Background: N-Reduction is catalyzed by a molybdenum-dependent three-component enzyme system.
Results: Essential components include mitochondrial but not microsomal cytochrome b5, and the mitochondrial amidoxime reducing components 1/2.
Conclusion: CYB5 heme is required for activity, and contribution of a particular mARC isoform to N-reduction is dependent on its expression level.
Significance: These findings contribute to the understanding of N-reductive pathway in detoxication and drug metabolism.
Keywords: Drug Metabolism, Heme, Mitochondria, Molybdenum, RNA Interference (RNAi), CYB5B, MOSC, N-reduction, mARC
Abstract
The mitochondrial amidoxime reducing component mARC is a recently discovered molybdenum enzyme in mammals. mARC is not active as a standalone protein, but together with the electron transport proteins NADH-cytochrome b5 reductase (CYB5R) and cytochrome b5 (CYB5), it catalyzes the reduction of N-hydroxylated compounds such as amidoximes. The mARC-containing enzyme system is therefore considered to be responsible for the activation of amidoxime prodrugs. All hitherto analyzed mammalian genomes code for two mARC genes (also referred to as MOSC1 and MOSC2), which share high sequence similarities. By RNAi experiments in two different human cell lines, we demonstrate for the first time that both mARC proteins are capable of reducing N-hydroxylated substrates in cell metabolism. The extent of involvement is highly dependent on the expression level of the particular mARC protein. Furthermore, the mitochondrial isoform of CYB5 (CYB5B) is clearly identified as an essential component of the mARC-containing N-reductase system in human cells. The participation of the microsomal isoform (CYB5A) in N-reduction could be excluded by siRNA-mediated down-regulation in HEK-293 cells and knock-out in mice. Using heme-free apo-CYB5, the contribution of mitochondrial CYB5 to N-reductive catalysis was proven to strictly depend on heme. Finally, we created recombinant CYB5B variants corresponding to four nonsynonymous single nucleotide polymorphisms (SNPs). Investigated mutations of the heme protein seemed to have no significant impact on N-reductive activity of the reconstituted enzyme system.
Introduction
The mitochondrial amidoxime reducing component mARC,2 together with the electron transport proteins cytochrome b5 (CYB5) and NADH-cytochrome b5 reductase (CYB5R), is part of the N-reductive enzyme system that is known to be responsible for oxygen-insensitive reduction of several N-hydroxylated compounds (1) (Fig. 1). In 2006, mARC was isolated from porcine liver mitochondria for the first time and was identified as a so far unknown molybdenum-containing protein (2). Beside sulfite oxidase, aldehyde oxidase and xanthine oxidoreductase, mARC is the fourth molybdenum-containing enzyme found in mammals with all of them binding the molybdenum cofactor (Moco) (3). Mutations in the highly conserved Moco biosynthetic pathway lead to the combined loss of activities of all molybdenum enzymes. Humans suffering from this Moco deficiency exhibit severe neurological abnormalities, ultimately resulting in the death of affected patients.
FIGURE 1.
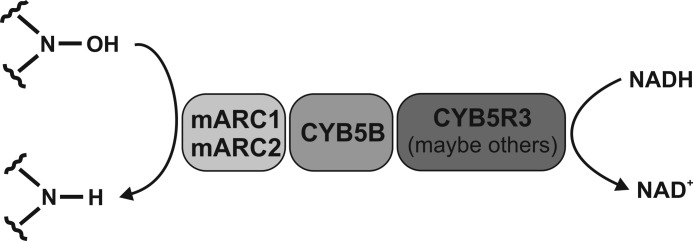
N-Reductive enzyme system in human cells.
All analyzed mammalian genomes contain two mARC genes (MOSC1 and MOSC2), which share a high degree of sequence similarity to each other. Classification of the terms “mARC1” and “mARC2” in different species are made on the basis of sequence alignments to the human proteins.
It is well accepted that the N-reductive enzyme system plays a major role in the activation of N-hydroxylated prodrugs such as amidoximes, N-hydroxy-guanidines, or sulfohydroxamic acids (4–6). The mARC-containing enzyme system is the counterpart of cytochrome P450 (P450) and flavin monooxygenase-catalyzed oxidations at nitrogen centers and can reduce back all N-oxygenated structures. Although their physiological substrates, and therefore their physiological functions, are as yet unknown, mARC proteins are assumed to be involved in detoxification of mutagenic and toxic aromatic hydroxylamines such as N-hydroxylated DNA base analogs (7). This hypothesis is supported by a recent study that describes significantly down-regulated expression of human mARC2 in colon tumors of human tissue samples (8). As the nitric oxide (NO) precursor N4-hydroxy-l-arginine is described as another putative physiological substrate, it has been suggested that the N-reductive enzyme system could also act as a regulator of l-arginine-dependent biosynthesis of NO (9). Furthermore, mARC2 expression has been found to be up-regulated in diabetic kidneys and by glucose in renal cells (10). Moreover, an association of mARC1 with lipid metabolism was recently postulated (11–13).
In previous studies, we have shown that both human mARC proteins are capable of reducing N-hydroxylated substrates in vitro when reconstituted with their electron transport proteins CYB5 and CYB5R and that all three components are necessary for N-reduction (2, 4–7, 9, 14). However, it is still unknown whether both mARC proteins are involved in the N-reductive metabolic pathway in vivo. The existence of two mARC genes in nearly all hitherto annotated genomes suggests the evolutionary need for the function of each isoform. In all investigated mammals, at least one mARC protein was identified in mitochondria (2, 14–16). Besides this eponymous localization, proteomic studies and immunofluorescence studies suggest an alternative peroxisomal localization for mARC2 (17, 18). This could be a hint at functions of mARC beyond the N-reductive system. To explore differences in the functions of the two mARC proteins and to clarify whether in cell metabolism both or only one mARC homolog is involved in the N-reductive pathway, siRNA-mediated knockdown studies of mARC in human cells were performed in this work.
In addition, involvement of the heme protein CYB5 in N-reductive metabolism was further characterized. Like the flavoprotein CYB5R, CYB5 can be found as an integral membrane protein in the endoplasmic reticulum and the outer mitochondrial membrane (OMM), as well as a soluble form in erythrocytes (19, 20). In contrast to CYB5R, two separate genes encode the different isoforms of CYB5, with CYB5A encoding the microsomal and soluble isoform and CYB5B encoding the mitochondrial isoform. Because mARC was identified in the OMM (2, 11, 16), it is likely that the mitochondrial isoform of CYB5 is the physiological partner of mARC. However, reduction studies of a two-component microsomal system consisting of CYB5A and its reductase have been described (21–24). By siRNA-mediated down-regulation of CYB5A or CYB5B in HEK-293 and by analyzing CYB5A knock-out mice, we demonstrate that only the mitochondrial heme protein is involved in the N-reductive metabolic pathway. In a continuation of our studies, we investigated the effects of known nonsynonymous single nucleotide polymorphisms (SNPs) of the Single Nucleotide Polymorphism Database (dbSNP) (www.ncbi.nlm.nih.gov/snp) on N-reduction. For this purpose, the recombinant CYB5B protein variants carrying the respective amino acid substitution were expressed in Escherichia coli and reconstituted with CYB5R and mARC.
