Background: Morphogenesis of diatom biosilica depends on a silaffin-dependent organic matrix inside intracellular vesicles (SDVs).
Results: Silaffin-derived 12–14-mer peptides containing five modified lysines and several phosphoserines (pentalysine clusters) are sufficient for silica targeting in vivo.
Conclusion: Pentalysine clusters function as address tags for SDV targeting of silaffins.
Significance: Elucidating the molecular mechanisms for biogenesis of mineral-forming vesicles is essential for understanding biomineral morphogenesis.
Keywords: Biomineralization, Cell Wall, Intracellular Trafficking, Morphogenesis, Post-translational Modification, Protein Motifs, Protein Targeting, Subcellular Organelles, Diatoms, Silica
Abstract
The biological formation of inorganic materials (biomineralization) often occurs in specialized intracellular vesicles. Prominent examples are diatoms, a group of single-celled eukaryotic microalgae that produce their SiO2 (silica)-based cell walls within intracellular silica deposition vesicles (SDVs). SDVs contain protein-based organic matrices that control silica formation, resulting in species specifically nanopatterned biosilica, an organic-inorganic composite material. So far no information is available regarding the molecular mechanisms of SDV biogenesis. Here we have investigated by fluorescence microscopy and subcellular membrane fractionation the intracellular transport of silaffin Sil3. Silaffins are a group of phosphoproteins constituting the main components of the organic matrix of diatom biosilica. We demonstrate that the N-terminal signal peptide of Sil3 mediates import into a specific subregion of the endoplasmic reticulum. Additional segments from the mature part of Sil3 are required to reach post-endoplasmic reticulum compartments. Further transport of Sil3 and incorporation into the biosilica (silica targeting) require protein segments that contain a high density of modified lysine residues and phosphoserines. Silica targeting of Sil3 is not dependent on a particular peptide sequence, yet a lysine-rich 12–14-amino acid peptide motif (pentalysine cluster), which is conserved in all silaffins, strongly promotes silica targeting. The results of the present work provide the first insight into the molecular mechanisms for biogenesis of mineral-forming vesicles from an eukaryotic organism.
Introduction
Biomineralization is a process by which organisms assemble inorganic materials of defined chemical composition into characteristic structures with specific functions (e.g. bone and teeth in mammals, mollusk shells, sea urchin spines, coral skeletons). Detailed studies in various model systems including prokaryotes and single-celled and multicellular eukaryotes have revealed common underlying principles in biomineralization, which include the utilization of intracellular vesicles and protein-based organic templates. The inorganic components of the biomineral are accumulated within the vesicles and then transformed into a bioinorganic precursor phase (1–4) or even a structurally fully developed building block (5–8) of the biomineral. Despite the importance of these intracellular organelles, hardly any information is available on the molecular composition of eukaryotic biomineralization vesicles and the mechanisms of their biogenesis.
One of the best studied eukaryotic systems for biomineralization is the formation of SiO2 (silica)-based cell walls of diatoms, a large group of single-celled aquatic microorganisms (6, 9). The cell wall of each diatom species exhibits characteristic nano- and micropatterned features demonstrating that silica formation is genetically controlled. Diatom silica is produced from silicic acid that the cells take up from the environment through specific silicic acid transporter proteins (10). Silica formation takes place inside the diatom cell within silica deposition vesicles (SDVs),5 which appear to be acidic compartments comparable in pH with the lysosomes (11, 12). SDVs are shaped by the cytoskeleton (13–17) and contain silica-forming organic components that are believed to control the deposition and patterning of the silica (6, 14). Previously, silica-forming organic components have been studied most thoroughly in the model diatom Thalassiosira pseudonana and include unique proteins (silaffins, cingulins, silacidins), chitin and linear long-chain polyamines (6, 18–20).
The discovery of silica-forming proteins has enabled the study of the molecular mechanism of SDV biogenesis through analyses of the intracellular transport pathways of silica-forming proteins. All diatom silica-forming proteins known to date, silaffins, silacidins, and cingulins, are derived from precursor polypeptides containing N-terminal signal peptides for co-translational import into the endoplasmic reticulum (ER) (6, 18, 19). Silaffins and silacidins are heavily phosphorylated (19, 21–23), and silaffins contain several additional post-translational amino acid modifications including O-glycosylation, alkylation of lysine residues, and hydroxylation of proline and lysine residues (22–24). Currently, no information is available regarding the post-translational amino acid modifications of cingulins. The activity of kinases that catalyze the phosphorylation of silaffin- and silacidin-derived unphosphorylated model peptides has been discovered in membrane fractions containing ER and Golgi membranes (25). The glycan moieties present in silaffins (e.g. rhamnose, xylose, glucuronic acid) (22, 23) are expected to be attached to the polypeptide backbone through glycosyltransferases present in the ER or Golgi apparatus (26). Therefore, it is assumed that intracellular transport of silica-forming proteins to the SDV involves vesicular trafficking from the ER to the cis-Golgi followed by intra-Golgi transport and sorting into vesicles that bud from the trans-Golgi network (TGN) and fuse with the SDV (see Fig. 1A). This scenario is analogous to intracellular transport of proteins to the lysosome in mammalian cells (27) and to the lytic vacuole in yeast (28). In these well studied examples, specific receptor proteins, which bind only to proteins that are destined for transport to the lysosome/vacuole (“cargo proteins”), are located in the TGN. The receptor-cargo complex is then incorporated into TGN-derived vesicles that fuse with the vacuole/lysosome rather than the plasma membrane or any other intracellular organelle. The specificity of cargo-receptor interaction is crucial for correct targeting and requires the presence of a specific peptide sequence for vacuolar targeting and a specific amino acid modification (mannose 6-phosphate) for lysosomal targeting (29). We hypothesized that the delivery of proteins to the SDV requires a specific targeting motif for interaction with yet unknown receptor proteins that mediate vesicular transport to the SDV. Therefore, in the present study, we have investigated by in vivo fluorescence microscopy and subcellular membrane fractionation whether silaffins contain such a targeting motif.
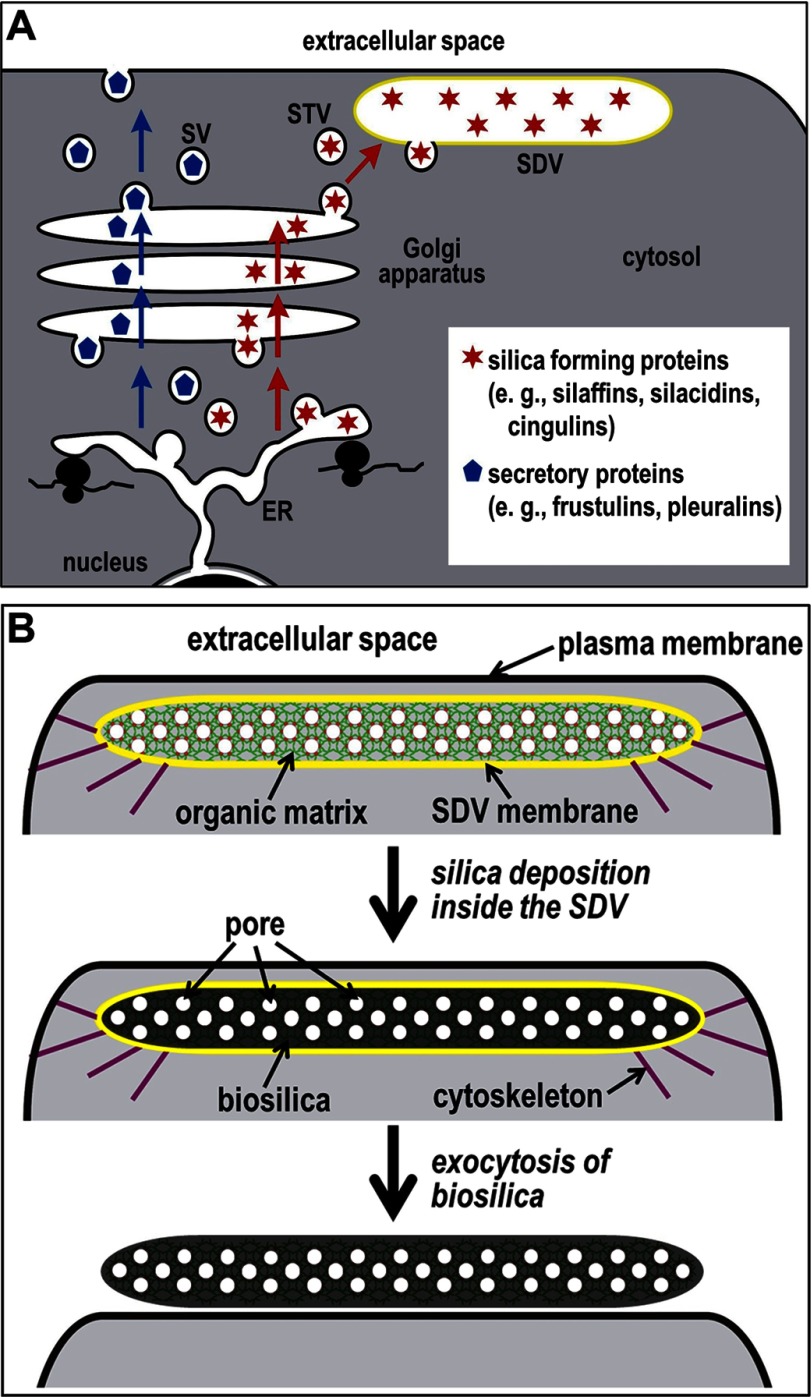
FIGURE 1.
