteins: MGMT (clone MT3.1; Millipore, CA), p53(clone DO-1; Santa Cruz Biotechnology, Inc); c-flip (clone NF6; Alexis Biochemicals, San Diego, CA). Antibodies specific for human GAPDH(clone 14C10,), p21 Waf1/Cip1(clone DCS60), H2AX(Ser139), total AKT (AKT1/2/3-his136), and AKT1/2/3ser473 were purchased from Cell Signaling Technology, Inc. The next day, the membrane was washed and incubated with goat anti-mouse or anti-rabbit HRP-conjugated secondary antibody according to the manufacturer’s instructions (Pierce Biotechnology, Inc., Rockford, IL) followed by detection of immunoreactive proteins using the Supersignal West Femto Maximum Sensitivity Substrate (Pierce) and exposure to x-ray film (Midwest Scientific, St. Louis, MO).
Caspase-8 and caspase-9 Colorimetric Activity Assay
To measure caspase-8 or caspase-9 activity following treatment, the Caspase-8 and Caspase-9 Colorimetric Activity Assay Kits (Chemicon/Millipore, Billerica, MA) were used according to manufacturer’s instructions. After treatment, cells were collected and resuspended in 100 μL of chilled 1X cell lysis buffer, incubated on ice for 10 minutes followed by centrifugation. Equal amounts of supernatant protein were used in the absence or presence of caspase-8 or caspase-9 inhibitors. Lysates were pre-incubated with caspase inhibitors for 10 minutes before adding caspase-8 or caspase-9 substrate solution. All samples were incubated for 90 mins at 37°C and then read at 405 nm in a microtiter plate reader. Fold-increase in caspase-8 or caspase-9 activity was determined by comparing the OD reading from the treated sample with the level of the untreated control.
Determination of mitochondrial DNA damage
Quantitative alkaline southern blotting was performed as described previously. (13) Following treatment, DNA was isolated and digested with BamHI. Sodium hydroxide (NaOH) was included in the loading dye, agarose gel, and electrophoresis buffers. The ethidium bromide-stained gel served as an internal control for equal loading between the lanes. After blotting BamHI-digested total DNA, the membrane was hybridized with a mitochondrial-specific probe which recognizes the CoII-ATPase 6 sequences; this probe recognizes a major BamHI restriction band that is larger than the sequence of the probe and allows for analysis of damage over a longer stretch of DNA than just the CoII-ATPase 6 sequence. After hybridization, membranes were exposed to an imaging screen to determine band intensity. A decrease in the hybridization of the mitochondrial-specific probe correlates with increased DNA damage. The number of pixels per band was determined by encompassing bands with identical rectangular regions of interest and subtracting the background. The break frequency was determined using the Poisson expression (s=−ln P0, where s is the number of breaks per fragment and P0 is the fraction of fragments free of breaks).
Statistical Analysis
All numerical data are represented as the mean and standard deviation of triplicate data points for each group. Statistical analyses were conducted using Student’s t-test and p < 0.05 was considered statistically significant.
Results
Generation and characterization of human myeloid precursor (MP) cells
Human CD34+ cells were incubated with human G-CSF and SCF to generate large numbers of MP cells so that sufficient amounts of DNA, RNA, and protein could be obtained for analysis. By days 10-12 post-expansion, MP cells represented the majority of the cell population (see Supplemental Figure 1).
Induction of DNA-damage response following exposure to myelosuppressive doses of chemotherapy
MP cells were exposed to 200 μM of TMZ which, based on pharmacokinetic studies in patients, would most likely exceed the maximal tolerated dose (MTD) of TMZ and therefore potentially lead to a myelosuppressive or myeloablative condition. For example, in a phase I trial in which submyelosuppressive doses of TMZ were used, a single dose of TMZ at 200 mg/m2 resulted in peak TMZ levels (Cmax) in the plasma of ~11 μg/ml or 57 μM; when TMZ was administered daily for 5 consecutive days, peak levels of TMZ in the plasma were ~16 μg/ml or 82 μM. (14) Additionally Quinn et al conducted a phase I clinical trial in adult patients with malignant glioma to determine the MTD of single-dose TMZ in combination with 6BG. When combined with 6BG, the MTD of temozolomide was found to be 472 mg/m2. (6) The plasma levels associated with a dose of TMZ at 472 mg/m2 have been reported in a phase I pediatric trial; the range of peak TMZ levels was 0.6.3-26.5 μg/ml or approximately 3 μM-136 μM in the plasma. (15)
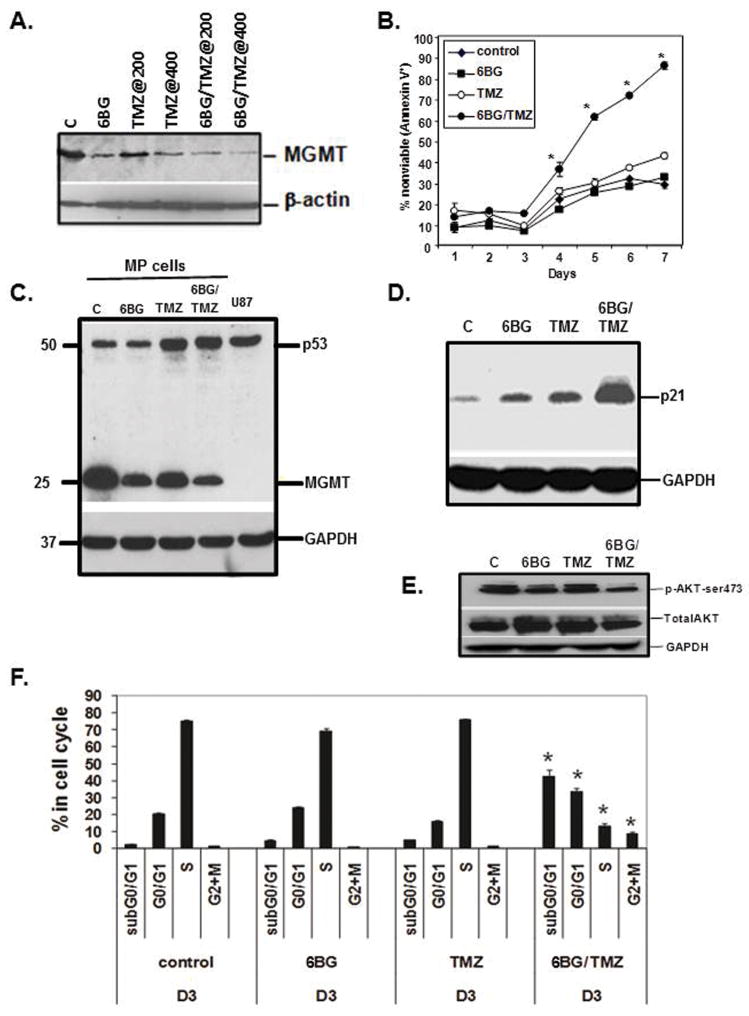
To understand how chemotherapy-mediated myelosuppression occurs when using 6BG in combination with TMZ, we first examined the impact of 6BG/TMZ exposure in MP cell viability assays. MGMT-mediated repair of TMZ-induced adducts is initiated by the transfer of the methyl group from the O6 position of guanine to an internal cysteine residue in the active site of MGMT, which results in ubiquitination and proteosome-mediated degradation of MGMT. In the presence of the MGMT inhibitor 6BG, MGMT is subsequently ubiquitinated and degraded due to transfer of the benzyl group from 6BG to the same cysteine residue in the active site.(9) 6BG/TMZ-based regimens have been used to downregulate MGMT expression in tumors and increase sensitivity to TMZ.(9) MP cells were exposed to 6BG/TMZ one time and compounds were not washed out. As expected, exposure of MP cells to 6BG and/or TMZ resulted in decreased MGMT levels (Fig 1A) by day 1 post-exposure. Consistent with depletion of MGMT, time-course viability studies of MP cells indicated that the combination of 6BG and TMZ resulted in a significant increase in cell death by day 4. By day 7 post-treatment, 90% of the 6BG/TMZ-treated cells were nonviable(Fig 1B). In TMZ-treated cultures, ~40% of the cells were nonviable by day 7 post-treatment. In the control and 6BG-treated cultures, cell death increased from ~10% to 20-25% over time which is typically seen in this culture system as the population moves toward a more differentiated phenotype.