CYB5 is considered to accept electrons from CYB5R and to pass them via its heme to the Moco of mARC, thereby functioning as electron transport protein in the N-reductive system (1). However, several P450-catalyzed reactions have been shown to be stimulated by both CYB5 and apo-CYB5 (25–27). These findings suggest that CYB5 does not function as an electron donor in these reactions, but instead exerts an allosteric effect facilitating productive electron transfer from P450 reductase to P450. To explore the mechanism by which CYB5B takes part in the mARC-containing N-reductive system, we determined whether the N-reductive system can also be reconstituted with apo-CYB5, which lacks the heme and hence cannot participate in electron transfer.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
HEK-293 human embryonic kidney cells and ZR-75-1 human breast carcinoma cells were purchased from Cell Lines Service (Eppelheim, Germany). Opti-MEM, Minimum Essential Medium, RPMI 1640, sodium pyruvate solution, sodium bicarbonate, minimum Eagle's medium nonessential amino acids, Lipofectamine® RNAiMAX, Stealth Select RNAi® siRNA sets of each three oligonucleotides targeting human CYB5A, human CYB5B, and human MOSC1, and Stealth RNAi® siRNA negative controls were obtained from Invitrogen. FBS, trypsin, l-glutamine, penicillin/streptomycin, and PBS were purchased from PAA Laboratories GmbH (Pasching, Austria). ON-TARGETplus SMARTpool siRNA targeting human MOSC2 was purchased from Thermo Scientific. Complete protease inhibitor mixture was acquired from Roche Applied Science (Mannheim, Germany). All other chemicals were purchased from Sigma-Aldrich (Munich, Germany), Merck KGaA (Darmstadt, Germany), or Roth (Karlsruhe, Germany). Methanol HPLC grade was purchased from J.T. Baker (Deventer, Netherlands). Anti-MOSC2, anti-CYB5B, and anti-GAPDH antibodies were obtained from Sigma. Anti-MOSC1 antibody was purchased from Abgent (San Diego, CA), anti-CYB5A was from Santa Cruz Biotechnology (Santa Cruz, CA), and anti-calnexin was from Acris Antibodies GmbH (Herford, Germany).
Cell Culture
HEK-293 cells were maintained in Minimum Essential Medium supplemented with 10% FBS, 2 mm l-glutamine, 0.1 mm nonessential amino acids, 1 mm sodium pyruvate, 1.5 g/liter sodium bicarbonate, and 1% penicillin/streptomycin. ZR-75-1 cells were maintained in RPMI 1640 supplemented with 10% FBS, 2 mm l-glutamine, and 1 mm sodium pyruvate. Cell lines were incubated at 37 °C in 5% CO2.
siRNA Transfection and Design of Knockdown Experiments
HEK-293 or ZR-75-1 cells were forward transfected according to the manufacturer's transfection protocol from Invitrogen. Briefly, the cells were trypsinized, counted, and plated in antibiotic-free culture medium in 24-well plates 24 h before transfection so that cells reached 30–50% confluence at the time of transfection. For transfection, siRNA oligonucleotides were diluted in Opti-MEM and incubated with transfection reagent to form liposome-siRNA complexes, which were then added to the cells. To ensure that all observed silencing was due to a specific effect of siRNA on its target gene and not to off-target effects, at least three different siRNA sequences to the target gene were tested, and optimal concentrations of siRNA were evaluated. The siRNA-mediated inhibition at protein level also depends on the half-life of the protein. To optimize the time required for highest knockdown, effects were examined on different days after transfection. Negative controls included a nontargeting siRNA control (scrambled siRNA) and transfection reagents without siRNA. Down-regulation of protein expression was verified by Western blot analysis.
Determination of N-Reductive Activity in Human Cell Lines
N-Reductive activity in HEK-293 or ZR-75-1 was determined by measuring reduction of the model substrate benzamidoxime to benzamidine. For N-reduction studies, culture medium was removed, and cells were carefully washed and subsequently preincubated with substrate-free incubation buffer (Hanks' balanced salt solution containing 10 mm HEPES, pH 7.4) at 37 °C for 10 min. Then 250 μl of incubation buffer containing 5 mm benzamidoxime was added to each well simultaneously, and cells were incubated at 37 °C for 120 min. After the designated time, incubation medium was carefully removed and centrifuged to eliminate cellular contaminants and debris. The supernatant was analyzed by HPLC. Samples were separated isocratically on a Phenomenex® Gemini (150 × 4.6 mm) 5-μm C18 column with a Phenomenex C18 4 mm × 3.0-mm guard column (Phenomenex, Aschaffenburg, Germany). The mobile phase was composed of 50 mm ammonium acetate, pH 7.0, and 10% (v/v) methanol. Flow rate was kept at 1 ml/min, and detection was carried out at 229 nm. The retention times were 5.2 ± 0.1 min (benzamidine) and 12.0 ± 0.2 min (benzamidoxime).
Total Cellular Protein Extraction
To harvest cellular protein, the medium was removed, and cells were washed with ice-cold PBS and collected by centrifugation. After removing PBS, cells were resuspended in ice-cold lysis buffer (1% (v/v) Nonidet P-40, 150 mm NaCl, 50 mm Tris, pH 8.0, protease inhibitor mixture) and shaken for 60 min at 4 °C. Then cell lysates were cleared by centrifugation. The supernatant contained the total cellular protein extract.
Determination of Protein Concentrations
Protein concentrations were measured using the bicinchoninic acid (BCA) protein assay kit from Pierce according to the manufacturer's instructions.
Western Blot Analysis
Samples were mixed with Laemmli sample buffer (28) and incubated for 5 min at 100 °C. Same amounts of protein for each sample of cell lysate or subcellular fraction were loaded and separated by SDS-PAGE. After electrophoresis, the proteins were transferred onto Hybond-P PVDF membranes (GE Healthcare, Chalfont St Giles, UK). The membranes were then blocked in TBS containing Tween 20 (TBST) and 5% (m/v) milk powder, incubated with primary antibodies, and washed with TBST. Additionally, they were incubated with HRP-labeled goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, Suffolk, UK) at 4 °C overnight. After washing with TBST, the membranes were developed with the ECL Plus Western blotting detection system (GE Healthcare). The protein levels of calnexin or GAPDH were used as loading controls.
Preparation of Crude Subcellular Fractions of CYB5A Knock-out and Wild Type Mouse Liver
Homozygous microsomal cytochrome b5 complete knock-out mice (KO, CYB5A−/−) and control (wild type, CYB5A+/+) mice were generated as described previously (29). Mouse liver tissues were fractionated into homogenate, mitochondria, and microsomes by differential centrifugation. Briefly, crude mitochondria were sedimented by centrifugation at 6300 × g, and microsomes were separated from cytosol by centrifugation at 14,000 × g. All steps were performed in 10 mm phosphate buffer, pH 7.4, containing 0.25 m sucrose, 1 mm EDTA, 1 mm DTT, and protease inhibitor mixture at 4 °C.
Expression and Purification of Recombinant Human Proteins
Expression and purification of mARC1 (reference sequence NP_073583), mARC2 (reference sequence NP_060368), CYB5B (reference sequence NP_085056), and CYB5R isoform 2 (reference sequence NP_015565) were performed according to Wahl et al. (14). Recombinant CYB5A was purchased from MoBiTec (Göttingen, Germany).
Expression of CYB5B Variants
Variants c.5C>T (rs117949766), c.41A>G (rs79522540), c.46A>G (rs79729055), and c.64A>G (rs77374917) of CYB5B were generated by PCR mutagenesis using primers carrying the desired mutation. Accuracy of PCR mutagenesis was confirmed by DNA sequencing (GATC Biotech, Konstanz, Germany). The four protein variants (S2F, D14G, K16E, and T22A) of CYB5B were expressed in E. coli.
Determination of Cytochrome b5 Reductase FAD Content
The FAD content of human recombinant CYB5R was determined by absorption at 450 nm as described before (9).
Determination of Cytochrome b5 Heme Content
The heme content of CYB5 was estimated by recording the difference spectrum of oxidized and NADH-reduced protein (30).