Development of the SDV. A, proposed pathways for intracellular transport of diatom cell wall-associated proteins. Frustulins and pleuralins (blue pentagons) are proteins that are not involved in silica formation and become associated with the silica surface in the extracellular space (6). It is assumed that such proteins become secreted through the standard secretory pathway (blue arrows) involving secretory vesicles (SV). In contrast, it is hypothesized that the pathway for proteins involved in silica formation (red stars) departs from the standard secretory pathway in the TGN (red arrows). Through receptor-mediated sorting, such proteins would become packaged into SDV transport vesicles (STV) that are targeted for fusion with the SDV. B, the shape and positioning of the SDV (SDV membrane is shown in yellow) close to the plasma membrane (black) are believed to be controlled by the cytoskeleton (purple) (14). It is assumed that the biomolecules in the SDV lumen constitute a nanopatterned organic matrix that templates the deposition of silica, thus generating a porous silica nanopattern. During this process, the organic matrix becomes entrapped inside the silica (6). After silica morphogenesis is complete, the biosilica (silica + organic matrix) is deposited on the cell surface through exocytosis.
EXPERIMENTAL PROCEDURES
Culture Conditions
T. pseudonana (Hustedt) Hasle et Heimdal clone CCMP1335 was grown in an enriched artificial seawater medium (ESAW) according to the North East Pacific Culture Collection protocol (Canadian Center for the Culture of Microorganisms (CCCM) ESAW Recipe) at 18 °C under constant light at 5,000–10,000 lux. ESAW contains 0.55 mm NaNO3 as the sole nitrogen source (nitrate medium). Where indicated, nitrate was replaced by 0.55 mm NH4Cl (ammonium medium).
Construction of Genes Encoding GFP-tagged Sil1 and Sil4
The Sil1 gene (hybrid genomic DNA/cDNA gene fusion: genomic DNA amino acids 1–179; cDNA amino acids 180–501)6 was amplified by PCR using the oligonucleotides 5′-AAC AAA ATG AAA GTT ACC ACG TCA ATC-3′ and 5′-GAA TCG CGG CCG CTC AAT TCA GAA AGA AGG AC-3′ that introduced a NotI restriction site (underlined). The resulting 2630-bp PCR product was digested with NotI and introduced into the EcoRV and NotI sites of pTpfcp (30) in which the SmaI site had been destroyed, generating pTpfcp/Sil1. The enhanced green fluorescent protein (eGFP) gene was amplified by PCR using the sense oligonucleotide 5′-GAT GGT GAG CAA GGG CGA GG-3′ and the antisense oligonucleotide 5′-CCC TTG TAC AGC TCG TCC ATG C-3′ and then introduced blunt into the SmaI site of pTpfcp/Sil1. The resulting plasmid, pTpfcp/Sil1-GFPint, generated a GFP fusion protein where the GFP is located internal of the Sil1 after amino acid position 399.
The Sil4 gene was PCR-amplified from genomic DNA of T. pseudonana using the oligonucleotides 5′-GATT GAT ATC ATA ATC ATG AAG ATC ATT TTT CCA GCA CT-3′ (EcoRV site underlined) and 5′-GTT ACG AGA AGA GTA TCT TTG GAG GTA CCG ATC-3′ (KpnI site underlined). The resulting 1601-bp PCR product was digested with EcoRV and KpnI and inserted into the corresponding restriction sites of vector pTpNR-GFP (30).
To construct an expression vector for nitrate-inducible expression of Sil3-GFP, the plasmid pTpfcp/sil3-gfp (31) was used as a template for PCR using the sense primer 5′-ACC AAA ATG AAG ACT TCT GCC ATT G-3′ and antisense primer 5′-GAA TGC GGC CGC TTA CTT GTA CAG CTC GTC-3′, which amplified the complete tpSil3-egfp fusion gene and introduced a NotI restriction site at the 3′-end (bold). The resulting 1184-bp PCR product was digested with NotI and incorporated into the EcoRV and NotI sites of vector pTpNR (30), generating pTpNR/sil3-gfp.The sequences of all constructs were confirmed by DNA sequencing.
Construction of Genes Encoding GFP-tagged Sil-derived Fragments and Pentalysine Clusters
The gene construction procedures are described in the supplemental text (see supplemental Tables S1, S2 and S3 for Sil3-derived gene segments and oligonucleotide sequences). Genes were incorporated into the pTpfcp/ctGFP vector (25). The sequences of all constructs were confirmed by DNA sequencing.
Genetic Transformation of T. pseudonana
Introduction of genes into T. pseudonana and selection of transformants were performed as described previously (30).
Cell Wall Isolation
To determine whether the resulting GFP fusion proteins were incorporated into the biosilica, cells were extracted at 55 °C for 1 h with a buffer containing 1% (w/v) SDS, 0.1 m EDTA (pH 8.0), and 1 mm PMSF. The extracted cell walls were then washed by centrifugation and resuspension three times with water, once with acetone, and again three times with water.
Fluorescence Microscopy
Confocal fluorescence microscopy imaging was performed using an inverted Zeiss laser scanning microscope LSM 510 (Jena, Germany). GFP fluorescence (Argon laser, 488 nm) was detected using a 505/550-nm bandpass filter. For dual imaging, chloroplast autofluorescence (HeNe laser, 543 nm) was detected with a 585-nm long pass filter in the multitrack mode of the microscope. The imaging of transformants expressing GFP fusion proteins of Sil4 and PLCart and Sil3 (under control of nitrate reductase promoter) was performed using a Zeiss LSM 780 confocal equipped an argon laser (488 nm) and a 32-channel GaAsP spectral detector (acquisition between 473 and 668 nm). The GFP signal was separated from chloroplast autofluorescence by linear spectral unmixing using Zen2010 software (Zeiss). Isolated biosilica was viewed using a Zeiss Axioplan 200 epifluorescence microscope equipped with a GFP long pass filter set (excitation, 450–490 nm, beam splitter FT 510 nm; emission, LP515, Zeiss).
Fractionation of Subcellular Membranes
Preparation of subcellular membrane fractions from T. pseudonana strains expressing GFP fusion proteins of Sil3-derived segments T1 and T6 was performed according to a previously described method (25). Western blots were performed in duplicate and probed with both an anti-eGFP antibody (Living Colors full-length A.v. antibody, Clontech) and anti-tpSTK1 antibodies (25).
