Background: Dact1 and Vangl2 regulate the planar cell polarity (PCP) pathway during development.
Results: Sestd1 genetically and biochemically interacts with Dact1 and Vangl2 and mediates PCP.
Conclusion: Sestd1 cooperates with Dact1 to regulate Vangl2 and PCP.
Significance: The discovery of PCP pathway regulators and components uncovers molecular mechanisms contributing to neural tube defects, other birth defects, and disease processes in humans.
Keywords: Cell Migration, Cell Polarity, Mouse Genetics, Rho, Transgenic Mice, Wnt Signaling, Planar Cell Polarity (PCP) Pathway
Abstract
The planar cell polarity (PCP) pathway is a conserved non-canonical (β-catenin-independent) branch of Wnt signaling crucial to embryogenesis, during which it regulates cell polarity and polarized cell movements. Disruption of PCP components in mice, including Vangl2 and Dact1, results in defective neural tube closure and other developmental defects. Here, we show that Sestd1 is a novel binding partner of Vangl2 and Dact1. The Sestd1-Dact1 interface is formed by circumscribed regions of Sestd1 (the carboxyl-terminal region) and Dact1 (the amino-terminal region). Remarkably, we show that loss of Sestd1 precisely phenocopies loss of Dact1 during embryogenesis in mice, leading to a spectrum of birth malformations, including neural tube defects, a shortened and/or curly tail, no genital tubercle, blind-ended colons, hydronephrotic kidneys, and no bladder. Moreover, as with Dact1, a knock-out mutation at the Sestd1 locus exhibits reciprocal genetic rescue interactions during development with a semidominant mutation at the Vangl2 locus. Consistent with this, examination of Wnt pathway activities in Sestd1 mutant mouse embryonic tissue reveals disrupted PCP pathway biochemistry similar to that characterized in Dact1 mutant embryos. The Sestd1 protein is a divergent member of the Trio family of GTPase regulatory proteins that lacks a guanine nucleotide exchange factor domain. Nonetheless, in cell-based assays the Sestd1-Dact1 interaction can induce Rho GTPase activation. Together, our data indicate that Sestd1 cooperates with Dact1 in Vangl2 regulation and in the PCP pathway during mammalian embryonic development.
Introduction
Wnt signaling plays important roles in embryonic development and in adult physiology (1, 2). It can be divided conceptually into a “canonical” β-catenin-dependent pathway and multiple β-catenin-independent pathways. β-Catenin-dependent Wnt signaling is initiated by binding of Wnts to a transmembrane receptor complex consisting of Frizzled proteins and low density lipoprotein receptor-related protein 5/6 (LRP5/6). Through inhibition of glycogen synthase kinase 3, this leads to cytoplasmic/nuclear β-catenin stabilization and β-catenin-mediated transcriptional changes in gene expression (3). Evolutionarily conserved β-catenin-independent pathways have also been described downstream of some Wnt ligands and Frizzled receptors, including the Wnt/Ca2+ pathway that regulates calcium flux and calcium-activated intracellular pathways, and the planar cell polarity (PCP)2 pathway that regulates cell polarity and cell migration during development (4–6). In mice mutant for any one of several core PCP molecules, including Dishevelled 2 (Dvl2), Scribble (Scrib), cadherin-EGF-LAG-seven-pass-G-type-receptor-1 (Celsr1), and Vang-like-2 (Vangl2), one major developmental consequence is defective neural tube closure resulting in spina bifida and related neural tube defects (NTDs) (7–10). Mutations in these genes are also a cause of NTDs in humans (11–14), and this is one of the most common birth defects in our species (15, 16). The Rho family of small GTPases are effectors downstream of β-catenin-independent Wnt pathways, including PCP (17, 18). These GTPases affect polarized cell morphology and movements via regulation of actin dynamics (19–21).
The Dapper-antagonist-of-catenin (Dact) family of scaffold proteins was initially identified by virtue of conserved interactions with the Dvl proteins central to virtually all forms of Wnt signaling (22–24). Three Dact family members (Dact1–3) have been identified in vertebrates (25), where based on a variety of in vitro and in vivo assays, they appear to have both overlapping and divergent signaling functions (26–31). In studies using genetically engineered mice, we have shown previously that Dact1 is required for normal post-translational regulation of Vangl2 during the epithelial-mesenchymal transition at the primitive streak, for normal PCP in posterior embryonic tissues, and that defects in these developmental processes in Dact1 null embryos lead to complex birth malformations, including NTDs (29). Consistent with the notion that this reflects conserved developmental functions for this protein, several rare mutations at this locus have been identified in human patients with NTDs (32).
In this study, we identify SEC14 and spectrin domains-1 (Sestd1) as a Dact-interacting protein, initially through a two-hybrid screen in yeast and a proteomic approach in immortalized mammalian cell lines. Sestd1 exhibits moderate sequence conservation with the Trio family proteins in an amino-terminal SEC14 domain and three carboxyl-terminal spectrin-like domains (33). To probe Sestd1 function in vivo, we generated a Sestd1 knock-out allele in mice and show that homozygosity for this allele phenocopies homozygosity for the Dact1 knock-out allele, causing an identical spectrum of posterior malformations. Moreover, mice that are null for both Sestd1 and Dact1 display the same spectrum of phenotypes as mice that are null for either locus alone, suggesting that these two proteins function in a unitary pathway during embryonic development. Consistent with the above findings, in cell-based assays loss of Sestd1 specifically disrupts PCP but not Wnt/β-catenin pathway activity, and Sestd1 cooperates with Dact1 to activate the PCP effector, Rho GTPase. Finally, we show that mutations at the Sestd1 and Vangl2 loci exhibit reciprocal rescue interactions in mice, highly reminiscent of genetic interactions between Dact1 and Vangl2. Together, our findings strongly support that the Sestd1 and Dact1 proteins closely cooperate during mammalian embryonic development in the regulation of Vangl2 and the PCP pathway.
MATERIALS AND METHODS
Generation of Sestd1−/− Mice
Approximately 8.4 kb of Sestd1 genomic DNA from the C57BL/6J mouse strain was inserted into PGKneoF2L2DTA2 to create the Sestd1 targeting vector. This targeting vector allows for Cre-mediated excision of exon 4, intron 4, and exon 5. Homologous recombination at the 5′ and 3′ arms of the targeting construct in G4 ES cells (34) was carried out under a service contract by the University of Washington Transgenic Resources Program and was confirmed by PCR in the Cheyette laboratory at University of California at San Francisco. Note that the G4 ES cell line is genetically distinct from the ES cell line previously used for Dact1 locus targeting (29). Mice carrying the targeted allele were generated by standard mouse embryo manipulation and chimera founder breeding techniques. To discriminate between Sestd1 KO and wild type (WT) alleles in mouse (tail) tissue extracts, specific PCR primer pairs were designed as follows. A primer pair (5′-GGATTAGATGTGGCGCATACAAGCTTTCAGTC-3′ and 5′-GACTTTCCTCATAACCCTGAAGGGAGAAGGC-3′) flanking the first LoxP site in the targeted allele was separated by 498 bases of genomic sequence in WT. Similarly, a primer pair (5′-GGATTAGATGTGGCGCATACAAGCTTTCAGTC-3′ and 5′-TCTTGGCTCAGCTTCTTGGGATGTGGTTTTCC-3′) flanking the first and second LoxP site was separated by 630 bases of genomic sequence in the Sestd1 Cre-excised allele.
Reagents
The following reagents were used: p3×FLAG-CMV-10 (Sigma); pcDNA3.1(−) and pcDNA 5/FRT/TO (Invitrogen); SRE.Luciferase reporter (provided by K. Kaibuchi, Nagoya University, Nagoya, Japan); pSuperTOP luciferase reporter and pRenilla (provided by Randall T. Moon, University of Washington); pC3-transferase (provided by Louis F. Reichardt, University of California at San Francisco); mouse IgG-agarose bead, anti-FLAG M2-conjugated agarose beads, and anti-HA-agarose beads (Sigma). The following antibodies were used for immunoblotting: FLAG (Sigma); HA (Roche Applied Science); Active β-catenin “ABC,” phospho-Mypt1 (Thr-696) (Millipore); β-catenin, Mypt1 (BD Biosciences), and Sestd1 (Thermo Scientific).
