Background: KEEP ON GOING (KEG) is required to maintain low levels of the nuclear-localized transcription factor ABI5.
Results: KEG-mediated degradation of ABI5 occurs in the cytoplasm/trans-Golgi network/early endosome and requires a C-terminal lysine residue.
Conclusion: Cytoplasmic degradation prevents nuclear accumulation of ABI5 so as to prohibit ABA signaling.
Significance: Knowing how ABI5 stability is controlled is crucial to understanding ABA signaling and plant survival of environmental stress.
Keywords: Abscisic Acid, Arabidopsis, Plant Hormones, Protein Degradation, Protein Turnover, Ubiquitin Ligase, Ubiquitination, ABI5 Transcription Factor, RING-type E3, Abiotic Stress
Abstract
To mitigate the effects of environmental stress the abscisic acid (ABA)-responsive transcription factor ABI5 is required to delay growth of germinated seedlings. In the absence of stress, KEEP ON GOING (KEG) E3 is required to maintain low levels of ABI5. However, the mechanism underlying KEG-dependent turnover of ABI5 is not known. In addition, localization studies place KEG at the trans-Golgi network, whereas ABI5 is nuclear. Here we show that KEG interacts directly with ABI5 via its conserved C3 region. Interactions between KEG and ABI5 were observed in the cytoplasm and trans-Golgi network only when the RING domain of KEG was inactivated or when ABI5 was stabilized via mutations. Deletion of the C-terminal region of ABI5 or substituting lysine 344 for alanine (K344A) prohibited protein turnover. Furthermore, ABI5 is observed in the cytoplasm of Arabidopsis thaliana root cells when the K344A mutation is combined with the deletion of a nuclear localization signal. Other lysine mutations (K353A, K364A, and K376A) in conjunction with the nuclear localization signal deletion did not result in cytoplasmic accumulation of ABI5. Loss of lysine 344 did not affect the ability of ABI5 to promote ABA responses, which demonstrates that the mutant transcription factor is still functional. Based on the results, a model is suggested where KEG targets ABI5 for degradation in the cytoplasm, thus reducing nuclear accumulation of the transcription factor in the absence of ABA.
Introduction
Ubiquitination is responsible for marking regulatory and abnormal proteins for degradation by the 26 S proteasome. The selective removal of specific targets via the covalent attachment of ubiquitin molecules represents an efficient and rapid strategy to control many cellular processes such as response to external stimuli, cell growth, and division. Formation of ubiquitin conjugates on a specific protein requires the sequential action of three enzymes, E1 (ubiquitin activating), which activates ubiquitin molecules forming an E1-ubiquitin intermediate; E2 (ubiquitin conjugating), which accepts the activated ubiquitin from the E1 forming an E2-ubiquitin intermediate; and E3 (ubiquitin ligase), which facilitates the transfer of ubiquitin from the E2-ubiquitin intermediate to the target protein. Ubiquitination of a specific substrate is mainly regulated through modulation of its degradation signal (degron) and through control of the activity of its cognate E3. The E3-dependent recognition of the various primary degradation signals leads to the enzymatic addition of a polyubiquitin chain that serves as a secondary signal for targeting of the substrates to the proteasome (1–3).
Ubiquitin is usually attached via an isopeptide bond to an internal lysine on the substrate. Most target proteins have multiple lysine residues, but only one or a few can be efficiently ubiquitinated. This implies that for these substrates, the position of the ubiquitin acceptor site or the local structure surrounding it serves as a determinant for degron function. The molecular basis for the specificity of a E3 ligase toward a particular lysine in a substrate is poorly understood. Structural studies suggest that specificity can be explained by the relative spatial relationship of the lysine to the E3 ligase (4, 5).
