Abstract
Secondary structure within the downstream region of mammalian polyadenylation signals has been proposed to perform important functions. The simian virus 40 late polyadenylation signal (SVLPA) forms alternate secondary structures in equilibrium. Their formation correlates with cleavage-polyadenylation efficiency (H. Hans and J. C. Alwine, Mol. Cell. Biol. 20:2926-2932, 2000; M. I. Zarudnaya, I. M. Kolomiets, A. L. Potyahaylo, and D. M. Hovorun, Nucleic Acids Res. 3:1375-1386, 2003), and oligonucleotides that disrupt the secondary structure inhibit in vitro cleavage. To define the important features of downstream secondary structure, we first minimized the SVLPA by deletion, forming a downstream region with fewer, and more stable, stem-loop structures. Specific mutagenesis showed that both stem stability and loop size are important functional features of the downstream region. Stabilization of the stem, thus minimizing alternative structures, decreased cleavage efficiency both in vitro and in vivo. This was most deleterious when the stem was stabilized at the base of the loop, constraining loop size by inhibiting breathing of the stem. The significance of loop size was supported by mutants that showed increased cleavage efficiency with increased loop size and vice versa. A loop of at least 12 nucleotides promoted cleavage; U richness in the loop also promoted cleavage and was particularly important when the stem was stabilized. A mutation designed to eliminate downstream secondary structure still formed many relatively weak alternative structures in equilibrium and retained function. The data suggest that although the downstream region is very important, its structure is quite malleable and is able to tolerate significant mutation within a wide range of primary and secondary structural features. We propose that this malleability is due to the enhanced ability of GU- and U-rich downstream elements to easily form secondary structures with surrounding sequences.
Polyadenylation is the process by which the 3′ ends of most mammalian mRNAs are formed. In a tightly coupled set of reactions, the precursor RNA is endonucleolytically cleaved at a specific site and then approximately 250 adenosine residues are polymerized to the cleaved end, forming the poly(A) tail. Nearly all mammalian cellular mRNAs are so processed in the nucleus. In addition, the mammalian DNA viruses which replicate in the nucleus utilize the cellular polyadenylation mechanisms. It has been clearly established that the poly(A) tail is essential for the stability, transport, and translation of most mRNAs (reviewed in references 9, 25, 26, 33, and 34).
The polyadenylation signal defines the site of polyadenylation through specific binding of a complex of proteins that orchestrate cleavage and polyadenylation of the precursor RNA. Binding specificity is provided by elements in the RNA. The central, nearly invariant, feature of mammalian polyadenylation signals is the six-nucleotide consensus element AAUAAA, located between 11 and 25 nucleotides upstream of the actual cleavage-and-polyadenylation site. The AAUAAA element is the binding site for the 160-kDa subunit of the cleavage-and-polyadenylation specificity factor (15, 21). Since AAUAAA sequences are also found within the coding regions of many genes, mechanisms must exist to specify the correct AAUAAA sequence to be utilized. This is provided by elements located downstream of the AAUAAA sequences (downstream elements [DSEs]), which greatly increase the efficiency of utilization of the preceding AAUAAA sequence (1-3, 5, 10, 17-19, 24, 27, 28, 35, 36, 38) and the coupling of splicing and polyadenylation (13). DSEs do not have a clear consensus but tend to be variable lengths (6 to 20 nucleotides) of GU- or U-rich sequences. These GU-rich elements serve as binding sites for the 64-kDa subunit of the cleavage-stimulatory factor (CStF) (9, 31). The optimal position for the DSE appears to be downstream of the cleavage site and 14 to 70 nucleotides downstream of the AAUAAA element. The position of the DSE relative to the AAUAAA element appears to be important; DSEs located either closer or farther away result in reduced polyadenylation efficiency (7; our unpublished observations). In cases in which efficient polyadenylation signals have their DSEs located far downstream of the AAUAAA element, secondary structure in the RNA brings the DSE to a more optimal position relative to the AAUAAA element (4).
We have previously postulated that secondary structure involving the sequences in the downstream region may have an important functional role in many, if not all, mammalian polyadenylation signals (14). U- or GU- rich regions are ideal for forming secondary structures with surrounding sequences, since G's and U's each have a choice of two bases to pair with (G's with C's and U's, U's with A's and G's). While the resulting secondary structures may not be exceptionally stable, they may exist long enough to allow polyadenylation factors to function, thus enhancing processing efficiency.
We have previously demonstrated extensive secondary structure in the downstream region of the simian virus 40 late polyadenylation signal (SVLPA) and that the ability to form structured downstream regions correlates with cleavage efficiency (14). The SVLPA is particularly efficient (6), corresponding to its structure, which is more complex than that of most polyadenylation signals (Fig. 1). The consensus AAUAAA element is located 13 nucleotides upstream of the cleavage-and-polyadenylation site (An). In addition to the AAUAAA element, elements both upstream and downstream affect its efficiency of utilization (6, 10, 27-29). The downstream region contains the expected U-rich element (DSE-U, U in Fig. 1) (10), which is similar to the DSEs of most polyadenylation signals in that it is bound by the 64-kDa component of CStF (32). The SVLPA also contains two additional DSEs, a G-rich element and a second U-rich element. The G-rich element (DSE-G, G in Fig. 1) can be bound by hnRNP H (2, 3, 23); this binding may enhance polyadenylation efficiency (1, 3). The second U-rich element (DSE-U′, U′ in Fig. 1A) (27, 28) can function as an hnRNP C protein binding site (35). It can function as a DSE in synthetic polyadenylation signals (35, 36) and has been shown to enhance the coupling of splicing and polyadenylation (13).