Cell Culture
HEK293T cells were grown in Dulbecco's modified Eagle's medium high glucose (DMEM H21) supplemented with 10% fetal bovine serum (FBS), 1× penicillin/streptomycin at 37 °C under 5% CO2, 95% air. 3T3 cells were maintained in DMEM H21 supplemented with 10% calf bovine serum and 1× penicillin/streptomycin at 37 °C under 5% CO2, 95% air. Mouse embryonic fibroblasts (MEFs) were prepared from E13.5/14.5 embryos of the Sestd1+/+ (WT) and Sestd1−/− (mutant) genotypes by cross-breeding Sestd1 heterozygous parents. Primary MEFs were cultured as described previously (35). Briefly, embryos were decapitated, eviscerated, minced, and trypsinized. The dissociated cells were cultured in DMEM H21 containing glutamine, nonessential amino acids, penicillin/streptomycin, β-mercaptoethanol, and 10% FBS.
Yeast Two-hybrid Screen
The Matchmaker two-hybrid system (Clontech) was used with a Xenopus laevis unfertilized oocyte cDNA library. Full-length XDpr1 was used as bait to recover a cDNA corresponding to the carboxyl-terminal half of Xenopus Sestd1.
Proteomics
Human embryonic kidney 293T cells were grown to confluence in culture dishes and transfected with FLAG-Dact1 plasmid. Stable transfected cells were lysed and incubated with anti-FLAG M2 beads for 3 h. Beads were collected, washed, and eluted by 3×FLAG peptide. The eluted supernatant was digested with trypsin, and the resulting peptides were analyzed by mass spectrometry.
Luciferase Reporter Activity Assays
Luciferase vectors were co-transfected with a Renilla vector for normalization of transfection efficiency. 3T3 cells and MEF cells were seeded in 24-well plates the day before transfection. Cells were transfected with the indicated plasmids using Lipofectamine 2000. For the SRE.Luciferase reporter assay, transfected cells were maintained in low serum medium (0.5% FBS) to sustain a basal Rho GTPase activity. Luciferase activity was measured 24 h post-transfection using the Dual-Luciferase Reporter System (Promega) in a plate-reading luminometer (Veritas). The data are presented as the relative luciferase activity, calculated as the ratio of the firefly to Renilla luciferase activity (mean ± S.E. (n = 4)).
Immunoprecipitation
HEK293T cells were plated in maintenance medium without antibiotics for 12 h and transfected with Lipofectamine 2000 (Invitrogen). After 48 h, transfected cells were lysed in a buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100 supplemented with protease inhibitor tablets (Roche Applied Science). Cell lysates were pre-cleared and incubated with anti-FLAG M2 beads or anti-HA beads for 3 h. Beads were collected and washed as described previously (36). Protein complexes were separated by SDS-PAGE followed by detection using the other antibody as appropriate.
RESULTS
Sestd1 Is a Novel Binding Partner of all Dact Family Members
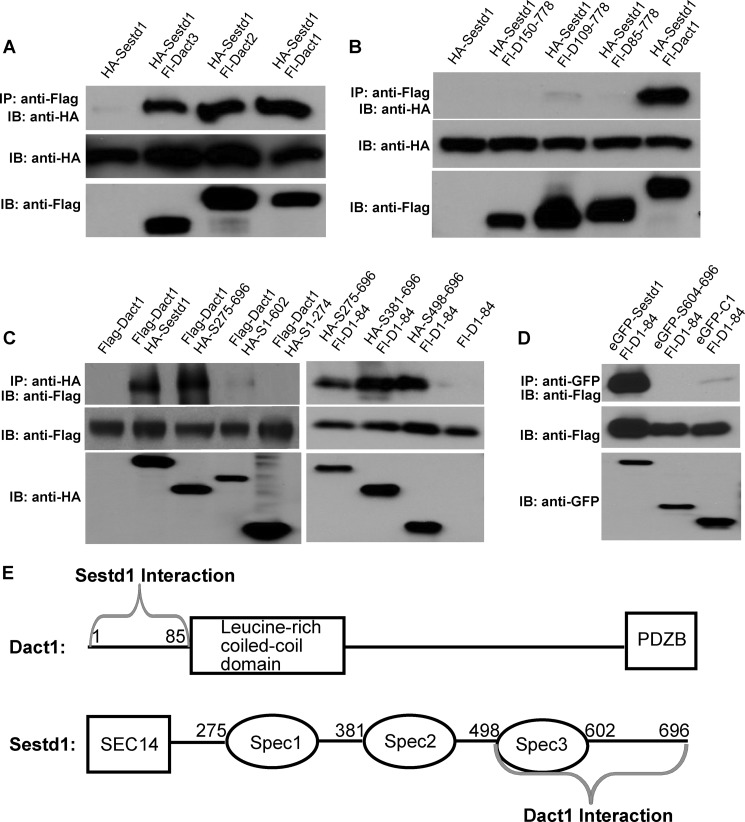
Dact1 is a scaffold protein that regulates Wnt signaling, and loss of Dact1 causes a phenotypic spectrum in mice resembling the lower mesodermal defects sequence in humans (29, 37). To dissect and elucidate the biochemical pathway involving Dact1, we sought to identify new binding partners of this protein. We used two complementary approaches to accomplish this using Dact1 as a molecular starting point. First, we performed a yeast two-hybrid screen using Xenopus Dapper (Dpr), a close homolog of mammalian Dact1, as bait. In a second strategy, we generated a HEK293T cell line that stably expressed mouse Dact1 with an amino-terminal FLAG epitope, allowing us to biochemically purify Dact1 along with associated proteins that could be identified through proteomic analysis (i.e. mass spectrometry). Both approaches independently identified Sestd1 as a binding partner of Dact1 (data not shown). To confirm this interaction, murine Sestd1 was recombinantly co-expressed in an immortalized cell line with each of the three murine Dact paralogs, and co-immunoprecipitation (co-IP) assays were performed. Sestd1 formed complexes with all three Dact family members in these assays (Fig. 1A).
FIGURE 1.
Sestd1 is a novel binding partner of Dact family members. A, Sestd1 interacts with all three Dact paralogs in HEK293T cells. HA-Sestd1 was expressed with or without FLAG-Dact1, FLAG-Dact2, and FLAG-Dact3 in HEK293T cells. Protein complexes were immunoprecipitated (IP) with anti-FLAG-agarose beads, and associated proteins were detected by immunoblot (IB) with anti-HA antibody. B–D, amino terminus of Dact1 interacts with the carboxyl terminus of Sestd1, including the third spectrin domain (Spec3). Appropriate FLAG-, HA-, or enhanced GFP-tagged proteins were recombinantly expressed in HEK293T cells. The co-IP assays were performed as in A. E, schematic summary of complex formation between Dact1 and Sestd1. Interacting regions are indicated by gray braces.
Identification of Binding Regions between Sestd1 and Dact1
Like Dvl, both Dact and Sestd1 are “scaffold” proteins, essentially meaning that they are intracellular proteins with conserved protein-protein interaction domains but whose primary structure reveals little else about their potential cellular functions, let alone about how they might cooperate mechanistically. As an initial step toward tackling this problem, we mapped the domains of their primary sequence that contribute to their interaction. Plasmids encoding full-length or truncated FLAG-Dact1 proteins were co-transfected with HA-Sestd1 in HEK293T cells and protein complexes analyzed by co-IP. Dact1 amino-terminal deletion mutants, including one lacking only the first 84 amino acids of the Dact1 protein (Dact1(150–778), Dact1(109–778), and Dact1(85–778)), failed to form complexes with full-length Sestd1 (Fig. 1B), whereas a polypeptide consisting of only the amino-terminal 84 amino acids of Dact1 (Dact1(1–84)) upstream of the leucine zipper domain could form complexes with Sestd1 (Fig. 1C). These data demonstrate that the amino-terminal 84 amino acids of Dact1 are both necessary and sufficient for complex formation with Sestd1 (Fig. 1E). Interestingly, this amino-terminal region of Dact1 is dispensable for interactions with all previously described Dact partnering proteins (24, 29, 38–41). The co-IP assays were similarly performed using a panel of Sestd1 truncations to identify the Dact-interacting region of Sestd1. Sestd1 mutant proteins lacking amino-terminal residues, including the first two spectrin domains (Sestd1(275–696), Sestd1(381–696), and Sestd1(498–696)), can form complexes with the N terminus of Dact1 (Dact1(1–84)), whereas a Sestd1 truncation mutant lacking only the carboxyl-terminal 94 residues (Sestd1(1–602)) forms complexes with Dact1 only very weakly (Fig. 1C). Nevertheless, this carboxyl-terminal polypeptide, which is unstable when expressed by itself (data not shown), is insufficient to mediate complex formation with Dact1(1–84) when expressed as a fusion with enhanced GFP (enhanced GFP-Sestd1(604–696)) (Fig. 1D). Taken together, these co-IP data suggest that the carboxyl-terminal 198 residues of Sestd1 (Sestd1(498–696)), including both the third spectrin domain and the C terminus, are required for complex formation with Dact1 (Fig. 1E). The fact that Dact1-Sestd1 complex formation is mediated by well circumscribed regions in each protein supports the notion that these proteins are uniquely allied in function.
