Abstract
The eukaryotic RFC clamp loader couples the energy of ATP hydrolysis to open and close the circular PCNA sliding clamp onto primed sites for use by DNA polymerases and repair factors. Structural studies reveal clamp loaders to be heteropentamers. Each subunit contains a region of homology to AAA+ proteins that defines two domains. The AAA+ domains form a right-handed spiral upon binding ATP. This spiral arrangement generates a DNA binding site within the center of RFC. DNA enters the central chamber through a gap between the AAA+ domains of two subunits. Specificity for a primed template junction is achieved by a third domain that blocks DNA, forcing it to bend sharply. Thus only DNA with a flexible joint can bind the central chamber. DNA entry also requires a slot in the PCNA clamp, which is opened upon binding the AAA+ domains of the clamp loader. ATP hydrolysis enables clamp closing and ejection of RFC, completing the clamp loading reaction.
Keywords: Clamp loader, Sliding clamp, DNA polymerase, Replisome, RFC, PCNA, AAA+ machine
14.1 Overview of Clamp Loaders and Sliding Clamps
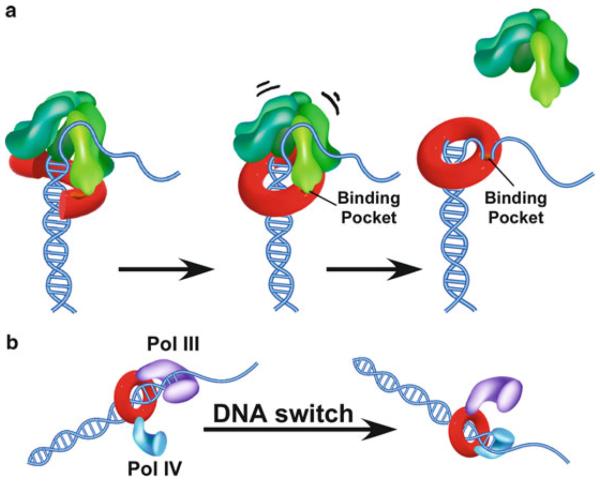
Clamp loaders are so called for their action in loading ring-shaped sliding clamps onto DNA (see Fig. 14.1a). Sliding clamps encircle DNA and slide along the duplex while binding DNA polymerases, tethering them to DNA for high processivity during chain extension (reviewed in O'Donnell and Kuriyan 2006). Clamps and clamp loaders are ubiquitous in all life forms and thus must have evolved in the progenitor ancestor cell from which all different cell types arose.
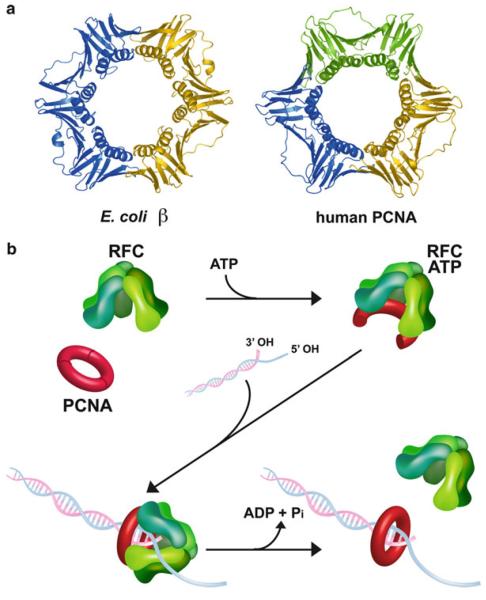
Fig. 14.1.
Scheme of clamp loader function. (a) The structures of the E. coli β and human PCNA sliding clamps (PDB: 2POL and PDB: AXC, respectively). (b) Schematic of clamp loader function using eukaryotic RFC and PCNA as the example. ATP binding to RFC enables RFC to bind and open PCNA. In the presence of a primed template, RFC places PCNA onto DNA and then hydrolyzes ATP to eject from the PCNA-DNA complex (Reproduced with permission from Figure 1a of Bowman et al. (2004))
Sliding clamps were the first type of protein known to function by encircling DNA (Kong et al. 1992; Stukenberg et al. 1991). Today, many DNA metabolic proteins are known to encircle DNA for performance of their function. The structures of the E. coli β clamp and the eukaryotic PCNA (proliferating cell nuclear antigen) clamp are shown in Fig. 14.1a (Gulbis et al. 1996; Krishna et al. 1994). The eukaryotic PCNA clamp is a homotrimer, while the bacterial clamp is a homodimer. Despite this difference in oligomeric state, the eukaryotic and bacterial clamps have essentially the same structure. In both cases, the clamps are six domain rings, and each domain has the same chain folding pattern. The domains have fused together during evolution in various ways giving rise to the different oligomerization states. In PCNA, the individual protomers of the trimer are composed of two domains each. In bacteria, the clamps are homodimers, and each protomer consists of three domains. The structure of sliding clamps from archaeal cells and bacteriophage T4 have also been solved; they share the same general chain fold and trimeric oligomerization state as PCNA (Dore et al. 2006; Matsumiya et al. 2001; Moarefi et al. 2000; Shamoo and Steitz 1999). The detailed structure and function of PCNA is the subject of the Chap. 15.
The crystal structures showed the sliding clamp rings were closed, implying that another factor was needed to crack these ring-shaped clamps open and then reclose them onto DNA. In fact, a second protein was known to be required for the clamp to function with DNA polymerase. In all systems, this “clamp loader” protein is a pentameric ATPase. The eukaryotic clamp loader was first identified as a necessary protein for SV40 replication in vitro (Fairman et al. 1988). The exact function was not known at the time and it was named replication factor C (RFC) (Waga and Stillman 1994) or activator-1 (Lee et al. 1989). The name RFC gained widespread use and is the term used today for eukaryotic and archaeal clamp loaders (Grabowski and Kelman 2003). The names of the clamp loader, clamp and replicative DNA polymerase in different cell types are given in Table 14.1.
Table 14.1.
Three component subunit structures of replicases from diverse organisms
| Organism | Polymerase | Clamp loader pentamer | Sliding clamp | |
|---|---|---|---|---|
| E. coli | Pol III |
|
β dimer | |
| T4 phage | gp43 | gp44/62 pentamer | gp45 trimer | |
| Archaea | Pol B | RFC pentamer | PCNA trimer | |
| Eukaryotes | Pol δ, Pol ε | RFC pentamer | PCNA trimer |
Clamp loaders are heteropentamers and they hydrolyze ATP to assemble their respective clamps onto a primed DNA site (see Fig. 14.1b) (O'Donnell and Kuriyan 2006). However, the clamp loader binds the same surface of the clamp as the DNA polymerase and prevents the interaction of DNA polymerase with the clamp (Jonsson et al. 1998; Naktinis et al. 1996). Thus, upon placing the clamp on DNA, the clamp loader must eject from the clamp to enable the polymerase access to the clamp. The hydrolysis of ATP accomplishes clamp loader ejection from the clamp as illustrated in the second step of Fig. 14.1b (Ason et al. 2003). The polymerase then binds to the clamp for highly processive function.
14.2 Clamp Loader Structure
The RFC clamp loader is composed of five essential “clamp loading” subunits referred to as RFC1 through 5 (Cullmann et al. 1995). With the exception of the large RFC1 subunit (approximately 128 kDa in human), the RFC2, 3, 4 and 5 subunits are approximately 38–41 kDa each. The five subunits contain a region of homology with one another (O'Donnell et al. 1993). This region of homology defines a large family of proteins referred to as AAA+ proteins (ATPases associated with a variety of cellular functions) (Erzberger and Berger 2006). The AAA+ region encodes two domains, the structure of which will be discussed shortly. AAA+ proteins generally perform protein remodeling reactions in a wide variety of cellular pathways (Neuwald et al. 1999). Many AAA+ proteins are circular hexamers, although other oligomerization states exist and in fact, the heteropentameric clamp loader is one of these exceptions. Numerous examples of AAA+ proteins participate in the replication process, including origin binding proteins (i.e. several subunits of the ORC complex, Cdc6/Cdc18, the Mcm2–7 helicase and of course, the RFC clamp loader) (Davey et al. 2002).
