Abstract
Integrin-mediated cell-extracellular matrix (ECM) interaction plays key roles in tissue morphogenesis and integrity. The LIM-domain only protein PINCH (the Particularly Interesting Cysteine- and Histidine-rich protein) functions as an adaptor essential for the assembly and function of the focal adhesion complex that links integrin signaling to the cytoskeleton and other intracellular signaling pathways and regulates diverse cellular processes such as cell adhesion, migration, growth, differentiation and survival. Recent biochemical and genetic studies have greatly advanced our knowledge surrounding the molecular interactions and functions of each component of the focal adhesion complex and revealed a requirement for PINCH in early embryogenesis, in morphogenesis of the neural crest and cardiac outflow, and in myocardial growth and remodeling. In this review article, we will provide an overview of the current knowledge of the molecular interactions of PINCH with other components of focal adhesions, highlighting recent discoveries of the in vivo role of PINCH and discuss its potential implication for human heart disease.
Keywords: PINCH, Focal adhesion, neural crest, myocardial remodeling, cardiomyopathy
Introduction
Cells communicate with their microenvironment and neighbors by several specialized cell membrane associated structures, which transduce diverse mechanical and biochemical signals across cell membrane and play essential roles in regulating morphogenesis and maintaining tissue structural and functional integrity and homeostasis. Cell and Extracellular Matrix (ECM) interaction is mediated mainly by integrin and its associated protein complexes, including integrin-linked kinase (ILK), Parvin and PINCH (Particularly Interesting Cysteine- and Histidine-rich protein) [1]. Engagement of integrins with the components of the ECM leads to recruitment and formation of a cytoplasmic focal adhesion complex, referred to as IPP complex (ILK, Parvin and PINCH) [1, 2]. Formation of the IPP complex is essential for targeting of ILK, Parvin and PINCH to focal adhesion sites and for stabilization of each component of the complex, preventing them from proteosomal degradation [3-6]. Signaling through integrins is bidirectional. Changes in intracellular signaling pathways and cytoskeletal organization modulates the binding of intracellular molecules to the integrin cytoplasmic tail, which in turn modifies integrin binding affinity to the ECM, and deposition and remodeling of the ECM [1, 2, 7]. Recent studies have provided an insight into the molecular interactions and functions of these proteins in cell adhesion, migration, proliferation, differentiation and survival and have revealed a central role for ILK and PINCH in mediating bidirectional integrin signaling. The focus of this review will be on the molecular interactions and in vivo functions of PINCH. We will discuss unresolved questions and future directions in dissecting the molecular mechanism of PINCH and highlight any potential clinical implications.
Molecular Interactions and Functions of PINCH
ILK contains an N-terminal ankyrin repeats domain, a pleckstrin homology (PH) domain and a C-terminal Ser/Thr kinase domain [8, 9]. ILK binds directly to the cytoplasmic tail of β1 and β3-integrin through its C-terminal kinase domain. ILK also binds to the LIM-domain-only protein PINCH [10-12] and a number of actin cytoskeletal associated proteins, such as parvin and paxillin, thus linking ECM-integrin to the actin cytoskeleton and other intracellular pathways [1, 13-17]. These interactions of ILK are fundamental to the establishment of the integrin-actin cytoskeletal network and for the accurate control of basic cellular functions such as cell migration, spreading, growth and survival. Disruption of these interactions by various experimental approaches targeting either PINCH, ILK or parvin, such dominant-negative overexpression, or siRNA gene knockdown or gene knockout, lead to defects in cell migration, spreading, survival and extracellular matrix assembly[3, 14, 15, 17-27].
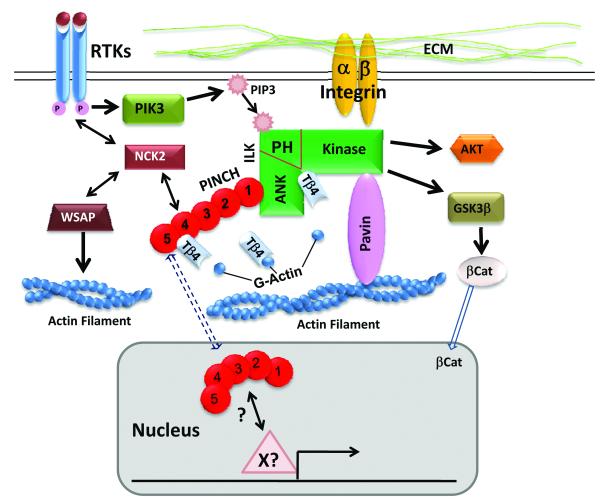
The kinase activity of ILK is regulated by cell-matrix adhesion and growth factors in a phosphoinositol-3 kinase (PI3K) dependent manner (Fig.1) [8, 9]. Cell adhesion and growth factor stimulation activate PI3K, which in turn increase the production of phosphatidylinositol 3,4,5-trisphosphate (PIP3) [8]. The 3-phosphoinoside lipid binds to the PH motif of ILK and activates its kinase activity, which in turn activates multiple signaling pathways involved in cell adhesion, migration, growth and survival [1, 2, 22, 27-32]. Overexpression of the PIP3 phosphatase, PTEN or treatment of cells with the PI3K inhibitors Wortmannin or Ly294002 inhibits ILK activation [8]. Activated ILK phosphorylates and activates PKB/Akt at Ser473, an event critical for cell growth and survival [8]. ILK can also phosphorylate and inhibit GSK3β, leading to the stabilization and translocation of β-catenin to the nucleus and activation of gene expression (Fig.1) [33].
Figure1.