Generation of Sestd1-deficient Mouse Lines
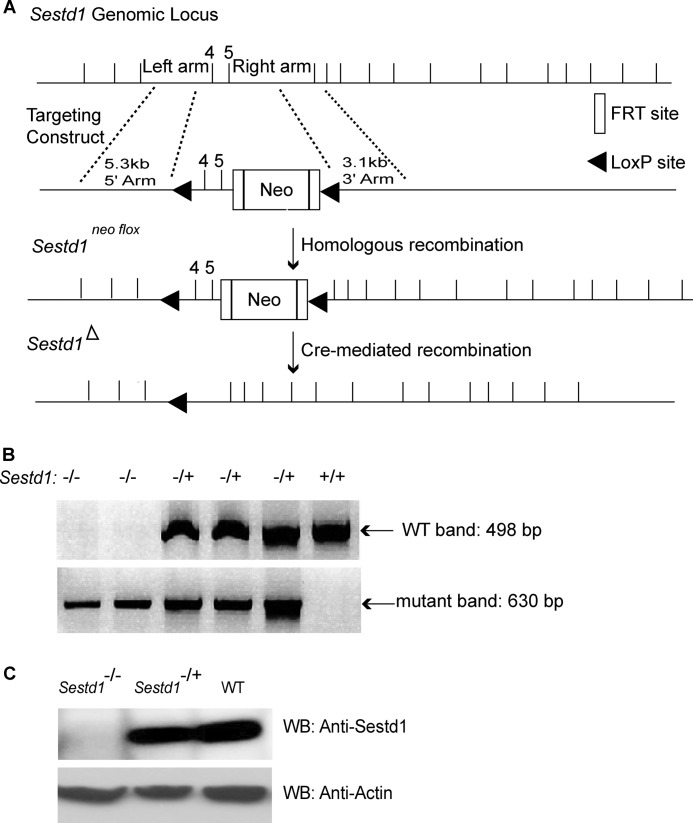
To further probe the biological functions of Sestd1, we created Sestd1 knock-out mice using standard homologous recombination, embryonic stem (ES) cell manipulation, and mouse genetic techniques. In brief, a Sestd1flox allele was engineered and introduced into mice by homologous recombination in ES cells followed by chimeric animal generation. Once this allele was established in a mouse line, a constitutive null allele was generated through mouse husbandry, including crossing to a transgenic mouse line expressing Cre recombinase in the germ line, resulting in Cre-mediated excision of exon 4, intron 4, and exon 5 to produce a constitutive null allele (Fig. 2A). Genotyping PCR (Fig. 2B) was employed to identify all targeted alleles in mice. Loss of the Sestd1 protein was verified by immunoblot in mice homozygous for the null allele (Fig. 2C), hereafter referred to as Sestd1 mutants or KO.
FIGURE 2.
Targeted inactivation of the Sestd1 gene. A, schema of the Sestd1 genomic locus and gene targeting intermediates. Exons 4 and 5 partially encode the functionally important SEC14 domain of Sestd1. Excision of exons 4 and 5 by Cre recombinase yields a truncated transcript followed by a frameshift. B, PCR genotyping of genomic tail DNA. Using the PCR primer pairs indicated under “Materials and Methods,” WT and Sestd1 mutant loci generate 498- and 630-bp fragments, respectively. C, immunoblot analysis of Sestd1 protein levels. A band at ∼79 kDa disappears in Sestd1−/− brain tissue lysates.
Loss of Sestd1 Phenocopies Loss of Dact1 in Mice with a Spectrum of Posterior Malformations
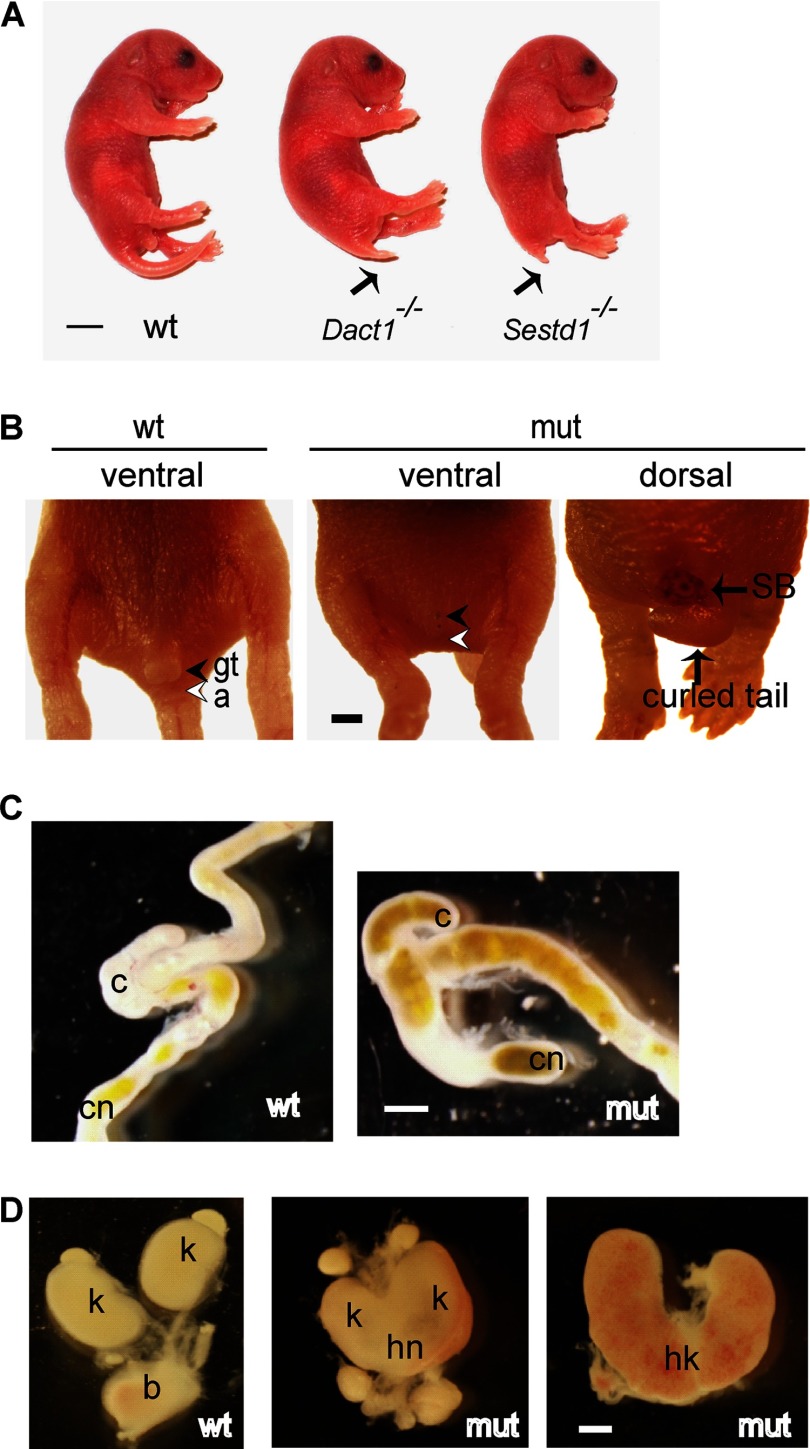
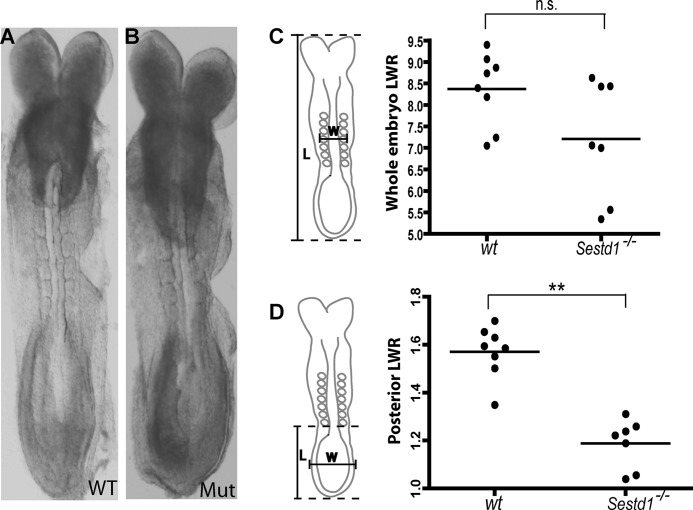
Sestd1 mutant mice survive to birth but typically die within 24 h. Remarkably, these mice display a spectrum of birth malformations, including a short tail, no genital tubercle, no anus, blind-ended colons, hydronephrotic kidneys, and no bladder (Fig. 3, A–D), that precisely phenocopies the spectrum of birth malformations in Dact1 mutant mice described previously by ourselves and independently by another group (29, 31). The amazing correspondence of this otherwise unique phenotypic spectrum in two mouse lines with mutations at separate loci (and also engineered in distinct ES cell lines; see under “Materials and Methods”) provides compelling evidence that Sestd1 and Dact1 closely cooperate during development. Like Dact1 mutants, Sestd1 mutants also display partly penetrant neural tube defects, including spina bifida (Fig. 3B), and some also have the curled tail common to many PCP mutants in mice, including mutants for Vangl2 and Scrib (7, 9). We traced birth defects in Sestd1 mutants back to the earliest defects observable during development at embryonic day 8.5 (E8.5). Compared with wild type (WT) embryos, Sestd1 mutants at this stage had a more rounded posterior contour (Fig. 4, A and B), very similar to Dact1 mutant embryos at the same stage (29). To quantify this morphological difference between wild type and Sestd1 mutant embryos, the whole embryo length-width ratio (LWR) and posterior LWR were measured. Compared with wild type, we could measure no significant difference in whole embryo LWR of Sestd1 mutant embryos (WT: 8.37 ± 0.29 versus KO: 7.21 ± 0.51, p = 0.066) (Fig. 4C). However, we measured a highly significant reduction in posterior LWR between WT and Sestd1 mutant embryos (WT: 1.57 ± 0.038 versus KO: 1.19 ± 0.038, p < 0.001) (Fig. 4D). Similar reductions in posterior LWR at this stage are detected in Dact1 mutant and Vangl2Lp/+ embryos, in which they are associated with abnormal convergent-extension movements in primary germ tissues (29). In brief, our phenotypic analyses suggest that the embryonic basis for posterior birth defects in Sestd1 mutants is identical to that in Dact1 mutants.
FIGURE 3.
Loss of Sestd1 causes a spectrum of posterior defects that phenocopies loss of Dact1. A, Sestd1 KO phenocopies Dact1 KO in mice. Sestd1 mutants (mut) die before or within 1 day of birth, and at postnatal day 0 they have a short tail (arrow). B, Sestd1 mutant neonates lack genital tubercle (gt) and anus (a) (filled arrowhead and empty arrowhead). Some Sestd1 mutant neonates also have spina bifida (SB) and curled tails. C, Sestd1 mutant neonates have blind-ended colons (cn). D, Sestd1 mutant neonates have hydronephrotic horseshoe kidneys (hk) and no bladder (b). Scale bar, 500 μm. Other abbreviations: wt, wild type; mut, Sestd1 mutant; c, cecum; k, kidney; hn, hydronephrosis (cf. Fig. 1 in Ref. 29).