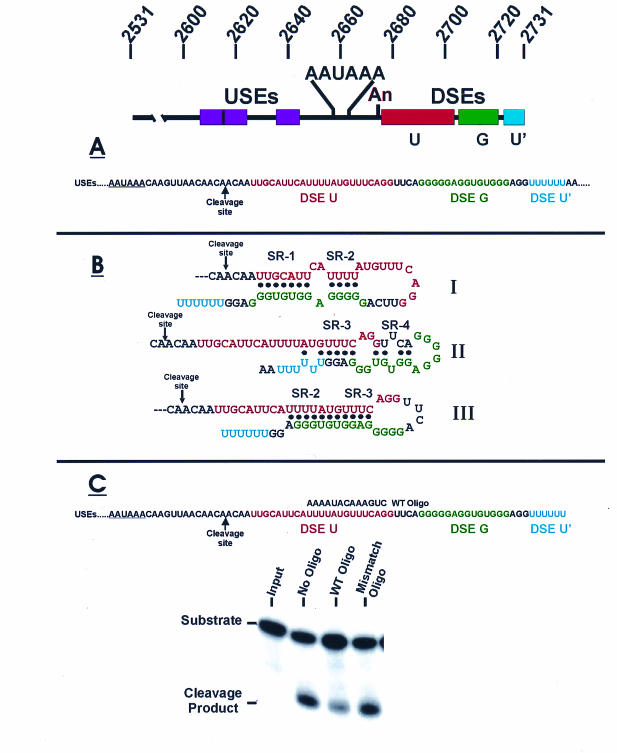
FIG. 1.
Linear and secondary structures of the SVLPA. (A) The linear structure of the SVLPA shows the AAUAAA element, the cleavage-polyadenylation site (An), the upstream efficiency elements (USEs; purple), and the defined downstream efficiency elements: a U-rich element in red (U, DSE U), a G-rich element in green (G, DSE G), and a second U-rich element in blue (U′, DSE U′). The linear sequence of the downstream region is shown below, beginning with the AAUAAA element; colors of the three DSEs are the same as above. (B) The three secondary structures of the SVLPA downstream sequence that can be predicted from previous data (14, 37). (C) Results of oligonucleotide (Oligo)-directed disruption of SVLPA secondary structure on in vitro cleavage efficiency.
We previously analyzed the secondary structure of the downstream region of the SVLPA by using nuclease sensitivity structure probing (14). Analysis of these data (37; our unpublished results) predicted that there are at least three structures (Fig. 1B) that exist in equilibrium. In the present work, we minimized the SVLPA by deletion to form a downstream region with fewer and simpler structures while retaining function. The resulting modified late polyadenylation signal (mLPA) was then specifically mutagenized to define the essential features of downstream secondary structure.
MATERIALS AND METHODS
Plasmids.
pGem2-UPAS contains the wild-type (WT) SVLPA (nucleotides 2531 to 2729) (29). By linker substitution mutagenesis, nucleotides 2618 and 2637 of pGem2-UPAS were replaced with an XbaI site to create pGem2-MLPA. Mutations were introduced into pGem2-MLPA by site-directed mutagenesis (Quick-Change; Stratagene) to generate the pGem2-MLPA mutant series. To test the polyadenylation efficiency of mLPA and its mutant forms in vivo by transfection analyses, they were introduced into pGL3-control (Promega). A SacII site was introduced at nucleotide 5095 in pGL3-control to eliminate the AAUAAA sequence in the synthetic polyadenylation signal. An EcoRI site was inserted at nucleotide 2185 to facilitate cloning of mLPA and mutant polyadenylation signals. The resulting plasmid was digested with HpaI and EcoRI and then ligated with polyadenylation signal-containing fragments from WT and mutant pGem2-MLPA plasmids.
RNA substrates.
Templates for in vitro transcription were obtained by linearizing WT and mutant pGem2-MLPA plasmids with DraI. For in vitro polyadenylation assays, RNA substrates were synthesized by T7 RNA polymerase (Promega) in accordance with the manufacturer's instruction in the presence of m7GpppG cap structure and [α-32P]UTP. Labeled RNAs were phenol-chloroform-isoamyl alcohol (50:49:1) extracted and purified through P-30 micro Bio-Spin columns (Bio-Rad) (11-13). RNA substrates used in structure probing were synthesized without the cap structure. After phenol extraction and ethanol precipitation, the RNAs were 3′ end labeled with cytidine 3′,5′-bis[α-32P]phosphate and T4 RNA ligase and then purified on a 5% acrylamide-7 M urea gel.