Cell adhesions play important roles in cell growth, differentiation, migration and survival. Engagement of integrin with extracellular matrix (ECM) leads to the recruitment and formation of a cytoplasmic focal adhesion complex composed of integrin-linked kinase (ILK), PINCH and Parvin, which links ECM-integrins to the actin cytoskeleton. PINCH, a five LIM domain (1-5) adaptor protein, interacts with the ankyrin domain (ANK) of ILK via its LIM1 domain and interacts with NCK2 via its LIM4 domain, thus Links integrin pathway to other intracellular pathways, especially growth factor-receptor tyrosine kinase (RTK) pathways. ILK kinase is activated in a PI3 kinase (PI3K) dependent manner, which in turn activated downstream PKB/AKT required for cell growth and survival. ILK also phosphorylates and inhibits GSK3b, leading to the stabilization and translocation of b-catenin to the nucleus and activation of gene expression. Thymosin β4 (Tβ4) formed a functional complex with PINCH and ILK via direct interactions with PINCH and ILK, resulting in activation of Akt and promoting myocardial cell migration, survival and repair. In the neural crest cells, PINCH is predominantly nuclear, suggesting a direct role of PINCH in regulating gene expression in complex with transcription factor(s) yet to be identified (X?) .
The N-terminal ankyrin domain of ILK binds directly to the LIM domain-only adaptor protein PINCH. Two PINCH proteins have been characterized in mammals, including PINCH1 (LIMS1) and PINCH2 (LIMS2). PINCH1 was originally identified in an antibody screen of a human cDNA library as a marker for senescent erythrocytes [34]. A yeast-two hybrid screen using the N-terminal ankyrin-domain of ILK as bait identified PINCH as a direct binding partner of ILK [11]. PINCH2, a close homolog of PINCH1, was identified by cDNA sequence-database mining. PINCH1 and PINCH2 share high sequence and structural homology and both are localized to focal adhesions and the nucleus [12, 35]. PINCH, through its LIM domain mediated protein interactions, functions as a molecular scaffold that supports the assembly of a multiprotein complex at sites of integrin enrichment.
PINCH1 is composed of five LIM domains (LIM1-5) and a short C-terminal sequence. It shares high homology to that of PINCH2, expect in the LIM5 domain and the C-terminal tail [10, 12, 35, 36]. The binding of the ankyrin repeat domain of ILK has been mapped to the first LIM domain (LIM1) of PINCH, which is required for localization and function of the ILK-PINCH complex (Fig.1) [36]. In addition to promoting ILK mediated phosphorylation and activation of PKB/Akt at Ser473, PINCH1 is also required for phosphorylation of PKB/Akt at Thr308 and survival even in cells with a constitutively active form of PKB/Akt [3]. These data suggest that PINCH1 activates PKB/Akt in an ILK-dependent and an ILK independent manner and functions both upstream and downstream of PKB/Akt [3]. Interestingly, the two PINCHs compete for binding to ILK and the PINCH1-ILK and PINCH2-ILK interactions are mutually exclusive [12]. Overexpression of PINCH2 inhibits the PINCH1-ILK interaction and reduces cell spreading and migration, suggesting an intriguing role for PINCH2 in fine tuning the PINCH1-ILK interaction in cell adhesion and migration [3, 12, 36]. In addition, the expression of a chimeric PINCH with PINCH1 LIM domains and PINCH2 C-terminal tail cannot rescue the spreading defect in PINCH1-knockdown HeLa cells [37], suggesting that the C-terminal of PINCH1 is required for its function. Nevertheless, expression of a full-length PINCH2 completely restores the adhesion and spreading defects of PINCH1-null fibroblasts [38], suggesting a redundant role of PINCH1 and PINCH2 in this context. Furthermore, global knockout of PINCH2 and single knockout of PINCH1 in the myocardium did not result in any basal cardiac phonotype, which has been attributed to the redundant role of PINCH1 and PINCH2 [39].
NCK2 is a Src Homology2/3 (SH2-SH3) adaptor protein implicated in various signaling pathways, including that of growth factors, cell adhesion receptors and the cytoskeleton [40]. It has been shown that integrin mediated signaling is required for potentiating growth factor signaling [1, 2, 40]. NCK2 has been shown to associate with receptor tyrosine kinases via its SH2 domain. NCK2, via its interactions with Rho effectors and other cytoskeletal associated proteins such as WASP, N-WASP, is implicated in linking receptor tyrosine kinases to actin cytoskeleton remodeling (Fig.1) [1, 41, 42]. The LIM4 domain of PINCH1 has been shown to bind to NCK2, but not its homolog NCK1 [40, 43], thus linking growth factor signaling pathways to integrin and cytoskeletal signaling. However the role of PINCH1-NCK2 interaction remains to be determined because the binding affinity of NCK2 to PINCH1 is very weak and NCK2 knockout mice did not present any phenotype [40, 44].
Rsu-1 is a highly conserved leucine rich repeat (LRR) protein, identified as a Ras suppressor based on its ability to inhibit transformation by Ras [45, 46]. Rsu-1 is co-localized with PINCH1 and ILK in focal adhesions [47]. Moreover studies have shown that the LIM5 domain of PINCH1, but not that of PINCH2, binds to Rsu-1 and this interaction plays a role in targeting PINCH1 to focal adhesions, stabilizing the IPP complex and inhibiting migration [10, 47, 48]. Ectopic expression of Rsu-1 inhibits anchorage independent growth of Ras-transformed cells and human tumor cell lines, in which both expression of ILK and PINCH are increased. Thus, Rsu-1 may represent an important crosstalk between Ras and integrin signaling pathways and play an important role in cell growth and tumoriogenesis [47-49].
The G-actin sequestrating peptide Thymosin-β4 binds to LIM4 and LIM5 domains of PINCH1 and forms a functional complex with PINCH1 and ILK, which activates PKB/Akt and promotes migration and survival of embryonic and postnatal cardiac cells in culture (Fig.1) [50]. In a mouse myocardial infarction model, Thymosin-b4 treatment results in upregulation of ILK and Akt activity in the heart, enhanced cardiomyocyte survival, and improved cardiac function [50]. Although ILK-Akt pathway has been implicated in these regeneration processes, the underlying molecular mechanisms remain to be determined, which may involve multiple cell types, alterations in metabolism and energy consumption and enhanced angiogenesis that promote cell survival. It is unknown whether PINCH2 also binds to Thymosin-β4; however given the redundant role of the two PINCHs in the myocardium, it is likely that PINCH2 may also bind to thymosin-β4 and act redundantly with PINCH1 in this context. Thus, Thymosin-β4, PINCH-ILK pathway may represent a promising therapeutic target for cardiac disease [51].