The AAA+ homologous region folds into two domains, and these domains bind ATP (Guenther et al. 1997). The P-loop and DEAD box ATP site motifs are located on the larger, N-terminal domain, and the smaller domain contains several residues that are important to binding and/or hydrolysis. In all clamp loader subunits there is at least one additional domain that is C-terminal to the AAA+ domains. The C-terminal domain that is outside of the region of AAA+ homology is mostly composed of α-helix and it mediates the strongest intersubunit interactions that hold the pentamer together. RFC1, the largest subunit of RFC, contains both N- and C- terminal extensions in addition to these three domains (Bunz et al. 1993). Although the N-terminal extension of RFC1 contains a region of homology to DNA ligases (i.e. the BRCT domain), it does not have ligase activity. The solution structure of the human RFC1 N-terminal BRCT domain has recently been solved and a model for BRCT DNA binding presented (Kobayashi et al. 2010). The N-terminal extension of RFC1 is not essential for cell viability (Gomes et al. 2000), nor is it required for in vitro clamp loading activity (Uhlmann et al. 1997), but removal of this region results in sensitivity to DNA damage in vivo (Gomes et al. 2000).
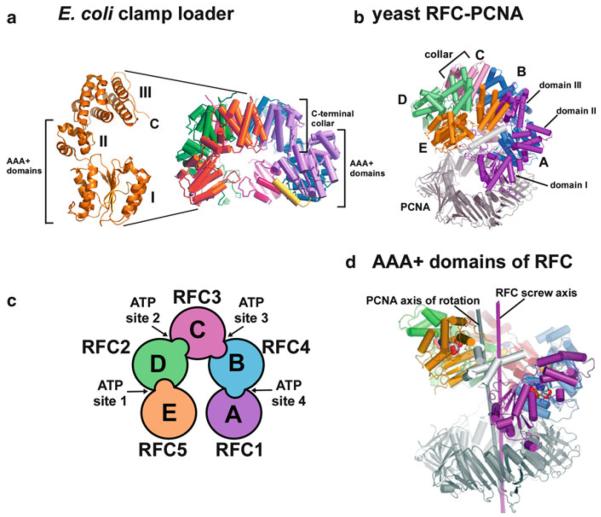
The subunits of RFC, like clamp loaders of all cell types, are arranged in a circular shape (Bowman et al. 2004; Jeruzalmi et al. 2001a). The crystal structures of the bacterial and eukaryotic clamp loaders, in Fig. 14.2a and b, respectively, show that the AAA+ domains of the five subunits are arranged in a spiral, while the C-terminal domains define a nearly planar circle, referred to as a “collar”. One subunit of the E. coli clamp loader is shown at the right of Fig. 14.2a, to illustrate the three domains structure of clamp loader subunits (Jeruzalmi et al. 2001a). By convention, clamp loaders are viewed from the “side” with the C-terminal domain at the top, and the N-terminal AAA+ domains at the bottom (Jeruzalmi et al. 2001a). Proceeding counterclockwise around the circle from the subunit at the far right, the subunit positions are referred to as the A, B, C, D and E subunits (see Fig. 14.2b and c). The C-terminal domains of the collar form a tightly closed circle with no gap and are the main connections that hold the complex together. In all clamp loaders, there is a gap between the AAA+ domains of subunits A and E (i.e. see Fig. 14.2a). In RFC this gap is present between RFC1 and RFC5 (Fig. 14.2b and c). In RFC, this gap is somewhat narrower than in the E. coli clamp loader because RFC1 has a C-terminal region that extends across the gap and protrudes down toward the N-terminal face of the clamp loader (colored white in Fig. 14.2b).
Fig. 14.2.
Architecture of the clamp loader. (a) E. coli minimal clamp loader heteropentamer of γ3δδ′ (PDB: 1JR3). The left part of the figure shows the three-domain architecture of each subunit. The right part of the figure shows the gap between the AAA+ domains of subunits δ (purple) and δ′ (orange). The yellow α helix of δ binds the β clamp at one end (see text for details) (Adapted with permission from Figure 1a of Jeruzalmi et al. (2001a)). (b) The yeast RFC-PCNA-ATP γ S complex (PDB: 1SXJ). RFC is in color and PCNA is in grey. The three domains of the subunits are as indicated, and subunits are noted according to their positions (a-e), which for yeast RFC are RFC1, RFC4, RFC3, RFC2 and RFC5, respectively (Adapted with permission from Figure 1b of Bowman et al. (2004)). (c) Cartoon of the arrangement of yeast RFC subunits. The location of ATP sites at subunit interfaces is indicated. ATP binding sites are within subunits RFC2 (ATP site 1), RFC3 (ATP site 2), RFC4 (ATP site 3) and RFC5 (ATP site 4). Subunits with arginine fingers that are needed for catalysis are in RFC5 (ATP site 1), RFC2 (ATP site 2), RFC3 (ATP site 3) and RFC4 (ATP site 4) (Adapted with permission from Figure 3b in O'Donnell and Kuriyan (2006)). (d) RFC–PCNA structure in which only the AAA+ domains of RFC are shown (color) and the collar is removed. The figure illustrates the spiral shape of the AAA+ domains. RFC1 (purple) forms the most extensive contact with PCNA (grey) (Reproduced with permission from Figure 2 of Bowman et al. (2004))
Although clamp loader subunits are homologous to AAA+ proteins, they do not all have a functional ATP binding site (e.g. lack the P-loop). Therefore, certain clamp loader subunits do not hydrolyze ATP. In the eukaryotic RFC clamp loader the RFC5 subunit lacks a P-loop. Thus the RFC pentamer has only four functional ATP sites. In fact, mutational studies show that only three of these sites are needed for PCNA clamp loading; the ATP site of RFC1 is not essential for clamp loading (Gomes and Burgers 2001). A common feature of AAA+ proteins is that the ATP site is located at the interface of two subunits, and catalysis requires residues from both subunits. Specifically, one subunit binds ATP, but residue(s) needed for catalysis of ATP are donated by the adjacent subunit. In RFC, one of these inter-subunit catalytic residues is an “arginine finger” set within a SRC motif that is conserved in clamp loader subunits of all cell types (Bowman et al. 2004; Jeruzalmi et al. 2001a; O'Donnell and Kuriyan 2006). Mutational analysis has shown that the arginine of the SRC motif is required for ATP catalysis (Johnson and O'Donnell 2003; Johnson et al. 2006; Snyder et al. 2004; Williams et al. 2004). The fact that the ATP sites of AAA+ oligomers are formed by residues of two neighboring subunits suggests that the AAA+ architecture may reflect an underlying necessity for intersubunit communication, consistent with the idea that cooperation of subunits within a large complex is important to performance of a complicated protein remodeling task.