Determination of Molybdenum Content in mARC
The molybdenum contents of human recombinant mARC proteins were determined by inductively coupled mass Spectrometry as described previously (9).
Preparation of Apo-Cytochrome b5
Apo-CYB5B was prepared by heme cleavage on the basis of Teale's methyl ethyl ketone method (31). Human recombinant CYB5B was diluted to ∼10 mg/ml protein with ice-cold distilled water. The enzyme solution was kept on ice during the whole procedure and adjusted to pH 2.5 by dropwise addition of 0.1 m HCl. An equal volume of ice-cold 2-butanone was added. The mixture was shaken for 30 s and allowed to stand at 0 °C for 1 min until separation of the upper red colored heme containing butanone phase and the colorless aqueous phase containing the apo-enzyme took place. The ketone phase was carefully removed. The buffer of the remaining aqueous solution was exchanged by ultrafiltration with Amicon Ultra-0.5 centrifugal filter devices (Merck Millipore, Billerica, MA) to a storage buffer (10 mm KH2PO4, 1 mm Na2EDTA, 0.1 mm DTT, 10% (m/v) glycerol). Loss of heme was confirmed by determination of CYB5 heme content.
Heme Incorporation into Apo-Cytochrome b5
The preparation of hemin chloride solution and its incorporation into apo-CYB5 were performed according to Mulrooney and Waskell (32). Briefly, solutions of heme were prepared by adding hemin chloride to a solution of 50% ethanol in water to give a final concentration of 1 mm. A small amount of 1 m NaOH was added and mixed to dissolve the hemin. The solution was then allowed to stand for 5 min so particles could settle. A 50-μl aliquot was removed and diluted into 950 μl of 20 mm Tris, 1 mm EDTA, pH 8.0, and the absorbance of the Tris-liganded heme was measured at 385 nm. The process was repeated until NaOH addition caused no further increase in absorbance at 385 nm. Apo-CYB5 prepared as above was diluted with 20 mm Tris, pH 8.0, containing 1 mm EDTA and 0.4% (m/v) sodium cholate, to yield a protein concentration of about 2 mg/ml. Aliquots of hemin chloride solution were added into the apo-CYB5 sample, and the reconstitution of apo-CYB5 with heme was monitored by recording absorbance spectra from 350 to 500 nm. The reconstitution was considered to be complete when the Soret peak at 413 nm of CYB5 turned into a plateau, caused by excess of free Tris-liganded hemin. A red-colored protein was obtained, which also indicated the binding of heme. The final heme content of the reconstituted CYB5 was determined as described above.
In Vitro N-Reduction Assays
N-Reductive activities of the in vitro reconstituted mARC-containing enzyme system with recombinant enzymes or subcellular fractions were determined by measuring reduction of the model substrate benzamidoxime to benzamidine. Incubations were carried out as described previously with slight modifications (9). Incubation mixtures contained 3 mm substrate, 1 mm NADH in a total volume of 150 μl of 20 mm MES buffer, pH 6.0 (in the case of recombinant proteins) or 100 mm phosphate buffer, pH 6.0 (in the case of subcellular fractions). The recombinant enzymes were applied according to their cofactors. Unless otherwise stated, the incubation mix contained 15 pmol of FAD of CYB5R, 150 pmol of heme of CYB5, and 150 pmol of Moco of mARC. Incubation mixtures with subcellular fractions contained 0.05 mg of total protein. Samples were analyzed by HPLC as described by Kotthaus et al. (9).
Data Analysis
Statistical analyses were carried out using the SigmaPlot 11 software (Systat Software Inc.). The significance of observed differences was evaluated by Student's unpaired t test or for multiple comparison by Bonferroni's test. A probability less than 5% was considered to be significant. All experimental values are given as means ± S.D.
RESULTS
Involvement of Human mARC1 and mARC2 in N-Reduction
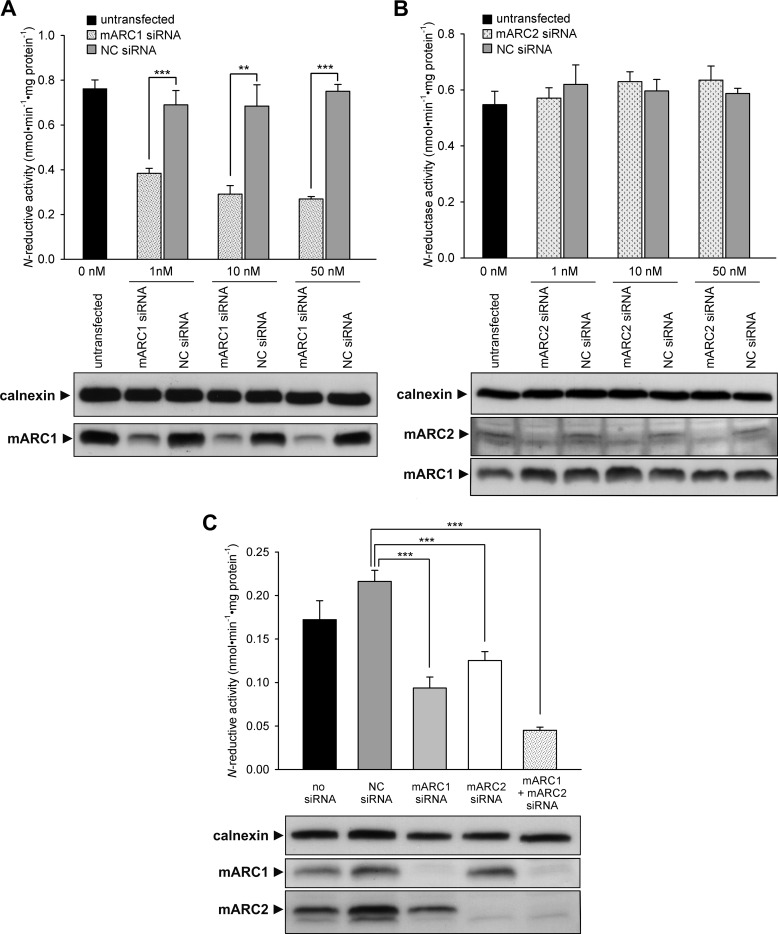
As already shown in our previous in vitro reconstitution assays, both recombinant human mARC proteins are capable of reducing N-hydroxylated substrates (7, 14). To find out whether this is also the case in living cells, siRNA-mediated mARC knockdown studies were carried out in human cells. Efficiency of protein down-regulation was verified by Western blot analysis (Fig. 2). Loss of protein as compared with nontargeting siRNA transfected cells or untransfected cells was shown in a dose- and time-dependent manner. The knockdown of one homolog did not affect the protein level of the other homolog, and maximal effect was seen between 3 and 5 days after transfection (Fig. 2, time dependence data not shown). mARC1 protein expression was almost completely recovered between days 8 and 11 after siRNA transfection. In HEK-293 cells, down-regulation of mARC1 resulted in an enormous reduction of N-reductive activity to about 35% (Fig. 2A). After recovery of mARC1 protein, N-reductive capacity increased simultaneously (data not shown). In contrast, knockdown of mARC2 in HEK-293 cells did not have any influence on N-reduction (Fig. 2B). However, mARC2 expression in HEK-293 cells seems to be quite low because protein can hardly be detected by Western blotting. To examine whether the missing effect of mARC2 knockdown on N-reductive activity is a result of low mARC2 expression or whether mARC2 is simply not involved in N-reduction in cell metabolism, ZR-75-1, a cell line with nearly equal mARC1 and mARC2 expression (33), was additionally tested. In our RNAi experiments, down-regulation of both mARC1 and mARC2 in ZR-75-1 resulted in a moderate but significant decrease of N-reductive activity. As a consequence, simultaneous knockdown of both homologs revealed an additive reductive effect on activity (Fig. 2C).