Roles of PINCH1 during early development stage and embryonic stem cell
Genetic studies in C. elegans and Drosophila have revealed an essential role for PINCH in mediating integrin-ILK-dependent signaling [35, 52]. The deletion of UNC-97, an orthologue of PINCH1, in C. elegans results in muscle detachment and an embryonic-lethal phenotype called PAT (paralyzed and arrested elongation at the twofold stage) [35] resembling that of β1-integrin/PAT-3 or ILK/PAT-4 [53, 54]. In Drosophila muscle, PINCH displays a completely overlapping expression pattern with ILK and βPS integrin, prominently enriched at the muscle attachment sites [52]. Flies deficient in PINCH1 (named stck in Drosophila) exhibit muscle detachment, similar to the phenotypes of ILK and PS-integrin [52, 55-57].
During early mouse embryogenesis, the inner cell mass (ICM) of the blastocyst develops into the primitive endoderm and the epiblast [58]. The primitive endoderm forms the surface of the ICM of the blastocyst and deposits a basement membrane. The basement membrane is required for adjacent ICM cells to polarize and to establish the columnar epiblast [59]. The importance of integrin-ILK-mediated cell-cell and cell-matrix interactions during early embryonic development is highlighted by genetic studies in mouse models [60-64]. In β1-integrin null embryos, the primitive endoderm fails to deposit laminin α1 and form the basement membrane [60, 62]. In ILK null mouse embryos, the primitive endoderm differentiates and produces a basement membrane but the epiblast fails to polarize or cavitate, and mutants die at the peri-implantation stage [63]. In contrast to that of invertebrates, two PINCH isoforms, PINCH1 and PINCH2, are expressed in mammals [10, 12, 34]. However, PINCH2 is not expressed until a later developmental stage from E14.5 onwards. PINCH1 is readily detectable in blastocysts at approximately E3.5 [6, 39]. PINCH null mouse embryos die at E5.5, exhibiting a disorganized egg cylinder, with decreased cell proliferation and excessive cell death, highlighting an important role of PINCH1 during early embryogenesis [5, 6]. In addition, studies from Fässler’ laboratory using a PINCH1 null embryoid body (EBs) model and comparing to that of an ILK null EBs highlighted an ILK-independent role of PINCH1 in endoderm survival and cell-cell adhesion [5].
Role of PINCH1 in Neural Crest and Outflow Tract Morphogenesis
Neural crest cells (NCCs) are a transient embryonic stem cell population that originate from the dorsal neural tube and migrate along well-defined migratory pathways to their final destinations, giving rise to a diversity of cell types and contributing to craniofacial and cardiac outflow tract morphogenesis, and formation of the entire peripheral nervous system [65-67]. Cardiac NCCs migrate into outflow tract and contribute to the smooth muscle component of the outflow tract and outflow tract septation, as well as endocardial cushion morphogenesis [66-72]. Perturbation of cardiac NCC development causes congenital heart defects in animal models and in humans, affecting the outflow tract and great vessels [70, 73-76].
Recent studies from our group have shown that PINCH1 is highly expressed in NCCs and that neural crest specific ablation of PINCH1 leads to severe cardiovascular defects, including an enlarged common arterial trunk, ventricular septal defects (VSDs), and defective cushion/valve maturation [77]. In addition to cardiovascular defects, mutants exhibited defects in craniofacial structures, such as hypoplastic thymus and craniofacial malformation. Interestingly, PINCH1-deficient NCCs did migrate correctly into the pharyngeal apparatus as demonstrated by fate mapping and by in situ hybridization using the neural crest marker CrbpI [77]. Importantly, we found that from E11.5 onwards, cardiac NCCs in the outflow tract continue to proliferate and fail to exit the cell cycle and undergo smooth muscle cell differentiation. PINCH1-deficient NCCs in the outflow tract cushion underwent markedly increased apoptosis at E11.5-E13.5, associated with an observed failure of cushion remodeling and valve maturation[77]. These observations demonstrate that PINCH1 plays important roles in proliferation, differentiation and survival of NCCs, but it appears dispensable for NCC migration into the outflow tract.
It has been shown that interaction of PINCH and ILK is essential for their stability, targeting to focal adhesion sites and function [3-6]. However, we found no significant change in ILK expression in cultured NCCs and in the outflow tract of PINCH1 mutants, suggesting that inactivation of PINCH1 in NCCs does not affect formation of the focal adhesion complex and cardiac phenotypes may be caused by an ILK independent mechanism. Supporting this, PINCH1 in NCCs was found to be predominantly nuclear and PINCH1 contains a presumed leucine-rich nuclear export signal and an overlapping basic nuclear localization signal, suggesting that it may act as a shuttling/signaling protein or directly involved in regulation of gene expression [77, 78].
A unique feature of the neural crest PINCH1 mutant is the aneurysmal arterial trunk. It is important to note that several human syndromes which include aortic and vascular aneurysms have been associated with alterations in TGFβ signaling [79-81]. TGFβ signaling plays an important role in specification, migration, survival, and differentiation of NCCs [65, 82, 83] and its expression in the PINCH1 mutant outflow tract was dramatically downregulated. It is tempting to speculate that PINCH1 mutation might be involved in human syndromes characterized by aortic and vascular aneurysms.