RESULTS
Expression and Localization of Silaffin-GFP Fusion Proteins
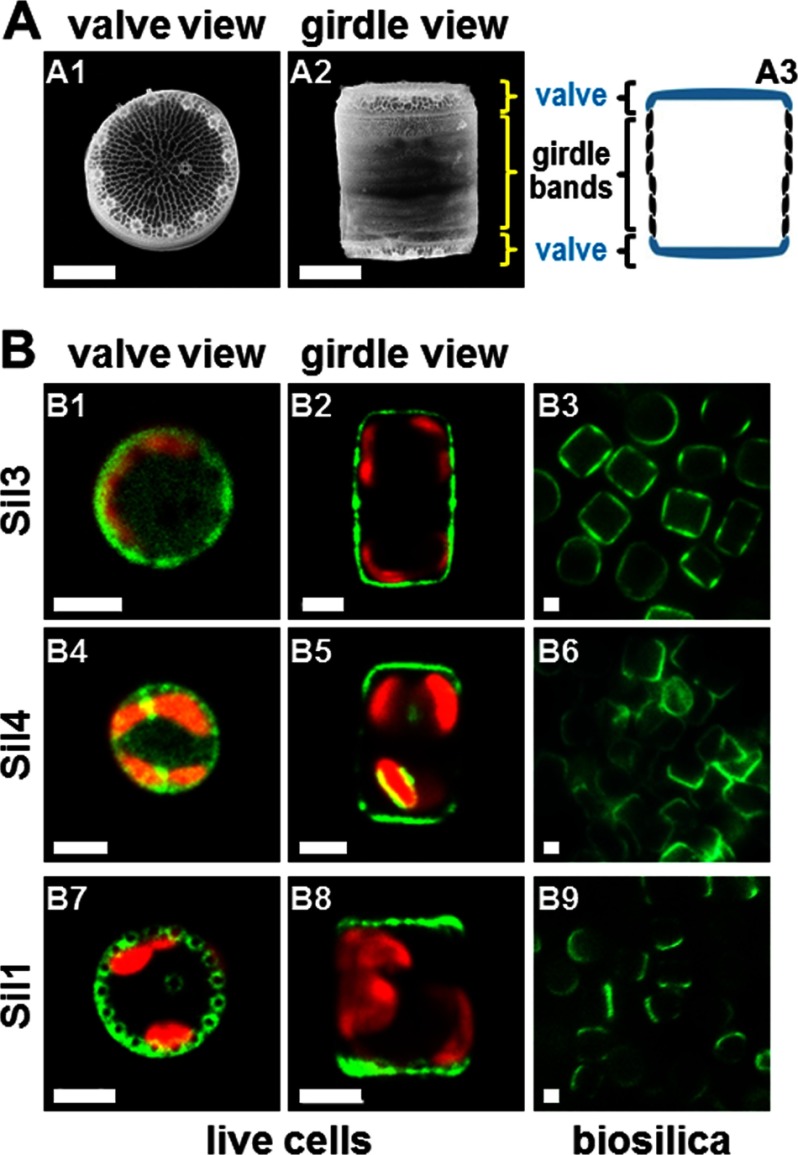
The genome of the diatom T. pseudonana encodes four silaffins (denoted Sil1–4), which are embedded in the biosilica of the cell wall (22). Silica embedding is likely a direct consequence of the mechanism of silica biogenesis because silaffins are believed to be part of a silica-forming organic matrix in the lumen of the SDV onto which silica becomes deposited. Subsequently, the organic matrix remains permanently trapped inside the silica after exocytosis and deposition on the cell surface (Fig. 1B). Previously, it has been demonstrated that expression of a Sil3-GFP fusion protein (GFP was fused to the C terminus of the silaffin) in T. pseudonana led to incorporation of the fusion protein into the cell wall (Fig. 2, B1 and B2) (31). The fusion protein was post-translationally modified, and like endogenous Sil3, could not be extracted from the biosilica by treatment with a hot solution of SDS (Fig. 2, B3) (31). This indicated that the GFP tag did not compromise processing, intracellular sorting, and the silica-forming activity of the silaffin domain. To investigate cell wall incorporation of the other silaffins, T. pseudonana expression vectors were constructed that encoded Sil1-GFP and Sil4-GFP fusion proteins (in both cases, GFP was fused to the C terminus of the silaffin). Expression of a GFP fusion protein of Sil2, which shares 91% amino acid sequence identity with Sil1, was not attempted as it was deemed redundant. The phenotype of transformant cells expressing Sil4-GFP resembled those of Sil3-GFP-expressing transformants with the GFP fusion protein being located in all parts of the cell wall. However, the localization of Sil4-GFP in girdle bands was substantially less pronounced than for Sil3-GFP (Fig. 2, B4 and B5). Treatment with a solution of SDS did not result in Sil4-GFP extraction from the cell walls, demonstrating its entrapment in the biosilica (Fig. 2, B6). None of the transformant clones carrying the Sil1-GFP encoding gene exhibited expression of the fusion protein when analyzed by fluorescence microscopy. We speculated that the C-terminal GFP tag may have become proteolytically removed during maturation of the silaffin domain because the C-terminal region of Sil1 contains an RXL tripeptide motif (RPL: amino acids 453–455), which is a recognition site for proteolytic cleavage in some diatom cell wall proteins (6, 18). Therefore, a new fusion gene was constructed, Sil1-GFPint, in which GFP was positioned upstream of the RXL motif. Introduction of the Sil1-GFPint encoding gene into T. pseudonana yielded GFP fluorescent transformant clones. The fusion protein was located in the two subsections of the cell wall, termed valves, and absent from all other regions of the cell wall (Fig. 2, B7 and B8). Treatment with a solution of SDS did not extract the GFP fluorescence from the biosilica, demonstrating biosilica entrapment of the fusion protein (Fig. 2, B9).
FIGURE 2.
Localization of silaffin-GFP fusion proteins in T. pseudonana. A, SEM micrographs (A1 and A2) and schematic (A3) of single biosilica cell walls in valve orientation (A1) and girdle band orientation (A2 and A3). T. pseudonana cell walls exhibit cylindrical shapes and thus appear as circles in valve view and as rectangles in girdle view. B, confocal fluorescence microscopy images of individual live cells in valve view (B1, B4, and B7) and girdle view (B2, B5, and B8) of T. pseudonana transformant strains expressing GFP fusion proteins (green color) of the indicated silaffins. Red color is caused by chloroplast autofluorescence. For each clone, the valve and girdle view images are from different cells. Epifluorescence microscopy images of isolated biosilica from multiple cells of the same transformant strains (B3, B6, and B9) are shown. White bars: 2 μm.
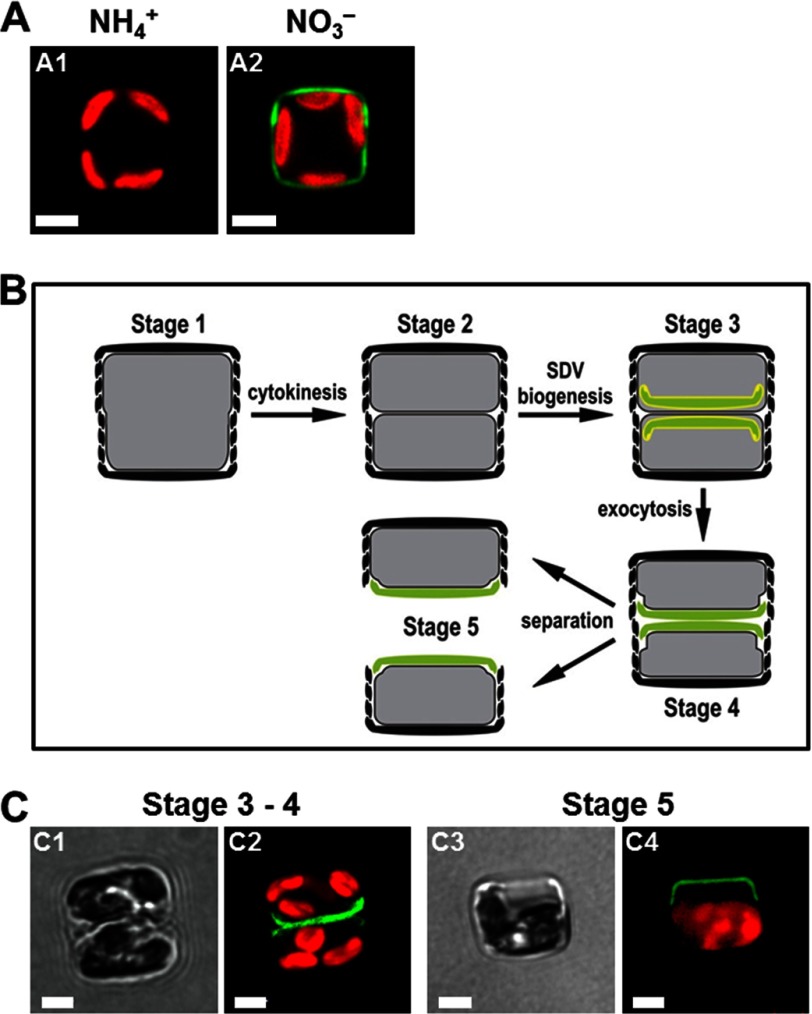
To date there is no direct information available about the intracellular transport pathway of silaffins. To investigate whether silaffins become incorporated into the biosilica via an SDV-dependent pathway as depicted in Fig. 1, the location of newly expressed Sil3-GFP during the cell division cycle was analyzed. This experiment required generating a T. pseudonana strain that carried the Sil3-GFP fusion gene under control of the nitrate reductase promoter, Pnr. This promoter prevents gene expression when ammonium ions are present in the growth medium and induces gene expression when cells are transferred into medium containing nitrate (30). As expected, cells harboring the Pnr-Sil3-GFP gene exhibited biosilica-associated GFP fluorescence only when grown in nitrate-containing medium and did not express Sil3-GFP in ammonium-containing medium (Fig. 3, A1 and A2). To investigate the fate of newly formed Sil3-GFP during silica biosynthesis, cells harboring the Pnr-Sil3-GFP gene were grown in ammonium medium thus lacking Sil3-GFP fluorescence. The cells were then transferred into silicic acid-free medium, which leads to an arrest predominantly in the G1 phase of the cell cycle (Fig. 3B, Stage 1) (32, 33). After 24 h, expression of Sil3-GFP was induced by transferring the G1-arrested cells into nitrate-containing medium, and 2 h later, silicic acid was added to induce cell division and silica biosynthesis. About 4 h after the addition of silicic acid, the cells commenced cell division (Fig. 3B, Stage 2). Each of the two resulting progeny cells produced a new silica element, termed the valve, which covered only one pole of the cell (Fig. 3B, Stages 3 and 4). Concomitantly with valve production, GFP fluorescence became visible in the same area as the newly produced valve, but was absent from all other regions of the cell (Fig. 3, C1 and C2). After separation of the progeny cells (Fig. 3B, Stage 5), GFP fluorescence continued to be visible only at one pole of each cell (Fig. 3, C3 and C4) until fluorescent girdle bands were produced during cell expansion in interphase, and new fluorescent valves were made in the next cell division. This experiment demonstrated that Sil3-GFP was incorporated into the cell wall together with newly formed biosilica, which is consistent with intracellular transport of silaffin-GFP to the SDV (Fig. 1).