14.3 RFC Clamp Loader Interaction with DNA
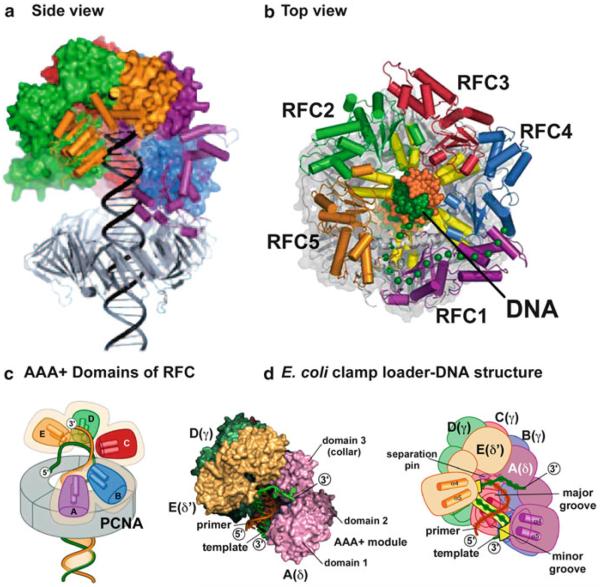
The structure of RFC in complex with PCNA and ATP γ S revealed an unanticipated mode by which clamp loaders bind to DNA, even though DNA was not present in the structure (Bowman et al. 2004, 2005). Modeling of DNA through the PCNA ring, located below the complex, indicated that DNA may reside inside the center of the clamp loader and be surrounded by all five subunits (see Fig. 14.3a). Surprisingly there is sufficient space inside the center of RFC to accommodate duplex DNA (Fig. 14.3b), but even more telling is the disposition of the AAA+ domains of all five subunits. The AAA+ domains are arranged in a right-handed spiral that closely match the pitch of the DNA duplex (Fig. 14.2d and illustration in Fig. 14.3c). This highly suggested that DNA binds inside the clamp loader. Furthermore, many positive charged residues present in each subunit are within hydrogen bonding distance to DNA modeled into the complex. These potential DNA interactive residues are conserved from bacteria to human, further suggesting their importance in bind to DNA (Bowman et al. 2005). These conserved residues are located on two α helices in each subunit that have positive dipoles pointed toward the direction of DNA modeled into the central chamber (illustrated in Fig. 14.3c). Although the AAA+ domains of the unliganded (no ATP) E. coli γ3δδ′ complex are also arranged in a spiral, the pitch is not as steep as that seen in the yeast RFC-PCNA-ATP γ S structure. This difference between unliganded E. coli clamp loader (no ATP) and ATP γ S bound RFC-PCNA suggests that ATP binding results in a conformational change that brings the AAA+ domains into the correct spiral shape to form a central DNA binding site. Subsequent mutational studies demonstrated that these positive charged residues are indeed required to bind DNA, both for the E. coli clamp loader and yeast RFC (Goedken et al. 2005; Yao et al. 2006).
Fig. 14.3.
Model of the RFC-PCNA-DNA complex. (a) Duplex DNA is modeled through PCNA and into the center of RFC (Reproduced with permission from Figure 3c in O'Donnell and Kuriyan (2006)). (b) View of the RFC-PCNA-DNA model looking from the “top” of RFC with the C-terminal “collar” removed. There exists space for DNA in the center of RFC, and each subunit contains residues on two α helices (yellow) that point toward the DNA (Adapted with permission from Figure 4b of Bowman et al. (2004)). (c) Cartoon of the spiral disposition of the AAA+ domains of RFC relative to DNA and PCNA (Reproduced with permission from Figure 4c of Bowman et al. (2004)). (d) Structure of the E. coli clamp loader bound to a primed DNA (left) and cartoon of the structure (right diagram) (PDB: 3GLF) (Simonetta et al. 2009). The template strand of DNA is colored green (left) or yellow (right) (Reproduced with permission from Figure 1c in Simonetta et al. (2009))
The proposal that DNA binds to the central chamber of the clamp loader provided an immediate answer to the question of why clamp loaders have a gap between two subunits. Since clamps are loaded at primed sites on template strands that do not have DNA “ends”, the DNA cannot simply enter the clamp loader through the “bottom”. Hence, the observed gap between two clamp loading subunits provide an entry port for an “endless” DNA strand to enter into the central cavity of the clamp loader. Entry of a long DNA strand would also require an open interface of the sliding clamp located under the clamp loader in which the clamp loader gap and the open interface of the clamp are aligned with one another for DNA entry.
Structural support for DNA binding to the central cavity of RFC was obtained from an electron microscopy study of an archaeal RFC-PCNA bound to DNA (Miyata et al. 2005). Although high resolution architectural details are not visible, the DNA appeared to reside inside the clamp loader. The electron microscopy study is described further below in the context of PCNA ring opening. A high resolution structure of DNA bound to the inside of a clamp loader has recently been obtained for the E. coli clamp loader in the presence of a primed template DNA (Simonetta et al. 2009). The structure, shown in Fig. 14.3d, confirms that duplex DNA resides within the central cavity of the γ3δδ′ clamp loader, and that each subunit interacts with the DNA backbone. Interestingly, the clamp loader only interacts with the template strand, not the primer strand. This finding was unanticipated and may reflect an important biological function. Specifically, the E. coli clamp loader must be capable of assembling the clamp onto an RNA primer made by primase, but it must also be capable of assembling the clamp onto a DNA primed site during various types of repair reactions outside the context of chromosome replication. RNA-DNA and DNA-DNA duplexes have different structures. RNA-DNA duplexes prefer the A-form, which has a much larger diameter than B-form duplex DNA. Interestingly, the fit of RNA-DNA modeled into the structure indicates that the template DNA strand interactions can be maintained with very little or no change. Furthermore, the central cavity has sufficient diameter to accommodate the greater diameter of A-form RNA-DNA relative to B-form DNA-DNA. Hence, the fact that the major interactions between the clamp loader and DNA occur through the template strand may facilitate binding to both A-form and B-form structures.
Primed sites are synthesized at nearly random positions during lagging strand synthesis ( Kornberg and Baker 1992), and thus clamp loaders must be capable of recognizing the structure of a primed site, not a specific sequence for clamp loading. Structure specific binding to a primed site is made possible by the tight packing of the C-terminal domains in the collar (Bowman et al. 2004, 2005). As discussed above, DNA will enter the central chamber of the clamp loader through the gap between the AAA+ domains of two subunits that are aligned with the open interface of the clamp, but the tightly packed C-terminal domains provide a “cap” that prevents DNA from going straight through the structure. This imposes the requirement that DNA must make a sharp bend in order to bind into the central chamber of the clamp loader. Duplex DNA is too rigid for this sharp bend, but the flexibility of single-strand DNA should allow this bend to occur. This is nicely apparent from the structure of the E. coli clamp loader bound to a primed template (Fig. 14.3d), in which the template strand bends out from the side of the clamp loader at the top of the central cavity, just below the “cap” formed by the C-terminal domains. Thus the clamp loader structure can be compared to a “screw cap” in which the screw binds duplex DNA and the cap imposes the sharp bend that provides specificity for a primed template junction that has a flexible template single strand.
14.4 ATP Binding and Opening of the Clamp
Studies in both E. coli and eukaryotes have shown that the clamp loader requires ATP binding to interact with the sliding clamp (Gomes and Burgers 2001; Naktinis et al. 1996). However, ATP does not need to be hydrolyzed for this function. Indeed, the E. coli clamp loader has been shown to open one interface of the β dimer upon binding ATP (or ATP γ S) (Turner et al. 1999). Therefore hydrolysis is not required for clamp binding or clamp opening, but is needed for completion of the reaction (clamp loader ejection/clamp closing).
Most proteins that bind the PCNA clamp contain a conserved motif, called the PIP (PCNA interacting peptide) motif (Warbrick 2000). The way that the PIP sequence motif binds to PCNA was determined from the structure of a peptide of this motif bound to the human PCNA clamp (Gulbis et al. 1996). The PCNA clamp has a hydrophobic pocket located between the two domains in each subunit. The PIP motif binds into this hydrophobic pocket. The PIP motif is required for most known protein-PCNA interactions. Subsequently, the structure of an E. coli clamp loader subunit (δ subunit) bound to the β clamp, and a peptide bound to the T4 clamp, revealed a very similar type of interaction in which a peptide sequence binds into a hydrophobic pocket located between the globular domains (Jeruzalmi et al. 2001b; Shamoo and Steitz 1999). A consensus sequence for bacterial clamp binding peptides has been described, in analogy to the PIP sequence (Dalrymple et al. 2001; Wijffels et al. 2004).