In vitro polyadenylation assay.
HeLa-S3 nuclear extract were prepared as previously described (20), from cells obtained from the National Cell Culture Center. 32P-labeled RNA substrates were analyzed for in vitro cleavage efficiency with cordycepin as previously described (12). The reaction product RNAs were phenol extracted, ethanol precipitated, separated on an 8% polyacrylamide-7 M urea gel, and then quantitated with a Molecular Dynamics PhosphorImager. For in vitro cleavage experiments with added oligonucleotides, the oligonucleotide and substrate RNA (25:1) were mixed without polyvinyl alcohol and nuclear extract, preincubated at 50°C for 10 min, and then slowly cooled to room temperature. The remaining components of the reaction mixture were added, and the mixture was assayed as described above. The sequence of the specific oligonucleotide used is shown in Fig. 1C; the mismatched oligonucleotide was 5′-CGGUACCUCCCGCGU-3′.
RNA structure probing.
3′-end-labeled RNAs prepared as described above were used in all experiments. RNA ladders were generated by partial hydrolysis of RNA in 50 mM sodium carbonate (pH 9.2) and 1 mM EDTA for 5 min at 95°C. RNase T1 digestion (0.01 U) was performed in 20 mM sodium citrate (pH 5)-1 mM EDTA-7 M urea for 15 min at 37°C. Nuclease S1 (0.1, 1, or 10 U), RNase V1 (0.5 × 10−3 or 1 × 10−3 U) or Pb2+ (0.2 or 0.5 mM lead acetate) digestion was performed in 10 mM Tris (pH 7)-100 mM KCl-10 mM MgCl2 for 15 min at room temperature. Reactions were stopped by adding 2 volumes of 95% formamide-18 mM EDTA-0.025% sodium dodecyl sulfate-0.025% xylene cyanol-0.025% bromophenol blue and loaded onto a 10% polyacrylamide-7 M urea gel (8, 16). RNase V1, under some conditions, can preferentially cleave the nucleotides on only one strand of a double-stranded region, thus showing only half of a structured region (16, 30). This has occurred with the SVLPA (14) and must be considered when interpreting structure analysis data.
Cell culture and transfection.
CV-1 cells were maintained in Dulbecco's modified Eagle's medium containing 5% fetal calf serum. Cells were plated on 12-well plates 1 day before transfection and reached 80% confluence on the day of transfection. Each well was transfected with 500 ng of DNA, 1.5 μl of Fugene 6 reagent (Roche), and serum-free medium to a final volume of 30 μl. After 18 h, luciferase expression was measured with the Promega Luciferase Assay System.
RESULTS
An oligonucleotide that disrupts downstream secondary structure inhibits in vitro polyadenylation.
To determine whether disruption of secondary structure in the downstream region of the SVLPA inhibits cleavage efficiency, we synthesized a 15-nucleotide RNA (Fig. 1C) complementary to sequences in the downstream region. We predicted that this oligonucleotide would significantly interfere with the formation of the natural secondary structures when annealed to the SVLPA substrate RNA. In vitro cleavage efficiency of SVLPA was significantly reduced in the presence of a 25-fold excess of the oligonucleotide (Fig. 1C), but not in the presence of a mismatched oligonucleotide. This result suggests that disruption of the natural secondary structure inhibits cleavage efficiency.
The mLPA.
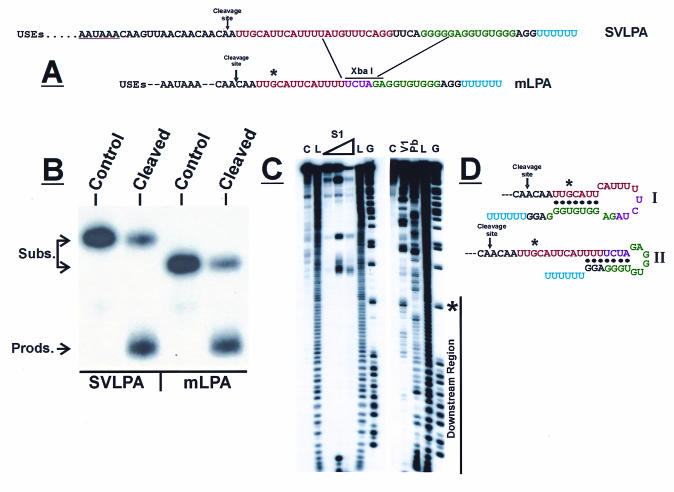
To determine whether a minimal functional secondary structure can be defined in the downstream region of the SVLPA, we constructed an mLPA that would minimize alternative secondary structures (Fig. 2A). Eighteen nucleotides of the WT SVLPA were replaced with four new nucleotides to form a new XbaI site. This deletion was chosen because it preserves, in large part, the base pairing predicted in SVLPA structure I (shown in Fig. 1B), which was predicted by previous data (14, 37) to be the predominant structure within the SVLPA. In vitro cleavage assays (Fig. 2B) show that the WT SVLPA and the mLPA have very similar cleavage efficiencies.
FIG. 2.