FIGURE 2.
mARC1 and mARC2 knockdown in human cells. A, mARC1 knockdown in HEK-293. B, mARC2 knockdown in HEK-293. C, mARC1 and mARC2 knockdown in ZR-75-1. For HEK-293, cells were transfected with various amounts of mARC1, mARC2, or nontargeting (NC) siRNA. For ZR-75-1, cells were transfected with 20 nm of NC, mARC1, or mARC1 siRNA. For simultaneous knockdown, 10 nm mARC1 and 10 nm mARC2 siRNA were transfected. N-Reductive activities in cells were determined as described under “Experimental Procedures” on day 5 (for HEK-293) or day 6 (for ZR-75-1) after transfection. Results are presented as means ± S.D. (n = 3). **, p < 0.01, ***, p < 0.001. The siRNA-mediated down-regulations of the proteins of interest were verified by Western blot using anti-mARC1, anti-mARC2, or anti-calnexin (loading control) antibody.
Participation of Mitochondrial Cytochrome b5 in the N-Reductive System
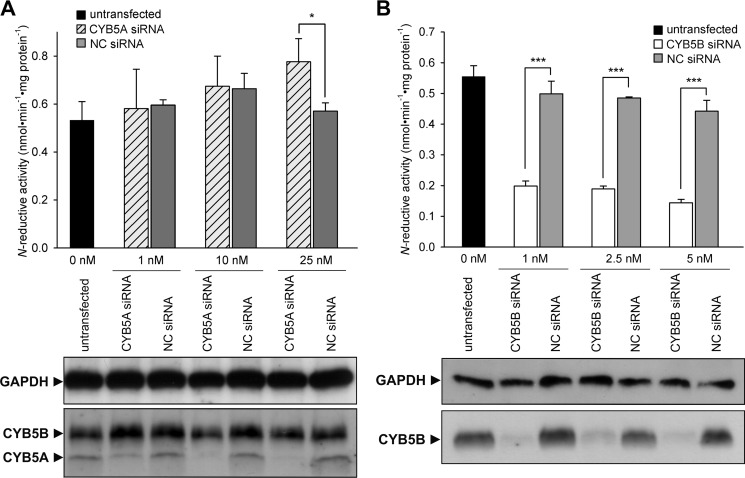
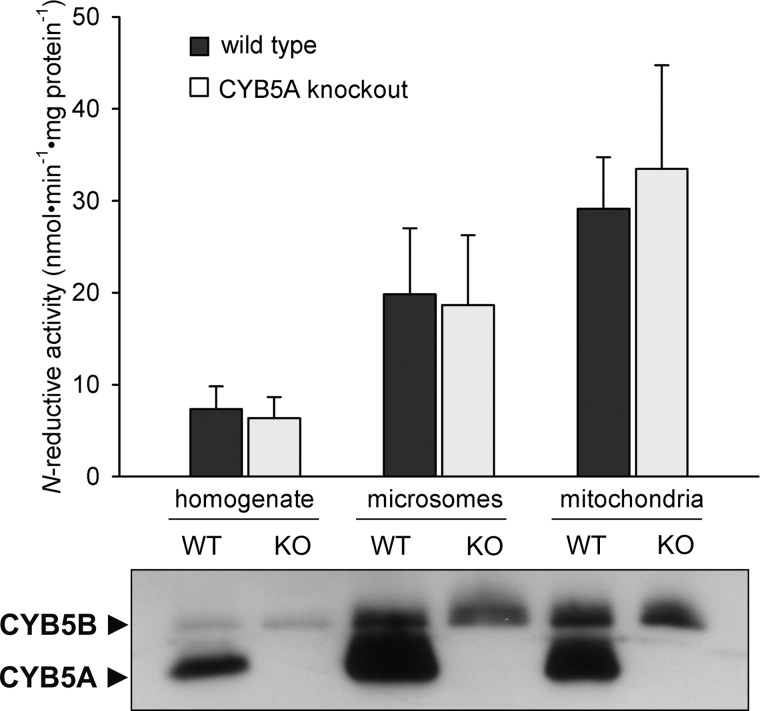
In vitro, the human N-reductive system can be reconstituted with both the microsomal and the mitochondrial isoform of CYB5 with nearly equal activity (data not shown). To investigate whether both or only one isoform is involved in N-reduction in cell metabolism, each CYB5 protein was down-regulated individually in HEK-293 cells by siRNA transfection in a dose- and time-dependent manner. Success of down-regulation as compared with nontargeting siRNA transfected cells or untransfected cells was shown by Western blot analysis (Fig. 3). Knockdown of one isoform did not affect the protein level of the other isoform, and maximal effect was seen between 3 and 5 days after transfection (Fig. 3, time dependence data not shown). Knockdown of microsomal CYB5A did not have any significant inhibitory effect on N-reductive activity or even a slightly stimulating effect with higher siRNA concentration (Fig. 3A). In contrast, down-regulation of mitochondrial CYB5B resulted in a tremendous decrease of N-reductive activity to about 35% as compared with negative control (Fig. 3B). In addition to our knockdown studies, subcellular fractions of CYB5A knock-out mouse livers were examined regarding their N-reductive activity. Western blot data confirmed the absence of microsomal CYB5A protein in knock-out samples, whereas expression of the mitochondrial isoform CYB5B was not affected by the CYB5A knock-out (Fig. 4). Due to cross-contamination, CYB5B was also detected in the crude microsomal fraction of wild type and knock-out mice, and CYB5A was detected in the crude mitochondrial fraction of wild type mice. Reductase activity was enriched in the mitochondrial fraction of wild type and knock-out mouse livers. Supporting the results found in CYB5A knockdown experiments, no significant difference in N-reductive activity was observed between wild type and CYB5A knock-out samples (Fig. 4).
FIGURE 3.
CYB5A and CYB5B knockdown in HEK-293 cells. A, CYB5A knockdown. B, CYB5B knockdown. Cells were transfected with various amounts of CYB5A, CYB5B, or nontargeting (NC) siRNA. N-Reductive activities in HEK-293 cells were determined as described under “Experimental Procedures” on day 5 after transfection. Results are presented as means ± S.D. (n = 3). *, p < 0.05, ***, p < 0.001. The siRNA-mediated down-regulations of the proteins of interest were verified by Western blot using anti-CYB5A, anti-CYB5B, or anti-GAPDH (loading control) antibody.
FIGURE 4.
N-Reductive activities of subcellular liver fractions of CYB5A knock-out mice. N-Reductive activities of subcellular fractions of wild type and CYB5A knock-out mouse livers were determined as described under “Experimental Procedures.” Assays were performed on a pool of two liver preparations and one individual liver sample of wild type or knock-out mice (n = 3). Incubations were performed in duplicates. Results are presented as means ± S.D. The absence of CYB5A protein in the individual knock-out samples was verified by Western blot analysis using anti-CYB5A antibody.