It is important to note that there appear to be additional binding sites to which proteins can interact with the clamp, in addition to the interaction with the hydrophobic pocket of the clamp. For example, one such secondary site is observed in the crystal structure of Pol IV bound to the bacterial clamp (Bunting et al. 2003). Secondary sites of interaction of protein binding to PCNA are suggested by mutational and genetic studies (Ayyagari et al. 1995; Eissenberg et al. 1997; Gomes and Burgers 2000; Johansson et al. 2004). Despite these additional interactions with the clamp, interaction with the hydrophobic pocket in the clamp is thought to be the major source of binding energy between the clamp and the proteins that it binds.
PCNA is a homotrimer and therefore has three identical protein binding sites, one in each subunit. Thus multiple proteins may attach to the PCNA clamp at the same time through binding to these identical sites. This aspect of clamp biology, in which multiple proteins bind the clamp at the same time, is referred to as the “tool belt” hypothesis. This very interesting subject will be briefly mentioned again later, and is expanded upon in Chap. 15.
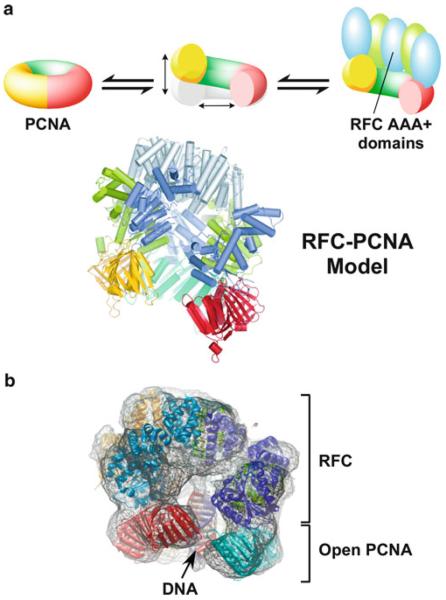
The N-terminal, or bottom surface, of RFC binds to PCNA (Fig. 14.2b). As described above, this surface is composed of the N-terminal AAA+ domains that are arranged in a right-handed spiral. The spiral shape of the N-terminal bottom surface of RFC can only accommodate binding of two to three RFC subunits, as a spiral geometry is inconsistent with complete interaction with a spiral (i.e. all five subunits of RFC) with a flat closed circular PCNA ring. However, if an open PCNA clamp adopts a right-handed spiral to match the right-handed spiral surface of the clamp loader, all the RFC subunits could bind the open PCNA clamp. Molecular simulations of PCNA in fact indicate that once an interface of PCNA is broken, the clamp spontaneously opens into a right-handed spiral (Kazmirski et al. 2005). Furthermore, the open PCNA spiral closely matches the N-terminal surface of RFC (illustrated in Fig. 14.4a). The electron micrographic reconstruction of an archaeal RFC-PCNADNA also reveals that the clamp is open in a right-handed spiral (Miyata et al. 2005). One problem with both of these studies is that the opening in the clamp (<15 Å) is less than the distance needed for passage of a DNA duplex (Fig. 14.4b) (Kazmirski et al. 2005; Miyata et al. 2005). The clamp must open at least 20 Å to accommodate passage of duplex DNA. Thus the mechanism of clamp opening is still an active issue for future research.
Fig. 14.4.
The PCNA clamp opens in a right-handed spiral. (a) Molecular simulations indicate that PCNA opens in a right-handed spiral that matches the shape of the RFC AAA+ domains (top diagram); the model of open PCNA with the structure of RFC is shown below (Kazmirski et al. 2005) (Reproduced with permission from Figure 5 of Kazmirski et al. (2005)). (b) Electron micro-graph particle reconstruction of an archaeal RFC-PCNA in the presence of DNA and ATP (Miyata et al. 2005) (Reproduced with permission from Figure 3a in Miyata et al. (2005))
The PCNA clamp is closed in the yeast RFC-PCNA-ATPγS structure, yet ATP binding should result in an open PCNA clamp when bound to RFC. Indeed, a fluorescent assay for clamp opening indicates that the PCNA clamp is open upon ATP binding to RFC (Zhuang et al. 2006). A likely reason for the closed clamp in the crystal structure is that RFC subunits were mutated in the “arginine finger” active site residue of the SRC motif to prevent any possible hydrolysis of ATPγS over the long time frames needed for crystal growth. These active site mutations probably prevented the conformation change required to open the clamp. The fact that ATP binding to wild type (i.e. non-mutated) RFC yields an open PCNA clamp is further supported by electron microscopy studies of a archaeal RFC-PCNA-DNA-ATP complex showing the clamp in an open form (Fig. 14.4b) (Miyata et al. 2005).
Substantial insight into the mechanism of clamp opening has been obtained from crystal structure and biochemical analysis of the E. coli β clamp in complex with the δ subunit of the clamp loader (i.e. δ is located in position A) (Jeruzalmi et al. 2001b). Biochemical studies demonstrated that the δ subunit of the clamp loader, by itself, opens the clamp (Turner et al. 1999). The δ subunit cannot load clamps on DNA, as the other subunits of the clamp loader are needed to perform the organized tasks of binding DNA, placing the clamp on a primed site, ejection of the clamp loader and clamp closing.
Only one δ subunit binds to a β dimer and surprisingly, the δ subunit binds much more tightly to a monomeric β clamp, mutated at the interface (a half clamp) (Stewart et al. 2001). This observation suggests that the binding energy of δ subunit to β is used to perform “work” on the circular β clamp, presumably to open the clamp on its own. This “work” of clamp opening is manifested in a lower affinity of δ to a β dimer compared to the β monomer mutant. In other words, the β monomer “half clamp” is already open, so δ does not need to expend binding energy to open it and thus binds the β monomer tighter then the β dimer. Biochemical studies have demonstrated that the clamp is not dismantled into monomers during clamp loading, and that only one interface opens during clamp loading onto DNA (Turner et al. 1999).
The crystal structure of δ subunit bound to the monomeric mutant β clamp provided much deeper insight into the clamp opening mechanism (Jeruzalmi et al. 2001b). Specifically, the structure of δ bound to a β monomer revealed that the β monomer adopts a much shallower crescent shape than the β protomers within a β ring. This shallower crescent shape was due to rigid body motions of the three domains of the β protomer. Interestingly, the major rigid body change that contributed to the shallower crescent shape of the β monomer was distant from the δ -β binding site. The implication of these observations for clamp loading is that the protomers within the clamp are under spring tension (i.e. the protomers bend inward because the force of the dimer interfaces are strong), but when one interface of the ring is broken (i.e. by δ), then each protomer can “relax” to a shallower crescent shape, thereby providing a gap for DNA strand passage.
The δ - β structure also indicates a specific mechanism by which the δ subunit forces one β interface open. The connection between δ and the hydrophobic pocket in β is mediated by residues near one end of a long α helix in δ (colored yellow in Fig. 14.2a). The opposite end of the same α helix extends to the β dimer interface and causes a distortion at the interface (Jeruzalmi et al. 2001a). This distortion may destabilize the β dimer interface and allow it to open.
14.5 ATP Hydrolysis and Closing of the Clamp
Clamp closure around DNA is the step at which ATP is hydrolyzed. In this regard, it is important to note that each subunit of the RFC heteropentamer is encoded by a different gene, and therefore each ATP site is structurally distinct (Cullmann et al. 1995). There are two extremes in which one may view the role of the different ATP sites. At one extreme, all the ATPs are hydrolyzed at once, and the purpose is to simply close the clamp and eject the clamp loader. In this view the clamp loader is a simple switch, either opening or closing the clamp, and DNA is the trigger for the switch in which primed template binding brings all catalytic residues into register at once and hydrolysis is essentially simultaneous at all sites regardless of the structural differences between the sites. At the other extreme, the ATP sites have individual functions enabled by their different molecular structures. In this view, the clamp loader is still a switch, but a more complicated one, where each ATP drives a different step along the path of clamp closure and clamp loader ejection.