FIGURE 4.
Sestd1 mutant embryonic phenotype. A and B, early Sestd1 mutant embryos have a more rounded posterior contour than WT littermate embryos. C and D, length-width ratio (LWR) measurements in wild type and Sestd1 mutants for whole (C) and posterior (D) embryo. n.s., not significant (p > 0.05); **, p < 0.01.
To genetically test the hypothesis that Sestd1 and Dact1 operate in a single pathway during embryonic development as opposed to different pathways that just happen to produce the same phenotypic outcome, an intercross was made between Sestd1−/+;Dact1−/+ (double heterozygous) mice and the phenotypes of all newborn pups, which include both double and single homozygous mutant animals, examined. Sestd1−/−;Dact1−/− (double homozygous mutant) offspring showed phenotypes identical to their littermates that were homozygous mutant at either locus alone (Table 1). In other words, the two loci are phenotypically epistatic, a genetic result most consistent with the interpretation that the two gene products operate in a linear biochemical pathway. Taken together, our protein interaction data plus phenotypic observations in Sestd1 and Dact1 mutant animals suggest that Sestd1 closely cooperates with Dact1 in a single pathway during embryonic development in mice.
TABLE 1.
Sestd1−/−;Dact1−/− combination mutants show the same phenotypes as either Sestd1−/− or Dact1−/− single mutants
An intercross was made between Sestd1−/+;Dact1−/+ mice. Genotype and phenotype (tail length and genital tubercle (GT)) of each newborn pup were determined.
| Genotype |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| WT | Dact1−/+ | Sestd1−/+ | Sestd1−/+ Dact1−/+ | Dact1−/− | Sestd1−/− | Sestd1−/+ Dact1−/− | Sestd1−/− Dact1−/+ | Sestd1−/− Dact1−/− | |
| Number | 5 | 3 | 4 | 7 | 3 | 3 | 8 | 3 | 4 |
| Phenotype | Normal | Normal | Normal | Normal | Short tail, no GT | Short tail, no GT | Short tail, no GT | Short tail, no GT | Short tail, no GT |
PCP Pathway Abnormalities in Sestd1 Mutant Embryonic Tissue
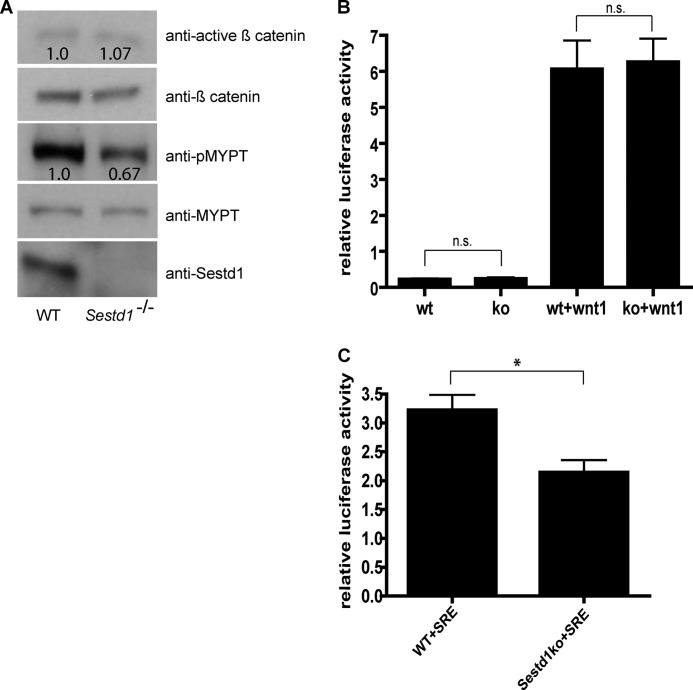
We previously showed that the Dact1 mutant phenotypic spectrum, which we also observe in Sestd1 mutant embryos, closely correlates with altered PCP but not altered Wnt/β-catenin signaling in affected tissues of mutant embryos (29). To extend these findings while using an independent methodology to determine which Wnt pathways are most altered in the absence of the Sestd1 protein, we isolated MEFs from the Sestd1 mutant and WT littermate embryos and analyzed pathway biochemistry in these cells. We first quantified relative levels of activated β-catenin and of phosphorylated Mypt1 (a substrate of Rho associated kinase) as endogenous biochemical correlates of the Wnt/β-catenin and Wnt/PCP pathways, respectively. In comparison with WT, there were no significant differences in levels of activated β-catenin in Sestd1 mutant MEFs, whereas phosphorylated Mypt1 protein was significantly decreased in Sestd1 mutant MEFs, supporting an effect on Rho GTPase but not on Wnt/β-catenin signaling in mutant embryonic cells (Fig. 5A). To confirm these findings more quantitatively, we employed plasmid-based luciferase reporter assays for both the Wnt/β-catenin and Rho GTPase pathways. For Wnt/β-catenin signaling, we used a well established luciferase reporter assay based on transient transfection of cells with the SuperTOP reporter plasmid containing a multimer of an optimal T cell factor-binding site upstream of the luciferase cDNA (42, 43). For Rho GTPase, we used the SRE.L reporter, containing a mutant serum-response element (SRE) upstream of the luciferase cDNA, and which is thereby responsive to activation downstream of Rho (44). These luciferase-based assays confirmed that although Wnt/β-catenin signaling is unaffected in the Sestd1 mutant MEFs (WT: 0.22 ± 0.02 versus KO: 0.24 ± 0.03, p = 0.68; WT+Wnt1: 6.06 ± 0.79 versus KO+Wnt1: 6.26 ± 0.65, p = 0.85) (Fig. 5B), Rho GTPase activity is significantly decreased compared with WT (WT: 3.22 ± 0.26 versus KO: 2.14 ± 0.21 p = 0.032) (Fig. 5C). Our signaling pathway assays in Sestd1 mutant MEFs closely parallel and strongly support our prior findings in Dact1 mutant embryos, demonstrating that the principal Wnt pathway affected by deletion of either of these proteins in mouse embryonic tissues activates Rho GTPase, in this context most likely representing PCP.