Figure 1. Correlation of MGMT status, cell viability, and DNA damage response in MP cells following exposure to 6BG/TMZ.
(A) Human MP cells were exposed to 6BG in the absence or presence of TMZ (200 uM and 400 uM) and 6BG/TMZ for one day and probed for MGMT expression. Beta-actin served as loading (B) MP cells were treated in triplicate and viability of myeloid cultures determined over time. (C-E). Analysis of 6BG/TMZ-induced p53 stabilization, p21, AKT phosphorylation, and cell cycle arrest at Day 3 post-treatment. GAPDH served as a loading control. *p < 0.05, 6BG/TMZ vs. control, 6BG, or TMZ. Data are representative of 3 independent experiments.
To delineate signaling pathways operative following DNA damage, gene expression was analyzed in MP cells treated with 6BG/TMZ for 18 hours. Microarray analysis was performed on treated MP cells using the PIQOR Cell-Death microarray which contains 494 probes specific for RNAs whose protein products are involved in apoptosis and include stress-like caspases, TNF-receptor family members, Bcl-2 family members, and heat shock proteins. Most of the increased RNA transcripts were targets of p53-mediated signaling. The 6BG/TMZ combination had the strongest impact on changes in gene expression in the MP cells compared to control, 6BG alone, or TMZ alone (see supplementary information, Table 1). The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (16) and are accessible through GEO Series accession number GSE44122 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE44122). Based on the microarray data obtained from the MP cells, protein expression was evaluated based on selected RNAs that were increased (p21 and FAS) or showed no change (p53) following 6BG/TMZ treatment. TMZ and 6BG/TMZ exposure resulted in a stabilization of p53 (Fig 1C), and increased levels of the p53-downstream target p21 (Fig 1D). Exposure of MP cells to 6BG/TMZ resulted in no changes in total AKT protein, but decreased phosphorylation of AKT at serine 473 compared to control, 6BG, or TMZ (Fig 1E). Decreased phosphorylation of AKT at serine 473 has been shown previously to correlate with decreased AKT activity.(17) These DNA-damage responses correlated with increased cell-cycle arrest in G0/G1 and G2/M; the subGo/G1 fraction also increased significantly by day 3 post-treatment with 6BG/TMZ (Fig 1F).
Myelosuppressive chemotherapy-mediated DNA damage and cell death
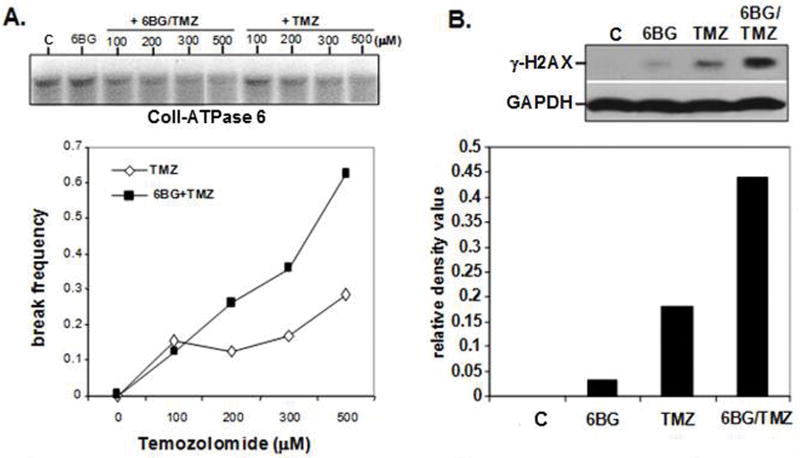
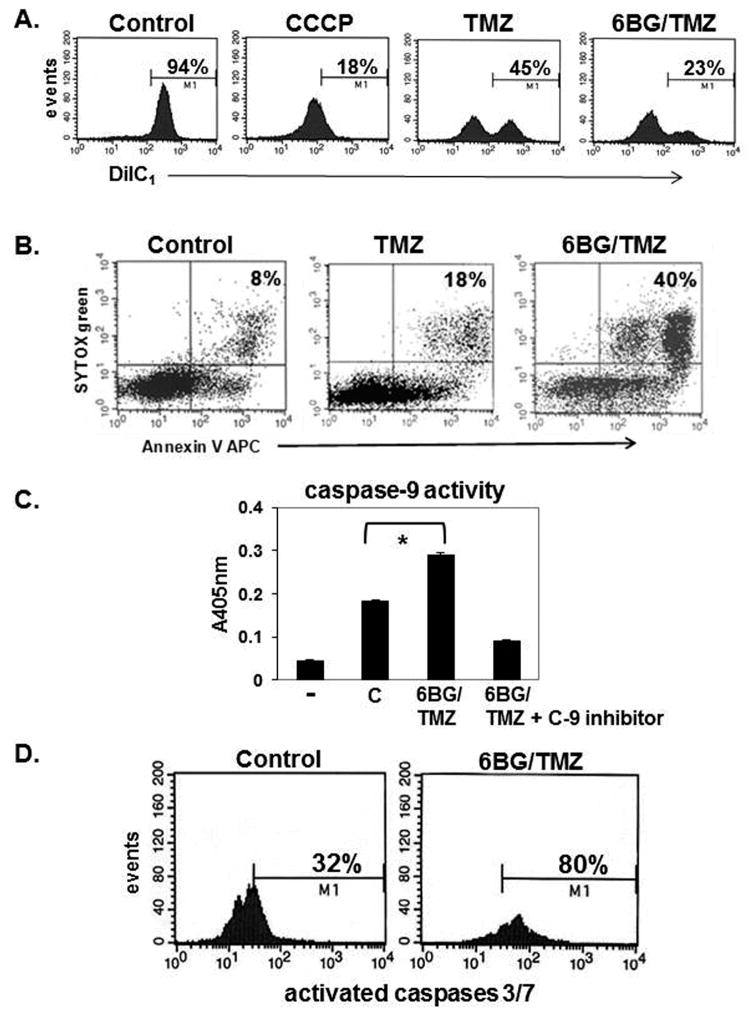
While the nuclear genome is commonly examined for DNA damage, the mitochondrial genome plays an essential role in regulating cell survival since this genome encodes for essential proteins involved in oxidative phosphorylation and respiration. Moreover, the extent to which myelosuppressive chemotherapy actually damages the mitochondrial genome of hematopoietic cells, and how damage correlates with cell death, is not known. The degree of mitochondrial-specific DNA-strand breaks was determined in MP cells (control, 6BG, TMZ and 6BG/TMZ) by alkaline Southern blot analysis using a mitochondrial-specific CoII-ATPase 6 DNA probe.