Study of ATP binding and ATP site mutations in RFC indicates that different ATP sites have different functions and thus favors a more complicated switch (Gomes and Burgers 2001; Gomes et al. 2001; Johnson et al. 2006; Schmidt et al. 2001). One report demonstrates that the four ATP sites of RFC fill in a specific fashion (Gomes and Burgers 2001). Binding of the first two ATP enable RFC to bind PCNA, followed by a third ATP when it binds PCNA, while the fourth ATP binds upon association with the primed template (Gomes et al. 2001). Mutational studies of ATP site P-loops in different RFC subunits demonstrate that mutation of the P-loop of any single subunit has an effect on activity, although defects due to mutation of the RFC1 P-loop can be overcome by increasing the concentration of ATP (Cai et al. 1998; Podust et al. 1998; Schmidt et al. 2001). Interestingly, P-loop mutants of RFC still bind PCNA but are defective in binding DNA (Cai et al. 1998). This may be explained by inability of the RFC mutants to open PCNA, precluding DNA from the central chamber of RFC.
Studies that mutate the arginine finger in different RFC subunits suggest an order to ATP hydrolysis and further support the proposal that different ATPase sites have distinct functions in the clamp loading mechanism (Johnson et al. 2006). These experiments suggested that ATP binding was sensed by RFC3 to promote DNA association. ATP hydrolysis in RFC2 was speci fi cally stimulated by PCNA, and thus may be the fi rst ATP to be hydrolyzed and coupled to PCNA ring closure around DNA. Remaining ATP appears to be hydrolyzed in an ordered fashion around the clamp loader ring, starting from subunit D (RFC2), then subunits in positions C (RFC3), B (RFC4) and A (RFC1). The RFC-PCNA-ATPγS structure indicates that upon closure of PCNA, contacts between PCNA to RFC2 and RFC5 will be severed (i.e. these subunit do not bind the closed ring in the structure). This would also disable ATP induced conformations in RFC needed for RFC binding to DNA. Hence, these differential effects conspire to achieve the same goal – specifically to disconnect RFC from PCNA and DNA, allowing ring closure and clamp loader ejection.
Studies of the E. coli clamp loader also suggest that different ATP sites may have distinct functions. The E. coli clamp loader pentamer (γ3δδ′) has three ATP binding sites; only the γ subunits bind ATP while δ and δ′ do not. Even though the three ATP sites are each in a γ subunit, they are not identical due to their formation by the union of two subunits. Thus only two sites are structurally similar (those formed by γ-γ junctions at positions B–C and C–D), while the third site (position D–E) involves an arginine finger from an SRC motif within δ′. A mutation in the arginine finger of δ′ results in deficient clamp binding, while mutations in the arginine fingers of γ disrupt DNA binding (Johnson and O'Donnell 2003; Snyder et al. 2004). These results further support the idea that different ATP sites have distinct functions in clamp loader action. Interestingly, after ATP hydrolysis and clamp loader ejection from β, the E. coli clamp loader remains inactive for a short time, possibly due to slow ADP release (Ason et al. 2003; Bertram et al. 2000). This may serve to prevent the clamp loader from unloading clamps, an observation for both RFC and bacterial clamp loaders (discussed later in this chapter).
A recent structure of the E. coli β clamp bound to a primed DNA site has implications for the clamp loading mechanism (Georgescu et al. 2008) and the recent structure of PCNA-DNA indicates these fi ndings may generalize to RFC (McNally et al. 2010). Both structures show that DNA is highly tilted as it passes through the ring. The electron density of DNA is too sparse in the PCNA-DNA structure to locate the template strand. But the β clamp–DNA structure revealed that the clamp binds the single-strand DNA template strand at the same hydrophobic pocket that is used to bind to proteins (Georgescu et al. 2008). Considering the substantial similarities in clamps and clamp loaders of bacteria and eukaryotes, this structural feature may generalize to PCNA. Both the PCNA and E. coli β clamp have several contacts to the duplex DNA. Most notably are large loops that extend from the ring and bind each of the two strands of duplex DNA through conserved residues in the loops. Upon mutation of the duplex DNA interactive residues, replication activity is significantly reduced, but the reduction specifically resides in the clamp loading reaction and not the function of the clamp with DNA polymerase (Georgescu et al. 2008).
Clamp-DNA interactions are proposed to function at the step that draws the clamp closed around DNA during the clamp loading reaction (Georgescu et al. 2008). The residues on the loops that extend from the β clamp and bind DNA result in a pronounced tilt of the clamp on DNA, and this tilt may help disconnect the clamp from some of the subunits of the clamp loader (the first step in Fig. 14.5a). One may presume that the last connection between the clamp loader and clamp to be disrupted is the tight interaction between δ and the β clamp. The δ subunit is located in position A, analogous to the RFC1 subunit which, like δ, binds the clamp tighter than any of the other clamp loading subunits (Yao et al. 2003). The δ subunit binds to the same hydrophobic pocket to which template single-strand DNA binds, and therefore single-strand DNA may compete δ from β as illustrated in the second step of Fig. 14.5b, thereby facilitating ejection of the clamp loader from the clamp-DNA complex.
Fig. 14.5.
Function of clamp-DNA interactions. (a) Proposed role of DNA-clamp interactions in the clamp loading mechanism. PCNA and β (shown) clamps bind duplex DNA with a high degree of tilt. The first arrow suggests that when the clamp closes, the tilt of DNA through the clamp may sever connections to some clamp loader subunits. The second arrow suggests that the template single-strand DNA competes with δ (analogous position to RFC1) to eject the clamp loader from the clamp-DNA complex. (b) The tilt of DNA through PCNA and β may help DNA switch among different proteins bound to the same clamp. Illustrated here is DNA switching among two different polymerases attached to one clamp (Adapted with permission from Figure 7 of Georgescu et al. (2008))
Another proposed function of clamp-DNA interaction is to hold the clamp at a primed template junction after clamp loading. In the absence of these interactions, the clamp could conceivably slide on duplex DNA and be lost from the primed template junction. Hence, the clamp-DNA interactions may hold the clamp at the primed template junction after it is loaded but before the polymerase has associated with it, keeping the clamp where it is needed for function with the polymerase. This hypothesis has support in single-molecule studies of clamp sliding on primed DNA (Laurence et al. 2008).
The two protomers of the β clamp are identical, and therefore any particular site is duplicated. DNA may be presumed to rapidly isomerize between the two identical sites, tilting one way and then tilting the other. When two different DNA polymerases bind one clamp, the DNA tilt likely favors binding one polymerase, but when DNA isomerizes and tilts in the opposite direction, it may favor binding to the other DNA polymerase (see Fig. 14.5b). Ability of the primed terminus to isomerize between different polymerases becomes quite important upon encounter with a template lesion that requires a translesion DNA polymerase to bypass the lesion. Indeed, it has been shown that both DNA polymerase III and translesion DNA polymerase IV can bind to the β dimer at the same time (Benkovic et al. 2001). Studies have shown that the high fidelity DNA polymerase III gains control of the primed DNA in preference to the translesion polymerase IV, but upon DNA polymerase III stalling (e.g. at a template lesion), the translesion polymerase IV gains control of the DNA (Benkovic et al. 2001). Thus DNA isomerization between different DNA polymerases bound to one clamp could enable replication forks that stall at a lesion to rapidly switch to the translesion polymerase for lesion bypass, then switch back to a high fidelity DNA polymerase to continue replication, thereby preventing fork collapse.
14.6 Clamp Loaders also Unload Clamps After Replication
Bacterial systems have demonstrated that the leading and lagging strand DNA polymerases are associated with the helicase, clamp loader and sliding clamps to form a “replisome” machine (reviewed in Benkovic et al. 2001; Johnson and O'Donnell 2005). Eukaryotic replisomes are anticipated to contain both leading and lagging polymerases as well. The presence of both polymerases in one replisome machine implies formation of DNA loops on the lagging strand, due to the antiparallel structure of duplex DNA (Sinha et al. 1980). Specifically, the leading strand proceeds in the direction of helicase unwinding, but the lagging strand must be copied in the opposite direction of fork movement. These opposed directions are made possible by formation of a DNA loop for each Okazaki fragment (Sinha et al. 1980). Upon discovery that both leading and lagging strand polymerases are held to DNA by a sliding clamp for high processivity, it became important to understand how a processive polymerase-clamp complex could dissociate from DNA upon completing each short Okazaki fragment.