In vitro cleavage efficiency of the mLPA is equivalent to that of WT SVLPA. (A) Linear sequence diagram of the construction of mLPA from SVLPA. (B) In vitro cleavage efficiency of mLPA in comparison to that of SVLPA. (C) RNA structure analysis of mLPA: C, control lane with substrate only; L, alkaline hydrolysis nucleotide ladder; S1, structure analysis with nuclease S1; V1, structure analysis with RNase V1; Pb, structure analysis with Pb2+; G, G sequence analysis with RNase T1. (D) Predicted alternative structures in the downstream region of mLPA. USEs, upstream efficiency elements; Subs., substrate; Prods., products.
RNA secondary-structure analysis of mLPA is shown in Fig. 2C; the downstream region is indicated. The asterisks shown in Fig. 2A, C, and D denote the position of a G nucleotide in the G sequence lane that serves as a reference for reading the structure analysis lanes in Fig. 2C. Increasing amounts of nuclease S1 were used for digestion of single-stranded regions (S1; Fig. 2C, left side). Most of the nucleotides in the downstream region were not accessible to nuclease S1, suggesting significant secondary structure and possible tertiary structure. RNase V1 digestion of double-stranded regions (V1) and Pb2+ metal ion cleavage of single-stranded nucleotides (Pb) show that several regions are sensitive to both Pb2+ and RNase V1 (Fig. 2C, right side). This result indicates that different structures exist in equilibrium. During the course of the structure analysis, some bases vary between single- and double-stranded states and thus are susceptible to both single- and double-stranded cleavage reagents. Figure 2D shows two structures consistent with the data. Structure I was used to model the mutational analysis for the mutant mLPAs described below, since structure II utilizes nucleotides of the inserted XbaI linker that are not naturally occurring sequences in the SVLPA.
Structural analysis of mutations in mLPA.
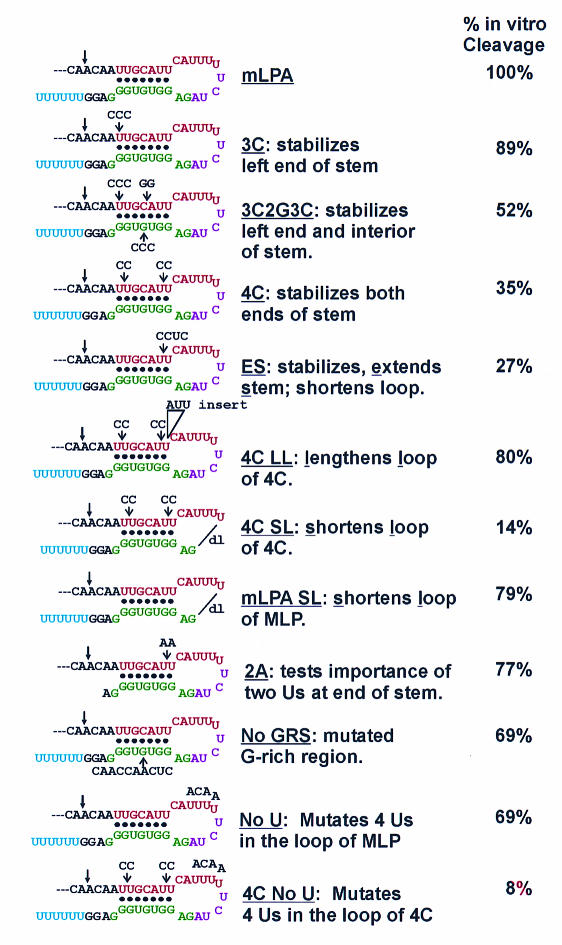
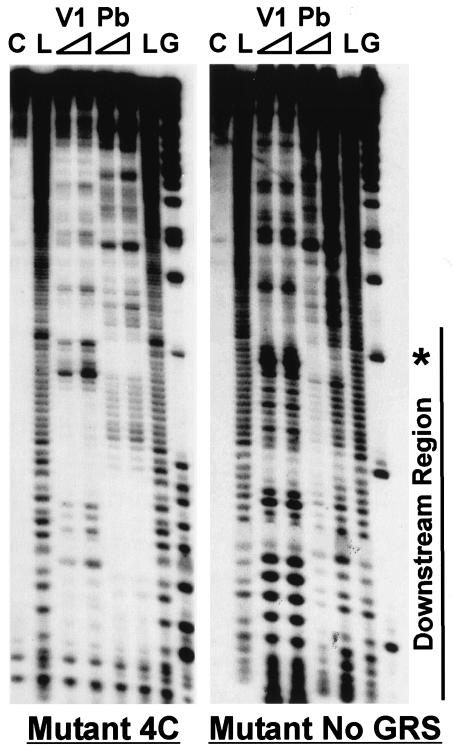
Several mutations designed to alter either the stability of the stem or the size of the loop were introduced into the mLPA (Fig. 3). Structure analysis of each mutant RNA was performed with Pb2+ and RNase V1. Mutants 3C2G3C, ES, 4C, 4C LL, 4C SL, and 4C No U were specifically designed to stabilize the stem structure in mLPA, and structural analysis showed that they each have one stable structure. For example, structural analysis of mutant 4C (Fig. 4) displays very good agreement, and little overlap, between the single-stranded Pb2+ cleavage and double-stranded RNase V1 cleavage. This result strongly suggests that the single structure shown in Fig. 3 forms stably in the downstream region of mutant 4C RNA.