FIGURE 5.
Disruption of the PCP pathway in Sestd1 mutant embryos. A, levels of phosphorylated MYPT (pMYPT; a target of Rho-associated kinase), but not of activated β-catenin, are reduced in Sestd1−/− MEFs. Cell lysates of WT and Sestd1−/− MEFs were subjected to immunoblot with the indicated antibodies. The relative density of active β-catenin was normalized to total β-catenin in the same sample, and the relative density of pMYPT was normalized by total MYPT. B, Sestd1−/− MEFs do not show reduced basal activation nor Wnt1-responsive activation of a Wnt/β-catenin-dependent luciferase reporter. MEF cells were transfected with Top flash, Renilla, or Wnt1 plasmids, and luciferase activity was measured 24 h post-transfection. n.s., not significant (p > 0.05). C, Sestd1−/− MEFs do show reduced basal activation of the Rho GTPase-dependent SRE luciferase reporter. MEF cells were transfected with SRE.L and Renilla plasmids in the presence of 0.5% serum for 24 h before luciferase activity was measured. *, p < 0.05.
Sestd1 and Dact1 Cooperatively Activate Rho GTPase in Cells
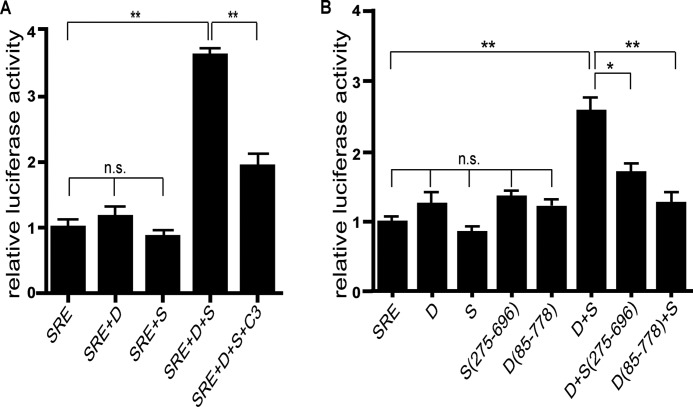
Sestd1 shares partial sequence similarity with Trio family members in its amino-terminal SEC14 domain and spectrin-like domains (33), but it lacks a GEF domain typically required for activating Rho GTPases. On this basis, we hypothesized that Sestd1 and Dact1 might regulate Rho GTPases via recruitment of a separate GEF protein. To test whether Sestd1 and Dact1 can indeed cooperatively regulate Rho GTPase activity in cells, we further employed the SRE.L reporter assay, but now in the context of recombinant expression of either wild type or mutant proteins. As achieving consistent relative levels of each of the two proteins was essential to test the hypothesis of cooperative activation, we molecularly engineered a panel of FLAG-Dact1–2A-HA-Sestd1 co-expression plasmids. The 2A sequence inserted between cDNAs in these plasmids impairs normal peptide bond formation between final glycine and proline codons of the 2A sequence via ribosomal skipping; the result is co-translation of distinct gene products from a single transcript, leading to equal levels of both translation products. Dact1, Sestd1, or Dact1–2A-Sestd1 plasmid was transfected in 3T3 cells along with the SRE.L and Renilla control plasmids, and luciferase levels were assessed 1 day later. Using this system, we found that Dact1 and Sestd1 by themselves had no effect on Rho GTPase activity (SRE: 1.0 ± 0.08 versus SRE+Dact1: 1.16 ± 0.15, p = 0.38; SRE: 1.0 ± 0.08 versus SRE+Sestd1:, 0.86 ± 0.05, p = 0.20) (Fig. 6A, 1st to 3rd bars). However, significant activation above base line was observed in cells co-expressing full-length Dact1 and Sestd1 (SRE: 1.0 ± 0.08 versus SRE+Dact1+Sestd1: 3.63 ± 0.06, p < 0.0001) (Fig. 6A, 4th bar). Moreover, when cells were co-transfected with C3 transferase, an ADP-ribosyltransferase that specifically inactivates Rho activity by catalyzing ribosylation of RhoA (45), SRE.L reporter induction by Dact1 and Sestd1 was sharply reduced (SRE+Dact1+Sestd1: 3.63 ± 0.06 versus SRE+Dact1+Sestd1+C3: 1.93 ± 0.18, p = 0.0009) (Fig. 6A, 5th bar). This confirmed that cooperative induction of luciferase activity by Dact1 and Sestd1 in this assay occurs primarily through Rho GTPase activation.
FIGURE 6.
Sestd1 cooperates with Dact1 in the PCP pathway. A, Dact1 and Sestd1 synergistically activate the Rho GTPase. The SRE.L reporter and Renilla plasmids were co-transfected with Dact1, Sestd1, or Dact1–2A-Sestd1 plasmids in 3T3 cells, and luciferase activity was measured 24 h later. Specificity of reporter activation for Rho GTPase activity is confirmed with the Rho-specific inhibitor, C3 exoenzyme (last lane). B, activation of Rho requires Dact1-Sestd1 complex formation and the Sestd1 SEC14 domain. The SRE.L reporter and Renilla plasmids were co-transfected with Dact1, Sestd1, Dact1–2A-Sestd1, Dact1–2A-Sestd1(275–696), or Dact1(85–778)-2A-Sestd1 and luciferase assayed as in A. n.s., not significant (p > 0.05); *, p < 0.05; **, p < 0.01.
Analysis of SRE.L reporter activation by co-expressed Dact1 and Sestd1 truncation mutants further demonstrated that the SEC14 domain of Sestd1 is important for Dact/Sestd1-mediated Rho activation (Dact1+Sestd1: 2.62 ± 0.19 versus Dact1+Sestd1(275–696): 1.72 ± 0.09, p = 0.014) (Fig. 6B, 2nd bar from right). In addition, the previously mapped amino-terminal Dact1 domain necessary for the Dact1-Sestd1 interaction is required (Dact1+Sestd1: 2.62 ± 0.19 versus Dact1(85–778)+Sestd1: 1.27 ± 0.11, p = 0.0039) (Fig. 6B, right-most bar). These data strongly support that Dact1 and Sestd1 cooperate in a complex to activate Rho GTPase and further show that this activation also requires the well conserved SEC14 domain in Sestd1.