The interesting question of how the lagging strand polymerase recycles from the end of one Okazaki fragment to begin extension of the next fragment is made possible by regulated attachment of the polymerase to the clamp. For example, yeast polymerase θ, the lagging strand polymerase (see Chap. 12, this volume), binds tightly to PCNA during processive synthesis but upon completing a substrate rapidly releases from PCNA and dissociates from the DNA (Langston and O'Donnell 2008). The polymerase then transfers to a new PCNA clamp at another primed site (assembled there by RFC) for extension of the next Okazaki fragment. This lagging strand polymerase recycling mechanism, whereby the polymerase hops from one clamp to the next, results in stoichiometric use of clamps, one per Okazaki fragment. This mechanism has been demonstrated to occur in bacterial systems as well (Johnson and O'Donnell 2005; Sinha et al. 1980).
The lagging strand mechanism whereby PCNA clamps are left behind on each completed Okazaki fragment is useful for the process by which RNA is removed at the 5′ terminus of Okazaki fragments. For example, PCNA functions with FEN1, the nuclease that removes RNA primers, and stimulates its action (Ayyagari et al. 2003; Kao and Bambara 2003). PCNA also interacts with ligase which seals Okazaki fragments together (Song et al. 2009). However, there are 10- to 100-fold more Okazaki fragments than there are clamps in both bacterial and eukaryotic systems and therefore clamps must be recycled on and off DNA many times during genome replication. However, PCNA is highly stabile on DNA, requiring over half an hour to dissociate from DNA at 37°C (Yao et al. 1996). Therefore clamps must be actively removed from DNA to enable their reuse during duplication of a genome.
Studies have demonstrated that the RFC clamp loader can remove clamps from DNA (Yao et al. 1996). ATP binding is needed, but hydrolysis is not required. However, subassemblies of RFC can also open PCNA and remove it from DNA (Yao et al. 2006), similar to the θ subunit of the E. coli clamp loader opening β and unloading it from DNA. Specifically, a complex of RFC2/RFC5 is fully capable of removing PCNA, as is a complex of RFC2/3/4/5. Intracellular concentrations of individual yeast RFC subunits suggest that RFC2/RFC5 complex is in excess over other subunits and thus may be present inside cells, much as E. coli contains a fivefold excess of θ for β clamp recycling (Leu et al. 2000). Subassemblies of RFC cannot load PCNA because they are lacking one or more subunits required for clamp loading. Hence, they are capable of unidirectional action in unloading clamps, rather than both loading and unloading as is the case with RFC. It is also interesting to note that certain alternative RFCs (yeast Ctf18-RFC and yeast Rad17-RFC) can also unload PCNA from DNA (Bylund and Burgers 2005; Yao et al. 2006). Alternative RFC's are the subject of the next section.
14.7 Alternative RFCs
The subunit composition of RFC can be altered by replacement of the RFC1 subunit with another protein. Examples include Elg1 (Bellaoui et al. 2003; Ben-Aroya et al. 2003; Kanellis et al. 2003) and Ctf18 (Bellaoui et al. 2003; Mayer et al. 2001). These alternative RFC complexes are thought to load (or unload) PCNA onto DNA for specific processes involved in genome stability (Elg1) and in cohesion (Ctf18). The specific role of these alternative clamp loaders is not yet elucidated. Perhaps the best understood alternative clamp loader is the Rad17-RFC, in which RFC1 is replaced by Rad17 (Rad24 in S. cerevisiae) (Green et al. 2000; Lindsey-Boltz et al. 2001). The Rad17-RFC (i.e. subunits RFC 2, 3, 4 and 5, along with Rad17) is involved in the DNA damage checkpoint response and it loads a novel clamp onto DNA. The novel clamp that Rad17-RFC functions with is a heterotrimer of Rad9/Rad1/Hus1 (reviewed in Sancar et al. 2004). This clamp is often referred to as the “911” clamp. The 911 clamp appears to activate a kinase instead of function with a DNA polymerase (Majka et al. 2006b). Phosphorylation of other proteins by the kinase then signals the cell that DNA damage has occurred thereby communicating that an S phase checkpoint is required. Interestingly, the Rad17-RFC appears to load the 911 clamp onto the 5¢ terminus of a primed template junction, the opposite polarity of RFC loading PCNA (Ellison and Stillman 2003; Majka et al. 2006a).
14.8 Conclusions
Clamp loaders are ubiquitous in all cellular life forms. The eukaryotic clamp loader is called RFC. Like other clamp loaders, RFC is a heteropentamer and harnesses the energy of ATP hydrolysis to assemble the ring-shaped PCNA processivity factor onto a primed site. The function of RFC has been aided by biochemical studies and structure determination, and also by the large body of research on the clamp loader of E. coli, which is also a heteropentamer like RFC and functions in a similar way. The studies to date answer many important questions, including the subunit organization that specifies how DNA is recognized, where clamps are bound to the clamp loader, and how clamp loading is targeted to a primed template. There still exist many important questions about the details of the clamp loading reaction. Multiple ATP molecules are involved for optimum activity, and whether the individual ATP sites perform individual functions is still unresolved. Also uncertain is the way that clamps are opened. It is not certain that RFC binds and then opens the PCNA clamp, or whether it waits for a PCNA clamp to open and then binds and stabilizes the open form of the clamp. The order of ATP hydrolysis in the individual subunits is also understudied and needs clarification. Despite these deficiencies in our knowledge, the RFC clamp loader remains one of the most understood of the AAA+ machines, which are involved in numerous cellular processes. Future research will enable a better grasp of the mechanism and help fill in our gaps in how AAA+ proteins, and clamp loaders in particular, carry out their function.