PINCH in Myocardial Growth, Maturation and Remodeling
Myocardial cells interact with each other and the matrix at specialized membranous structures, referred to as the intercalated discs and costameres, respectively [1, 31, 61, 84-86], which provide mechanical and electrical coupling between myocytes and enable the myocardium to function as a syncytium. During late development and early postnatal life, cardiomyocytes undergo physiological hypertrophic growth, realign cytoskeletal components and acquire a mature cytoarchitecture to meet a dramatically increased hemodynamic load. One feature of these postnatal myocardial remodeling and maturation is the redistribution and segregation of distinct cell-matrix and cell-cell adhesions [84, 86-89]. During embryonic development, myocytes interact with each other extensively via the cadherin-mediated adherens junction, which appears to play a dominant role in mediating myocyte adhesion and function [90]. During perinatal development, the myocytes continue to growth, elongate and interact with each other only at the bipolar ends by intercalated discs. Intercalated discs are disassembled from lateral cell membranes and reassembled at the bipolar ends of the cells, whereas costameres remain in the lateral cell membranes [91, 92]. However, molecular mechanisms regulating the segregation and redistribution of cell adhesions during postnatal myocardium remodeling and maturation remain largely unknown.
The Integrin signaling pathway has been shown to be important for maintenance of cardiac structure and function [93-95]. Cardiac specific deletion of integrinβ1 or ILK in mice resulted in disruption of the focal adhesion complex and development of cardiomyopathy and heart failure [93-95]. Deletion of PINCH1 in mouse myocardium did not lead to any basal cardiac phenotype, In addition, global PINCH2 knockout did not result in any phenotype [5, 6, 39].
However, analyses of mice which were doubly homozygous null for PINCH1 and PINCH2 in the myocardium (CDKO) have revealed a redundant yet essential role for PINCH in postnatal myocardial remodeling and maturation and in maintaining myocardial integrity [39]. Hearts of CDKO mutants were dilated and ventricular wall thickness was highly irregular with abnormal trabeculation, and mutant mice developed cardiomyopathy and heart failure within 4 weeks [39]. Consistent with the observed role of PINCH in mediating cell-matrix interaction, PINCH CDKO myocytes in culture failed to attach and spread and exhibited disrupted focal adhesions. Moreover, expression of ILK, Parvin, integrinβ1 and phosphorylation of Akt were significantly reduced, and there was markedly increased cell death in hearts of PINCH CDKO mice [39]. In addition, electron microscopy analysis revealed disruption of costameric structures and intercalated discs in the PINCH CDKO myocardium [39].
PINCH also plays a critical role in modulating the stability and polarized distribution of intercalated disc proteins during postnatal myocardial maturation and remodeling [39]. The expression of proteins that function in adherens junctions and gap junctions of the intercalated discs are markedly affected in the PINCH mutant myocardium. In CDKO myocardium, Connexin43, α-E-catenin and ZO-1 were significantly reduced and retained a disperse expression pattern throughout the lateral membrane comparable to that seen at earlier stages of development, rather than being expressed at the intercalated discs. Similarly, expression of N-cadherin, β-catenin and Vinculin remained largely in the lateral cell membrane of PINCH mutant heart; however, their expression at the intercalated discs were lost despite the overall amounts of these three proteins were unchanged. Taken together, our study demonstrates that PINCH proteins play essential roles in myocardial growth, maturation, remodeling and function, and highlights the importance of studying the role of PINCH proteins in human cardiac injury and cardiomyopathy.
Conclusions
Despite significant progress in our understanding of the molecular regulation of cell adhesions by integrin and its associated complex, many outstanding questions remain. ILK and PINCH are ubiquitously expressed in all cell types. However, it is not clear how one specific function in a given cellular context is achieved over others, such as survival vs. growth, differentiation and migration. PINCH1 plays a critical role in the neural crest in a seemingly ILK independent manner, and ILK expression is not affected in the PINCH1 null neural crest. In addition, PINCH1 in NCCs was found to be predominantly nuclear and hence, it will be important to determine whether PINCH1 functions as a shuttling/signaling protein or is directly involved in the regulation of gene expression (Fig.1). It will also be interesting to determine the potential role of ILK in neural crest cells in order to establish how PINCH and ILK function inside and outside of focal adhesion complexes.
Cardiac integrity and function are maintained by dynamic interactions of multiple cell types within the heart, including myocytes, fibroblasts and endothelial cells, and therefore it would be interesting to define the role for PINCH in these contexts. Given the important role thymosinβ4 in mediating cardiomyocyte survival, it is important to determine the detailed molecular mechanisms by which the interactions of thymosinβ4, ILK, and PINCH promote cardiomyocyte survival. Answers to these questions will increase our understanding of fundamental ECM-integrin signaling as well as facilitate the development of novel therapies to human heart disease.
Acknowledgements
We thank Drs. Indroneal Banerjee and Robert Lyon for critical reading of the manuscript. The work cited in the authors’ lab was supported by NIH grants for JC.
Footnotes
Portions of this work were supported by the National Institutes of Health (NIH) via grants to JC.