(13) The lack of hybridization to the mitochondrial-specific probe is indicative of mitochondrial DNA damage. In cultures exposed to 6BG, there was no increase in mitochondrial DNA damage (Fig 2A). When myeloid cells were exposed to increasing doses of TMZ in the presence of 6BG, there was a dramatic increase in the number of mitochondrial DNA-strand breaks compared to TMZ alone, indicating that the rate of repair was diminished in the 6BG/TMZ-treated cultures (Fig 2A). The increase in mitochondrial DNA damage correlated with doses of 6BG/TMZ resulting in cell death by day 4 post-treatment as shown in Figure 1B. The levels of γ-H2A-X which are indicative of double-strand DNA-strand breaks were increased in total cellular lysates following 6BG/TMZ treatment (Fig 2B). By day 4 post-treatment with 6BG/TMZ, mitochondrial membrane depolarization (Fig 3A) and apoptotic cells were increased in cultures compared to TMZ or control alone (Fig 3B). To examine if the intrinsic pathway is activated in response to 6BG/TMZ, we evaluated caspase-9 activity following treatment. Levels of activated caspase-9 activity increased significantly following exposure to 6BG/TMZ (Fig 3C) and correlated with activation of Caspases 3 and 7 (Fig 3D), indicating that mitochondrial-mediated cell death is activated and plays a prominent role in chemotherapy-mediated myeloid cell death.
Figure 2. Determination of mitochondrial-genome specific and global DNA damage exposure to 6BG/TMZ.
(A) Human MP cells were exposed to increasing doses of TMZ (100-500 uM) in absence or presence of 6BG for 2 hours and the level of mitochondrial DNA strand breaks determined by alkaline Southern Blot. A mitochondrial-specific probe recognizing the CoII-ATPase 6 sequences was used to detect mitochondrial DNA damage (B) Human MP cells were exposed to 6BG in the absence or presence of 200 uM TMZ for 3 days and the levels of γ-H2AX determined by Western analysis. GAPDH served as a loading control. Data are representative of 2 independent experiments.
Figure 3. Analysis of mitochondrial membrane depolarization and apoptosis following exposure to myelosuppressive chemotherapy.
Human MP cells were treated with 6BG +/- TMZ for 4 days. (A) Mitochondrial membrane depolarization was determined by DiIC1 staining. Increased depolarization correlates with decreased DiC1 stain. Insert on control histogram is DiIC1 staining of myeloid cells treated with the membrane depolarization agent, CCCP. (B) Apoptotic cell death was determined by Annexin V and Sytox- green stain. (C) Caspase-9 activity was determined in the +/- of the caspase-9 specific inhibitor. (D) Caspase-3/7 activation was determined by flow cytometry. Data are representative of 3 independent experiments for A-C and 2 independent experiments for D.