Functional Analysis of Cytochrome b5 Type B SNP Variants
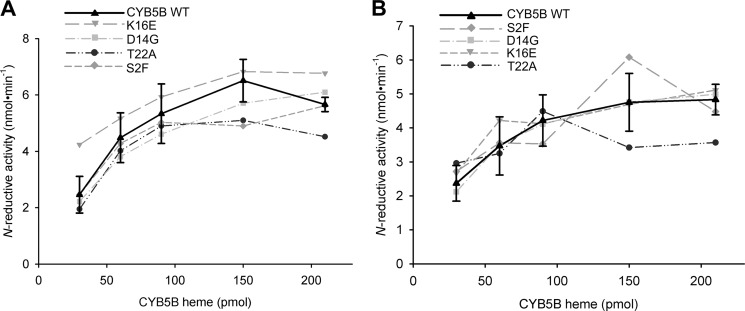
The corresponding protein variants (S2F, D14G, K16E, and T22A) of published SNPs in dbSNP build 132 were successfully expressed in E. coli. Expression levels and heme contents of CYB5B variants were comparable with those of wild type (data not shown). N-Reductive activities of variants and wild type were determined in incubations that contained different concentrations of heme (30–210 pmol). In the incubation mixture, when CYB5B proteins were used in excess over CYB5R, maximal activities were always achieved by all variants. Moreover, after reconstituting the enzyme system with the various CYB5B variants, N-reductive activities of each complete system were found to lie in the activity range of wild type CYB5B (Fig. 5).
FIGURE 5.
Reduction of benzamidoxime using cytochrome b5 type B wild type and variants. A, assay carried out with mARC1. B, assay carried out with mARC2. 120 pmol of molybdenum (mARC), 30–210 pmol of heme (CYB5B), and 6 pmol of FAD (CYB5R) were incubated with 3 mm benzamidoxime in 20 mm MES, pH 6.0, for 15 min under aerobic conditions. Data are means ± S.D. of one batch (CYB5B variants) or of four batches (CYB5B WT) each incubated in duplicates and measured twice by HPLC.
Involvement of Cytochrome b5 Heme in the N-Reductive System
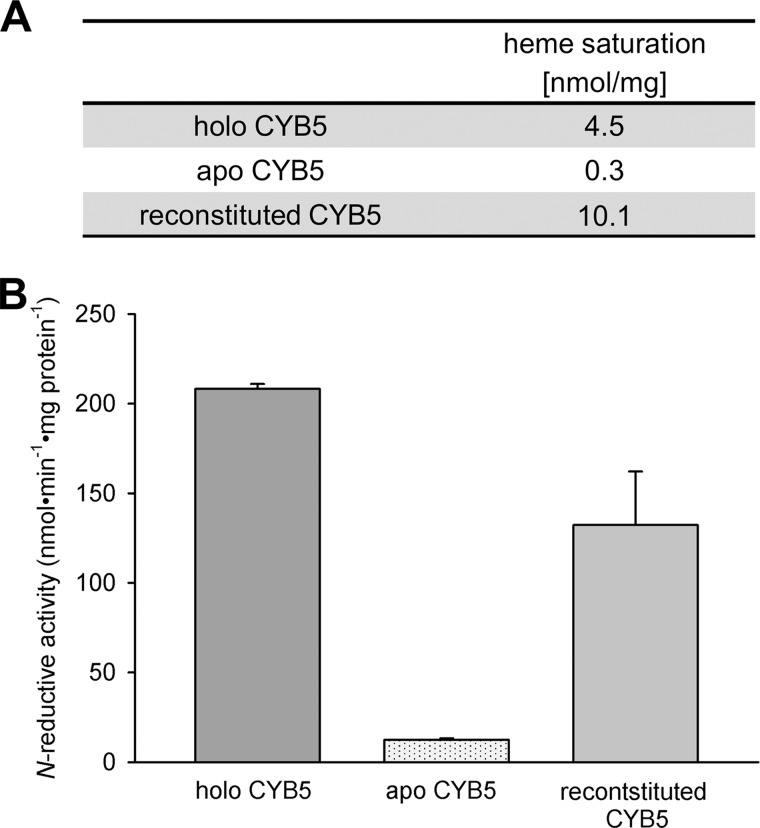
To explore the mechanism by which CYB5 takes part in the N-reductive system, we examined N-reductive activity of the reconstituted three-component enzyme system completed with either holo-CYB5B or apo-CYB5B. For this purpose, we prepared apo-CYB5 from recombinantly expressed holo-CYB5 by heme removal with the 2-butanone method (see “Experimental Procedures”). To prove the functionality of apo-CYB5 after butanone treatment, we afterward reconstituted the protein by heme incorporation. Heme saturation of apo-, holo-, and heme-reincorporated CYB5 was measured to ensure the loss and the regain of CYB5 heme (Fig. 6A). Subsequently, activities of the N-reductive system reconstituted with either form of CYB5 (holo-, apo-, and reconstituted CYB5) were determined. As shown in Fig. 6B, N-reductive activity was obtained with holo-CYB5 but was almost totally lost with apo-CYB5. The remaining activity can be attributed to a small fraction of heme-loaded CYB5 left in the apo-CYB5 preparation. By reinsertion of heme into CYB5, activity could be recovered, supporting the necessity of CYB5-bound heme for N-reductive activity.
FIGURE 6.
Involvement of cytochrome b5 heme in N-reduction. Recombinant CYB5 (holo-CYB5) was gained as described under “Experimental Procedures.” Apo-CYB5 was prepared from holo-CYB5 by heme cleavage, and afterward the same batch was reconstituted by heme incorporation. The human recombinant N-reductive enzyme system was then reconstituted with mARC1, CYB5R, and either form of CYB5. N-Reductive activities were measured with benzamidoxime as a model substrate. A, heme saturations of holo-CYB5, apo-CYB5, and reconstituted CYB5 were determined to confirm the success of heme cleavage and heme incorporation. B, N-reductive activities of holo-CYB5, apo-CYB5, and reconstituted CYB5 were determined as described under “Experimental Procedures.” The incubation mix contained 75 pmol of mARC, 7.5 pmol of CYB5R, and 75 pmol of CYB5 in the case of holo and reconstituted CYB5 or the same amount of protein as holo-CYB5 in the case of apo-CYB5. Incubations were performed in duplicates. Results are presented as means ± S.D.
DISCUSSION
The mARC-containing three-component enzyme system is responsible for reduction of several N-hydroxylated compounds and therefore plays a major role in N-reductive drug metabolism (1). In previous in vitro assays, we have shown that both human mARC proteins are capable of reducing N-hydroxylated substrates when reconstituted with the electron transport proteins CYB5 and CYB5R (6, 9, 14). Omitting one component in the incubation mixture leads to a nearly complete loss of activity (6, 9, 14). In vitro, both mARC proteins act on the same N-hydroxylated substrates (1). Nevertheless, for some substrates, differences in specific activities can be seen. The fact that all mammalian genomes analyzed up to now harbor two mARC genes suggests the evolutionary need of each protein. However, it still remains to be shown whether differences exist between mARC1 and mARC2 in vivo, for instance with respect to tissue-specific localizations, subcellular localization, or substrate specificity, respectively. Besides its mitochondrial localization, a peroxisomal co-localization for mARC2 has been suggested (17, 18). Thus, functions of mARC outside the described mitochondrial N-reductive system are also conceivable. To examine whether both or only one mARC protein is involved in N-reductive pathway in vivo, RNAi studies with human cell lines were performed. Our investigations demonstrate that in HEK-293, hardly any mARC2 protein can be detected, whereas mARC1 protein expression is considerably higher. In support of this, database researches (33) indicate that this trend is reflected in mRNA levels in almost all human cell lines and tissues listed in BioGPS (gene numbers 64757 and 54996). In accordance with the observed expression levels of each mARC protein, siRNA-mediated knockdown of mARC1 in HEK-293 cells lowers N-reductive activity tremendously, whereas knockdown of mARC2 does not affect N-reduction (Fig. 2). In contrast to HEK-293 cells, protein expression levels of mARC1 and mARC2 in ZR-75-1 cells are nearly the same. In fact, in this human cell line, N-reductive activity is decreased by both mARC1 and mARC2 knockdown, with the lowest activity being seen when knocking down both isoforms simultaneously. These results prove for the first time that not only in vitro but also in cell metabolism mARC1 and mARC2 have the capacity to reduce N-hydroxylated substrates with the extent of involvement in N-reduction being highly dependent on the expression level of the particular mARC isoform. In contrast to the present study, other siRNA-mediated knockdown studies in murine cells could only prove the involvement of mARC2 in N-reduction (11). This discrepancy is likely to be attributed to the variance of examined species. In contrast to human cells and tissues, for almost all murine cell lines, including the examined cell line of Neve et al. (11), high gene expression levels for mARC2 and extremely low gene expression levels for mARC1 are described (33). Therefore, the missing influence of mARC1 on N-reductive activity in murine cells is supposedly due to the small protein expression of mARC1 as compared with mARC2. These observations indicate that depending on the species, usually only one isoform, either mARC1 or mARC2, is predominantly expressed. Consequently, the mARC protein that is predominantly responsible for the N-reductive pathway varies from species to species. This hypothesis is supported by the finding that independent from the investigated species, N-reductive activity is consistently found to be enriched in the OMM (2, 9, 11, 34), but up to now, either mARC1 or mARC2 protein and never both proteins could be attributed to the OMM. For instance, in pig and rat mARC2, but no mARC1 was found in the OMM (2, 11), whereas human OMM was demonstrated to contain mARC1 rather than mARC2 (16). The physiological reasons for the existence of two mARC proteins and why one mARC protein seems to be subordinated to the other remains to be shown. However, mARC2 was reported to be down-regulated in human colon tumors (8) and up-regulated by glucose treatment in human cells (10). Furthermore, ZR-75-1, a human breast cancer cell line, is an exceptional example for a human cell line with equal mARC1 and mARC2 protein expression levels. It thus appears to be conceivable that the expression of the subordinated mARC protein is regulated by special circumstances, whereas expression of the respective other isoform remains constant, eventually to fulfill housekeeping functions.