FIG. 3.
Mutants that alter the secondary structure in the downstream region of mLPA alter in vitro cleavage efficiency. Each mutant was tested for in vitro cleavage efficiency, shown as a percentage of the activity of mLPA (100%).
FIG. 4.
RNA structure analysis of mutants 4C and No GRS. Analysis was similar to that described in the legend to Fig. 2.
Structure analysis of the other mutants (3C, mLPA SL, 2A, No GRS, and No U) suggested that, like mLPA, they formed more than one secondary structure by utilizing the sequences in the downstream region (not shown). Mutant mLPAs lacking downstream structure have not been found. Mutant No GRS (Fig. 3), which replaces the G-rich region, was predicted to have minimal ability to form structures involving only sequences downstream of the AAUAAA element. However, the structure analysis (Fig. 4) showed extensive cleavage throughout the downstream region by both Pb2+ and RNase V1. This result indicates that many relatively weak structures form and dissociate throughout the downstream region, resulting in most nucleotides being alternatively double and single stranded and thus susceptible to both Pb2+ and RNase V1. The potential for forming structures involving only the downstream sequences of mutant No GRS has probably been eliminated; thus, the structures that do form are likely the result of the downstream sequences pairing with sequences from the region upstream of the AAUAAA element.
In vitro cleavage analysis of mutant mLPAs shows that stem stability and loop size are important for function.
In Fig. 3, the vitro cleavage efficiency of each mutant substrate is presented relative to the cleavage efficiency of mLPA, which is set at 100%. Each mutant showed accurate utilization of the known cleavage site in mLPA, and no evidence of the utilization of alternative sites was noted (data not shown). Mutants 3C, 3C2G3C, 4C, and ES were designed to increase the base-pairing stability in the stem by replacing GU base pairs with GC base pairs or otherwise mutating to increasing the GC base pairing in the stem. The data suggest that stabilizing the stem reduces cleavage efficiency, and stabilization of the right end (at the base of the loop) is most inhibitory. Specifically, mutant 3C, which stabilizes the left end of the stem, had little effect (89% of the efficiency of mLPA), whereas mutant 4C, which stabilizes both the left and right ends of the stem, was very deleterious (35% of the efficiency of mLPA). In agreement, mutant 3C2G3C, which stabilizes the left end and center of the stem, showed intermediate activity (52% of the efficiency of mLPA).
Stabilization of the right end of the stem, as in mutant 4C, may restrict the size of the loop by preventing breathing of the stem at the base of the loop. The significance of loop size was shown by constructing mutants ES and 4C LL (Fig. 3). Mutant ES extends the stem with additional base pairing, further constricting loop size. This resulted in even lower polyadenylation efficiency (27% of the efficiency of mLPA). In contrast, polyadenylation efficiency was dramatically increased in mutant 4C LL (80% of the efficiency of mLPA), in which three nonpairing nucleotides (AUU) were inserted to increase loop size in the context of the stable stem of mutant 4C.
We further tested the need for a critical loop size by deleting four bases in the loops of mLPA and mutant 4C, creating mutants mLPA SL and 4C SL. In the case of mutant 4C SL, containing the stable stem, the further reduction of loop size was even more detrimental to polyadenylation efficiency (14% of the efficiency of mLPA). The shortening of the loop in the context of the mLPA was less detrimental (79% of the efficiency of mLPA). We believe that the milder effect of loop shortening in mLPA is due either to the ability of the stem in mLPA to breathe and provide expansion of the loop or to the ability of mLPA to form alternative structures. Taken together, these data suggest that increased stem stability is detrimental to cleavage efficiency because it limits the effective loop size.
It is possible that the two U's at the right end of the stem, mutated to two C's in mutant 4C, are critical in the nucleotide sequence in ways that do not involve stem-loop formation. To rule out this possibility, mutant 2A was constructed, in which the two U's were replaced with two A's, which do not have the stem-stabilizing effect of two C's. Although there is some diminution of cleavage efficiency (77% of the efficiency of mLPA), the data suggest that two U's in this position are not essential features of the linear nucleotide sequence since they can be replaced with A's. However, altering the nucleotides of this region in ways that stabilize the stem, and constrain the loop, significantly affects polyadenylation efficiency.
In mutant No GRS, the G-rich region was replaced with nucleotides that would minimize secondary structure involving the downstream nucleotides. However, as described above, the data suggest that extensive structure exists in the form of many relatively weak structures involving the downstream sequences. The in vitro cleavage analysis shows that loss of the G-rich region was tolerated and that the alternative structures formed could support accurate cleavage at 69% of the efficiency of mLPA.