Reciprocal Genetic Rescue between Sestd1 and Vangl2 Mutant Alleles in Mice
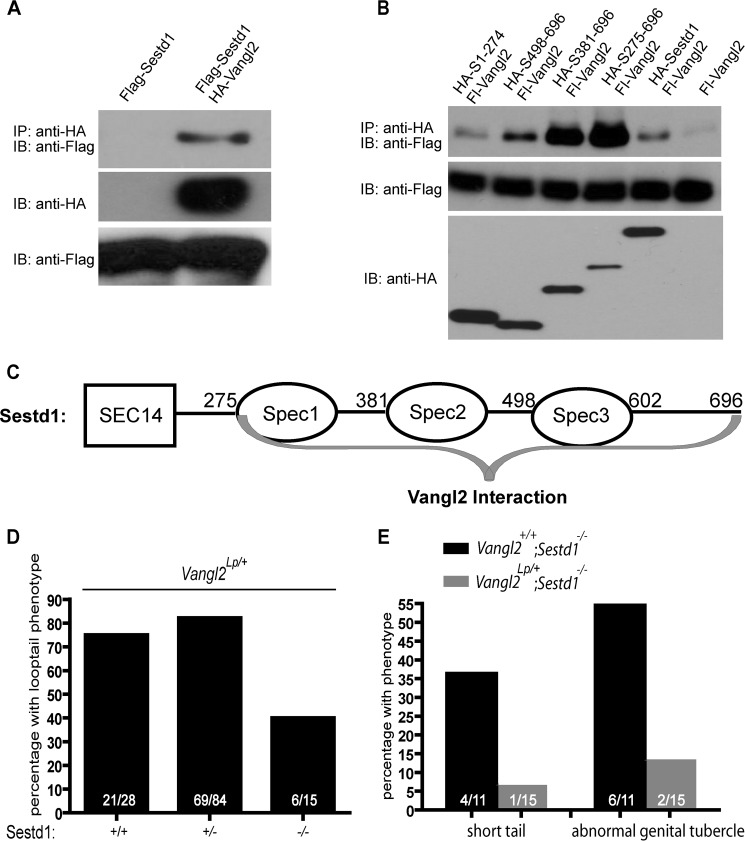
Dact1 biochemically and genetically interacts with Vangl2 (29). Because Sestd1 and Dact1 work in the same pathway to activate Rho GTPase presumably as part of PCP signaling, we speculated that Sestd1 might also interact with Vangl2. To test this hypothesis biochemically, we used co-IP to assess whether the Sestd1 and Vangl2 proteins form a complex when expressed in cells. This confirmed that Sestd1 can form a complex with Vangl2 (Fig. 7A). To map the domains of their primary sequence that contribute to Sestd1-Vangl2 interaction, plasmids encoding full-length or truncated HA-Sestd1 proteins were co-transfected with FLAG-Vangl2 in HEK293T cells, and protein complexes analyzed by co-IP. A Sestd1 carboxyl-terminal deletion mutant lacking all three spectrin domains (Sestd1(1–274)) failed to form complexes with Vangl2 (Fig. 7B). Sestd1 deletion mutants lacking one or two spectrin domains (Sestd1(381–696) and Sestd1(498–696)) recruited less Vangl2 protein than a truncation mutant that had all three spectrin repeats but no SEC14 domain (Sestd1(275–696)) (Fig. 7B). These data demonstrate that all three spectrin domains in Sestd1 contribute to complex formation with Vangl2 (Fig. 7C). Interestingly, compared with full-length Sestd1, Sestd1 truncation mutants that lack the SEC14 domain (Sestd1(275–696), Sestd1(381–696), and Sestd1(498–696)) consistently recruit more Vangl2 protein in these co-IP experiments (Fig. 7B). This suggests that the presence of the SEC14 domain somehow inhibits complex formation with Vangl2 in cells. Moreover, because the region of Sestd1 responsible for Vangl2 complex formation (Fig. 7C) overlaps but does not precisely coincide with the region responsible for Dact1 complex formation (Fig. 1E), we conclude that Sestd1-Vangl2 complex formation is not mediated exclusively and indirectly through Dact1, but it must include other intermolecular interactions involving the first two spectrin domains, which are negatively regulated by presence of the amino-terminal SEC14 domain.
FIGURE 7.
Sestd1 is a novel binding partner of Vangl2 and mutations in Sestd1 and Vangl2 reciprocally rescue. A, Sestd1 interacts with Vangl2 in HEK293T cells. FLAG-Sestd1 was expressed with or without HA-Vangl2 in HEK293T cells. Protein complexes were immunoprecipitated (IP) with anti-HA-agarose beads and associated proteins detected by immunoblot (IB) with anti-FLAG antibody. B, carboxyl terminus of Sestd1, including all three spectrin domains, interacts with Vangl2. Appropriate FLAG- or HA-tagged proteins were recombinantly expressed in HEK293T cells. The co-IP assays were performed as in Fig. 1A. C, schematic summary of complex formation between Sestd1 and Vangl2: Interacting regions are indicated by gray braces. D, loss of Sestd1 partly rescues the semidominant Vangl2Lp phenotype. An intercross was made between Sestd1+/−;Vangl2+/+ and Sestd1+/−;Vangl2Lp/+ mice, and neonatal offspring were genotyped and phenotyped. In the bar graph, the numbers of offspring corresponding to each phenotype/genotype are shown in white. E, heterozygosity for the Vangl2Lp allele partly rescues the Sestd1 null phenotype, graphed as in D.
Having shown that Sestd1 biochemically interacts with Vangl2, we next asked whether the Sestd1 knock-out mutation we engineered in mice genetically interacts with the semidominant Looptail (Lp) mutation in Vangl2, as we previously found for knock-out mutations at the Dact1 locus (29). The Vangl2Lp allele corresponds to a missense point mutation at the Vangl2 locus that semidominantly impairs function of both Vangl family members (Vangl1 and Vangl2) in mice (46). We crossed Sestd1+/−;Vangl2+/+ to Sestd1+/−;Vangl2Lp/+ mice and examined resultant phenotypes in newborn littermate offspring corresponding to the different genotypic combinations of these alleles. With regard to the semidominant curly “Loop-” tail phenotype, 40% (6/15) of Vangl2Lp/+;Sestd1−/− pups had curly tails as compared with 75% (21/28) of Vangl2Lp/+;Sestd1+/+ pups and 82% (69/84) of Vangl2Lp/+;Sestd1+/− pups (Fig. 7D). These genetic outcomes demonstrate that complete loss of Sestd1 partly rescues the Vangl2Lp/+ phenotype. Conversely, 36% (4/11) of Sestd1−/−;Vangl2+/+ pups from this cross displayed reduced tail length, and 54% (6/11) displayed genitourinary defects, but the penetrance of these phenotypes dropped to <7% (1/15) and ∼13% (2/15), respectively, in Sestd1−/−;Vangl2Lp/+ pups (Fig. 7E). These genetic outcomes demonstrate that the Sestd1 null phenotype is substantially rescued by the semidominant Vangl2Lp allele.
To summarize, genetic experiments demonstrate that mutations at the Sestd1 and Vangl2 loci mutually rescue, mimicking our genetic results for the Dact1 and Vangl2 loci. These data provide support for the conclusion that Sestd1 cooperates closely with Dact1 in the post-translational regulation of Vangl2 during embryonic development in the mouse, as we have previously shown for Dact1 (29).
DISCUSSION
Rho GTPases have emerged as key effectors of the PCP pathway and of other β-catenin independent branches of Wnt signaling, altering cytoskeletal dynamics and leading to changes in cell shape and migration during embryonic development (21). Dact1 null embryos have defects in biochemical correlates of PCP, including reduced activity of the Rho-associated kinase and elevated activity of c-Jun amino-terminal kinase (JNK) (29). In this report, we identify Sestd1 as a close functional partner of Dact1 during development. The two proteins can form a complex via circumscribed domains that have not been reported to be involved in other protein-protein interactions. Loss of Sestd1 in mice closely phenocopies loss of Dact1; in fact, loss of Sestd1 produces a neonatal phenotypic spectrum indistinguishable from that caused by loss of Dact1, including the highly penetrant truncation of posterior mesoderm and endoderm derivatives, along with less penetrant NTDs and the curled tails characteristic of other mouse PCP mutants. In addition, Sestd1 mutant embryonic tissues, as exemplified here by MEFs, have reduced basal Rho GTPase activity but do not have reductions in activated β-catenin either in the presence or absence of exogenous Wnt1. In genetic epistasis experiments, Sestd1;Dact1 double mutant mice show the same phenotypic spectrum and severity as either Sestd1 or Dact1 single mutants, and like the Dact1 null allele, the Sestd1 null allele reciprocally rescues the semidominant Lp mutation in Vangl2. Together, the biochemical, genetic, and signaling data presented here provide compelling evidence that the Sestd1 and Dact1 proteins function tightly together in the PCP pathway upstream of Rho GTPase activation in mouse embryos. Our prior findings in Dact1 mutant animals combined with our genetic data here further indicate that these proteins participate in the post-translational regulation of Vangl2 during epithelial-mesenchymal transition and associated mesendodermal cell movements at the primitive streak in the posterior embryo (29).