FIGURE 3.
Expression of Sil3-GFP in T. pseudonana under control of the nitrate reductase promoter/terminator cassette. Cells are shown in girdle view. Green color is indicative of Sil3-GFP, and red color is caused by chloroplast autofluorescence. White bars: 2 μm. A, confocal fluorescence microscopy images of individual live cells from a T. pseudonana strain grown in medium containing ammonium (NH4+) (A1) or nitrate (NO3−) (A2) as the sole nitrogen source. B, schematic showing a cell in girdle band view in subsequent stages in during cell division. The newly formed valves are shown in green, and SDV membranes are depicted in yellow. C, bright field (C1 and C3) and corresponding confocal fluorescence microscopy images (C2 and C4) of individual live cells. The cells were synchronized by the silicon starvation-replenishment method according to an established protocol (33). Sil3-GFP expression was induced by transferring cells from ammonium medium to nitrate medium 2 h prior to silicon replenishment.
Search for a Silica-targeting Motif
According to the working model for intracellular sorting of biosilica-embedded proteins (Fig. 1A), it would be reasonable to assume that silaffins may be endowed with a specific peptide region (in the following termed silica-targeting motif) that is required for receptor-mediated transport from the Golgi apparatus to the SDV. To investigate whether silaffins carry a silica-targeting motif, fusion genes were synthesized that encode selected segments of the Sil3 polypeptide chain flanked by a signal peptide for ER translocation at the N terminus (SP) and GFP at the C terminus (Fig. 4A). The encoded fusion proteins, denoted T1–T12, were expressed in T. pseudonana under control of the constitutive Pfcp promoter, and their intracellular locations were analyzed by confocal fluorescence microscopy (Fig. 4B). The results of these experiments are summarized in the following.
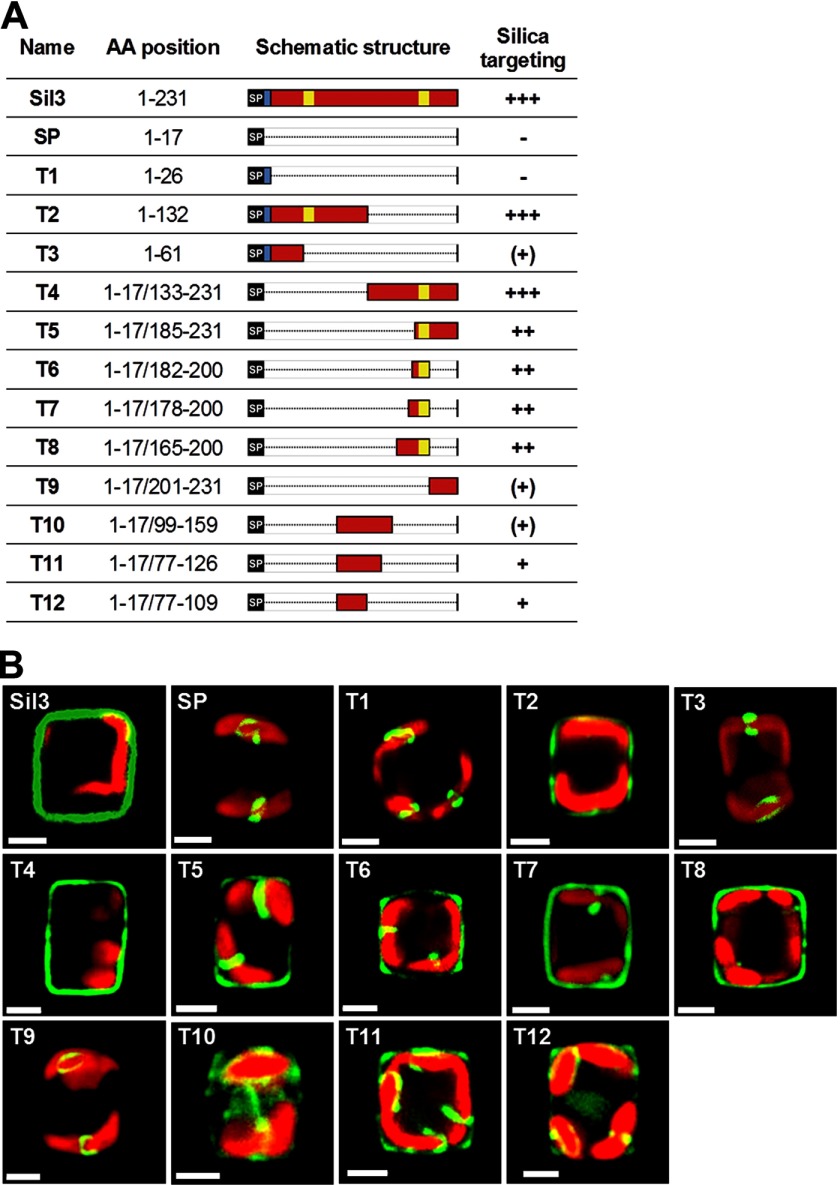
FIGURE 4.
Silica-targeting efficacy of various polypeptide segments derived from Sil3. A, list of Sil3-derived polypeptide segments used for expression as GFP fusion proteins in T. pseudonana. The AA position column indicates the amino acid positions of the Sil3-derived polypeptide segments in the Sil3 precursor polypeptide sequence according to Ref. 22. Note that each fusion protein contains the N-terminal signal peptide (amino acid positions 1–17). The Schematic structure column shows schematic renderings of the primary structures of the Sil3-derived polypeptide segments. Red bars: segments from the region of mature Sil3; black bars labeled SP: N-terminal signal peptide; blue bar: pro-peptide; yellow bars: pentalysine clusters. The Silica targeting column indicates the approximate relative silica-targeting efficacies of the Sil3-derived segments. The greater the number of + signs, the greater the targeting efficacy. (+) indicates silica targeting with low efficacy, and − indicates lack of silica targeting. B, confocal fluorescence microscopy images of individual live cells in girdle view expressing the indicated Sil3-derived GFP fusion proteins. Each cell is a representative example of the cells in the corresponding transformant strain. Green color is indicative of Sil3-GFP, and red color is caused by chloroplast autofluorescence. White bars: 2 μm.
Neither the signal peptide alone (SP) nor the fusion protein T1 that also contains the pro-peptide (i.e. amino acids 16–26, which are absent from the N terminus of mature Sil3 (22)) was capable of targeting GFP for incorporation into the biosilica (Fig. 4B, SP and T1, supplemental Fig. S1, SP and T1). In these cases, the GFP fusion proteins were located inside the cell within clamp-like structures closely associated with the plastids (Fig. 4B, SP and T1). These structures likely represent subcompartments of the ER as similar structures have been observed when GFP fusion proteins were expressed that carried the signal peptide of the ER resident kinase, tpSTK1, fused to the C-terminal ER retention signal, DDEL (25). Fusion protein T3, which is an extension of T1 containing the first 36 amino acids of mature Sil3, was also present in the plastid-associated ER subcompartment (Fig. 4B, T3). However, very low amounts of T3 were also detected in isolated biosilica, indicating successful albeit rather inefficient silica targeting (supplemental Fig. S1, T3). Fusion protein T2, which contained the N-terminal half of the Sil3 polypeptide (amino acids 1–132), was incorporated into the biosilica with high efficiency (Fig. 4B, T2, supplemental Fig. S1, T2). This demonstrated the presence of a strong silica-targeting motif in the N-terminal half of Sil3. Surprisingly, a fusion protein containing the C-terminal half of the Sil3 polypeptide (T4) was also efficiently incorporated into the biosilica (Fig. 4B, T4, supplemental Fig. S1, T4). Therefore, it was concluded that the N-terminal half and the C-terminal half of Sil3 each contain a strong silica-targeting motif that can function independently of each other.
To identify the strong silica-targeting motif of the C-terminal half of Sil3, the GFP fusion proteins T5–T9 (Fig. 4A) were expressed in T. pseudonana, and their locations were analyzed. T5, T6, T7, and T8 were incorporated into the biosilica with high efficacy, but significant amounts of the fusion proteins were also present in the clamp-like structures (Fig. 4B, T5–T8, supplemental Fig. S1, T5–T8). In contrast, only very low amounts of T9 (amino acids 201–231) were incorporated into biosilica (supplemental Fig. S1, T9) as this GFP fusion protein was mainly located in the plastid-associated ER compartment (Fig. 4B, T9). These data suggested that a motif for strong silica targeting is located within amino acids 133–200 of Sil3.