Lack of FAS-mediated signaling during exposure of cells to myelosuppressive chemotherapy
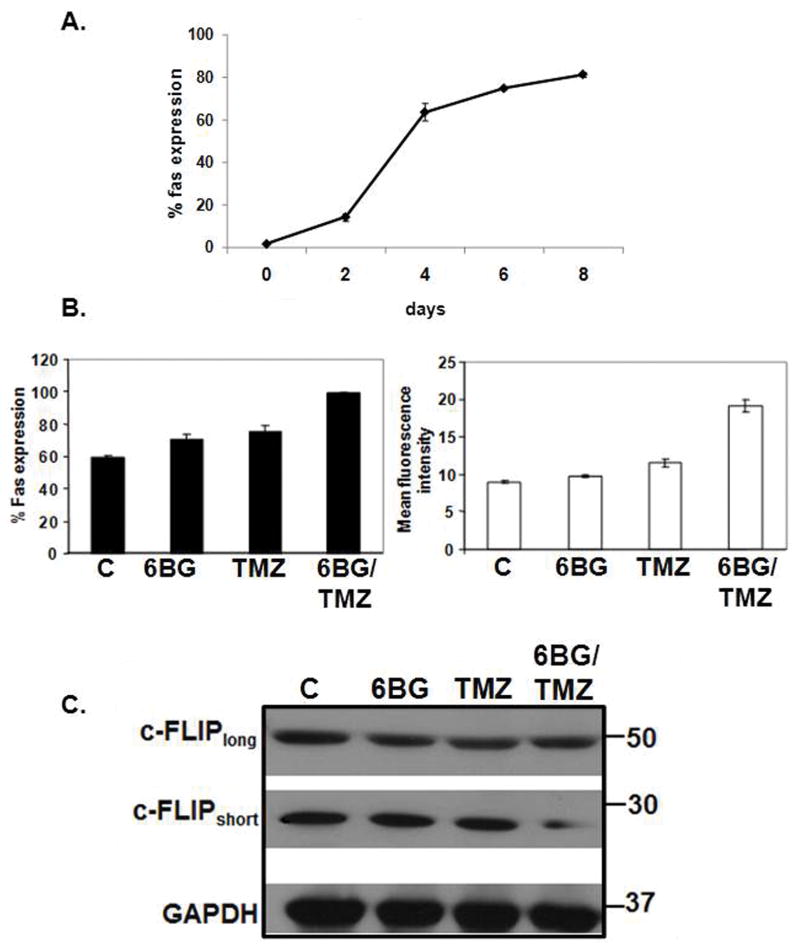
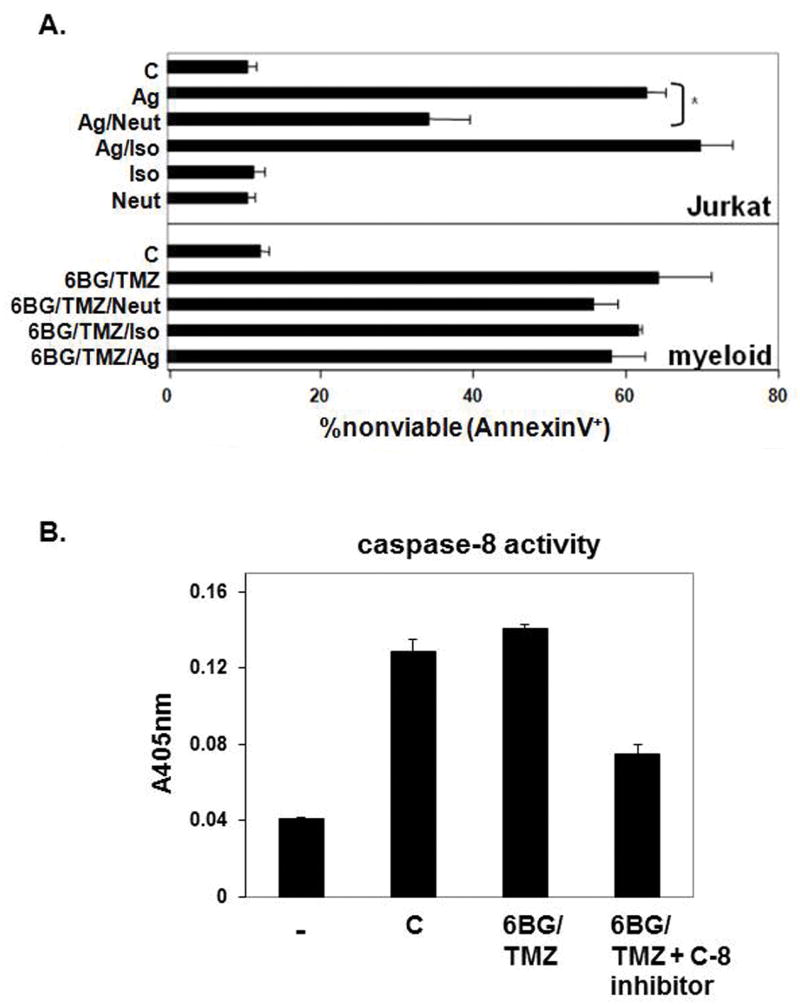
Time-course studies indicated that FAS expression increased as the human CD34+ cells were pushed via cytokine exposure to proliferate and differentiate into MP cells (Fig 4A). The percentage of FAS+ MP cells as well as the density of FAS per cell increased following 6BG/TMZ treatment compared to control, 6BG or TMZ treatment (Fig. 4B). FAS-mediated signaling typically results in activation of caspase-8 and caspase-9 leading to subsequent apoptosis. To investigate the relevance of FAS-mediated signaling in more detail, the levels of c-Fliplong and c-Flipshort in MP cells were determined since both c-Flip isoforms can inhibit FAS-mediated signaling in other cell systems.(18, 19) Both c-Fliplong and c-Flipshort were expressed in MP cells. No changes in the level of c-Fliplong were observed following treatment with 6BG alone, TMZ alone, or 6BG/TMZ (Fig 4C). However, in comparison with control cells or cells treated with 6BG or TMZ, there was a decrease in c-Flipshort in MP cells exposed to 6BG/TMZ. We next determined if decreases in c-flipshort correlated with activation of FAS-mediated signaling (Fig 5). To directly determine if FAS-mediated signaling is operative during 6BG/TMZ-mediated cell death, agonistic (CH-11) and neutralizing (ZB4) anti-FAS antibodies were used. Jurkat cells served as a positive control since FAS-mediated signaling is operative when these cells undergo apoptosis. (20) Treatment of Jurkat cells with agonistic anti-FAS antibodies resulted in cell death. This could be prevented by pre-incubation of Jurkat cells with the neutralizing anti-FAS antibodies followed by incubation with the agonistic anti-FAS antibodies (Fig 5A). When 6BG/TMZ-treated MP cells were incubated with agonistic anti-FAS in the absence or presence of isotype control or neutralizing anti-FAS antibodies for 3 days, there were no significant changes in cell death, indicating that at least under these conditions, FAS-mediated signaling was not operative in MP cells treated with 6BG/TMZ. Caspase-8 has been previously reported to be involved in myeloid cell differentiation (21, 22) and bioactivity analyses indicated detectable levels of caspase-8 activity in the myeloid cultures; however, the levels of active caspase-8 did not change following exposure to 6BG/TMZ (Fig 5B). These data show that while the extrinsic, FAS-mediated cell-death pathway is upregulated molecularly in response to 6BG/TMZ treatment, it is the intrinsic, mitochondrial-mediated pathway that is fully activated and correlates with increased apoptosis of MP cells.
Figure 4. Modulation of FAS and c-FLIP expression in MP cells exposed to 6BG/TMZ.
(A) Time course of FAS expression was performed during the expansion period. FAS expression was analyzed in triplicate via flow cytometry on cells in the early expansion and differentiation phase - days 0-8 post-expansion. (B) At 10 days post-expansion, control MP cells (C) or MP cells were exposed to 6BG, TMZ, or combination of 6BG/TMZ for 3 days. The percent of FAS positive and the relative density of FAS expression was determined via flow cytometry. (C) MP cells were exposed to 6BG, TMZ, or 6BG/TMZ for 3 days and cellular lysates analyzed via Western blot for c-FLIPshort and c-FLIP long. Flow cytometric data are expressed as the mean and standard deviation of triplicate data points for each group. Flow cyotmetric data and Western analysis were repeated twice with similar results.