Mutant No U was designed to determine the significance of the cluster of U's in the loop of mLPA. Their mutation decreases cleavage efficiency to 69% of that of mLPA, suggesting that while the U's are not strictly essential, they augment the efficiency of the reaction. When the No U mutation was inserted into the mutant 4C context, with the stable stem (4C No U), we noted that nearly all of the remaining activity was lost (8% of the efficiency of mLPA). Thus, in mutant 4C, the remaining in vitro cleavage activity depends on the U cluster. This result suggests that the necessity for a single-stranded U cluster increases with increased stem stability and constricted variability of the downstream structure.
In vivo effects of mutant mLPAs correlate well with in vitro cleavage efficiencies.
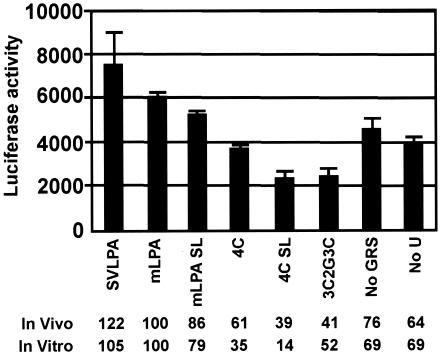
To determine whether the in vitro cleavage activities of WT and mutant mLPAs correlated with in vivo polyadenylation efficiency, luciferase reporter plasmids were constructed containing WT or mutant mLPAs as the polyadenylation signals. These plasmids were transfected into CV-1 cells, and luciferase activity was used as an indication of in vivo polyadenylation efficiency. The in vivo data correlate well with the in vitro data (Fig. 5). For example, mutant 3C2G3C, with the stabilized stem, has 41% of the luciferase activity of mLPA, which compares well with 52% in vitro cleavage activity. Mutant 4C showed the most variation between in vivo and in vitro data, showing 61% of the mLPA luciferase activity and 35% in vitro cleavage activity. However, both the in vivo and in vitro activities of 4C were significantly reduced when the loop size was shortened in mutant 4C SL. The fact that the effects of these mutations are not as severe in vivo as in vitro is likely due to the presence of RNA helicases in vivo that help open the stem and increase loop size.
FIG. 5.
In vivo polyadenylation efficiency of mLPA and its mutant forms corresponds well to in vitro cleavage efficiencies. WT mLPA, various mutant mLPAs, and the SVLPA were used as the polyadenylation signals in luciferase reporter plasmids. The levels of luciferase activity were compared to the activity of mLPA, which was set at 100%. The in vitro cleavage activities from Fig. 3 are shown for comparison.
Mutant No-GRS, in which the entire G-rich region is replaced, remained quite functional both in vitro and in vivo, suggesting that the more random, and less stable, secondary structures that form (Fig. 4) are operable.
DISCUSSION
Previous analysis of the secondary structure of the SVLPA showed that the downstream region forms stem-loop structures in solution (14, 37), and the ability to form these structures correlates with increased in vitro cleavage efficiency (14). Although the SVLPA signal is more efficient and structurally more complex than most mammalian polyadenylation signals, it is likely that downstream secondary structure is a common feature of mammalian polyadenylation signals, since all contain DSEs that tend to be U or GU rich. Since G's and U's each have a choice of two bases to pair with (G's with C's and U's, U's with A's and G's), the ability of GU- and U-rich elements to enter into secondary structure with surrounding sequences is heightened. In addition, many nonclassical base pairings can occur in RNA under the proper sequence and folding conditions. While the resulting secondary structures may not be exceptionally stable, they may exist long enough to allow polyadenylation factors to function. Thus, the stability of the secondary structure may be a determinant of polyadenylation efficiency.
In the present studies, we have characterized the functional downstream secondary structure of the SVLPA to establish the general features of downstream secondary structure. Our approach was to minimize the SVLPA to a form that retains function as well as stable stem-and-loop structures and then characterize the contributions of various elements by specific mutagenesis. Mutations that stabilized the stem generally decreased in vitro cleavage efficiency (Fig. 3). The effect was most deleterious when mutations stabilizing the stem were near the base of the loop, which appears to constrain loop size by minimizing breathing of the stem. The importance of loop size was further demonstrated by mutants that either decreased loop size in the presence of a stable stem, resulting in decreased cleavage efficiency, or increased loop size in the presence of a stable stem, resulting in increased cleavage efficiency. The data suggest that a loop size of at least 12 nucleotides promotes efficient polyadenylation.
One possible reason for the requirement of a loop of this minimum size may be the stable binding of CStF-64. It is possible that the stem and loop function to bind and position factors such as CStF and cleavage factors CFI and CFII near the cleavage site. In analogy with the splicing reaction, it is interesting to speculate that the RNA in the stem-and-loop region, in conjunction with the polyadenylation factors, may be a catalytic participant in the cleavage reaction.
A significant functional feature of the loop in mutant 4C is the cluster of U's. Recently, the RNA binding domain of CStF-64 has been resolved by nuclear magnetic resonance analysis (22). The structure suggests that it binds to UU dinucleotides; thus, the U's in the loop and the surrounding single-stranded loop nucleotides may make up the binding site. This may account for the critical nature of the U cluster when it is mutated in the context of mutant 4C, where the loop size is constrained.