References
- 1.Wu C. PINCH, N(i)ck and the ILK: network wiring at cell-matrix adhesions. Trends Cell Biol. 2005;15:460–466. doi: 10.1016/j.tcb.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Legate KR, Montanez E, Kudlacek O, et al. ILK, PINCH and parvin: the tIPP of integrin signalling. Nat Rev Mol Cell Biol. 2006;7:20–31. doi: 10.1038/nrm1789. [DOI] [PubMed] [Google Scholar]
- 3.Fukuda T, Chen K, Shi X, et al. PINCH-1 is an obligate partner of integrin-linked kinase (ILK) functioning in cell shape modulation, motility, and survival. J Biol Chem. 2003;278:51324–51333. doi: 10.1074/jbc.M309122200. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Chen K, Tu Y, et al. Distinct roles of two structurally closely related focal adhesion proteins, alpha-parvins and beta-parvins, in regulation of cell morphology and survival. J Biol Chem. 2004;279:41695–41705. doi: 10.1074/jbc.M401563200. [DOI] [PubMed] [Google Scholar]
- 5.Li S, Bordoy R, Stanchi F, et al. PINCH1 regulates cell-matrix and cell-cell adhesions, cell polarity and cell survival during the peri-implantation stage. J Cell Sci. 2005;118:2913–2921. doi: 10.1242/jcs.02422. [DOI] [PubMed] [Google Scholar]
- 6.Liang X, Zhou Q, Li X, et al. PINCH1 plays an essential role in early murine embryonic development but is dispensable in ventricular cardiomyocytes. Mol Cell Biol. 2005;25:3056–3062. doi: 10.1128/MCB.25.8.3056-3062.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coppolino MG, Dedhar S. Bi-directional signal transduction by integrin receptors. Int J Biochem Cell Biol. 2000;32:171–188. doi: 10.1016/s1357-2725(99)00043-6. [DOI] [PubMed] [Google Scholar]
- 8.Delcommenne M, Tan C, Gray V, et al. Phosphoinositide-3-OH kinase-dependent regulation of glycogen synthase kinase 3 and protein kinase B/AKT by the integrin-linked kinase. Proc Natl Acad Sci U S A. 1998;95:11211–11216. doi: 10.1073/pnas.95.19.11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hannigan GE, Leung-Hagesteijn C, Fitz-Gibbon L, et al. Regulation of cell adhesion and anchorage-dependent growth by a new beta 1-integrin-linked protein kinase. Nature. 1996;379:91–96. doi: 10.1038/379091a0. [DOI] [PubMed] [Google Scholar]
- 10.Braun A, Bordoy R, Stanchi F, et al. PINCH2 is a new five LIM domain protein, homologous to PINCHand localized to focal adhesions. Exp Cell Res. 2003;284:239–250. doi: 10.1016/s0014-4827(02)00039-3. [DOI] [PubMed] [Google Scholar]
- 11.Tu Y, Li F, Goicoechea S, et al. The LIM-only protein PINCH directly interacts with integrin-linked kinase and is recruited to integrin-rich sites in spreading cells. Mol Cell Biol. 1999;19:2425–2434. doi: 10.1128/mcb.19.3.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Chen K, Guo L, et al. Characterization of PINCH-2, a new focal adhesion protein that regulates the PINCH-1-ILK interaction, cell spreading, and migration. J Biol Chem. 2002;277:38328–38338. doi: 10.1074/jbc.M205576200. [DOI] [PubMed] [Google Scholar]
- 13.Olski TM, Noegel AA, Korenbaum E. Parvin, a 42 kDa focal adhesion protein, related to the alpha-actinin superfamily. J Cell Sci. 2001;114:525–538. doi: 10.1242/jcs.114.3.525. [DOI] [PubMed] [Google Scholar]
- 14.Tu Y, Huang Y, Zhang Y, et al. A new focal adhesion protein that interacts with integrin-linked kinase and regulates cell adhesion and spreading. J Cell Biol. 2001;153:585–598. doi: 10.1083/jcb.153.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamaji S, Suzuki A, Sugiyama Y, et al. A novel integrin-linked kinase-binding protein, affixin, is involved in the early stage of cell-substrate interaction. J Cell Biol. 2001;153:1251–1264. doi: 10.1083/jcb.153.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikolopoulos SN, Turner CE. Actopaxin, a new focal adhesion protein that binds paxillin LD motifs and actin and regulates cell adhesion. J Cell Biol. 2000;151:1435–1448. doi: 10.1083/jcb.151.7.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikolopoulos SN, Turner CE. Integrin-linked kinase (ILK) binding to paxillin LD1 motif regulates ILK localization to focal adhesions. J Biol Chem. 2001;276:23499–23505. doi: 10.1074/jbc.M102163200. [DOI] [PubMed] [Google Scholar]
- 18.Attwell S, Mills J, Troussard A, et al. Integration of cell attachment, cytoskeletal localization, and signaling by integrin-linked kinase (ILK), CH-ILKBP, and the tumor suppressor PTEN. Mol Biol Cell. 2003;14:4813–4825. doi: 10.1091/mbc.E03-05-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terpstra L, Prud’homme J, Arabian A, et al. Reduced chondrocyte proliferation and chondrodysplasia in mice lacking the integrin-linked kinase in chondrocytes. J Cell Biol. 2003;162:139–148. doi: 10.1083/jcb.200302066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vouret-Craviari V, Boulter E, Grall D, et al. ILK is required for the assembly of matrix-forming adhesions and capillary morphogenesis in endothelial cells. J Cell Sci. 2004;117:4559–4569. doi: 10.1242/jcs.01331. [DOI] [PubMed] [Google Scholar]
- 21.Tan C, Cruet-Hennequart S, Troussard A, et al. Regulation of tumor angiogenesis by integrin-linked kinase (ILK) Cancer Cell. 2004;5:79–90. doi: 10.1016/s1535-6108(03)00281-2. [DOI] [PubMed] [Google Scholar]
- 22.Persad S, Attwell S, Gray V, et al. Inhibition of integrin-linked kinase (ILK) suppresses activation of protein kinase B/Akt and induces cell cycle arrest and apoptosis of PTEN-mutant prostate cancer cells. Proc Natl Acad Sci U S A. 2000;97:3207–3212. doi: 10.1073/pnas.060579697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Attwell S, Roskelley C, Dedhar S. The integrin-linked kinase (ILK) suppresses anoikis. Oncogene. 2000;19:3811–3815. doi: 10.1038/sj.onc.1203711. [DOI] [PubMed] [Google Scholar]