By various enzyme purification studies and reconstitution assays (2, 14, 35–37), the heme protein CYB5 is known to be part of the N-reductive metabolic pathway. There are two membrane-bound isoforms of CYB5, the microsomal (CYB5A) and the mitochondrial (CYB5B) isoform, which both derive from two different genes (19, 20). CYB5A and CYB5B show high sequence and structural similarities (20, 38). Therefore, it is not surprising that in vitro, both recombinant proteins are able to complete the reconstituted three-component enzyme system. To investigate which isoform is involved in N-reduction in vivo, RNAi experiments were carried out. Our siRNA-mediated knockdown studies in human cells show that only the mitochondrial isoform is part of the N-reductive system in cell metabolism (Fig. 3) because knockdown of the microsomal isoform does not affect N-reduction. These results are in accordance with the results of RNAi studies in murine cells obtained by Neve et al. (11). In addition, subcellular fractions of livers from CYB5A knock-out mouse livers were examined regarding their N-reductive activity. Supporting the results found with CYB5A knockdown, no significant difference in activity is seen between wild type fractions and CYB5A knock-out fractions (Fig. 4). These results undoubtedly demonstrate that only the mitochondrial, but not the microsomal, isoform of CYB5 is required for N-reduction. Therefore, the mitochondrial isoform represents the physiological part of the mARC-containing enzyme system. As CYB5B is exclusively associated with the OMM (11, 39) and N-reductive activity is found to be enriched in this compartment (2, 9, 16, 34, 40), a microsomal N-reductive system (21–24) is highly unlikely.
In continuation of our studies, we examined whether nonsynonymous SNPs of CYB5B have a functional effect on N-reductive activity. At the beginning of our studies, allele frequencies of nonsynonymous SNPs were reviewed in dbSNP. The dbSNP (build 132) published four nonsynonymous SNPs in CYB5B. The corresponding recombinant protein variants (S2F, D14G, K16E, and T22A) were successfully expressed in E. coli, and each variant was able to complete the N-reductive enzyme system. Our results suggest that the tested amino acid substitutions do not affect the function of CYB5B in the N-reductive pathway. However, a recent release of dbSNP (build 137) published three additional nonsynonymous SNPs, which in principle cannot be excluded to affect N-reduction (www.ncbi.nlm.nih.gov/snp). However, the protein variant encoded by the SNP with the highest frequency of the variant allele (c.5C>T) was already included in our investigation. These findings therefore suggest that SNPs in CYB5B are allowed to establish in populations as long as they do not affect important enzymatic or structural properties.
In vitro reconstitution studies with human apo-mARCs confirmed that N-reductive activity is clearly dependent on Moco (14). Likewise, we examined the reductase activity of the human enzyme system reconstituted with either holo-CYB5B or apo-CYB5B (Fig. 6). The removal of heme with the 2-butanone method results in a nearly total loss of N-reductive activity. To prove functionality of apo-CYB5 after butanone treatment, we afterward reconstituted the protein by heme reincorporation. By reinsertion of heme, the recovered holo-CYB5B is able to complete N-reductive activity of the mARC-containing enzyme system. Based on these results, it is reasonable to state that the heme of CYB5B is necessary for catalysis, and therefore, heme of CYB5B is likely to be involved in electron transfer within the N-reductive system.
Characterization of the involvement of CYB5R, the third component of the N-reductive system, remains the subject of further investigations. As for mARC and CYB5B, RNAi experiments and studies on cofactor involvement should be performed analogously for a more detailed understanding of the functional and mechanistic properties of this protein within the N-reductive three-component system.
In this study, we demonstrate that the mARC-containing enzyme system is responsible for the N-reduction of amidoximes in whole cell metabolism. mARC1 and/or mARC2 and CYB5B could be identified as essential components of the N-reductive system in cells. These results confirm that the N-reductive system plays a major role in prodrug activation. Amidines and guanidines are very often elements of new drug candidates as the cations formed after protonation interact with carboxylates of the target protein. However, these functional groups have negative pharmacokinetic properties. Replacements of amidines and guanidines usually result in a high loss of affinity. Using the prodrug principle amidoximes instead of amidines allows the maintenance of the amidine function and oral bioavailability at the same time. The prodrug activation is not dependent on P450 enzymes. Thus, P450-dependent interactions are avoided. Additionally, the rapid reduction of N-oxygenated compounds, which often have toxic properties, to the corresponding nitrogen functions by the N-reductive system provides an effective way of detoxication. Accordingly, a detailed understanding of the mARC-mediated prodrug activation and detoxication of N-oxygenated compounds is of high relevance for further prodrug developments and for the understanding of detoxication pathways.
Acknowledgments
We thank Petra Köster, Ulrike Westernströer, and Sven Wichmann for technical assistance.
This work was supported by Deutsche Forschungsgemeinschaft (DFG) Grants CL56/9-1 and ME 1266/24-1.
- mARC
- mitochondrial amidoxime reducing component
- CYB5
- cytochrome b5
- CYB5R
- NADH-cytochrome b5 reductase
- Moco
- molybdenum cofactor
- OMM
- outer mitochondrial membrane
- P450
- cytochrome P450
- TBST
- TBS containing Tween 20.