Figure 5. Relevance of FAS-mediated signaling in MP cells exposed to 6BG/TMZ.
(A) As a control for antibody-mediated signaling through FAS, Jurkat T cells were incubated in triplicate with agonistic (Ag), neutralizing (Neut), isotype (Iso) or combinations of antibodies for 24 hours and the percent of nonviable cells determined via Annexin V staining and flow cytometry. MP cells that had been expanded in culture for 10 days were treated with 6BG/TMZ for 3 days in the presence of combinations of agonistic (Ag), neutralizing (Neut), and isotype (Iso) antibodies. Antibody was added each day of the 3-day culture and nonviable cells determined via Annexin V staining and flow cytometry on day 4 post-treatment. (B) Caspase-8 activity was determined in lysates from control and 6BG/TMZ-treated cells in the +/- of a caspase-8 specific inhibitor. Data are representative of two independent experiments (A) or 3 independent experiments (B).
DISCUSSION
In the present MP ex vivo model, we determined the extent to which the mitochondrial-versus FAS-mediated cell-death pathways are activated in 6BG/TMZ-treated MP cells. We focused on DNA-damage responses elicited via a TMZ-based regimen previously used in the clinic that can be myelosuppressive.(6) In 6BG/TMZ-treated MP cells, p53-mediated signaling was activated and the intrinsic, mitochondrial-mediated cell death pathway predominated following exposure to 6BG/TMZ. Interestingly, damage to both mitochondrial and nuclear DNA persisted following exposure to 6BG/TMZ. Increases in mitochondrial membrane depolarization and caspase-9 activity following 6BG/TMZ exposure were observed. Decreased phosphorylation of AKT at serine 473, an indicator of decreased AKT activity, was observed in 6BG/TMZ-treated MP cells. (23) AKT blocks apoptosis through phosphorylation and inactivation of pro-apoptotic proteins such as Bad,(24) forkhead transcription factors,(25) and caspase-9. (26) Decreased AKT activity correlated with increased caspase-9 activity in treated MP cells. In comparison to control, 6BG, or TMZ, cell-surface FAS expression increased in 6BG/TMZ-treated MP cells. Caspase-8 inhibitors, c-FLIPlong and c-FLIPshort, which can associate with the FAS-receptor complex were constitutively expressed in MP cells, and c-FLIPshort expression decreased in the presence of 6BG/TMZ. However, this did not correlate with increases in caspase-8 activity. In addition, experiments with agonistic and neutralizing anti-FAS antibodies further confirmed that FAS-mediated signaling resulting in cell death was not operative.
The response of mammalian cells to DNA-damaging agents is dependent on complex intracellular signaling networks and the interplay of these networks can be cell-type specific. Depending on the cell type studied, the extrinsic, death receptor-mediated versus intrinsic, mitochondrial-mediated apoptotic pathways induced by chemotherapy can be preferentially activated. (27-30) While O6MeG adducts induce FAS-mediated apoptosis in primary human T-lymphocytes(29), formation of O6MeG adducts induced mitochondrial-mediated cell death in primary human fibroblasts (28) and B-lymphoblastoid cells. (27) In glioblastoma multiforme cells, Roos et al previously showed that relative rates of cell proliferation, double-strand breaks, MGMT levels, and p53 status could dictate whether GBM cells died predominantly via a mitochondrial-versus FAS-mediated apoptotic cell death. (30)
In MP cells, although FAS was expressed and c-FLIPshort levels were decreased in 6BG/TMZ-treated cells, MP cells were not sensitive to signals delivered through the FAS receptor complex. There are 3 isoforms of c-FLIP that can be detected in mammalian cells – c-FLIPlong, c-FLIPRAJI (R), and c-FLIPshort. (19) Both the c-FLIPlong and c-FLIPshort were constitutively expressed on the MP cells and only the c-FLIPshort isoform was modulated by 6BG/TMZ exposure. Others have shown that both of these isoforms can be found at the FAS death-inducing signaling complex (DISC) and can block procaspase-8 activation and death-receptor-mediated apoptosis.(19) However, the precise function of c-FLIPlong – pro-apoptotic versus anti-apoptotic- can be based on cellular context. For example, cFLIPlong exhibits pro-apoptotic function when there is strong receptor activation or when high levels of short c-FLIP isoforms – c-FLIPshort or c-FLIPR - are present.(31) Poukkula et al previously demonstrated that c-FLIPshort in contrast to c-FLIPlong was more susceptible to ubiquitination and proteasomal degradation and was partially attributable to two lysine residues in the C-terminal 20 amino acids that are unique to c-FLIPshort. (32) Whether FAS-mediated signaling plays a different functional role or becomes active once the MP cells fully mature is possible. Bauer et al recently reported that TMZ-mediated apoptosis of human peripheral blood monocytes induced both the mitochondrial- and FAS-mediated apoptotic pathways. (33)
Maintenance of both nuclear and mitochondrial genome stability is essential for normal cell survival and also prevents the emergence of pre-cancerous cells. Mitochondria function not only as the primary energy producer but also as a key regulator of cell survival and death.(34) Other laboratories have shown that mitochondrial DNA can incur greater levels of damage than nuclear DNA following treatment with alkylating or oxidizing agents. (35-37) Indeed, various disease states such as diabetes, ischemic heart disease, Parkinson’s disease, Alzheimer’s disease and the normal aging process have been correlated with increased mutations in mitochondrial DNA.(38, 39) Studies of mitochondrial DNA repair demonstrate the presence of base excision repair (39, 40) and mismatch repair in the mitochondria,(41, 42) but no nuclear excision repair. There is some evidence for reversal of O6-methylguanine lesions by direct repair in mitochondria.(43, 44) However, MGMT is most likely not found in the mitochondria since it does not contain a mitochondrial-localization signal. Additionally, we previously could not detect endogenous MGMT protein in the mitochondria via confocal microscopy in human hematopoietic cells. (45) The importance of mitochondrial DNA damage was previously documented by our group. In this study, we demonstrated that forced localization of MGMT via fusion of a mitochondrial-localization signal to MGMT, resulted in detectable MGMT in the mitochondria and that expression of MGMT in the mitochondria correlated with increased resistance to alkylator exposure in primary hematopoietic progenitor cells and in the leukemic cell line K562.(45) In our current study, we directly measure DNA damage and repair kinetics in the mitochondria of MP cells. Significant levels of DNA damage in the mitochondrial genome of 6BG/TMZ-treated MP cells were evident. We also demonstrate that depletion of a DNA repair protein (i.e. MGMT) that is normally only localized in the nucleus can have a profound effect on mitochondrial DNA damage status and function. Additionally, lack of repair to the mitochondrial DNA can lead to a secondary generation of ROS which also triggers the intrinsic pathway of apoptosis.