- 24.Gary DS, Milhavet O, Camandola S, et al. Essential role for integrin linked kinase in Akt-mediated integrin survival signaling in hippocampal neurons. J Neurochem. 2003;84:878–890. doi: 10.1046/j.1471-4159.2003.01579.x. [DOI] [PubMed] [Google Scholar]
- 25.Friedrich EB, Liu E, Sinha S, et al. Integrin-linked kinase regulates endothelial cell survival and vascular development. Mol Cell Biol. 2004;24:8134–8144. doi: 10.1128/MCB.24.18.8134-8144.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yau CY, Wheeler JJ, Sutton KL, et al. Inhibition of integrin-linked kinase by a selective small molecule inhibitor, QLT0254, inhibits the PI3K/PKB/mTOR, Stat3, and FKHR pathways and tumor growth, and enhances gemcitabine-induced apoptosis in human orthotopic primary pancreatic cancer xenografts. Cancer Res. 2005;65:1497–1504. doi: 10.1158/0008-5472.CAN-04-2940. [DOI] [PubMed] [Google Scholar]
- 27.Edwards LA, Thiessen B, Dragowska WH, et al. Inhibition of ILK in PTEN-mutant human glioblastomas inhibits PKB/Akt activation, induces apoptosis, and delays tumor growth. Oncogene. 2005;24:3596–3605. doi: 10.1038/sj.onc.1208427. [DOI] [PubMed] [Google Scholar]
- 28.Brakebusch C, Bouvard D, Stanchi F, et al. Integrins in invasive growth. J Clin Invest. 2002;109:999–1006. doi: 10.1172/JCI15468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 30.Giancotti FG, Tarone G. Positional control of cell fate through joint integrin/receptor protein kinase signaling. Annu Rev Cell Dev Biol. 2003;19:173–206. doi: 10.1146/annurev.cellbio.19.031103.133334. [DOI] [PubMed] [Google Scholar]
- 31.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 32.Wu C, Dedhar S. Integrin-linked kinase (ILK) and its interactors: a new paradigm for the coupling of extracellular matrix to actin cytoskeleton and signaling complexes. J Cell Biol. 2001;155:505–510. doi: 10.1083/jcb.200108077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Novak A, Hsu SC, Leung-Hagesteijn C, et al. Cell adhesion and the integrin-linked kinase regulate the LEF-1 and beta-catenin signaling pathways. Proc Natl Acad Sci U S A. 1998;95:4374–4379. doi: 10.1073/pnas.95.8.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rearden A. A new LIM protein containing an autoepitope homologous to “senescent cell antigen”. Biochem Biophys Res Commun. 1994;201:1124–1131. doi: 10.1006/bbrc.1994.1822. [DOI] [PubMed] [Google Scholar]
- 35.Hobert O, Moerman DG, Clark KA, et al. A conserved LIM protein that affects muscular adherens junction integrity and mechanosensory function in Caenorhabditis elegans. J Cell Biol. 1999;144:45–57. doi: 10.1083/jcb.144.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li F, Zhang Y, Wu C. Integrin-linked kinase is localized to cell-matrix focal adhesions but not cell-cell adhesion sites and the focal adhesion localization of integrin-linked kinase is regulated by the PINCH-binding ANK repeats. J Cell Sci. 1999;112(Pt 24):4589–4599. doi: 10.1242/jcs.112.24.4589. [DOI] [PubMed] [Google Scholar]
- 37.Xu Z, Fukuda T, Li Y, et al. Molecular dissection of PINCH-1 reveals a mechanism of coupling and uncoupling of cell shape modulation and survival. J Biol Chem. 2005;280:27631–27637. doi: 10.1074/jbc.M504189200. [DOI] [PubMed] [Google Scholar]
- 38.Stanchi F, Bordoy R, Kudlacek O, et al. Consequences of loss of PINCH2 expression in mice. J Cell Sci. 2005;118:5899–5910. doi: 10.1242/jcs.02686. [DOI] [PubMed] [Google Scholar]
- 39.Liang XQ, Sun YF, Ye MQ, Scimia MC, Cheng HQ, Martin J, Wang G, Rearden A, Wu C, Peterson K, Powell HC, Evans SM, Chen J. Targeted ablation of PINCH1 and PINCH2 from murine myocardium results in dilated cardiomyopathy and early postnatal lethality. Circulation. 2009 doi: 10.1161/CIRCULATIONAHA.109.864686. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tu Y, Li F, Wu C. Nck-2, a novel Src homology2/3-containing adaptor protein that interacts with the LIM-only protein PINCH and components of growth factor receptor kinase-signaling pathways. Mol Biol Cell. 1998;9:3367–3382. doi: 10.1091/mbc.9.12.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buday L, Wunderlich L, Tamas P. The Nck family of adapter proteins: regulators of actin cytoskeleton. Cell Signal. 2002;14:723–731. doi: 10.1016/s0898-6568(02)00027-x. [DOI] [PubMed] [Google Scholar]
- 42.Li W, Fan J, Woodley DT. Nck/Dock: an adapter between cell surface receptors and the actin cytoskeleton. Oncogene. 2001;20:6403–6417. doi: 10.1038/sj.onc.1204782. [DOI] [PubMed] [Google Scholar]
- 43.Velyvis A, Vaynberg J, Yang Y, et al. Structural and functional insights into PINCH LIM4 domain-mediated integrin signaling. Nat Struct Biol. 2003;10:558–564. doi: 10.1038/nsb938. [DOI] [PubMed] [Google Scholar]
- 44.Bladt F, Aippersbach E, Gelkop S, et al. The murine Nck SH2/SH3 adaptors are important for the development of mesoderm-derived embryonic structures and for regulating the cellular actin network. Mol Cell Biol. 2003;23:4586–4597. doi: 10.1128/MCB.23.13.4586-4597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cutler ML, Bassin RH, Zanoni L, et al. Isolation of rsp-1, a novel cDNA capable of suppressing v-Ras transformation. Mol Cell Biol. 1992;12:3750–3756. doi: 10.1128/mcb.12.9.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsuda T, Marinetti MR, Masuelli L, et al. The Ras suppressor RSU-1 localizes to 10p13 and its expression in the U251 glioblastoma cell line correlates with a decrease in growth rate and tumorigenic potential. Oncogene. 1995;11:397–403. [PubMed] [Google Scholar]