ABSCISIC ACID INSENSITIVE 5 (ABI5),2 a member of the Arabidopsis thaliana (Arabidopsis) basic leucine zipper (bZIP) transcription factor family, has been shown to play an important role in controlling abscisic acid (ABA)-dependent post-germinative growth arrest as well as late phases of seed maturation (6, 7). The efficiency of the ABA-induced growth arrest is dependent on ABI5 protein accumulation through transcriptional activation and enhanced protein stability (7, 8). ABI5 protein accumulates in seedlings treated with 26 S proteasome inhibitors and in RPN10 (a subunit of the 26 S proteasome) mutant plants, suggesting that ABI5 turnover is dependent on the 26 S proteasome pathway (7, 9). A number of E3 ligases have been shown to be involved in modulating ABI5 stability (10). KEEP ON GOING (KEG), a multidomain really interesting new gene (RING)-type E3 ligase, is required to maintain low levels of ABI5 in the absence of the hormone. This is based on the fact that keg mutants undergo growth arrest immediately after germination, accumulate extremely high levels of ABI5, and display hypersensitivity to ABA, whereas overexpression of KEG leads to ABA insensitivity (11, 12). Furthermore, complementation studies demonstrate that KEG containing a nonfunctional E3 ligase domain is not able to rescue the keg-1 phenotype, whereas an intact KEG is able to fully rescue the KEG mutant and restore the levels of ABI5 to that observed for wild type plants (12). In addition, KEG is capable of attaching ubiquitin to ABI5 in biochemical assays. Overall, these studies demonstrate that KEG negatively regulates ABI5 abundance to prohibit activation of ABA responses in the absence of the hormone or stress stimulus.
Although KEG has been clearly demonstrated to negatively regulate the abundance of ABI5, the ability of KEG to directly interact with and mediate the turnover ABI5 is inconsistent with previously described cellular localization patterns of ABI5 and KEG. Recent reports suggest that KEG localizes to the trans-Golgi network/early endosome (TGN/EE) vesicles of transiently transformed tobacco epidermal cells (13). This is in contrast to ABI5, which has been shown to be constitutively localized in the nucleus via the use of a promoter-β-glucuronidases reporter system (14). Under the control of the cauliflower mosaic virus 35S promoter, ABI5 was only observed in the nucleus of both transiently transformed Arabidopsis and onion epidermal cells (15). In light of the apparent spatial separation of the E3 ligase and substrate, the outstanding question of how KEG directly regulates ABI5 turnover remains to be addressed. Here we show that KEG interacts directly with ABI5 in the cytoplasm and TGN/EE via ABI5 conserved C3 region. ABI5 mutations that prohibit KEG-mediated turnover lead to the stabilization and accumulation of ABI5 in the cytoplasm. Overall our results suggest a model where in the absence of ABA, KEG targets ABI5 for degradation in the cytoplasm to maintain low levels of the transcription factor.
EXPERIMENTAL PROCEDURES
Sequence Analysis and Alignment
To identify potential nuclear localization and export signal, the complete amino acid sequence of ABI5 was examined by using the WoLF PSORT computer program (16). Alignments were generated with the ClustalX program (17) and revised using the Se-Al sequence editor (Evolutionary Biology Group, University of Oxford).
Cloning and Mutagenesis
The full-length wild type KEG (11), the RING domain mutant of KEG (KEGAA; C29A, H31A), and the ABI5 cDNAs in the gateway entry vector pDONR201 (Invitrogen) were obtained as previously described (12). To be used for C-terminal fusion expression, these cDNAs were amplified again using Phusion polymerase (Finnzymes) to remove the STOP codon and introduced back into pDONR201 as per the manufacturer's instructions. The partial cDNA regions of KEG encoding the RING, kinase domain, and ankyrin repeats (KEG1–829; 1–2487 nucleotides (nt)), the RING and kinase domain (KEG1–442; 1–1326 nt), and the ankyrin repeats (KEG497–829; 1489–2487 nt) were introduced into pDONR201 as per the manufacturer's instructions. Deletion mutants and point mutations of ABI5 were generated using the Phusion site-directed mutagenesis kit (Finnzymes). Primers used for making these mutants are listed under supplemental Table S1. Nucleotide sequences were confirmed by DNA sequencing (McGill University and Génome Québec Innovation Centre).
Yeast Two-hybrid Assay
Gateway-compatible vectors pGBKT7-DEST (bait) and pGADT7-DEST (prey) modified from the pGBKT7 and pGADT7-Rec vectors, respectively, were gifts from Dr. Yuhai Cui (Agriculture and Agri-Food Canada, London, ON). The full-length open reading frame of KEG as well as KEG1–829, KEG1–442, and KEG497–829 were introduced into the activation domain vector pGADT7-DEST. The full-length open reading frames of ABI5, deletion mutants, and point mutations of ABI5 were cloned into the DNA-binding domain vector pGBKT7-DEST. Both bait and prey constructs were co-transformed into yeast strain AH109 using the lithium acetate method according to the manufacturer's instructions (Clontech). Transformants were selected for on medium lacking tryptophan and leucine (SD/-Trp/-Leu). To select for interactions, equal amounts of diluted AH109 carrying the bait and prey constructs was spotted onto selective medium lacking tryptophan, leucine, histidine, and adenine (SD/-Trp/-Leu/-His/-Ade) and incubated at 28 °C for up to 5 days. Empty pGBKT7 and pGADT7-Rec vectors were used as negative controls.