To investigate the location of T8 at different stages of the cell cycle, the T8-GFP fusion protein was expressed under control of the inducible nitrate reductase promoter. After induction of T8-GFP expression, the location of the fusion protein was restricted to the valve during and shortly after cell division (supplemental Fig. S2A, F and BF). After progression through two cell cycles, T8-GFP was present in all parts of the cell wall (supplemental Fig. S2B, F and BF). This location is identical to the location of T8-GFP that was expressed using the constitutive fcp promoter (Fig. 4B, T8). Therefore, it reasonable to assume that targeting of the Sil3 segments follows the same SDV-dependent route as the full-length protein.
Altogether, the results obtained from expression of the GFP fusion proteins SP and T1–T9 indicate that silica targeting of Sil3 can be mediated by various non-overlapping segments of mature Sil3. Furthermore, silica targeting of Sil3 does not operate in an “all-or-nothing” fashion as polypeptide segments with high silica-targeting efficacy and polypeptide segments with low silica-targeting efficacy are present.
Structure-Function Correlation in Silica-targeting Motifs
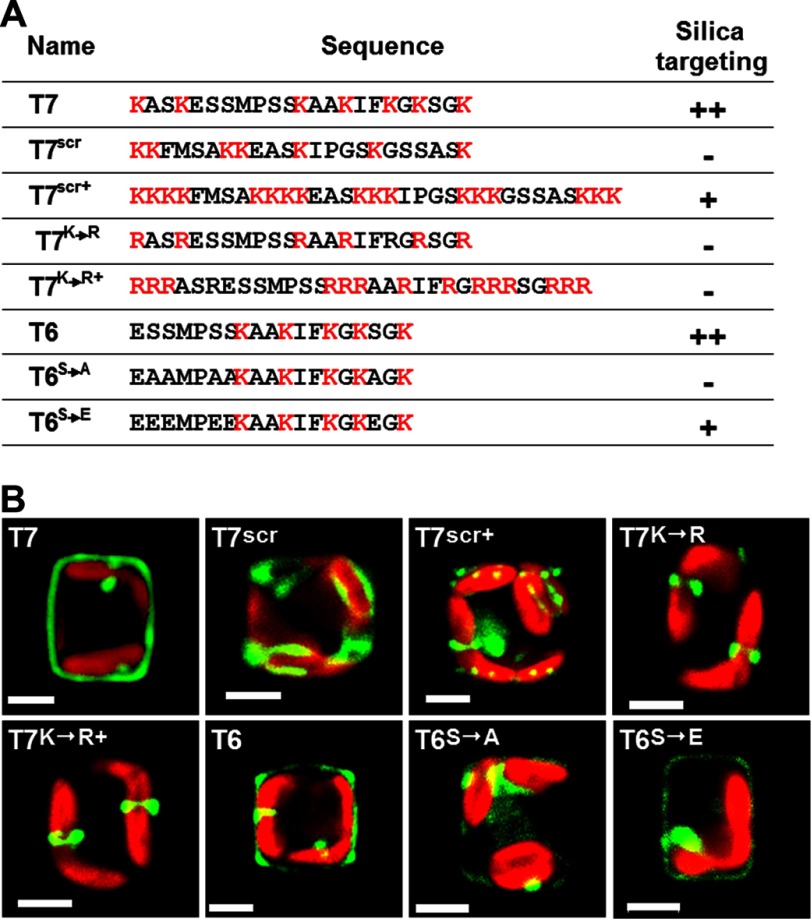
Segments T6 and T7 were the shortest peptides investigated that exhibited high silica-targeting efficacy and were therefore chosen to investigate the structure-function correlation in silica-targeting motifs. When the T7 sequence was scrambled (Fig. 5A, T7scr) silica targeting was completely abolished, and the fusion protein was entirely trapped in the clamp-like structure (Fig. 5B, T7scr). It is possible that the scrambling of the T7 sequence prevented modifications of lysine (methylation, polyamines) (22, 34) and/or serine (phosphorylation, glycosylation) (22) residues. Therefore, the observed abolishment of silica targeting could also be a consequence of incorrect peptide modification. To investigate this, a GFP fusion protein harboring sequence T7scr+ was expressed. The T7scr+ sequence was derived from T7scr, but 10 additional lysine residues were introduced according the expected positive charge conferred by the lysine modifications that are present in this peptide segment of Sil3 (34) (Fig. 5A, T7scr+). In case the polyamine modifications were missing, the additional lysine residues were meant to increase the number of amino groups within the peptide to the same level as in the T7 wild type sequence. The T7scr+ fusion protein was incorporated into the biosilica, although with relatively low efficiency as most of the fusion protein remained inside the cell (Fig. 5B, T7scr+, supplemental Fig. S1, T7scr+). Interestingly, the T7scr+ fusion protein exhibited a highly restricted distribution within the cell wall, being predominantly located in a spot-like pattern at the rim of the valves (Fig. 5B, T7scr+). The incorporation of T7scr+ but not T7scr into the cell wall indicated that a high density of amino groups is a crucial factor in silica targeting. The peptide sequence appears to have an indirect influence through determining the post-translational modifications of the lysine residues.
FIGURE 5.
Effects of amino acid exchanges on the silica-targeting efficacies of Sil3-derived polypeptide segments T6 and T7. A, list of the wild type and mutated polypeptide sequences used for expression as GFP fusion proteins in T. pseudonana. The Silica targeting column indicates the approximate relative silica-targeting efficacies of the Sil3-derived segments. The higher the number of + signs, the higher the targeting efficacy. − indicates lack of silica targeting. B, confocal fluorescence microscopy images of individual live cells in girdle view expressing the indicated GFP fusion proteins. Each cell is a representative example of the cells in the corresponding transformant strain. Green color is indicative of GFP fusion proteins, and red color is caused by chloroplast autofluorescence. White bars: 2 μm.
To further investigate the importance of amino groups in silica targeting, all lysine residues in wild type segment T7 were exchanged against arginine residues (Fig. 5A, T7K→R), or 10 arginine residues were added (Fig. 5A, T7K→R+). The additional arginine residues were introduced to compensate for the loss of positive charges due to the expected lack of the polyamine modifications (note: attachment of polyamines to arginine residues is unknown to date). Fluorescent microscopy analyses demonstrated that neither the T7K→R nor the T7K→R+ segment exhibited silica-targeting activity (Fig. 5B, T7K→R and T7K→R+; supplemental Fig. S1, T7K→R and T7K→R+). This demonstrated that arginines cannot substitute for the lysines in silica targeting and suggests that the function of lysines (and the polyamines attached to them) extends beyond the presentation of positive charges.
To address the role of serine residues in silica targeting, site-directed mutations were introduced into segment T6, which contains fewer amino acids than T7 but exhibits a similar targeting efficacy. When all five serine residues in T6 were exchanged against alanine residues (Fig. 5A, T6S→A), no biosilica incorporation of GFP was observed (Fig. 5B, T6S→A). In contrast, when all serines in T6 were exchanged against glutamate, a mimic of phosphoserine (35) (Fig. 5A, T6S→E), silica targeting was achieved (Fig. 5B, T6S→E, supplemental Fig. S1, T6S→E). These data indicate that one or several negative charges in segment T6 are crucial for silica targeting, thus suggesting that at least one of the serine residues is phosphorylated.
The Role of Pentalysine Clusters
As demonstrated above (Fig. 4), the N-terminal and C-terminal halves of the mature silaffin Sil3 each contain an independently functional silica-targeting motif. Therefore, we analyzed whether the N-terminal half contains a sequence region that is similar to the T6 segment (i.e. the shortest C-terminal segment with high silica-targeting efficacy). The 12-mer peptide KAAKLFKPKASK in the N-terminal half (amino acids 62–73) shows striking similarity regarding the positions of the lysine residues and the amino acid composition with the terminal 12 amino acids of the T6 segment, KAAKIFKGKSGK (amino acids 189–200). These two peptides exhibit the highest density of five lysine residues within the Sil3 sequence and therefore were termed pentalysine clusters. Interestingly, the other three silaffins of T. pseudonana also contain one (Sil1 and Sil2) or two (Sil4) pentalysine clusters (Table 1).
TABLE 1.