That secondary structure is a functional feature of the downstream region is also suggested by the inability to create a mutant mLPA containing no downstream structure. Mutant No GRS was designed to do this, but the data suggest that extensive structure still forms in its downstream region as many relatively weak structures in equilibrium. Since mutant No GRS cleaves accurately and relatively efficiently, this result suggests that an equilibrium of relatively unstable structures can function. In contrast, the mutant 4C data suggest that a stable structure may be inefficient.
These findings suggest that there may be an optimal stability of the secondary structure in the downstream region, and variations from this optimum may be a means of controlling gene expression by affecting polyadenylation efficiency. For example, in a simple model, more stable structures with small loops would result in weaker gene expression, whereas a marginally stable secondary structure may be more efficient and easily regulated. However, the utilization of downstream secondary structure could be more dynamic; for example, a stable structure may result in inefficient polyadenylation under some cellular conditions, but upon the induction of additional processing factors or RNA helicases, polyadenylation and gene expression may increase. Alternatively, control of polyadenylation efficiency may arise from the balance of marginally stable alternative secondary structures and how these alternative structures are affected by trans-acting factors. In this regard, it is interesting that the SVLPA retains sequences able to form three competing secondary structures (Fig. 1B); the mLPA also forms alternative structures (Fig. 2D). Since each of these polyadenylation signals is efficient, both in vitro and in vivo, the ability to form alternative structures correlates with increased efficiency. Thus, it is possible that each alternative structure, or changes between the structures during the polyadenylation reaction, plays an important role in the polyadenylation reaction and its regulation.
Our data also reassess the functions of the DSEs of the SVLPA. The mLPA signal retains in vitro and in vivo activities similar to those of the SVLPA signal, despite the deletion of a substantial portion of DSE-U and part of DSE-G (Fig. 2). This suggests that there is redundancy in the SVLPA. Interestingly, the mutant No GRS (Fig. 3) consists of the mLPA signal with DSE-G completely replaced. This resulted in only a 30% loss of in vitro activity and a 25% loss of in vivo activity in comparison with the mLPA signal. In contrast, when DSE-G was completely replaced in the context of the intact SVLPA, there was a 75 to 80% decrease in cleavage efficiency (2, 3). Thus, the loss of all of DSE-G is better tolerated in the context of the mLPA than in that of the SVLPA. This may be due to disruption of the balance between the alternative secondary structures of the SVLPA as discussed above.
Our findings suggest that downstream structural features of polyadenylation signals are quite malleable, are able to tolerate significant mutation, and function within a wide range of primary and secondary structural features. We believe that GU- and U-rich elements, with their heightened ability to nucleate secondary structure with surrounding sequences, provide such malleability.
Acknowledgments
We thank the members of the Alwine laboratory for helpful discussion and critical evaluation of the data and Sherri Adams for critical evaluation of the manuscript.
This work was supported by NIH grant GM45773 provided to J.C.A. by the Public Health Service.
REFERENCES
- 1.Arhin, G. K., M. Boots, P. S. Bagga, C. Milcarek, and J. Wilusz. 2002. Downstream sequence elements with different affinities for the hnRNP H/H′ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res. 30:1842-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagga, P. S., G. K. Arhin, and J. Wilusz. 1998. DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res. 26:5343-5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagga, P. S., L. P. Ford, F. Chen, and J. Wilusz. 1995. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-factor. Nucleic Acids Res. 23:1625-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bar-Shira, A., A. Panet, and A. Honigman. 1991. An RNA secondary structure juxtaposes two remote genetic signals for human T-cell leukemia virus type I RNA 3′-end processing. J. Virol. 65:5165-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhat, B. M., and W. S. M. Wold. 1985. ATTAAA as well as downstream sequences are required for RNA 3′-end formation in the E3 complex transcription unit of adenovirus. Mol. Cell. Biol. 5:3183-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carswell, S., and J. C. Alwine. 1989. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol. Cell. Biol. 9:4248-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, F., C. C. MacDonald, and J. Wilusz. 1995. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 23:2614-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciesiolka, J., D. Michalowski, J. Wrzesinski, J. Krajewski, and W. J. Krzyzosiak. 1998. Patterns of cleavages induced by lead ions in defined RNA secondary structure motifs. J. Mol. Biol. 275:211-220. [DOI] [PubMed] [Google Scholar]
- 9.Colgan, D. F., and J. L. Manley. 1997. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11:2755-2766. [DOI] [PubMed] [Google Scholar]