Although Sestd1 exhibits moderate sequence conservation with Trio family members in its amino-terminal SEC14 domain and carboxyl-terminal spectrin-like domains (33), it lacks a GEF domain and exhibits no GEF activity toward Rho GTPases based on cellular Rho activation assays. Instead, we demonstrate here that Sestd1 and Dact1 cooperate to activate Rho GTPase activity in cells and that this cooperation requires both the Sestd1 SEC14 domain and the amino-terminal domain of Dact1 necessary for physical interaction with Sestd1. On the basis of these data, we infer that activation of Rho GTPase downstream of these proteins may occur via recruitment of another GEF protein into a Dact1-Sestd1 complex. Interestingly, in ongoing proteomic studies we have identified just such a GEF-containing protein, rho/rac-guanine nucleotide exchange factor-2 (Arhgef2), as a potential Sestd1 binding partner (data not shown). Future work in our laboratory will be aimed at addressing whether Arhgef2 is directly involved in Sestd1 and Dact1-mediated Rho activation.
Although there are many members of the Trio protein family, Sestd1 is among the most divergent and is unique in lacking its own GTPase regulatory domain. The single copy of Sestd1 in the mammalian genome is highly conserved; the mouse and human Sestd1 proteins are >98% identical, consistent with a critical nonredundant physiological function. Notwithstanding its apparent importance, little has been previously established about the function of Sestd1. To our knowledge, the only prior investigation of this protein focused on its role in vascular tissues where it was implicated in nonselective cation channel regulation downstream of phospholipid signaling (47). Combined with the work presented here, this suggests the intriguing possibility that Sestd1 could be part of a signaling node linking the regulation of cation-dependent pathways (e.g. the Wnt/Ca2+ pathway) with the regulation of Rho GTPases downstream of Vangl2 (i.e. the Wnt/PCP pathway). Intriguingly, the Sestd1 SEC14 domain required for co-activation with Dact1 of Rho GTPase activity and that may also regulate Sestd1 interactions with Vangl2 has previously been implicated in the regulation of cation channels (47). Given the distribution of Sestd1 during embryonic development and in mature vascular, neural, and other tissues, studies in mice with the conditional Sestd1flox allele that we have generated will enable a molecular dissection of cross-talk between these important signaling pathways and an exploration of consequences of postnatal Sestd1 gene disruption, including contributions to cardiovascular, neuropsychiatric, and other pathophysiology.
Acknowledgments
We are deeply indebted to Dr. Randall T. Moon and his laboratory at the Howard Hughes Medical Institute and the Department of Pharmacology at the University of Washington for generous assistance, including conduct of the Xenopus Dapper yeast two-hybrid screen and mass spectrometry of our samples.
This work was supported, in whole or in part, by National Institutes of Health Grant R01HD055300 (to B. N. R. C.) and Grant T32 MH089920 from the National Institute of Mental Health (to X. Y. Y.). This work was also supported by the Department of Psychiatry and the Center for Neurobiology and Psychiatry at the University of California San Francisco.
This article was selected as a Paper of the Week.
- PCP
- planar cell polarity
- co-IP
- co-immunoprecipitation
- Dact
- Dapper antagonist of catenin
- Dvl
- Dishevelled
- GEF
- guanine nucleotide exchange factor
- MEF
- mouse embryonic fibroblasts
- NTD
- neural tube defect
- Sestd1
- SEC14 and spectrin domains 1
- SRE
- serum-response element
- Vangl2
- Van Gogh-like protein 2
- LWR
- length-width ratio
- SRE.L
- SRE.Luciferase.
REFERENCES
- 1. Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. (2004) WNT and β-catenin signalling: diseases and therapies. Nat. Rev. Genet. 5, 691–701 [DOI] [PubMed] [Google Scholar]
- 2. Logan C. Y., Nusse R. (2004) The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781–810 [DOI] [PubMed] [Google Scholar]
- 3. Huang H., He X. (2008) Wnt/β-catenin signaling: new (and old) players and new insights. Curr. Opin. Cell Biol. 20, 119–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Veeman M. T., Axelrod J. D., Moon R. T. (2003) A second canon. Functions and mechanisms of β-catenin-independent Wnt signaling. Dev. Cell 5, 367–377 [DOI] [PubMed] [Google Scholar]
- 5. Barrow J. R. (2006) Wnt/PCP signaling: a veritable polar star in establishing patterns of polarity in embryonic tissues. Semin. Cell Dev. Biol. 17, 185–193 [DOI] [PubMed] [Google Scholar]
- 6. Gao B. (2012) Wnt regulation of planar cell polarity (PCP). Curr. Top. Dev. Biol. 101, 263–295 [DOI] [PubMed] [Google Scholar]
- 7. Kibar Z., Vogan K. J., Groulx N., Justice M. J., Underhill D. A., Gros P. (2001) Ltap, a mammalian homolog of Drosophila Strabismus/Van Gogh, is altered in the mouse neural tube mutant Loop-tail. Nat. Genet. 28, 251–255 [DOI] [PubMed] [Google Scholar]
- 8. Curtin J. A., Quint E., Tsipouri V., Arkell R. M., Cattanach B., Copp A. J., Henderson D. J., Spurr N., Stanier P., Fisher E. M., Nolan P. M., Steel K. P., Brown S. D., Gray I. C., Murdoch J. N. (2003) Mutation of Celsr1 disrupts planar polarity of inner ear hair cells and causes severe neural tube defects in the mouse. Curr. Biol. 13, 1129–1133 [DOI] [PubMed] [Google Scholar]
- 9. Murdoch J. N., Henderson D. J., Doudney K., Gaston-Massuet C., Phillips H. M., Paternotte C., Arkell R., Stanier P., Copp A. J. (2003) Disruption of scribble (Scrb1) causes severe neural tube defects in the circletail mouse. Hum. Mol. Genet. 12, 87–98 [DOI] [PubMed] [Google Scholar]
- 10. Wang J., Hamblet N. S., Mark S., Dickinson M. E., Brinkman B. C., Segil N., Fraser S. E., Chen P., Wallingford J. B., Wynshaw-Boris A. (2006) Dishevelled genes mediate a conserved mammalian PCP pathway to regulate convergent extension during neurulation. Development 133, 1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lei Y. P., Zhang T., Li H., Wu B. L., Jin L., Wang H. Y. (2010) VANGL2 mutations in human cranial neural-tube defects. N. Engl. J. Med. 362, 2232–2235 [DOI] [PubMed] [Google Scholar]
- 12. Kibar Z., Salem S., Bosoi C. M., Pauwels E., De Marco P., Merello E., Bassuk A. G., Capra V., Gros P. (2011) Contribution of VANGL2 mutations to isolated neural tube defects. Clin. Genet. 80, 76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Juriloff D. M., Harris M. J. (2012) A consideration of the evidence that genetic defects in planar cell polarity contribute to the etiology of human neural tube defects. Birth Defects Res. A Clin. Mol. Teratol. 94, 824–840 [DOI] [PubMed] [Google Scholar]
- 14. Robinson A., Escuin S., Doudney K., Vekemans M., Stevenson R. E., Greene N. D., Copp A. J., Stanier P. (2012) Mutations in the planar cell polarity genes CELSR1 and SCRIB are associated with the severe neural tube defect craniorachischisis. Hum. Mutat. 33, 440–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Detrait E. R., George T. M., Etchevers H. C., Gilbert J. R., Vekemans M., Speer M. C. (2005) Human neural tube defects: developmental biology, epidemiology, and genetics. Neurotoxicol. Teratol. 27, 515–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Marco P., Merello E., Cama A., Kibar Z., Capra V. (2011) Human neural tube defects: genetic causes and prevention. Biofactors 37, 261–268 [DOI] [PubMed] [Google Scholar]