- 47.Dougherty GW, Chopp T, Qi SM, et al. The Ras suppressor Rsu-1 binds to the LIM 5 domain of the adaptor protein PINCH1 and participates in adhesion-related functions. Exp Cell Res. 2005;306:168–179. doi: 10.1016/j.yexcr.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 48.Dougherty GW, Jose C, Gimona M, et al. The Rsu-1-PINCH1-ILK complex is regulated by Ras activation in tumor cells. Eur J Cell Biol. 2008;87:721–734. doi: 10.1016/j.ejcb.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kadrmas JL, Smith MA, Clark KA, et al. The integrin effector PINCH regulates JNK activity and epithelial migration in concert with Ras suppressor 1. J Cell Biol. 2004;167:1019–1024. doi: 10.1083/jcb.200408090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bock-Marquette I, Saxena A, White MD, et al. Thymosin beta4 activates integrin-linked kinase and promotes cardiac cell migration, survival and cardiac repair. Nature. 2004;432:466–472. doi: 10.1038/nature03000. [DOI] [PubMed] [Google Scholar]
- 51.Srivastava D, Saxena A, Michael Dimaio J, et al. Thymosin beta4 is cardioprotective after myocardial infarction. Ann N Y Acad Sci. 2007;1112:161–170. doi: 10.1196/annals.1415.048. [DOI] [PubMed] [Google Scholar]
- 52.Clark KA, McGrail M, Beckerle MC. Analysis of PINCH function in Drosophila demonstrates its requirement in integrin-dependent cellular processes. Development. 2003;130:2611–2621. doi: 10.1242/dev.00492. [DOI] [PubMed] [Google Scholar]
- 53.Gettner SN, Kenyon C, Reichardt LF. Characterization of beta pat-3 heterodimers, a family of essential integrin receptors in C. elegans. J Cell Biol. 1995;129:1127–1141. doi: 10.1083/jcb.129.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mackinnon AC, Qadota H, Norman KR, et al. C. elegans PAT-4/ILK functions as an adaptor protein within integrin adhesion complexes. Curr Biol. 2002;12:787–797. doi: 10.1016/s0960-9822(02)00810-2. [DOI] [PubMed] [Google Scholar]
- 55.Leptin M, Bogaert T, Lehmann R, et al. The function of PS integrins during Drosophila embryogenesis. Cell. 1989;56:401–408. doi: 10.1016/0092-8674(89)90243-2. [DOI] [PubMed] [Google Scholar]
- 56.MacKrell AJ, Blumberg B, Haynes SR, et al. The lethal myospheroid gene of Drosophila encodes a membrane protein homologous to vertebrate integrin beta subunits. Proc Natl Acad Sci U S A. 1988;85:2633–2637. doi: 10.1073/pnas.85.8.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zervas CG, Gregory SL, Brown NH. Drosophila integrin-linked kinase is required at sites of integrin adhesion to link the cytoskeleton to the plasma membrane. J Cell Biol. 2001;152:1007–1018. doi: 10.1083/jcb.152.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tam PP, Beddington RS. Establishment and organization of germ layers in the gastrulating mouse embryo. Ciba Found Symp. 1992;165:27–41. doi: 10.1002/9780470514221.ch3. discussion 42-29. [DOI] [PubMed] [Google Scholar]
- 59.Coucouvanis E, Martin GR. Signals for death and survival: a two-step mechanism for cavitation in the vertebrate embryo. Cell. 1995;83:279–287. doi: 10.1016/0092-8674(95)90169-8. [DOI] [PubMed] [Google Scholar]
- 60.Aumailley M, Pesch M, Tunggal L, et al. Altered synthesis of laminin 1 and absence of basement membrane component deposition in (beta)1 integrin-deficient embryoid bodies. J Cell Sci. 2000;113(Pt 2):259–268. doi: 10.1242/jcs.113.2.259. [DOI] [PubMed] [Google Scholar]
- 61.Fassler R, Meyer M. Consequences of lack of beta 1 integrin gene expression in mice. Genes Dev. 1995;9:1896–1908. doi: 10.1101/gad.9.15.1896. [DOI] [PubMed] [Google Scholar]
- 62.Li S, Harrison D, Carbonetto S, et al. Matrix assembly, regulation, and survival functions of laminin and its receptors in embryonic stem cell differentiation. J Cell Biol. 2002;157:1279–1290. doi: 10.1083/jcb.200203073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sakai T, Li S, Docheva D, et al. Integrin-linked kinase (ILK) is required for polarizing the epiblast, cell adhesion, and controlling actin accumulation. Genes Dev. 2003;17:926–940. doi: 10.1101/gad.255603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stephens LE, Sutherland AE, Klimanskaya IV, et al. Deletion of beta 1 integrins in mice results in inner cell mass failure and peri-implantation lethality. Genes Dev. 1995;9:1883–1895. doi: 10.1101/gad.9.15.1883. [DOI] [PubMed] [Google Scholar]
- 65.Le Douarin NM, Dupin E. Multipotentiality of the neural crest. Curr Opin Genet Dev. 2003;13:529–536. doi: 10.1016/j.gde.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 66.Kirby ML, Waldo KL. Neural crest and cardiovascular patterning. Circ Res. 1995;77:211–215. doi: 10.1161/01.res.77.2.211. [DOI] [PubMed] [Google Scholar]
- 67.Jiang X, Rowitch DH, Soriano P, et al. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- 68.Kirby ML, Gale TF, Stewart DE. Neural crest cells contribute to normal aorticopulmonary septation. Science. 1983;220:1059–1061. doi: 10.1126/science.6844926. [DOI] [PubMed] [Google Scholar]
- 69.Miyagawa-Tomita S, Waldo K, Tomita H, et al. Temporospatial study of the migration and distribution of cardiac neural crest in quail-chick chimeras. Am J Anat. 1991;192:79–88. doi: 10.1002/aja.1001920109. [DOI] [PubMed] [Google Scholar]