Plant Material and Growth Conditions
Arabidopsis ecotype Columbia-0 (Col-0) wild type, mutant, and transgenic seeds were surface-sterilized with 50% (v/v) bleach and 0.1% Triton X-100. After cold treatment at 4 °C for 48 h, seeds were germinated and grown solid on 0.5× Murashige and Skoog (MS) medium containing 0.8% agar and 1% sucrose under continuous light at 22 °C. For plants grown in soil, 7-day-old seedlings were transferred from MS medium to soil and grown under photoperiodic cycles of 16 h light and 8 h dark at 22 °C in a growth chamber. keg-1 (Salk_049542) seeds obtained from the Arabidopsis Biological Resource Center (ABRC) (19) were a gift from the laboratory of Dr. Judy Callis (University of California, Davis, CA (11)).
Transient Protein Expression in Nicotiana benthamiana
Five-week-old N. benthamiana (tobacco) plants used for transient protein expression were grown under a photoperiod of 16 h light and 8 h dark at 23 °C. Agrobacterium tumefaciens (Agrobacterium) strain GV3101 transformed with the appropriate binary plasmids were grown and prepared for transient expression as described previously (20). Agrobacterium cultures were resuspended in infiltration solution (5 mg/ml of d-glucose, 50 mm MES, 2 mm Na3PO4, 0.1 mm acetosyringone) at an optical density at 600 nm of 0.8. For co-expression of multiple constructs, suspensions were mixed in equal ratios. Agrobacterium suspension mixtures were infiltrated using a needleless syringe into leaves of 5-week-old tobacco plants. Forty-eight hours after Agrobacterium infiltration, samples were collected for protein extraction and microscopic imaging. For proteasome inhibitor treatments, tobacco leaves were injected with 50 μm MG132 32 h after Agrobacterium infiltration, excised, and incubated in 50 μm MG132 for another 16 h.
Subcellular Localization and Bimolecular Fluorescence Complementation (BiFC) Assay
For subcellular localization, KEG and KEGAA cDNAs in the entry vector pDONR201 were introduced into the pEarleyGate101 plant transformation vector (21) using Gateway cloning (Invitrogen) to produce in-frame fusions with the C-terminal yellow fluorescence protein (YFP) and hemagglutinin (HA) tag (KEG-YFP) under the control of the cauliflower mosaic virus 35S promoter. Similarly, full-length ABI5, deletion mutants, and point mutations of ABI5 were cloned into the pEarleyGate102 vector to generate the C-terminal-tagged cyan fluorescent protein (CFP) and HA fusions (ABI5-CFP). For simplification, the HA tag was not included in the names of the fusion proteins throughout the text. Constructs were then introduced into Agrobacterium strain GV3101 and transformed into Arabidopsis using the flower-dipping method (22) or infiltrated into tobacco leaves as described above.
BiFC vectors pEarleyagte201-YN, containing the N-terminal portion (1–174 amino acids) of enhanced YFP (eYFP) and a HA tag and pEarleygate202-YC containing the C-terminal portion (175–239 amino acids) of eYFP and a FLAG tag were created based on the Gateway-compatible vectors pEarleygate201 and pEarleygate202, respectively, by the laboratory of Dr. Yuhai Cui (Agriculture and Agri-Food Canada, London, United Kingdom (18)). KEG and ABI5 (including deletions and point mutations) cDNAs in the gateway entry vector pDONR201 were recombined into pEarleygate201-YN and pEarleygate202-YC, respectively (Invitrogen). KEG is expressed as a fusion with the N-terminal portion of eYFP and a HA tag (KEG-YN). ABI5 is expressed as a fusion with the C-terminal portion of eYFP and a FLAG tag (ABI5-YC). For simplification, the HA and FLAG tags were not included in the names of the fusion proteins throughout the text. Each plasmid DNA was transformed into Agrobacterium strain GV3101. Equal volumes of Agrobacterium cultures with cDNAs in the pEarleygate201-YN and pEarleygate202-YC vectors were mixed and used for infiltrating tobacco leaves as described above. Leaves were assayed for fluorescence 48 h after infiltration for both subcellular localization and BiFC assay. Each localization experiment and BiFC assay was repeated at least three times.