An inverse correlation existed between mitochondrial DNA damage and the presence of MGMT in MP cells. A tight interconnection between mitochondrial and nuclear genome integrity most likely accounts for our observations. The persistence of nuclear DNA damage caused by 6BG-mediated MGMT downregulation and activation of p53-mediated signaling is most likely linked to mitochondrial dysfunction. Further, a large number of genes encoding mitochondria-localized proteins are found in the nuclear genome and damage to the nuclear genome could affect transcription of numerous nuclear-encoded genes.(46) Additionally, increases in oxidative damage, abasic sites, methylated adducts, and DNA-strand breaks in the mitochondrial DNA could all lead to mitochondrial dysfunction and apoptosis. Consistent with a close relationship between integrity of mitochondrial and nuclear genomes, Martin et al previously demonstrated that the PTEN-induced putative kinase 1 (PINK1), a nuclear-encoded protein that is specifically found in the mitochondria, has a profound effect on nuclear DNA integrity.(47) In this study, both nuclear and mitochondrial oxidative DNA lesions were enhanced in cells that did not express mitochondrial-localized PINK1. These findings suggest that modulation of mitochondrial-specific DNA damage could affect nuclear-specific DNA damage and that regulation of mitochondrial and nuclear integrity can be closely linked. Our study is the first to document the presence of mitochondrial DNA damage following exposure to TMZ and 6BG/TMZ in primary human MP cells. Additionally, we found that the degree of nuclear DNA damage in MP cells depleted of MGMT via 6BG could influence mitochondrial function.
Basic research investigations into understanding how non-transformed cells respond to and repair pro-mutagenic DNA lesions will be important for future screening and refinement of new therapeutics. Adequate detection of intracellular signaling proteins and modulation of these pathways can be challenging when working with primary culture systems. In this model system, generation of primary human MP cells required only 2 cytokines: G-CSF and SCF. Large numbers of cells adequate for most molecular analyses could be obtained. Different hematopoietic lineages can have differential sensitivities to genotoxic agents. For example, others have demonstrated that human monocytes but not macrophages or monocyte-derived dendritic cells were highly sensitive to the killing effect of TMZ and other methylating agents, but had similar sensitivities to the cross-linking chemotherapeutics fotemustine, mafosfamide, and cisplatin.(48) (33) While hematopoietic colony-forming unit assays (49) and humanized mouse models(11) have merit as screening tools for compound-induced toxicity, MP cells can be used not only for screening new therapeutics but also for comparing the pharmacodynamic molecular profiles of how normal hematopoietic versus cancer cells respond to different chemotherapeutic compounds. It is important to emphasize, however, that alkylating agents such as TMZ can be toxic to hematopoietic stem cells and toxicity can be more pronounced when given in combination with O6-benzylguanine. While the MP model cannot be used to probe mechanisms of alkylating agent-mediated toxicity to the hematopoietic stem-cell compartment, it does present a feasible in vitro model to understand how primary myeloid cells respond at the molecular level to therapy-induced DNA damage. Due to differences in cell cycling, DNA repair capacity, and bone marrow microenvironmental cues, it is highly likely that the kinetics and molecular mechanisms of DNA damage responses in progenitors versus stem cells are different. For example, in studies by Milyavsky et al, human hematopoietic stem cells derived from umbilical cord blood exhibited slower repair of ionizing radiation-induced DNA damage compared to progenitor cells; this correlated with increased apoptosis of stem cells compared to the progenitor cells. (50) It would be of interest in the future studies to investigate molecular responses of nucleotide analogs such as cytarabine and vidarabine that specifically target more rapidly cycling hematopoietic progenitor cells, as studied in the present model. The MP cell model can also be used to explore at the molecular level the sensitivity to different modes of DNA damage, signaling mechanisms operative under different types of DNA stress, and for testing strategies that improve DNA repair and maintain genome integrity in normal cells.
Supplementary Material
Translational relevance.
In response to chemotherapeutic treatment, cancer patients can experience adverse side effects due to myelosuppression as well as increased propensity to develop secondary hematological malignancies. However, model systems designed to study molecular events initiated by chemotherapy-mediated toxicity in immature hematopoietic cells are lacking. To address this important clinical issue, we examined the DNA-damage response of human myeloid precursor cells exposed to a temozolomide-based regimen known to cause myelosuppression in patients. Activation of the FAS/CD95/APO1- versus the mitochondrial-mediated cell-death pathway was investigated. When temozolomide-induced DNA damage persisted in the nuclear and mitochondrial genomes, apoptosis of myeloid precursor cells was induced by the mitochondrial- but not the FAS-mediated cell-death pathway. Primary human MP cells represent a clinically relevant approach to investigate how non-transformed primary hematopoietic cells respond to chemotherapy-induced DNA damage. Understanding the molecular basis of myelosuppression will aid in the development and design of more effective chemotherapies for cancer treatment.
Acknowledgments
The authors wish to thank Mary Murray and Dr. Harlan Shannon for their helpful discussions and critical reading of this manuscript. We thank the nursing staff at the St. Vincent Women’s Hospital for donation of their time for collection of umbilical cord blood products. We also acknowledge the continuing support of the Riley Children’s Foundation and the Indiana University Simon Cancer Center.
GRANT SUPPORT
This work was supported by RO1 CA138798 (K.E. Pollok); KO8 HL75253 (W.S. Goebel); the Riley Children’s Foundation (H. Wang, L.D. Mayo, W.S. Goebel, and K. E. Pollok), Jeff Gordon Research Foundation (H. Wang, L.D. Mayo, and K.E. Pollok), CA109262 (L.D. Mayo) VA-CDA2 (A. Suvannasankhah), and the NIH/NIDDK P30DK090948 CEMH (K.E. Pollok).
Footnotes
Conflict of Interest: A perceived conflict of interest exists for Dr. W. Scott Goebel. He holds the following positions: Medical Director, General BioTechnology, LLC; Medical Director and ownership interest, The Genesis Bank, LLC.; and, Medical Director and ownership interest, RenovoCyte, LLC. There are no conflicts of interest to declare for all other authors.