- 70.Conway SJ, Henderson DJ, Copp AJ. Pax3 is required for cardiac neural crest migration in the mouse: evidence from the splotch (Sp2H) mutant. Development. 1997;124:505–514. doi: 10.1242/dev.124.2.505. [DOI] [PubMed] [Google Scholar]
- 71.Epstein JA, Li J, Lang D, et al. Migration of cardiac neural crest cells in Splotch embryos. Development. 2000;127:1869–1878. doi: 10.1242/dev.127.9.1869. [DOI] [PubMed] [Google Scholar]
- 72.Brand T. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev Biol. 2003;258:1–19. doi: 10.1016/s0012-1606(03)00112-x. [DOI] [PubMed] [Google Scholar]
- 73.Lipson AH, Yuille D, Angel M, et al. Velocardiofacial (Shprintzen) syndrome: an important syndrome for the dysmorphologist to recognise. J Med Genet. 1991;28:596–604. doi: 10.1136/jmg.28.9.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brannan CI, Perkins AS, Vogel KS, et al. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- 75.Brown CB, Boyer AS, Runyan RB, et al. Requirement of type III TGF-beta receptor for endocardial cell transformation in the heart. Science. 1999;283:2080–2082. doi: 10.1126/science.283.5410.2080. [DOI] [PubMed] [Google Scholar]
- 76.Stoller JZ, Epstein JA. Cardiac neural crest. Semin Cell Dev Biol. 2005;16:704–715. doi: 10.1016/j.semcdb.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 77.Liang X, Sun Y, Schneider J, et al. Pinch1 is required for normal development of cranial and cardiac neural crest-derived structures. Circ Res. 2007;100:527–535. doi: 10.1161/01.RES.0000259041.37059.8c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martinsen BJ, Neumann AN, Frasier AJ, et al. PINCH-1 expression during early avian embryogenesis: implications for neural crest and heart development. Dev Dyn. 2006;235:152–162. doi: 10.1002/dvdy.20616. [DOI] [PubMed] [Google Scholar]
- 79.Loeys BL, Schwarze U, Holm T, et al. Aneurysm syndromes caused by mutations in the TGF-beta receptor. N Engl J Med. 2006;355:788–798. doi: 10.1056/NEJMoa055695. [DOI] [PubMed] [Google Scholar]
- 80.Mizuguchi T, Collod-Beroud G, Akiyama T, et al. Heterozygous TGFBR2 mutations in Marfan syndrome. Nat Genet. 2004;36:855–860. doi: 10.1038/ng1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Neptune ER, Frischmeyer PA, Arking DE, et al. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat Genet. 2003;33:407–411. doi: 10.1038/ng1116. [DOI] [PubMed] [Google Scholar]
- 82.Wurdak H, Ittner LM, Lang KS, et al. Inactivation of TGFbeta signaling in neural crest stem cells leads to multiple defects reminiscent of DiGeorge syndrome. Genes Dev. 2005;19:530–535. doi: 10.1101/gad.317405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brown CB, Boyer AS, Runyan RB, et al. Antibodies to the Type II TGFbeta receptor block cell activation and migration during atrioventricular cushion transformation in the heart. Dev Biol. 1996;174:248–257. doi: 10.1006/dbio.1996.0070. [DOI] [PubMed] [Google Scholar]
- 84.Forbes MS, Sperelakis N. Intercalated discs of mammalian heart: a review of structure and function. Tissue Cell. 1985;17:605–648. doi: 10.1016/0040-8166(85)90001-1. [DOI] [PubMed] [Google Scholar]
- 85.Wiesner S, Legate KR, Fassler R. Integrin-actin interactions. Cell Mol Life Sci. 2005;62:1081–1099. doi: 10.1007/s00018-005-4522-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jamora C, Fuchs E. Intercellular adhesion, signalling and the cytoskeleton. Nat Cell Biol. 2002;4:E101–108. doi: 10.1038/ncb0402-e101. [DOI] [PubMed] [Google Scholar]
- 87.Drees F, Pokutta S, Yamada S, et al. Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell. 2005;123:903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aberle H, Schwartz H, Kemler R. Cadherin-catenin complex: protein interactions and their implications for cadherin function. J Cell Biochem. 1996;61:514–523. doi: 10.1002/(SICI)1097-4644(19960616)61:4%3C514::AID-JCB4%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 89.Perriard JC, Hirschy A, Ehler E. Dilated cardiomyopathy: a disease of the intercalated disc? Trends Cardiovasc Med. 2003;13:30–38. doi: 10.1016/s1050-1738(02)00209-8. [DOI] [PubMed] [Google Scholar]
- 90.Radice GL, Rayburn H, Matsunami H, et al. Developmental defects in mouse embryos lacking N-cadherin. Dev Biol. 1997;181:64–78. doi: 10.1006/dbio.1996.8443. [DOI] [PubMed] [Google Scholar]
- 91.Angst BD, Khan LU, Severs NJ, et al. Dissociated spatial patterning of gap junctions and cell adhesion junctions during postnatal differentiation of ventricular myocardium. Circ Res. 1997;80:88–94. doi: 10.1161/01.res.80.1.88. [DOI] [PubMed] [Google Scholar]
- 92.Hirschy A, Schatzmann F, Ehler E, et al. Establishment of cardiac cytoarchitecture in the developing mouse heart. Dev Biol. 2006;289:430–441. doi: 10.1016/j.ydbio.2005.10.046. [DOI] [PubMed] [Google Scholar]
- 93.Shai SY, Harpf AE, Babbitt CJ, et al. Cardiac myocyte-specific excision of the beta1 integrin gene results in myocardial fibrosis and cardiac failure. Circ Res. 2002;90:458–464. doi: 10.1161/hh0402.105790. [DOI] [PubMed] [Google Scholar]
- 94.White DE, Coutu P, Shi YF, et al. Targeted ablation of ILK from the murine heart results in dilated cardiomyopathy and spontaneous heart failure. Genes Dev. 2006;20:2355–2360. doi: 10.1101/gad.1458906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ieda M, Tsuchihashi T, Ivey KN, et al. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev Cell. 2009;16:233–244. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]