Sequence comparison of pentalysine clusters from T. pseudonana silaffins
The numbering of the amino acid positions is according to Ref. 22 for silaffins Sil1, -2, and -3, and according to Ref. 43 for Sil4. The artificial pentalysine cluster, PLCart, has been designed to contain only lysine (bolded) and serine residues and to match the spacing of lysine residues of PLC1. AA, amino acid.
| Silaffin | AA position | PLC sequence | Name |
|---|---|---|---|
| Sil3 | 62–73 | KAAKLFKP--KASK | PLC3nt |
| Sil3 | 189–200 | KAAKIFKG--KSGK | PLC3ct |
| Sil1 | 430–442 | KATKTLKVS-KSGK | PLC1 |
| Sil2 | 420–432 | KVSKSGKSS-KSSK | PLC2 |
| Sil4 | 47–58 | KAAKLFKP--KSSK | PLC4a |
| Sil4 | 213–226 | KATKVFKSSGKSGK | PLC4b |
| KSSKSSKSS-KSSK | PLCart |
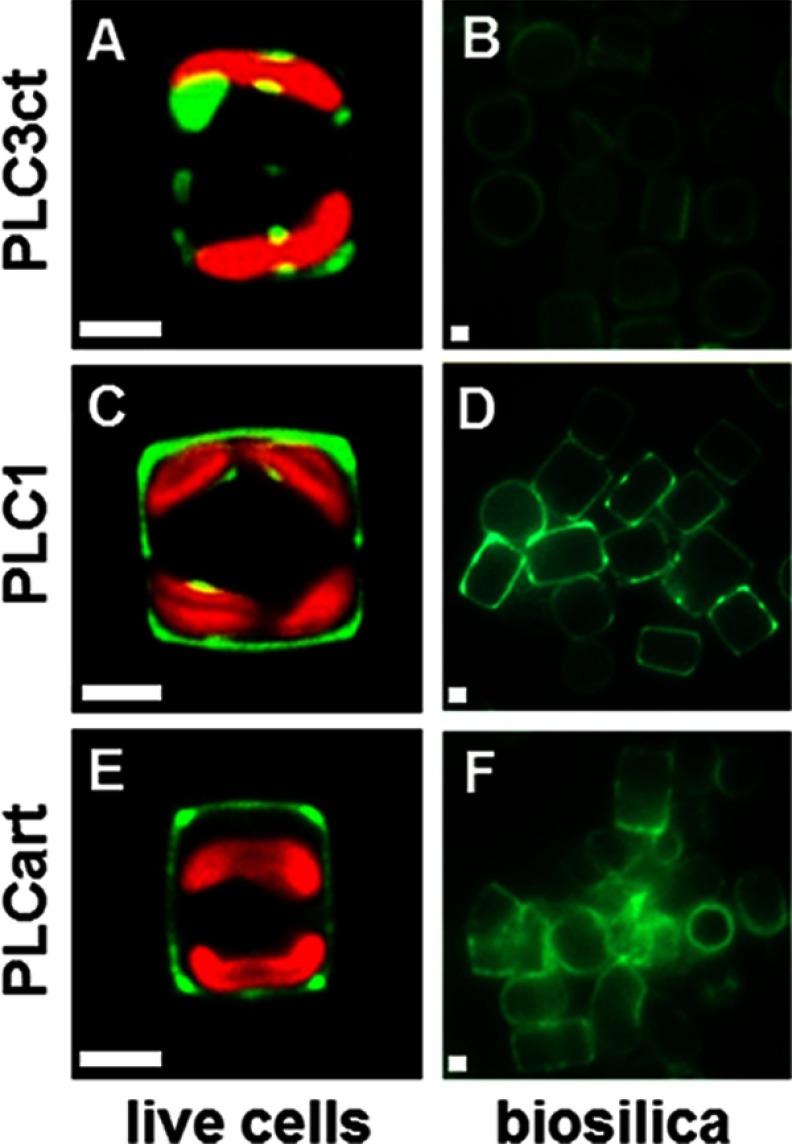
To investigate the silica-targeting efficacies of pentalysine clusters, two GFP fusion genes were constructed (GFP tags on 3′-ends) that encoded the C-terminal pentalysine cluster from Sil3 (PLC3ct) or the pentalysine clusters from Sil1 (PLC1) flanked by the N-terminal signal peptide from Sil3 or Sil1, respectively. The vast majority of the PLC3ct-containing fusion protein remained trapped within the cell (Fig. 6A), yet fluorescence microscopy of isolated biosilica clearly demonstrated that a small fraction of the fusion protein was also incorporated into the biosilica (Fig. 6B). This demonstrated that PLC3ct exhibited silica-targeting activity, albeit with very low efficacy. In contrast, PLC1 acted as a very efficient silica-targeting signal as judged from (i) the strong GFP fluorescence associated with biosilica (Fig. 6, C and D) and (ii) the fact that only a minor fraction of the fusion protein was associated with the clamp-like structure inside the cell (Fig. 6C). This result indicated that a pentalysine cluster is sufficient for silica targeting and suggested that the targeting efficacy is dependent on the structural characteristic of the pentalysine cluster. The striking differences between PLC1 and PLC3ct are the additional amino acid present in PLC1 (13 amino acids) and the higher number of hydroxylated amino acids (4 versus 1) in PLC1. Interestingly, T6, which is the shortest Sil3-derived segment with high silica-targeting efficacy, contains a high amount of serine residues (4 out of 7 amino acids) outside the PLC3ct motif. Therefore, it was hypothesized that a polypeptide consisting of a PLC-like arrangement of lysine residues and interspaced serine residues may be sufficient for efficient silica targeting. To test this hypothesis, an artificial 13-amino acid-containing pentalysine cluster, PLCart (Table 1), was generated and expressed as a fusion protein with the N-terminal signal peptide of Sil3 and a C-terminal GFP tag. In agreement with the hypothesis, the fusion protein proved to become incorporated into the biosilica with an efficacy that was intermediate between that of PLC1 and PLC3ct (Fig. 6, E and F).
FIGURE 6.
Silica-targeting efficacy of pentalysine clusters. A, C, and E, confocal fluorescence microscopy images of individual live cells in girdle view expressing the indicated pentalysine cluster-GFP fusion proteins. B, D, and F, epifluorescence microscopy images of isolated biosilica from multiple cells of the same transformant strains. Green color is indicative of Sil3-GFP, and red color is caused by chloroplast autofluorescence. White bars: 2 μm.
These results prompted the question as to whether pentalysine clusters are essential components of silica-targeting motifs. To investigate this, GFP fusion proteins containing the pentalysine cluster-free segments T10, T11, and T12 were expressed in T. pseudonana. Analysis by fluorescence microscopy revealed that each of the fusion proteins was incorporated into the biosilica, although with low (T11 and T12) or very low (T10) efficacy (Fig. 4B, T10–T12). In each case, relatively large amounts of the fusion proteins remained trapped inside the cell (Fig. 4B, T10–T12). These results demonstrated that pentalysine clusters are not essential for silica targeting, yet segments containing pentalysine clusters are particularly efficient in silica targeting. For example, the 19-amino acid-containing segment T6, which bears one pentalysine cluster, mediated efficient silica targeting, whereas the substantially longer segment T12 (33 amino acids) that lacks a pentalysine cluster exhibited only low silica-targeting efficacy (Fig. 4B, T6 and T12).
Intracellular Transport Route of Silaffins
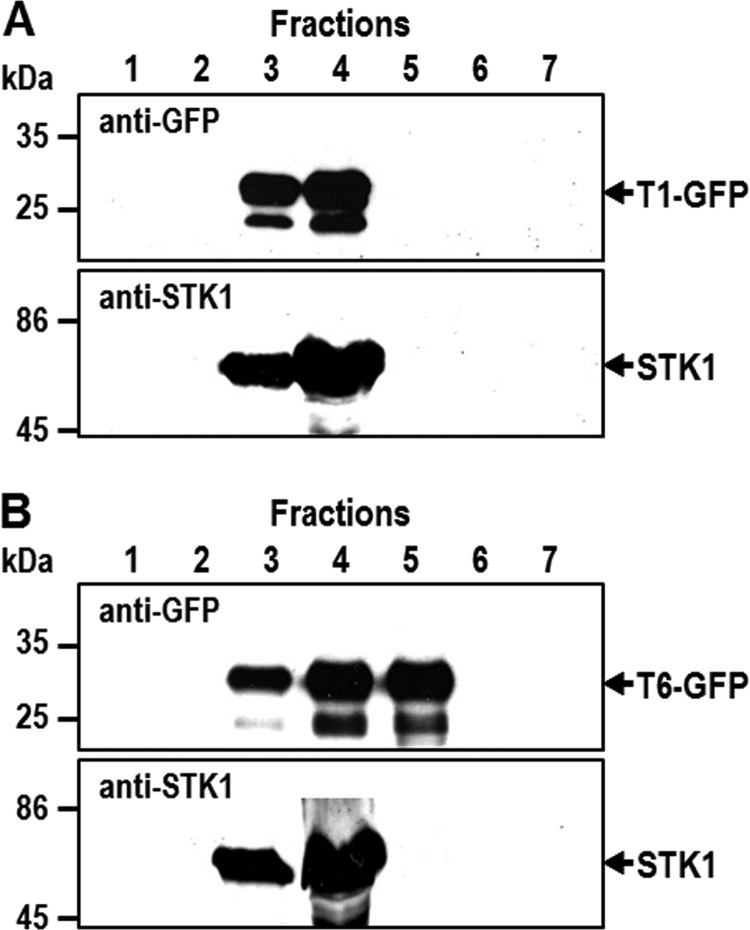
In fluorescence microscopy analysis of T. pseudonana transformants expressing various Sil3-derived GFP fusion proteins, only two subcellular locations could be discerned: the biosilica cell wall and the chloroplast-associated clamp-like structure (Figs. 2–6). It was unexpected that even those GFP fusion proteins that contained highly efficient silica-targeting motifs could not be detected in other subcellular compartments (e.g. Golgi apparatus, transport vesicles, SDV), which they are supposed to transit on their route to silica incorporation (Fig. 1A). It is possible that detection of GFP fusion proteins in such transit compartments was strongly hampered by low accumulation rates (i.e. rapid transit through post-ER compartments) and low luminal pH values resulting in quenching of GFP fluorescence (36). The fluorescence intensity of GFP fusion proteins inside the SDV, which is an acidic compartment (11), may be strongly reduced, and transport vesicles may not be readily visible due to the intense background fluorescence of the chloroplasts. To achieve a fluorescence-independent intracellular localization of silaffin-derived GFP fusion proteins, subcellular membrane fractionation according to a previously established protocol for T. pseudonana (25) was performed. In Western blot analysis of the subcellular membrane fractions using anti-GFP antibodies, the T1-GFP fusion protein was present exclusively in ER-containing fractions (Fig. 7A). This is consistent with the assumption that the clamp-like structure around the chloroplast observed in fluorescence microscopy images (Fig. 4B, T1) represents a subcompartment of the ER (note: in secondary endosymbionts like the diatoms, the outer membrane of the chloroplast is continuous with the ER). The T6-GFP fusion protein was present in both ER-containing and Golgi apparatus-containing fractions and absent from fractions containing plasma membranes and mitochondria (Fig. 7B). This result confirmed that intracellular transport of T6-GFP involves additional membrane-bound compartments other than the ER. It is likely that one of these compartments is the Golgi apparatus, but other subcellular membranes (e.g. transport vesicles, SDV membranes before silica deposition) may also be present in the Golgi membrane fraction.