References
- 1.Parent-Massin D, Hymery N, Sibiril Y. Stem cells in myelotoxicity. Toxicology. 2010;267:112–7. doi: 10.1016/j.tox.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 2.Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell stem cell. 2012;10:120–36. doi: 10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Casorelli I, Pelosi E, Biffoni M, Cerio AM, Peschle C, Testa U, et al. Methylation damage response in hematopoietic progenitor cells. DNA repair. 2007;6:1170–8. doi: 10.1016/j.dnarep.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 4.Gerson SL, Phillips W, Kastan M, Dumenco LL, Donovan C. Human CD34+ hematopoietic progenitors have low, cytokine-unresponsive O6-alkylguanine-DNA alkyltransferase and are sensitive to O6-benzylguanine plus BCNU. Blood. 1996;88:1649–55. [PubMed] [Google Scholar]
- 5.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 6.Quinn JA, Desjardins A, Weingart J, Brem H, Dolan ME, Delaney SM, et al. Phase I trial of temozolomide plus O6-benzylguanine for patients with recurrent or progressive malignant glioma. J Clin Oncol. 2005;23:7178–87. doi: 10.1200/JCO.2005.06.502. [DOI] [PubMed] [Google Scholar]
- 7.Strik HM, Marosi C, Kaina B, Neyns B. Temozolomide dosing regimens for glioma patients. Current neurology and neuroscience reports. 2012;12:286–93. doi: 10.1007/s11910-012-0262-y. [DOI] [PubMed] [Google Scholar]
- 8.Denny BJ, Wheelhouse RT, Stevens MF, Tsang LL, Slack JA. NMR and molecular modeling investigation of the mechanism of activation of the antitumor drug temozolomide and its interaction with DNA. Biochemistry. 1994;33:9045–51. doi: 10.1021/bi00197a003. [DOI] [PubMed] [Google Scholar]
- 9.Kaina B, Christmann M, Naumann S, Roos WP. MGMT: key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA repair. 2007;6:1079–99. doi: 10.1016/j.dnarep.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Karran P, Stephenson C. Mismatch binding proteins and tolerance to alkylating agents in human cells. Mutation research. 1990;236:269–75. doi: 10.1016/0921-8777(90)90010-3. [DOI] [PubMed] [Google Scholar]
- 11.Cai S, Wang H, Bailey B, Ernstberger A, Juliar BE, Sinn AL, et al. Humanized bone marrow mouse model as a preclinical tool to assess therapy-mediated hematotoxicity. Clin Cancer Res. 2011;17:2195–206. doi: 10.1158/1078-0432.CCR-10-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levenson R, Macara IG, Smith RL, Cantley L, Housman D. Role of mitochondrial membrane potential in the regulation of murine erythroleukemia cell differentiation. Cell. 1982;28:855–63. doi: 10.1016/0092-8674(82)90064-2. [DOI] [PubMed] [Google Scholar]
- 13.Driggers WJ, Holmquist GP, LeDoux SP, Wilson GL. Mapping frequencies of endogenous oxidative damage and the kinetic response to oxidative stress in a region of rat mtDNA. Nucleic Acids Res. 1997;25:4362–9. doi: 10.1093/nar/25.21.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammond LA, Eckardt JR, Baker SD, Eckhardt SG, Dugan M, Forral K, et al. Phase I and pharmacokinetic study of temozolomide on a daily-for-5-days schedule in patients with advanced solid malignancies. J Clin Oncol. 1999;17:2604–13. doi: 10.1200/JCO.1999.17.8.2604. [DOI] [PubMed] [Google Scholar]
- 15.Broniscer A, Gururangan S, MacDonald TJ, Goldman S, Packer RJ, Stewart CF, et al. Phase I trial of single-dose temozolomide and continuous administration of o6-benzylguanine in children with brain tumors: a pediatric brain tumor consortium report. Clin Cancer Res. 2007;13:6712–8. doi: 10.1158/1078-0432.CCR-07-1016. [DOI] [PubMed] [Google Scholar]
- 16.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, et al. Mechanism of activation of protein kinase B by insulin and IGF-1. The EMBO journal. 1996;15:6541–51. [PMC free article] [PubMed] [Google Scholar]
- 18.Krueger A, Schmitz I, Baumann S, Krammer PH, Kirchhoff S. Cellular FLICE-inhibitory protein splice variants inhibit different steps of caspase-8 activation at the CD95 death-inducing signaling complex. J Biol Chem. 2001;276:20633–40. doi: 10.1074/jbc.M101780200. [DOI] [PubMed] [Google Scholar]
- 19.Safa AR, Pollok KE. Targeting the Anti-Apoptotic Protein c-FLIP for Cancer Therapy. Cancers. 2011;3:1639–71. doi: 10.3390/cancers3021639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aronis A, Melendez JA, Golan O, Shilo S, Dicter N, Tirosh O. Potentiation of Fas-mediated apoptosis by attenuated production of mitochondria-derived reactive oxygen species. Cell death and differentiation. 2003;10:335–44. doi: 10.1038/sj.cdd.4401150. [DOI] [PubMed] [Google Scholar]
- 21.Kang TB, Ben-Moshe T, Varfolomeev EE, Pewzner-Jung Y, Yogev N, Jurewicz A, et al. Caspase-8 serves both apoptotic and nonapoptotic roles. J Immunol. 2004;173:2976–84. doi: 10.4049/jimmunol.173.5.2976. [DOI] [PubMed] [Google Scholar]
- 22.Pellegrini M, Bath S, Marsden VS, Huang DC, Metcalf D, Harris AW, et al. FADD and caspase-8 are required for cytokine-induced proliferation of hemopoietic progenitor cells. Blood. 2005;106:1581–9. doi: 10.1182/blood-2005-01-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheppard K, Kinross KM, Solomon B, Pearson RB, Phillips WA. Targeting PI3 kinase/AKT/mTOR signaling in cancer. Critical reviews in oncogenesis. 2012;17:69–95. doi: 10.1615/critrevoncog.v17.i1.60. [DOI] [PubMed] [Google Scholar]
- 24.Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, et al. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–41. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 25.Huang H, Tindall DJ. Dynamic FoxO transcription factors. Journal of cell science. 2007;120:2479–87. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 26.Cardone MH, Roy N, Stennicke HR, Salvesen GS, Franke TF, Stanbridge E, et al. Regulation of cell death protease caspase-9 by phosphorylation. Science. 1998;282:1318–21. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 27.Hickman MJ, Samson LD. Apoptotic signaling in response to a single type of DNA lesion, O(6)-methylguanine. Mol Cell. 2004;14:105–16. doi: 10.1016/s1097-2765(04)00162-5. [DOI] [PubMed] [Google Scholar]