Fluorescence Microscopy
Roots of 7-day-old transgenic Arabidopsis seedlings or pieces of infiltrated tobacco leaves excised 48 h after infiltration were imaged using a Zeiss LSM 510 META inverted confocal laser scanning microscope (Carl Zeiss MicroImaging GmbH) equipped with a ×25 oil immersion objective. YFP fluorescence was detected at an excitation wavelength of 514 nm, and emissions were collected between 530 and 600 nm. For CFP fluorescence, an excitation wavelength of 458 nm was used, and emissions were collected between 475 and 525 nm. To show the outline of each epidermal cell, a single slice of the fluorescence and bright-field images were overlaid. For those images showing only fluorescence, a series of Z-stack images were collected and then combined and processed using the Zeiss LSM Image Browser (Carl Zeiss MicroImaging GmbH).
Protein Extraction and Immunoblot Analysis
Forty-eight hours after Agrobacterium infiltration, tobacco leaf discs were collected from the same leaf used for microscopic imaging (see above), frozen in liquid nitrogen, and stored at −80 °C until needed. At least three independent repetitions were performed for each experiment. Total protein was extracted from tissues in protein extraction buffer (HEPES, pH 7.5, 5 mm EDTA, 5 mm EGTA, 10 mm Na3VO4, 10 mm NaF, 50 mm β-glycerophosphate, 10 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 5% glycerol and protease inhibitor mixture (Sigma)). The concentration of protein was determined using the Bradford reagent (Sigma) with bovine serum albumin as a standard. Total protein was mixed with 6× SDS loading buffer (300 mm Tris-HCl, pH 6.8, 30% glycerol, 12% SDS, 0.6% bromphenol blue) boiled for 5 min before loading on SDS-polyacrylamide gel (SDS-PAGE).
For immunoblot analysis, 30 μg of total protein per lane was separated on SDS-PAGE. After electrophoresis, proteins were electrotransferred to polyvinylidene fluoride (PVDF) membrane. After blocking for 1 h in TBST (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.05% Tween 20) with 5% nonfat dry milk at room temperature, membranes were incubated with mouse anti-FLAG (Sigma) or anti-HA (Sigma) at 1:10,000 dilution for 2 h. Following three washes with TBST, membranes were incubated with horseradish peroxidase-conjugated goat anti-mouse (Sigma) at 1:5000 dilution. After three washes with TBST, the membranes were visualized using an enhanced Lumi-Light Western blotting Substrate kit (Thermo Scientific) following the manufacturer's instructions.
Cell-free Degradation Assay
For recombinant proteins used in the assay, wild type and mutated ABI5 cDNAs were recombined into modified pDEST527 vectors (Addgene plasmid 11518; donated by Dominic Esposito, National Cancer Institute, Frederick, MD) to obtain fusion proteins with N-terminal FLAG-His tag (12). Recombinant proteins were expressed in Escherichia coli strain Rosetta (DE3) and purified using nickel-charged resin (Bio-Rad) according to the manufacturer's protocols.
Cell-free degradation assays were carried out as previously described (23). Total protein was extracted from 4-day-old Col-0 or 8-day-old keg-1 seedlings using extraction buffer (25 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 5 mm DTT, and protease inhibitor mixture). After centrifugation at 18,000 × g for 20 min at 4 °C, the supernatant was transferred to a new tube and the protein concentration was determined using the Bio-Rad protein assay kit. For the degradation assay, 1.6 μg of each recombinant ABI5 protein was incubated in 0.5 mg of total protein extract. MgCl2 (10 mm) and ATP (10 mm) were added to the reaction prior to incubation at 30 °C and the total reaction volume was adjusted to 240 μl with extraction buffer. 30 μl of the reaction mixture was taken at the indicated time points and 5 μl of 6× SDS loading buffer was added to stop the reaction. For proteasome inhibitor treatments, 30 μm MG132 was added to protein extracts 30 min prior to the start of the assay. ABI5 protein levels were determined by immunoblots using His antibodies (Sigma) at 1:5,000 dilution followed by horseradish peroxidase-conjugated anti-mouse IgG (Sigma) at 1:5,000 dilution.
Abscisic Acid Root Sensitivity Assay
Surface sterilized Col-0 and transgenic seedlings were germinated and grown vertically on solid MS medium for 3 days as described above. Seedlings were then transferred to solid MS medium with or without 2 μm ABA (Sigma) and grown vertically for another 5 days. Root growth after transfer was measured.