References
- Ason B, Handayani R, Williams CR, Bertram JG, Hingorani MM, O'Donnell M, Goodman MF, Bloom LB. Mechanism of loading the Escherichia coli DNA polymerase III β sliding clamp on DNA. Bona fide primer/templates preferentially trigger the γ complex to hydrolyze ATP and load the clamp. J Biol Chem. 2003;278:10033–10040. doi: 10.1074/jbc.M211741200. [DOI] [PubMed] [Google Scholar]
- Ayyagari R, Impellizzeri KJ, Yoder BL, Gary SL, Burgers PM. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol Cell Biol. 1995;15:4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyagari R, Gomes XV, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J Biol Chem. 2003;278:1618–1625. doi: 10.1074/jbc.M209801200. [DOI] [PubMed] [Google Scholar]
- Bellaoui M, Chang M, Ou J, Xu H, Boone C, Brown GW. Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. EMBO J. 2003;22:4304–4313. doi: 10.1093/emboj/cdg406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aroya S, Koren A, Liefshitz B, Steinlauf R, Kupiec M. ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc Natl Acad Sci U S A. 2003;100:9906–9911. doi: 10.1073/pnas.1633757100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annu Rev Biochem. 2001;70:181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- Bertram JG, Bloom LB, Hingorani MM, Beechem JM, O'Donnell M, Goodman MF. Molecular mechanism and energetics of clamp assembly in Escherichia coli. The role of ATP hydrolysis when γ complex loads β on DNA. J Biol Chem. 2000;275:28413–28420. doi: 10.1074/jbc.M910441199. [DOI] [PubMed] [Google Scholar]
- Bowman GD, O'Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- Bowman GD, Goedken ER, Kazmirski SL, O'Donnell M, Kuriyan J. DNA polymerase clamp loaders and DNA recognition. FEBS Lett. 2005;579:863–867. doi: 10.1016/j.febslet.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Bunting KA, Roe SM, Pearl LH. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the β-clamp. EMBO J. 2003;22:5883–5892. doi: 10.1093/emboj/cdg568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F, Kobayashi R, Stillman B. cDNAs encoding the large subunit of human replication factor C. Proc Natl Acad Sci U S A. 1993;90:11014–11018. doi: 10.1073/pnas.90.23.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund GO, Burgers PM. Replication protein A-directed unloading of PCNA by the Ctf18 cohesion establishment complex. Mol Cell Biol. 2005;25:5445–5455. doi: 10.1128/MCB.25.13.5445-5455.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Yao N, Gibbs E, Finkelstein J, Phillips B, O'Donnell M, Hurwitz J. ATP hydrolysis catalyzed by human replication factor C requires participation of multiple subunits. Proc Natl Acad Sci U S A. 1998;95:11607–11612. doi: 10.1073/pnas.95.20.11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullmann G, Fien K, Kobayashi R, Stillman B. Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:4661–4671. doi: 10.1128/mcb.15.9.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalrymple BP, Kongsuwan K, Wijffels G, Dixon NE, Jennings PA. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc Natl Acad Sci U S A. 2001;98:11627–11632. doi: 10.1073/pnas.191384398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MJ, Jeruzalmi D, Kuriyan J, O'Donnell M. Motors and switches: AAA+ machines within the replisome. Nat Rev Mol Cell Biol. 2002;3:826–835. doi: 10.1038/nrm949. [DOI] [PubMed] [Google Scholar]
- Dore AS, Kilkenny ML, Jones SA, Oliver AW, Roe SM, Bell SD, Pearl LH. Structure of an archaeal PCNA1-PCNA2-FEN1 complex: elucidating PCNA subunit and client enzyme specificity. Nucleic Acids Res. 2006;34:4515–4526. doi: 10.1093/nar/gkl623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Ayyagari R, Gomes XV, Burgers PM. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase δ and DNA polymerase ε. Mol Cell Biol. 1997;17:6367–6378. doi: 10.1128/mcb.17.11.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- Fairman M, Prelich G, Tsurimoto T, Stillman B. Identification of cellular components required for SV40 DNA replication in vitro. Biochim Biophys Acta. 1988;951:382–387. doi: 10.1016/0167-4781(88)90110-8. [DOI] [PubMed] [Google Scholar]
- Georgescu RE, Kim SS, Yurieva O, Kuriyan J, Kong XP, O'Donnell M. Structure of a sliding clamp on DNA. Cell. 2008;132:43–54. doi: 10.1016/j.cell.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedken ER, Kazmirski SL, Bowman GD, O'Donnell M, Kuriyan J. Mapping the interaction of DNA with the Escherichia coli DNA polymerase clamp loader complex. Nat Struct Mol Biol. 2005;12:183–190. doi: 10.1038/nsmb889. [DOI] [PubMed] [Google Scholar]
- Gomes XV, Burgers PM. Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 2000;19:3811–3821. doi: 10.1093/emboj/19.14.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes XV, Burgers PM. ATP utilization by yeast replication factor C. I. ATP-mediated interaction with DNA and with proliferating cell nuclear antigen. J Biol Chem. 2001;276:34768–34775. doi: 10.1074/jbc.M011631200. [DOI] [PubMed] [Google Scholar]
- Gomes XV, Gary SL, Burgers PM. Overproduction in Escherichia coli and characterization of yeast replication factor C lacking the ligase homology domain. J Biol Chem. 2000;275:14541–14549. doi: 10.1074/jbc.275.19.14541. [DOI] [PubMed] [Google Scholar]
- Gomes XV, Schmidt SL, Burgers PM. ATP utilization by yeast replication factor C. II. Multiple stepwise ATP binding events are required to load proliferating cell nuclear antigen onto primed DNA. J Biol Chem. 2001;276:34776–34783. doi: 10.1074/jbc.M011743200. [DOI] [PubMed] [Google Scholar]
- Grabowski B, Kelman Z. Archaeal DNA replication: eukaryal proteins in a bacterial context. Annu Rev Microbiol. 2003;57:487–516. doi: 10.1146/annurev.micro.57.030502.090709. [DOI] [PubMed] [Google Scholar]
- Green CM, Erdjument-Bromage H, Tempst P, Lowndes NF. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr Biol. 2000;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- Guenther B, Onrust R, Sali A, O'Donnell M, Kuriyan J. Crystal structure of the δ′ subunit of the clamp-loader complex of E. coli DNA polymerase III. Cell. 1997;91:335–345. doi: 10.1016/s0092-8674(00)80417-1. [DOI] [PubMed] [Google Scholar]
- Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. Structure of the C-terminal region of p21WAF1/CIP1 complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D, O'Donnell M, Kuriyan J. Crystal structure of the processivity clamp loader γ complex of E. coli DNA polymerase III. Cell. 2001a;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D, Yurieva O, Zhao Y, Young M, Stewart J, Hingorani M, O'Donnell M, Kuriyan J. Mechanism of processivity clamp opening by the δ subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell. 2001b;106:417–428. [PubMed] [Google Scholar]
- Johansson E, Garg P, Burgers PM. The Pol32 subunit of DNA polymerase δ contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J Biol Chem. 2004;279:1907–1915. doi: 10.1074/jbc.M310362200. [DOI] [PubMed] [Google Scholar]
- Johnson A, O'Donnell M. Ordered ATP hydrolysis in the γ complex clamp loader AAA+ machine. J Biol Chem. 2003;278:14406–14413. doi: 10.1074/jbc.M212708200. [DOI] [PubMed] [Google Scholar]
- Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- Johnson A, Yao NY, Bowman GD, Kuriyan J, O'Donnell M. The replication factor C clamp loader requires arginine finger sensors to drive DNA binding and proliferating cell nuclear antigen loading. J Biol Chem. 2006;281:35531–35543. doi: 10.1074/jbc.M606090200. [DOI] [PubMed] [Google Scholar]
- Jonsson ZO, Hindges R, Hubscher U. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 1998;17:2412–2425. doi: 10.1093/emboj/17.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellis P, Agyei R, Durocher D. Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr Biol. 2003;13:1583–1595. doi: 10.1016/s0960-9822(03)00578-5. [DOI] [PubMed] [Google Scholar]
- Kao HI, Bambara RA. The protein components and mechanism of eukaryotic Okazaki fragment maturation. Crit Rev Biochem Mol Biol. 2003;38:433–452. doi: 10.1080/10409230390259382. [DOI] [PubMed] [Google Scholar]
- Kazmirski SL, Zhao Y, Bowman GD, O'Donnell M, Kuriyan J. Out-of-plane motions in open sliding clamps: molecular dynamics simulations of eukaryotic and archaeal proliferating cell nuclear antigen. Proc Natl Acad Sci U S A. 2005;102:13801–13806. doi: 10.1073/pnas.0506430102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Ab E, Bonvin AM, Siegal G. Structure of the DNA-bound BRCA1 C-terminal region from human replication factor C p140 and model of the protein-DNA complex. J Biol Chem. 2010;285:10087–10097. doi: 10.1074/jbc.M109.054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong XP, Onrust R, O'Donnell M, Kuriyan J. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- Kornberg A, Baker TA. DNA replication. W. H. Freeman; New York: 1992. [Google Scholar]
- Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Langston LD, O'Donnell M. DNA polymerase δ is highly processive with proliferating cell nuclear antigen and undergoes collision release upon completing DNA. J Biol Chem. 2008;283:29522–29531. doi: 10.1074/jbc.M804488200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence TA, Kwon Y, Johnson A, Hollars CW, O'Donnell M, Camarero JA, Barsky D. Motion of a DNA sliding clamp observed by single molecule fluorescence spectroscopy. J Biol Chem. 2008;283:22895–22906. doi: 10.1074/jbc.M800174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Eki T, Hurwitz J. Synthesis of DNA containing the simian virus 40 origin of replication by the combined action of DNA polymerases α and δ. Proc Natl Acad Sci U S A. 1989;86:7361–7365. doi: 10.1073/pnas.86.19.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu FP, Hingorani MM, Turner J, O'Donnell M. The δ subunit of DNA polymerase III holoenzyme serves as a sliding clamp unloader in Escherichia coli. J Biol Chem. 2000;275:34609–34618. doi: 10.1074/jbc.M005495200. [DOI] [PubMed] [Google Scholar]
- Lindsey-Boltz LA, Bermudez VP, Hurwitz J, Sancar A. Purification and characterization of human DNA damage checkpoint Rad complexes. Proc Natl Acad Sci U S A. 2001;98:11236–11241. doi: 10.1073/pnas.201373498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka J, Binz SK, Wold MS, Burgers PM. Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J Biol Chem. 2006a;281:27855–27861. doi: 10.1074/jbc.M605176200. [DOI] [PubMed] [Google Scholar]
- Majka J, Niedziela-Majka A, Burgers PM. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell. 2006b;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumiya S, Ishino Y, Morikawa K. Crystal structure of an archaeal DNA sliding clamp: proliferating cell nuclear antigen from Pyrococcus furiosus. Protein Sci. 2001;10:17–23. doi: 10.1110/ps.36401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML, Gygi SP, Aebersold R, Hieter P. Identification of RFCCtf18p, Ctf8p, Dcc1p: an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol Cell. 2001;7:959–970. doi: 10.1016/s1097-2765(01)00254-4. [DOI] [PubMed] [Google Scholar]
- McNally R, Bowman GD, Goedken ER, O'Donnell M, Kuriyan J. Analysis of the role of PCNA-DNA contacts during clamp loading. BMC Struct Biol. 2010;10:3. doi: 10.1186/1472-6807-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Suzuki H, Oyama T, Mayanagi K, Ishino Y, Morikawa K. Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis. Proc Natl Acad Sci U S A. 2005;102:13795–13800. doi: 10.1073/pnas.0506447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moarefi I, Jeruzalmi D, Turner J, O'Donnell M, Kuriyan J. Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J Mol Biol. 2000;296:1215–1223. doi: 10.1006/jmbi.1999.3511. [DOI] [PubMed] [Google Scholar]
- Naktinis V, Turner J, O'Donnell M. A molecular switch in a replication machine defined by an internal competition for protein rings. Cell. 1996;84:137–145. doi: 10.1016/s0092-8674(00)81000-4. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- O'Donnell M, Kuriyan J. Clamp loaders and replication initiation. Curr Opin Struct Biol. 2006;16:35–41. doi: 10.1016/j.sbi.2005.12.004. [DOI] [PubMed] [Google Scholar]
- O'Donnell M, Onrust R, Dean FB, Chen M, Hurwitz J. Homology in accessory proteins of replicative polymerases: E. coli to humans. Nucleic Acids Res. 1993;21:1–3. doi: 10.1093/nar/21.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust VN, Tiwari N, Ott R, Fanning E. Functional interactions among the subunits of replication factor C potentiate and modulate its ATPase activity. J Biol Chem. 1998;273:12935–12942. doi: 10.1074/jbc.273.21.12935. [DOI] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Schmidt SL, Gomes XV, Burgers PM. ATP utilization by yeast replication factor C. III. The ATP-binding domains of Rfc2, Rfc3, and Rfc4 are essential for DNA recognition and clamp loading. J Biol Chem. 2001;276:34784–34791. doi: 10.1074/jbc.M011633200. [DOI] [PubMed] [Google Scholar]
- Shamoo Y, Steitz TA. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- Simonetta KR, Kazmirski SL, Goedken ER, Cantor AJ, Kelch BA, McNally R, Seyedin SN, Makino DL, O'Donnell M, Kuriyan J. The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell. 2009;137:659–671. doi: 10.1016/j.cell.2009.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha NK, Morris CF, Alberts BM. Efficient in vitro replication of double-stranded DNA templates by a purified T4 bacteriophage replication system. J Biol Chem. 1980;255:4290–4293. [PubMed] [Google Scholar]
- Snyder AK, Williams CR, Johnson A, O'Donnell M, Bloom LB. Mechanism of loading the Escherichia coli DNA polymerase III sliding clamp: II. Uncoupling the beta and DNA binding activities of the γ complex. J Biol Chem. 2004;279:4386–4393. doi: 10.1074/jbc.M310430200. [DOI] [PubMed] [Google Scholar]
- Song W, Pascal JM, Ellenberger T, Tomkinson AE. The DNA binding domain of human DNA ligase I interacts with both nicked DNA and the DNA sliding clamps, PCNA and hRad9-hRad1-hHus1. DNA Repair. 2009;8:912–919. doi: 10.1016/j.dnarep.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J, Hingorani MM, Kelman Z, O'Donnell M. Mechanism of β clamp opening by the δ subunit of Escherichia coli DNA polymerase III holoenzyme. J Biol Chem. 2001;276:19182–19189. doi: 10.1074/jbc.M100592200. [DOI] [PubMed] [Google Scholar]
- Stukenberg PT, Studwell-Vaughan PS, O'Donnell M. Mechanism of the sliding β-clamp of DNA polymerase III holoenzyme. J Biol Chem. 1991;266:11328–11334. [PubMed] [Google Scholar]
- Turner J, Hingorani MM, Kelman Z, O'Donnell M. The internal workings of a DNA polymerase clamp-loading machine. EMBO J. 1999;18:771–783. doi: 10.1093/emboj/18.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Cai J, Gibbs E, O'Donnell M, Hurwitz J. Deletion analysis of the large subunit p140 in human replication factor C reveals regions required for complex formation and replication activities. J Biol Chem. 1997;272:10058–10064. doi: 10.1074/jbc.272.15.10058. [DOI] [PubMed] [Google Scholar]
- Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- Warbrick E. The puzzle of PCNA's many partners. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Wijffels G, Dalrymple BP, Prosselkov P, Kongsuwan K, Epa VC, Lilley PE, Jergic S, Buchardt J, Brown SE, Alewood PF, Jennings PA, Dixon NE. Inhibition of protein interactions with the β2 sliding clamp of Escherichia coli DNA polymerase III by peptides from β2-binding proteins. Biochemistry. 2004;43:5661–5671. doi: 10.1021/bi036229j. [DOI] [PubMed] [Google Scholar]
- Williams CR, Snyder AK, Kuzmic P, O'Donnell M, Bloom LB. Mechanism of loading the Escherichia coli DNA polymerase III sliding clamp: I. Two distinct activities for individual ATP sites in the γ complex. J Biol Chem. 2004;279:4376–4385. doi: 10.1074/jbc.M310429200. [DOI] [PubMed] [Google Scholar]
- Yao N, Turner J, Kelman Z, Stukenberg PT, Dean F, Shechter D, Pan ZQ, Hurwitz J, O'Donnell M. Clamp loading, unloading and intrinsic stability of the PCNA, β and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells. 1996;1:101–113. doi: 10.1046/j.1365-2443.1996.07007.x. [DOI] [PubMed] [Google Scholar]
- Yao N, Coryell L, Zhang D, Georgescu RE, Finkelstein J, Coman MM, Hingorani MM, O'Donnell M. Replication factor C clamp loader subunit arrangement within the circular pentamer and its attachment points to proliferating cell nuclear antigen. J Biol Chem. 2003;278:50744–50753. doi: 10.1074/jbc.M309206200. [DOI] [PubMed] [Google Scholar]
- Yao NY, Johnson A, Bowman GD, Kuriyan J, O'Donnell M. Mechanism of proliferating cell nuclear antigen clamp opening by replication factor C. J Biol Chem. 2006;281:17528–17539. doi: 10.1074/jbc.M601273200. [DOI] [PubMed] [Google Scholar]
- Zhuang Z, Yoder BL, Burgers PM, Benkovic SJ. The structure of a ring-opened proliferating cell nuclear antigen-replication factor C complex revealed by fluorescence energy transfer. Proc Natl Acad Sci U S A. 2006;103:2546–2551. doi: 10.1073/pnas.0511263103. [DOI] [PMC free article] [PubMed] [Google Scholar]