FIGURE 7.
Biochemical analysis of the intracellular location of Sil3-derived GFP fusion proteins T1 and T6. Subcellular membrane fractions F1–F7 of T. pseudonana transformants were isolated by density sucrose gradient centrifugation according to an established protocol (25): F3/F4 = ER membranes, F5 = Golgi membranes, and F6/F7 = mitochondria and plasma membrane. Equal aliquots of the fractions were separated by SDS-PAGE and probed for the presence of T1-GFP (A) and T6-GFP (B) by Western blot analysis using anti-GFP antibodies. As a reference, the same membrane fractions from the T. pseudonana transformants were analyzed for the presence of the ER-resident kinase tpSTK1 using previously generated anti-tpSTK1 antibodies (25).
DISCUSSION
In the present work, the molecular mechanism for intracellular targeting of silica-forming proteins in diatoms has been addressed. It is demonstrated here that transport of silaffins into the silica cell wall (i.e. silica targeting) is not dependent on a particular peptide sequence. Instead, various non-overlapping segments throughout the mature part of Sil3 exhibit silica-targeting activity, but the efficacy of targeting can vary drastically between the different segments. The highest silica-targeting efficacy was always obtained with Sil3 segments containing a pentalysine cluster, which is characterized by five non-consecutive lysines (KXXXK, KXXK, or KXK) within a stretch of 12–14 amino acid residues. Each silaffin contains one or two pentalysine clusters, which is consistent with a crucial role of this peptide motif in silica targeting. Analysis by site-directed mutagenesis demonstrated that in pentalysine clusters, lysine residues cannot be functionally replaced by arginines. Furthermore, serine residues can be functionally replaced by glutamic acid, but not by alanines. These results indicate that the post-translational modification of the lysine residues (methyl groups, polyamine chains) (22, 34) and the serine residues (phosphoryl groups, carbohydrate moieties) (22) are essential for silica targeting. This situation is reminiscent of the intracellular targeting of proteins from the TGN to the lysosome, which is also dependent on a post-translational modification, mannose 6-phosphate (present on N-glycan moieties), rather than a specific peptide sequence (29). Selection of N-glycosylated proteins for mannose 6-phosphate addition does not depend on a particular peptide sequence of the transport substrate, which is analogous to the situation found with silica targeting of silaffins.
To date intracellular protein targeting in diatoms has been most thoroughly investigated for import of chloroplast proteins that are encoded in the nuclear genome. The outermost membrane of diatom chloroplasts is continuous with the ER, and nuclear encoded chloroplast proteins require a bipartite topogenic signal sequence at the N terminus for import into the chloroplast. The N-terminal half of the bipartite topogenic signal sequence is a typical signal peptide, and the second half is a transit peptide that mediates export from the ER and further transport into the chloroplast (37–39). As silaffins had previously been shown to contain a pro-peptide immediately following the N-terminal signal peptide (22, 24), we speculated that the pro-peptide may function as a signal for SDV targeting. However, the present data demonstrate that the pro-peptide of silaffin Sil3 (amino acids 18–26) is not required for silica targeting (Fig. 4B, SP). Therefore, the function(s) of the pro-peptide sequences of silaffins and the reason for their removal during silaffin maturation remain to be determined.
It is striking that the same structural features required for silaffin-mediated silica formation, namely serine phosphorylation and polyamine modification of lysines (6), are also crucial for silica targeting of silaffins. This concurrence may hint to a mechanistic connection between these two seemingly unrelated processes. Due to their strong zwitterionic nature, silaffins assemble into large supramolecular aggregates that become even larger when long-chain polyamines are added (21–23). It is hypothesized that these aggregates, rather than the monomeric constituents, are active in silica formation in vivo (6, 43). In other organisms, it has been shown that protein aggregation is crucial for the sorting of peptide hormones (e.g. insulin) to regulated secretory vesicles (40). The peptide hormones aggregate with each other and with other proteins (chromogranins A and B, secretogranin) in the TGN. Subsequent interaction with sorting receptors (e.g. carboxypeptidase E) triggers the budding of specialized vesicles from the TGN that then fuse with each other to form large secretory granules (40). Identification of targeting motifs in proteins destined for regulated secretory granules has been severely hampered by the difficulty of distinguishing whether a certain peptide segment from a cargo protein is required for interaction with the sorting receptor or for aggregation. Aggregation-dependent sorting could explain why we were unable in the present study to identify one specific silica-targeting motif for sorting of silaffins to the SDV. In this context, it is interesting to note that the SDV shares two important features with regulated secretory granules: (i) the large size (i.e. much larger than constitutive secretory vesicles) and (ii) secretion only upon a certain stimulus (in case of the SDV, the stimulus is unknown). Insight into a possible relationship between the machineries for the secretion of silica and that of peptide hormones may be obtained through a bioinformatics search in diatom genomes for homologues of components involved in biogenesis of regulated secretory granules.
In the present work, we have made a first step toward biochemical analysis of SDV biogenesis by demonstrating that silaffin transport intermediates can be detected in subcellular membrane fractions (Fig. 7). Future research on SDV targeting of proteins will require improvements in purification of subcellular membranes to enable biochemical characterization of transport vesicles and early (i.e. not silica-filled) stages of the SDV. Through such analysis in combination with the established tools for molecular genetic manipulation of diatoms (30, 41, 42), it should become possible to further increase the understanding of the molecular mechanisms of SDV biogenesis.
Supplementary Material
Acknowledgment
We thank Jennifer Klemm for technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) through Grant KR1853/3-1 (to N. K.) and Postdoctoral Fellowship SCHE1637/1-1 (to A. S.), by the United States Department of Energy (DOE) Office of Basic Energy Sciences through Grant DE-SC0002245 (to N. K.), and by the National Science Foundation (NSF) through Grant DMR-0845939 (to N. K.).
This article was selected as a Paper of the Week.

This article contains supplemental text and Figs. S1 and S2.
N. Poulsen, A. Scheffel, V. C. Sheppard, P. M. Chesley, and N. Kröger, unpublished data.
- SDV
- silica deposition vesicle
- ER
- endoplasmic reticulum
- TGN
- trans-Golgi network
- ESAW
- enriched artificial seawater medium
- SP
- signal peptide
- PLC
- pentalysine cluster.