- 10.Conway, L., and M. Wickens. 1985. A sequence downstream of AAUAAA is required for formation of simian virus 40 late mRNA 3′ termini in frog oocytes. Proc. Natl. Acad. Sci. USA 82:3949-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooke, C., and J. C. Alwine. 1996. The cap and the 3′ splice site similarly affects polyadenylation efficiency. Mol. Cell. Biol. 16:2579-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooke, C., and J. C. Alwine. 2002. Characterization of specific protein-RNA complexes associated with the coupling of polyadenylation and last-intron removal. Mol. Cell. Biol. 22:4579-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooke, C., H. Hans, and J. C. Alwine. 1999. Utilization of splicing elements and polyadenylation signal elements in the coupling of splicing and last-intron removal. Mol. Cell. Biol. 19:4971-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hans, H., and J. C. Alwine. 2000. Functionally significant secondary structure of the simian virus 40 late polyadenylation signal. Mol. Cell. Biol. 20:2926-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keller, W., S. Bienroth, K. M. Lang, and G. Christofori. 1991. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3′ processing AAUAAA. EMBO J. 10:4241-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowman, H. B., and D. E. Draper. 1986. On the recognition of helical RNA by cobra venom V1 nuclease. J. Biol. Chem. 261:5396-5403. [PubMed] [Google Scholar]
- 17.McDevitt, M. A., R. P. Hart, W. W. Wong, and J. R. Nevins. 1986. Sequences capable of restoring poly(A) site function define two distinct downstream elements. EMBO J. 5:2907-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDevitt, M. A., M. J. Imperiale, H. Ali, and J. R. Nevins. 1984. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell 37:992-999. [DOI] [PubMed] [Google Scholar]
- 19.McLauchlan, J., D. Gaffney, J. L. Whitton, and J. B. Clements. 1985. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3′ termini. Nucleic Acids Res. 13:1347-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore, C. L., and P. A. Sharp. 1984. Site-specific polyadenylation in a cell-free reaction. Cell 36:581-591. [DOI] [PubMed] [Google Scholar]
- 21.Murthy, K. G. K., and J. L. Manley. 1991. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J. Biol. Chem. 267:14804-14811. [PubMed] [Google Scholar]
- 22.Perez-Canadillas, J. M., and G. Varani. 2003. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J. 22:2821-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian, Z., and J. Wilusz. 1991. An RNA-binding protein specifically interacts with a functionally important domain of the downstream element of the simian virus 40 late polyadenylation signal. Mol. Cell. Biol. 11:5312-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryner, L. C., Y. Takagaki, and J. L. Manley. 1989. Sequences downstream of AAUAAA signals affect pre-mRNA cleavage and polyadenylation in vitro both directly and indirectly. Mol. Cell. Biol. 9:1759-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sachs, A., and E. Wahle. 1993. Poly(A) tail metabolism and function in eucaryotes. J. Biol. Chem. 268:22955-22958. [PubMed] [Google Scholar]
- 26.Sachs, A. B., and G. Varani. 2000. Eukaryotic translation initiation: there are (at least) two sides to every story. Nat. Struct. Biol. 7:356-361. [DOI] [PubMed] [Google Scholar]
- 27.Sadofsky, M., and J. C. Alwine. 1984. Sequences on the 3′ side of hexanucleotide AAUAAA affect efficiency of cleavage at the polyadenylation site. Mol. Cell. Biol. 4:1460-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sadofsky, M., S. Connelly, J. L. Manley, and J. C. Alwine. 1985. Identification of a sequence element on the 3′ side of AAUAAA which is necessary for simian virus 40 late mRNA 3′-end processing. Mol. Cell. Biol. 5:2713-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schek, N., C. Cooke, and J. C. Alwine. 1992. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal by using in vitro analyses. Mol. Cell. Biol. 12:5386-5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sperger, J. M., and T. R. Cech. 2001. A stem-loop of tetrahymena telomerase RNA distant from the template potentiates RNA folding and telomerase activity. Biochemistry 40:7005-7016. [DOI] [PubMed] [Google Scholar]
- 31.Takagaki, Y., C. C. MacDonald, T. Shenk, and J. L. Manley. 1992. The human 64-kDa polyadenylation factor contains a ribonucleoprotein-type RNA binding domain and unusual auxiliary motifs. Proc. Natl. Acad. Sci. USA 89:1403-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takagaki, Y., and J. L. Manley. 1997. RNA recognition by the human polyadenylation factor CstF. Mol. Cell. Biol. 17:3907-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wahle, E., and W. Keller. 1996. The biochemistry of polyadenylation. Trends Biochem. Sci. 27:247-250. [PubMed] [Google Scholar]
- 34.Wahle, E., and U. Ruegsegger. 1999. 3′-end processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev. 23:277-295. [DOI] [PubMed] [Google Scholar]
- 35.Wilusz, J., d. I. Feig, and T. Shenk. 1988. The C proteins of heterogeneous nuclear ribonucleoprotein complexes interact with RNA sequences downstream of polyadenylation cleavage sites. Mol. Cell. Biol. 8:4477-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilusz, J., and T. Shenk. 1990. A uridylate tract mediates efficient heterogeneous nuclear ribonucleoprotein C protein-RNA cross-linking and functionally substitutes for the downstream element of the polyadenylation signal. Mol. Cell. Biol. 10:6397-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zarudnaya, M. I., I. M. Kolomiets, A. L. Potyahaylo, and D. M. Hovorun. 2003. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 3:1375-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, F., and C. N. Cole. 1987. Identification of a complex associated with processing and polyadenylation in vitro of herpes simplex type 1 thymidine kinase precursor RNA. Mol. Cell. Biol. 7:3277-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]