- 17. Tada M., Kai M. (2012) Planar cell polarity in coordinated and directed movements. Curr. Top. Dev. Biol. 101, 77–110 [DOI] [PubMed] [Google Scholar]
- 18. Wallingford J. B. (2012) Planar cell polarity and the developmental control of cell behavior in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 28, 627–653 [DOI] [PubMed] [Google Scholar]
- 19. Hall A. (2005) Rho GTPases and the control of cell behaviour. Biochem. Soc. Trans. 33, 891–895 [DOI] [PubMed] [Google Scholar]
- 20. Ybot-Gonzalez P., Savery D., Gerrelli D., Signore M., Mitchell C. E., Faux C. H., Greene N. D., Copp A. J. (2007) Convergent extension, planar-cell-polarity signalling, and initiation of mouse neural tube closure. Development 134, 789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schlessinger K., Hall A., Tolwinski N. (2009) Wnt signaling pathways meet Rho GTPases. Genes Dev. 23, 265–277 [DOI] [PubMed] [Google Scholar]
- 22. Cheyette B. N., Waxman J. S., Miller J. R., Takemaru K., Sheldahl L. C., Khlebtsova N., Fox E. P., Earnest T., Moon R. T. (2002) Dapper, a Dishevelled-associated antagonist of β-catenin and JNK signaling, is required for notochord formation. Dev. Cell 2, 449–461 [DOI] [PubMed] [Google Scholar]
- 23. Gloy J., Hikasa H., Sokol S. Y. (2002) Frodo interacts with Dishevelled to transduce Wnt signals. Nat. Cell Biol. 4, 351–357 [DOI] [PubMed] [Google Scholar]
- 24. Zhang L., Gao X., Wen J., Ning Y., Chen Y. G. (2006) Dapper 1 antagonizes Wnt signaling by promoting dishevelled degradation. J. Biol. Chem. 281, 8607–8612 [DOI] [PubMed] [Google Scholar]
- 25. Fisher D. A., Kivimäe S., Hoshino J., Suriben R., Martin P. M., Baxter N., Cheyette B. N. (2006) Three Dact gene family members are expressed during embryonic development and in the adult brains of mice. Dev. Dyn 235, 2620–2630 [DOI] [PubMed] [Google Scholar]
- 26. Waxman J. S., Hocking A. M., Stoick C. L., Moon R. T. (2004) Zebrafish Dapper1 and Dapper2 play distinct roles in Wnt-mediated developmental processes. Development 131, 5909–5921 [DOI] [PubMed] [Google Scholar]
- 27. Jiang X., Tan J., Li J., Kivimäe S., Yang X., Zhuang L., Lee P. L., Chan M. T., Stanton L. W., Liu E. T., Cheyette B. N., Yu Q. (2008) DACT3 is an epigenetic regulator of Wnt/β-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell 13, 529–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meng F., Cheng X., Yang L., Hou N., Yang X., Meng A. (2008) Accelerated re-epithelialization in Dpr2-deficient mice is associated with enhanced response to TGFβ signaling. J. Cell Sci. 121, 2904–2912 [DOI] [PubMed] [Google Scholar]
- 29. Suriben R., Kivimäe S., Fisher D. A., Moon R. T., Cheyette B. N. (2009) Posterior malformations in Dact1 mutant mice arise through misregulated Vangl2 at the primitive streak. Nat. Genet. 41, 977–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Okerlund N. D., Kivimäe S., Tong C. K., Peng I. F., Ullian E. M., Cheyette B. N. (2010) Dact1 is a postsynaptic protein required for dendrite, spine, and excitatory synapse development in the mouse forebrain. J. Neurosci. 30, 4362–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wen J., Chiang Y. J., Gao C., Xue H., Xu J., Ning Y., Hodes R. J., Gao X., Chen Y. G. (2010) Loss of Dact1 disrupts planar cell polarity signaling by altering dishevelled activity and leads to posterior malformation in mice. J. Biol. Chem. 285, 11023–11030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shi Y., Ding Y., Lei Y. P., Yang X. Y., Xie G. M., Wen J., Cai C. Q., Li H., Chen Y., Zhang T., Wu B. L., Jin L., Chen Y. G., Wang H. Y. (2012) Identification of novel rare mutations of DACT1 in human neural tube defects. Hum. Mutat. 33, 1450–1455 [DOI] [PubMed] [Google Scholar]
- 33. Sato T., Mishina M. (2003) Representational difference analysis, high-resolution physical mapping, and transcript identification of the zebrafish genomic region for a motor behavior. Genomics 82, 218–229 [DOI] [PubMed] [Google Scholar]
- 34. George S. H., Gertsenstein M., Vintersten K., Korets-Smith E., Murphy J., Stevens M. E., Haigh J. J., Nagy A. (2007) Developmental and adult phenotyping directly from mutant embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 104, 4455–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Xu J. (2005) Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr. Protoc. Mol. Biol. Chapter 28, Unit 28.1 [DOI] [PubMed] [Google Scholar]
- 36. Kivimäe S., Yang X. Y., Cheyette B. N. (2011) All Dact (Dapper/Frodo) scaffold proteins dimerize and exhibit conserved interactions with Vangl, Dvl, and serine/threonine kinases. BMC Biochem. 12, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pauli R. M. (1994) Lower mesodermal defects: a common cause of fetal and early neonatal death. Am. J. Med. Genet. 50, 154–172 [DOI] [PubMed] [Google Scholar]
- 38. Hikasa H., Sokol S. Y. (2004) The involvement of Frodo in TCF-dependent signaling and neural tissue development. Development 131, 4725–4734 [DOI] [PubMed] [Google Scholar]
- 39. Brott B. K., Sokol S. Y. (2005) A vertebrate homolog of the cell cycle regulator Dbf4 is an inhibitor of Wnt signaling required for heart development. Dev. Cell 8, 703–715 [DOI] [PubMed] [Google Scholar]
- 40. Park J. I., Ji H., Jun S., Gu D., Hikasa H., Li L., Sokol S. Y., McCrea P. D. (2006) Frodo links Dishevelled to the p120-catenin/Kaiso pathway: distinct catenin subfamilies promote Wnt signals. Dev. Cell 11, 683–695 [DOI] [PubMed] [Google Scholar]
- 41. Gao X., Wen J., Zhang L., Li X., Ning Y., Meng A., Chen Y. G. (2008) Dapper1 is a nucleocytoplasmic shuttling protein that negatively modulates Wnt signaling in the nucleus. J. Biol. Chem. 283, 35679–35688 [DOI] [PubMed] [Google Scholar]
- 42. Korinek V., Barker N., Morin P. J., van Wichen D., de Weger R., Kinzler K. W., Vogelstein B., Clevers H. (1997) Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science 275, 1784–1787 [DOI] [PubMed] [Google Scholar]
- 43. DasGupta R., Kaykas A., Moon R. T., Perrimon N. (2005) Functional genomic analysis of the Wnt-wingless signaling pathway. Science 308, 826–833 [DOI] [PubMed] [Google Scholar]
- 44. Sah V. P., Seasholtz T. M., Sagi S. A., Brown J. H. (2000) The role of Rho in G protein-coupled receptor signal transduction. Annu. Rev. Pharmacol. Toxicol. 40, 459–489 [DOI] [PubMed] [Google Scholar]
- 45. Ridley A. J., Hall A. (1992) The small GTP-binding protein Rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70, 389–399 [DOI] [PubMed] [Google Scholar]
- 46. Song H., Hu J., Chen W., Elliott G., Andre P., Gao B., Yang Y. (2010) Planar cell polarity breaks bilateral symmetry by controlling ciliary positioning. Nature 466, 378–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miehe S., Bieberstein A., Arnould I., Ihdene O., Rütten H., Strübing C. (2010) The phospholipid-binding protein SESTD1 is a novel regulator of the transient receptor potential channels TRPC4 and TRPC5. J. Biol. Chem. 285, 12426–12434 [DOI] [PMC free article] [PubMed] [Google Scholar]