REFERENCES
- 1. Havemeyer A., Lang J., Clement B. (2011) The fourth mammalian molybdenum enzyme mARC: current state of research. Drug Metab. Rev. 43, 524–539 [DOI] [PubMed] [Google Scholar]
- 2. Havemeyer A., Bittner F., Wollers S., Mendel R., Kunze T., Clement B. (2006) Identification of the missing component in the mitochondrial benzamidoxime prodrug-converting system as a novel molybdenum enzyme. J. Biol. Chem. 281, 34796–34802 [DOI] [PubMed] [Google Scholar]
- 3. Hille R., Nishino T., Bittner F. (2011) Molybdenum enzymes in higher organisms. Coord Chem. Rev. 255, 1179–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gruenewald S., Wahl B., Bittner F., Hungeling H., Kanzow S., Kotthaus J., Schwering U., Mendel R. R., Clement B. (2008) The fourth molybdenum containing enzyme mARC: cloning and involvement in the activation of N-hydroxylated prodrugs. J. Med. Chem. 51, 8173–8177 [DOI] [PubMed] [Google Scholar]
- 5. Havemeyer A., Grünewald S., Wahl B., Bittner F., Mendel R., Erdélyi P., Fischer J., Clement B. (2010) Reduction of N-hydroxy-sulfonamides, including N-hydroxy-valdecoxib, by the molybdenum-containing enzyme mARC. Drug Metab. Dispos. 38, 1917–1921 [DOI] [PubMed] [Google Scholar]
- 6. Froriep D., Clement B., Bittner F., Mendel R. R., Reichmann D., Schmalix W., Havemeyer A. (February 4, 2013) Activation of the anti-cancer agent upamostat by the mARC enzyme system. Xenobiotica 10.3109/00498254.2013.767481 [DOI] [PubMed] [Google Scholar]
- 7. Krompholz N., Krischkowski C., Reichmann D., Garbe-Schönberg D., Mendel R. R., Bittner F., Clement B., Havemeyer A. (2012) The mitochondrial amidoxime reducing component (mARC) is involved in detoxification of N-hydroxylated base analogues. Chem. Res. Toxicol. 25, 2443–2450 [DOI] [PubMed] [Google Scholar]
- 8. Mikula M., Rubel T., Karczmarski J., Goryca K., Dadlez M., Ostrowski J. (2011) Integrating proteomic and transcriptomic high-throughput surveys for search of new biomarkers of colon tumors. Funct. Integr. Genomics 11, 215–224 [DOI] [PubMed] [Google Scholar]
- 9. Kotthaus J., Wahl B., Havemeyer A., Kotthaus J., Schade D., Garbe-Schönberg D., Mendel R., Bittner F., Clement B. (2011) Reduction of N(ω)-hydroxy-l-arginine by the mitochondrial amidoxime reducing component (mARC). Biochem. J. 433, 383–391 [DOI] [PubMed] [Google Scholar]
- 10. Malik A. N., Rossios C., Al-Kafaji G., Shah A., Page R. A. (2007) Glucose regulation of CDK7, a putative thiol related gene, in experimental diabetic nephropathy. Biochem. Biophys. Res. Commun. 357, 237–244 [DOI] [PubMed] [Google Scholar]
- 11. Neve E. P. A., Nordling A., Andersson T. B., Hellman U., Diczfalusy U., Johansson I., Ingelman-Sundberg M. (2012) Amidoxime reductase system containing cytochrome b5 type B (CYB5B) and MOSC2 is of importance for lipid synthesis in adipocyte mitochondria. J. Biol. Chem. 287, 6307–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aslibekyan S., Goodarzi M. O., Frazier-Wood A. C., Yan X., Irvin M. R., Kim E., Tiwari H. K., Guo X., Straka R. J., Taylor K. D., Tsai M. Y., Hopkins P. N., Korenman S. G., Borecki I. B., Chen Y.-D. I., Ordovas J. M., Rotter J. I., Arnett D. K. (2012) Variants identified in a GWAS meta-analysis for blood lipids are associated with the lipid response to fenofibrate. PLoS ONE 7, e48663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., Johansen C. T., Fouchier S. W., Isaacs A., Peloso G. M., Barbalic M., Ricketts S. L., Bis J. C., Aulchenko Y. S., Thorleifsson G., Feitosa M. F., Chambers J., Orho-Melander M., Melander O., Johnson T., Li X., Guo X., Li M., Shin Cho Y., Jin Go M., Jin Kim Y., Lee J.-Y., Park T., Kim K., Sim X., Twee-Hee Ong R., Croteau-Chonka D. C., Lange L. A., Smith J. D., Song K., Hua Zhao J., Yuan X., Luan J., Lamina C., Ziegler A., Zhang W., Zee R. Y., Wright A. F., Witteman J. C., Wilson J. F., Willemsen G., Wichmann H.-E., Whitfield J. B., Waterworth D. M., Wareham N. J., Waeber G., Vollenweider P., Voight B. F., Vitart V., Uitterlinden A. G., Uda M., Tuomilehto J., Thompson J. R., Tanaka T., Surakka I., Stringham H. M., Spector T. D., Soranzo N., Smit J. H., Sinisalo J., Silander K., Sijbrands E. J., Scuteri A., Scott J., Schlessinger D., Sanna S., Salomaa V., Saharinen J., Sabatti C., Ruokonen A., Rudan I., Rose L. M., Roberts R., Rieder M., Psaty B. M., Pramstaller P. P., Pichler I., Perola M., Penninx B. W., Pedersen N. L., Pattaro C., Parker A. N., Pare G., Oostra B. A., O'Donnell C. J., Nieminen M. S., Nickerson D. A., Montgomery G. W., Meitinger T., McPherson R., McCarthy M. I., McArdle W., Masson D., Martin N. G., Marroni F., Mangino M., Magnusson P. K., Lucas G., Luben R., Loos R. J., Lokki M.-L., Lettre G., Langenberg C., Launer L. J., Lakatta E. G., Laaksonen R., Kyvik K. O., Kronenberg F., König I. R., Khaw K.-T., Kaprio J., Kaplan L. M., Johansson A., Jarvelin M.-R., Janssens A. C., Ingelsson E., Igl W., Kees Hovingh G., Hottenga J.-J., Hofman A., Hicks A. A., Hengstenberg C., Heid I. M., Hayward C., Havulinna A. S., Hastie N. D., Harris T. B., Haritunians T., Hall A. S., Gyllensten U., Guiducci C., Groop L. C., Gonzalez E., Gieger C., Freimer N. B., Ferrucci L., Erdmann J., Elliott P., Ejebe K. G., Döring A., Dominiczak A. F., Demissie S., Deloukas P., de Geus E. J., de Faire U., Crawford G., Collins F. S., Chen Y.-D. I., Caulfield M. J., Campbell H., Burtt N. P., Bonnycastle L. L., Boomsma D. I., Boekholdt S. M., Bergman R. N., Barroso I., Bandinelli S., Ballantyne C. M., Assimes T. L., Quertermous T., Altshuler D., Seielstad M., Wong T. Y., Tai E.-S., Feranil A. B., Kuzawa C. W., Adair L. S., Taylor H. A., Borecki I. B., Gabriel S. B., Wilson J. G., Holm H., Thorsteinsdottir U., Gudnason V., Krauss R. M., Mohlke K. L., Ordovas J. M., Munroe P. B., Kooner J. S., Tall A. R., Hegele R. A., Kastelein J. J., Schadt E. E., Rotter J. I., Boerwinkle E., Strachan D. P., Mooser V., Stefansson K., Reilly M. P., Samani N. J., Schunkert H., Cupples L. A., Sandhu M. S., Ridker P. M., Rader D. J., van Duijn C. M., Peltonen L., Abecasis G. R., Boehnke M., Kathiresan S. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wahl B., Reichmann D., Niks D., Krompholz N., Havemeyer A., Clement B., Messerschmidt T., Rothkegel M., Biester H., Hille R., Mendel R. R., Bittner F., (2010) Biochemical and spectroscopic characterization of the human mitochondrial amidoxime reducing components hmARC-1 and hmARC-2 suggests the existence of a new molybdenum enzyme family in eukaryotes. J. Biol. Chem. 285, 37847–37859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Da Cruz S., Xenarios I., Langridge J., Vilbois F., Parone P. A., Martinou J.-C. (2003) Proteomic analysis of the mouse liver mitochondrial inner membrane. J. Biol. Chem. 278, 41566–41571 [DOI] [PubMed] [Google Scholar]