- 28.Ochs K, Kaina B. Apoptosis induced by DNA damage O6-methylguanine is Bcl-2 and caspase-9/3 regulated and Fas/caspase-8 independent. Cancer Res. 2000;60:5815–24. [PubMed] [Google Scholar]
- 29.Roos W, Baumgartner M, Kaina B. Apoptosis triggered by DNA damage O6-methylguanine in human lymphocytes requires DNA replication and is mediated by p53 and Fas/CD95/Apo-1. Oncogene. 2004;23:359–67. doi: 10.1038/sj.onc.1207080. [DOI] [PubMed] [Google Scholar]
- 30.Roos WP, Batista LF, Naumann SC, Wick W, Weller M, Menck CF, et al. Apoptosis in malignant glioma cells triggered by the temozolomide-induced DNA lesion O6-methylguanine. Oncogene. 2007;26:186–97. doi: 10.1038/sj.onc.1209785. [DOI] [PubMed] [Google Scholar]
- 31.Fricker N, Beaudouin J, Richter P, Eils R, Krammer PH, Lavrik IN. Model-based dissection of CD95 signaling dynamics reveals both a pro- and antiapoptotic role of c-FLIPL. The Journal of cell biology. 2010;190:377–89. doi: 10.1083/jcb.201002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poukkula M, Kaunisto A, Hietakangas V, Denessiouk K, Katajamaki T, Johnson MS, et al. Rapid turnover of c-FLIPshort is determined by its unique C-terminal tail. J Biol Chem. 2005;280:27345–55. doi: 10.1074/jbc.M504019200. [DOI] [PubMed] [Google Scholar]
- 33.Bauer M, Goldstein M, Heylmann D, Kaina B. Human Monocytes Undergo Excessive Apoptosis following Temozolomide Activating the ATM/ATR Pathway While Dendritic Cells and Macrophages Are Resistant. PloS one. 2012;7:e39956. doi: 10.1371/journal.pone.0039956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horbinski C, Chu CT. Kinase signaling cascades in the mitochondrion: a matter of life or death. Free Radic Biol Med. 2005;38:2–11. doi: 10.1016/j.freeradbiomed.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 35.LeDoux SP, Driggers WJ, Hollensworth BS, Wilson GL. Repair of alkylation and oxidative damage in mitochondrial DNA. Mutation research. 1999;434:149–59. doi: 10.1016/s0921-8777(99)00026-9. [DOI] [PubMed] [Google Scholar]
- 36.Grishko VI, Druzhyna N, LeDoux SP, Wilson GL. Nitric oxide-induced damage to mtDNA and its subsequent repair. Nucleic Acids Res. 1999;27:4510–6. doi: 10.1093/nar/27.22.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobson AW, Xu Y, Kelley MR, LeDoux SP, Wilson GL. Enhanced mitochondrial DNA repair and cellular survival after oxidative stress by targeting the human 8-oxoguanine glycosylase repair enzyme to mitochondria. J Biol Chem. 2000;275:37518–23. doi: 10.1074/jbc.M000831200. [DOI] [PubMed] [Google Scholar]
- 38.de Souza-Pinto NC, Bohr VA. The mitochondrial theory of aging: involvement of mitochondrial DNA damage and repair. Int Rev Neurobiol. 2002;53:519–34. doi: 10.1016/s0074-7742(02)53018-x. [DOI] [PubMed] [Google Scholar]
- 39.Bohr VA. Mitochondrial DNA repair. Prog Nucleic Acid Res Mol Biol. 2001;68:255–6. doi: 10.1016/s0079-6603(01)68104-2. [DOI] [PubMed] [Google Scholar]
- 40.LeDoux SP, Wilson GL, Beecham EJ, Stevnsner T, Wassermann K, Bohr VA. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis. 1992;13:1967–73. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- 41.de Souza-Pinto NC, Mason PA, Hashiguchi K, Weissman L, Tian J, Guay D, et al. Novel DNA mismatch-repair activity involving YB-1 in human mitochondria. DNA repair. 2009;8:704–19. doi: 10.1016/j.dnarep.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mason PA, Matheson EC, Hall AG, Lightowlers RN. Mismatch repair activity in mammalian mitochondria. Nucleic Acids Res. 2003;31:1052–8. doi: 10.1093/nar/gkg167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myers KA, Saffhill R, O’Connor PJ. Repair of alkylated purines in the hepatic DNA of mitochondria and nuclei in the rat. Carcinogenesis. 1988;9:285–92. doi: 10.1093/carcin/9.2.285. [DOI] [PubMed] [Google Scholar]
- 44.Satoh MS, Huh N, Rajewsky MF, Kuroki T. Enzymatic removal of O6-ethylguanine from mitochondrial DNA in rat tissues exposed to N-ethyl-N-nitrosourea in vivo. J Biol Chem. 1988;263:6854–6. [PubMed] [Google Scholar]
- 45.Cai S, Xu Y, Cooper RJ, Ferkowicz MJ, Hartwell JR, Pollok KE, et al. Mitochondrial targeting of human O6-methylguanine DNA methyltransferase protects against cell killing by chemotherapeutic alkylating agents. Cancer Res. 2005;65:3319–27. doi: 10.1158/0008-5472.CAN-04-3335. [DOI] [PubMed] [Google Scholar]
- 46.Neupert W. Protein import into mitochondria. Annual review of biochemistry. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 47.Martin SA, Hewish M, Sims D, Lord CJ, Ashworth A. Parallel high-throughput RNA interference screens identify PINK1 as a potential therapeutic target for the treatment of DNA mismatch repair-deficient cancers. Cancer Res. 2011;71:1836–48. doi: 10.1158/0008-5472.CAN-10-2836. [DOI] [PubMed] [Google Scholar]
- 48.Briegert M, Kaina B. Human monocytes, but not dendritic cells derived from them, are defective in base excision repair and hypersensitive to methylating agents. Cancer Res. 2007;67:26–31. doi: 10.1158/0008-5472.CAN-06-3712. [DOI] [PubMed] [Google Scholar]
- 49.Pessina A, Albella B, Bayo M, Bueren J, Brantom P, Casati S, et al. Application of the CFU-GM assay to predict acute drug-induced neutropenia: an international blind trial to validate a prediction model for the maximum tolerated dose (MTD) of myelosuppressive xenobiotics. Toxicol Sci. 2003;75:355–67. doi: 10.1093/toxsci/kfg188. [DOI] [PubMed] [Google Scholar]
- 50.Milyavsky M, Gan OI, Trottier M, Komosa M, Tabach O, Notta F, et al. A distinctive DNA damage response in human hematopoietic stem cells reveals an apoptosis-independent role for p53 in self-renewal. Cell stem cell. 2010;7:186–97. doi: 10.1016/j.stem.2010.05.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.