REFERENCES
- 1. Wilt F. H., Killian C. E., Hamilton P., Croker L. (2008) The dynamics of secretion during sea urchin embryonic skeleton formation. Exp. Cell Res. 314, 1744–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ali S. Y., Sajdera S. W., Anderson H. C. (1970) Isolation and characterization of calcifying matrix vesicles from epiphyseal cartilage. Proc. Natl. Acad. Sci. U.S.A. 67, 1513–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson H. C., Garimella R., Tague S. E. (2005) The role of matrix vesicles in growth plate development and biomineralization. Front. Biosci. 10, 822–837 [DOI] [PubMed] [Google Scholar]
- 4. Weiner S., Addadi L. (2011) Crystallization pathways in biomineralization. Annu. Rev. Mater. Res. 41, 21–40 [Google Scholar]
- 5. Wang X., Wiens M., Schröder H. C., Schlossmacher U., Pisignano D., Jochum K. P., Müller W. E. G. (2011) Evagination of cells controls bio-silica formation and maturation during spicule formation in sponges. PLoS One 6, e20523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kröger N., Poulsen N. (2008) Diatoms—From cell wall biogenesis to nanotechnology. Annu. Rev. Genet. 42, 83–107 [DOI] [PubMed] [Google Scholar]
- 7. Bäuerlein E. (2003) Biomineralization of unicellular organisms: An unusual membrane biochemistry for the production of inorganic nano- and microstructures. Angew Chem. Int. Ed. Engl. 42, 614–641 [DOI] [PubMed] [Google Scholar]
- 8. Jogler C., Schüler D. (2009) Genomics, genetics, and cell biology of magnetosome formation. Annu. Rev. Microbiol. 63, 501–521 [DOI] [PubMed] [Google Scholar]
- 9. Round F. E., Crawford R. M., Mann D. G. (1990) The Diatoms: Biology and Morphology of the Genera, Cambridge University Press, Cambridge [Google Scholar]
- 10. Hildebrand M., Wetherbee R. (2003) Components and control of silicification in diatoms. Prog. Mol. Subcell. Biol. 33, 11–57 [DOI] [PubMed] [Google Scholar]
- 11. Vrieling E. G., Gieskes W. W. C., Beelen T. P. M. (1999) Silicon deposition in diatoms: Control by the pH inside the silicon deposition vesicle. J. Phycol. 35, 548–559 [Google Scholar]
- 12. Desclés J., Vartanian M., El Harrak A., Quinet M., Bremond N., Sapriel G., Bibette J., Lopez P. J. (2008) New tools for labeling silica in living diatoms. New Phytol. 177, 822–829 [DOI] [PubMed] [Google Scholar]
- 13. Tesson B., Hildebrand M. (2010) Extensive and intimate association of the cytoskeleton with forming silica in diatoms: Control over patterning on the meso- and micro-Scale. PLoS One 5, e14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pickett-Heaps J. D., Schmid A. M., Edgar L. A. (1990) The cell biology of diatom valve formation. in Progress in Phycological Research (Round F. E., Chapman D. J., eds) pp. 1–168, Biopress Ltd., Bristol, UK [Google Scholar]
- 15. van de Meene A. M. L., Pickett-Heaps J. D. (2002) Valve morphogenesis in the centric diatom Proboscia alata Sundstrom. J. Phycol. 38, 351–363 [Google Scholar]
- 16. Van de Meene A. M. L., Pickett-Heaps J. D. (2004) Valve morphogenesis in the centric diatom Rhizosolenia setigera (Bacillariophyceae, Centrales) and its taxonomic implications. Eur. J. Phycol. 39, 93–104 [Google Scholar]
- 17. Schmid A. M. M. (1994) Aspects of morphogenesis and function of diatom cell-walls with implications for taxonomy. Protoplasma 181, 43–60 [Google Scholar]
- 18. Scheffel A., Poulsen N., Shian S., Kröger N. (2011) Nanopatterned protein microrings from a diatom that direct silica morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 3175–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wenzl S., Hett R., Richthammer P., Sumper M. (2008) Silacidins: Highly acidic phosphopeptides from diatom shells assist in silica precipitation in vitro. Angew Chem. Int. Ed. Engl. 47, 1729–1732 [DOI] [PubMed] [Google Scholar]
- 20. Brunner E., Richthammer P., Ehrlich H., Paasch S., Simon P., Ueberlein S., van Pee K.-H. (2009) Chitin-based organic networks: An integral part of cell wall biosilica in the diatom Thalassiosira pseudonana. Angewandte Chemie Int. Ed. 48, 9724–9727 [DOI] [PubMed] [Google Scholar]
- 21. Kröger N., Lorenz S., Brunner E., Sumper M. (2002) Self-assembly of highly phosphorylated silaffins and their function in biosilica morphogenesis. Science 298, 584–586 [DOI] [PubMed] [Google Scholar]
- 22. Poulsen N., Kröger N. (2004) Silica morphogenesis by alternative processing of silaffins in the diatom Thalassiosira pseudonana. J. Biol. Chem. 279, 42993–42999 [DOI] [PubMed] [Google Scholar]
- 23. Poulsen N., Sumper M., Kröger N. (2003) Biosilica formation in diatoms: Characterization of native silaffin-2 and its role in silica morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 100, 12075–12080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kröger N., Deutzmann R., Sumper M. (1999) Polycationic peptides from diatom biosilica that direct silica nanosphere formation. Science 286, 1129–1132 [DOI] [PubMed] [Google Scholar]
- 25. Sheppard V., Poulsen N., Kröger N. (2010) Characterization of an endoplasmic reticulum-associated silaffin kinase from the diatom Thalassiosira pseudonana. J. Biol. Chem. 285, 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Varki A., Esko J. D., Colley K. J. (2009) Cellular organization of glycosylation. in Essentials of Glycobiology, 2nd Ed (Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E., eds) pp. 37–61, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 27. McVey Ward D., Shiflett S. L., Kaplan J. (2005) Lysosome biogenesis and dynamics. in The Biogenesis of Cellular Organelles (Mullins C., ed) pp. 111–126, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 28. Graham T. R., Nothwehr S. F. (2002) Protein transport to the yeast vacuole. in Protein Targeting, Transport and Translocation (Dalbey R. E., von Heijne G. eds) pp. 322–357, Academic Press, London [Google Scholar]
- 29. Griffiths G., Hoflack B., Simons K., Mellman I., Kornfeld S. (1988) The mannose-6-phosphate receptor and the biogenesis of lysosomes. Cell 52, 329–341 [DOI] [PubMed] [Google Scholar]
- 30. Poulsen N., Chesley P. M., Kröger N. (2006) Molecular genetic manipulation of the diatom Thalassiosira pseudonana (Bacillariophyceae). J. Phycol. 42, 1059–1065 [Google Scholar]
- 31. Poulsen N., Berne C., Spain J., Kröger N. (2007) Silica immobilization of an enzyme through genetic engineering of the diatom Thalassiosira pseudonana. Angew Chem. Int. Ed. Engl. 46, 1843–1846 [DOI] [PubMed] [Google Scholar]
- 32. Frigeri L. G., Radabaugh T. R., Haynes P. A., Hildebrand M. (2006) Identification of proteins from a cell wall fraction of the diatom Thalassiosira pseudonana: Insights into silica structure formation. Mol. Cell. Proteomics 5, 182–193 [DOI] [PubMed] [Google Scholar]
- 33. Hildebrand M., Frigeri L. G., Davis A. K. (2007) Synchronized growth of Thalassiosira pseudonana (Bacillariophyceae) provides novel insights into cell-wall synthesis processes in relation to the cell cycle. J. Phycol. 43, 730–740 [Google Scholar]
- 34. Sumper M., Hett R., Lehmann G., Wenzl S. (2007) A code for lysine modifications of a silica biomineralizing silaffin protein. Angew Chem. Int. Ed. Engl. 46, 8405–8408 [DOI] [PubMed] [Google Scholar]
- 35. Dean A. M., Koshland D. E. (1990) Electrostatic and steric contributions to regulation at the active-site of isocitrate dehydrogenase. Science 249, 1044–1046 [DOI] [PubMed] [Google Scholar]
- 36. Llopis J., McCaffery J. M., Miyawaki A., Farquhar M. G., Tsien R. Y. (1998) Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc. Natl. Acad. Sci. U.S.A. 95, 6803–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kilian O., Kroth P. G. (2005) Identification and characterization of a new conserved motif within the presequence of proteins targeted into complex diatom plastids. Plant J. 41, 175–183 [DOI] [PubMed] [Google Scholar]
- 38. Gould S. B., Sommer M. S., Hadfi K., Zauner S., Kroth P. G., Maier U. G. (2006) Protein targeting into the complex plastid of cryptophytes. J. Mol. Evol. 62, 674–681 [DOI] [PubMed] [Google Scholar]
- 39. Felsner G., Sommer M. S., Maier U. G. (2010) The physical and functional borders of transit peptide-like sequences in secondary endosymbionts. BMC Plant Biol. 10, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tooze S. A., Martens G. J. M., Huttner W. B. (2001) Secretory granule biogenesis: rafting to the SNARE. Trends Cell Biol. 11, 116–122 [DOI] [PubMed] [Google Scholar]
- 41. Apt K. E., Kroth-Pancic P. G., Grossman A. R. (1996) Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol. Gen. Genet. 252, 572–579 [DOI] [PubMed] [Google Scholar]
- 42. Fischer H., Robl I., Sumper M., Kröger N. (1999) Targeting and covalent modification of cell wall and membrane proteins heterologously expressed in the diatom Cylindrotheca fusiformis (Bacillariophyceae). J. Phycol. 35, 113–120 [Google Scholar]
- 43. Sumper M., Brunner E. (2008) Silica biomineralisation in diatoms: The model organism Thalassiosira pseudonana. Chembiochem 9, 1187–1194 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.