- 16. Klein J. M., Busch J. D., Potting C., Baker M. J., Langer T., Schwarz G. (2012) The mitochondrial amidoxime-reducing component (mARC1) is a novel signal-anchored protein of the outer mitochondrial membrane. J. Biol. Chem. 287, 42795–42803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Islinger M., Lüers G. H., Li K. W., Loos M., Völkl A. (2007) Rat liver peroxisomes after fibrate treatment: a survey using quantitative mass spectrometry. J. Biol. Chem. 282, 23055–23069 [DOI] [PubMed] [Google Scholar]
- 18. Wiese S., Gronemeyer T., Ofman R., Kunze M., Grou C. P., Almeida J. A., Eisenacher M., Stephan C., Hayen H., Schollenberger L., Korosec T., Waterham H. R., Schliebs W., Erdmann R., Berger J., Meyer H. E., Just W., Azevedo J. E., Wanders R. J., Warscheid B. (2007) Proteomics characterization of mouse kidney peroxisomes by tandem mass spectrometry and protein correlation profiling. Mol. Cell Proteomics 6, 2045–2057 [DOI] [PubMed] [Google Scholar]
- 19. Borgese N., D'Arrigo A., De Silvestris M., Pietrini G. (1993) NADH-cytochrome b5 reductase and cytochrome b5 isoforms as models for the study of post-translational targeting to the endoplasmic reticulum. FEBS Lett. 325, 70–75 [DOI] [PubMed] [Google Scholar]
- 20. Altuve A., Silchenko S., Lee K. H., Kuczera K., Terzyan S., Zhang X., Benson D. R., Rivera M. (2001) Probing the differences between rat liver outer mitochondrial membrane cytochrome b5 and microsomal cytochromes b5. Biochemistry 40, 9469–9483 [DOI] [PubMed] [Google Scholar]
- 21. Kurian J. R., Longlais B. J., Trepanier L. A. (2007) Discovery and characterization of a cytochrome b5 variant in humans with impaired hydroxylamine reduction capacity. Pharmacogenet. Genomics 17, 597–603 [DOI] [PubMed] [Google Scholar]
- 22. Sacco J. C., Trepanier L. A. (2010) cytochrome b5 and NADH cytochrome b5 reductase: genotype-phenotype correlations for hydroxylamine reduction. Pharmacogenet. Genomics 20, 26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rhoads K., Sacco J. C., Drescher N., Wong A., Trepanier L. A. (2011) Individual variability in the detoxification of carcinogenic arylhydroxylamines in human breast. Toxicol. Sci. 121, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sacco J. C., Abouraya M., Motsinger-Reif A., Yale S. H., McCarty C. A., Trepanier L. A. (2012) Evaluation of polymorphisms in the sulfonamide detoxification genes NAT2, CYB5A, and CYB5R3 in patients with sulfonamide hypersensitivity. Pharmacogenet. Genomics 22, 733–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Auchus R. J., Lee T. C., Miller W. L. (1998) cytochrome b5 augments the 17,20-lyase activity of human P450c17 without direct electron transfer. J. Biol. Chem. 273, 3158–3165 [DOI] [PubMed] [Google Scholar]
- 26. Yamazaki H., Shimada T., Martin M. V., Guengerich F. P. (2001) Stimulation of cytochrome P450 reactions by apo-cytochrome b5: evidence against transfer of heme from cytochrome P450 3A4 to apo-cytochrome b5 or heme oxygenase. J. Biol. Chem. 276, 30885–30891 [DOI] [PubMed] [Google Scholar]
- 27. Kotrbová V., Aimová D., Ingr M., Borek-Dohalská L., Martínek V., Stiborová M. (2009) Preparation of a biologically active apo-cytochrome b5 via heterologous expression in Escherichia coli. Protein Expr. Purif. 66, 203–209 [DOI] [PubMed] [Google Scholar]
- 28. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 29. McLaughlin L. A., Ronseaux S., Finn R. D., Henderson C. J., Roland Wolf C. (2010) Deletion of microsomal cytochrome b5 profoundly affects hepatic and extrahepatic drug metabolism. Mol. Pharmacol. 78, 269–278 [DOI] [PubMed] [Google Scholar]
- 30. Estabrook R. W., Werringloer J. (1978) The measurement of difference spectra: application to the cytochromes of microsomes. Methods Enzymol. 52, 212–220 [DOI] [PubMed] [Google Scholar]
- 31. Teale F. W. (1959) Cleavage of the haem-protein link by acid methylethylketone. Biochim. Biophys. Acta 35, 543. [DOI] [PubMed] [Google Scholar]
- 32. Mulrooney S. B., Waskell L. (2000) High-level expression in Escherichia coli and purification of the membrane-bound form of cytochrome b5. Protein Expr. Purif. 19, 173–178 [DOI] [PubMed] [Google Scholar]
- 33. Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C. L., Haase J., Janes J., Huss J. W., 3rd, Su A. I. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Andersson S., Hofmann Y., Nordling A., Li X. Q., Nivelius S., Andersson T. B., Ingelman-Sundberg M., Johansson I. (2005) Characterization and partial purification of the rat and human enzyme systems active in the reduction of N-hydroxymelagatran and benzamidoxime. Drug Metab. Dispos. 33, 570–578 [DOI] [PubMed] [Google Scholar]
- 35. Bernheim M. L. (1972) The reduction of hydroxylamine and some aryl hydroxamates by liver mitochondria from mammals and birds. Enzymologia. 43, 167–176 [PubMed] [Google Scholar]
- 36. Kadlubar F. F., Ziegler D. M. (1974) Properties of a NADH-dependent N-hydroxy amine reductase isolated from pig liver microsomes. Arch. Biochem. Biophys. 162, 83–92 [DOI] [PubMed] [Google Scholar]
- 37. Kurian J. R., Bajad S. U., Miller J. L., Chin N. A., Trepanier L. A. (2004) NADH cytochrome b5 reductase and cytochrome b5 catalyze the microsomal reduction of xenobiotic hydroxylamines and amidoximes in humans. J. Pharmacol. Exp. Ther. 311, 1171–1178 [DOI] [PubMed] [Google Scholar]
- 38. Rodríguez-Marañón M. J., Qiu F., Stark R. E., White S. P., Zhang X., Foundling S. I., Rodríguez V., Schilling C. L., 3rd, Bunce R. A., Rivera M. (1996) 13C NMR spectroscopic and X-ray crystallographic study of the role played by mitochondrial cytochrome b5 heme propionates in the electrostatic binding to cytochrome c. Biochemistry 35, 16378–16390 [DOI] [PubMed] [Google Scholar]
- 39. D'Arrigo A., Manera E., Longhi R., Borgese N. (1993) The specific subcellular localization of two isoforms of cytochrome b5 suggests novel targeting pathways. J. Biol. Chem. 268, 2802–2808 [PubMed] [Google Scholar]
- 40. Clement B., Mau S., Deters S., Havemeyer A. (2005) Hepatic, extrahepatic, microsomal, and mitochondrial activation of the N-hydroxylated prodrugs benzamidoxime, guanoxabenz, and Ro 48-3656([[1-[(2s)-2-[[4-[(hydroxyamino)iminomethyl]benzoyl]amino]-1-oxopropyl]-4-piperidinyl]oxy]-acetic acid). Drug Metab. Dispos. 33, 1740–1747 [DOI] [PubMed] [Google Scholar]