RESULTS
KEG Interacts Directly with ABI5 in the Cytoplasm/TGN/EE
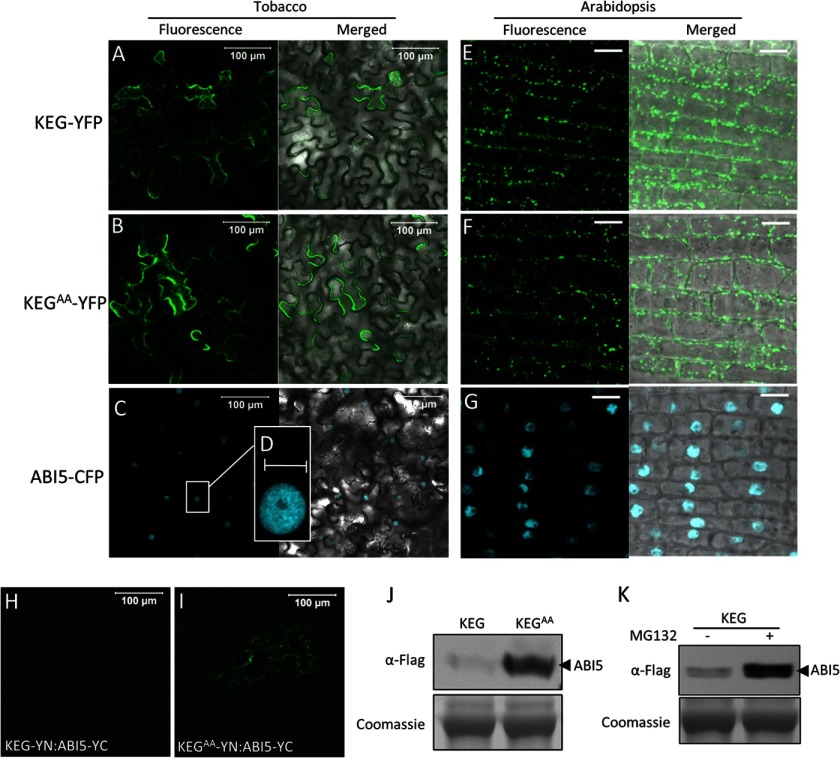
KEG has been shown to function as a negative regulator of ABA signaling by modulating the abundance of ABA-responsive transcription factor ABI5 (11, 12). Gu and Innes (13) recently showed that KEG is predominantly localized to the TGN/EE (13). This is in contrast to ABI5, which has been reported to be constitutively localized in the nucleus (14). The results from these subcellular localization studies raise the question of how KEG is able to regulate ABI5 abundance. Does KEG directly interact with ABI5 in plant cells? To address this question we first determined the subcellular localization of KEG and ABI5 in our experimental systems (Fig. 1, A–G). Full-length KEG and KEG containing a nonfunctional RING E3 ligase domain (KEG RING mutant; KEGAA) were expressed as C-terminal fusions with YFP under control of the cauliflower mosaic virus 35S promoter in transiently transformed tobacco leaf epidermal cells. Both KEG-YFP and KEGAA-YFP were observed in the cytoplasm as well as small punctate structures reminiscent of the previously observed localization (Fig. 1, A and B) (13). ABI5 was expressed as a CFP C-terminal fusion (ABI5-CFP) driven by the 35S promoter in transiently transformed tobacco leaf epidermal cells (Fig. 1, C and D). Unsurprisingly, in our experimental system, ABI5 was observed only in the nucleus (Fig. 1, C and D). We also generated Arabidopsis transgenic plants expressing ABI5-CFP, KEG-YFP, and KEGAA-YFP under the control of the 35S promoter. Localization for KEG-YFP, KEGAA-YFP, and ABI5-CFP in Arabidopsis root cells was similar to that observed in transiently transformed tobacco cells (Fig. 1, E–G).
FIGURE 1.
KEG interacts with ABI5 in the cytoplasm. A–D, localization of KEG-YFP (A), KEGAA-YFP (B), and ABI5-CFP (C and D) in transiently transformed tobacco leaf epidermal cells. Right panels show fluorescence images from a combined series of Z-stack. Left panels show transmitted light images merged with fluorescence images from a single optical section. Bars = 100 μm. D displays an enlargement of the section delimited by a white square in C. Bar = 10 μm. E–G, localization of KEG-YFP (E), KEGAA-YFP (F), and ABI5-CFP (G) in root cells of transgenic Arabidopsis seedlings. Right panels show fluorescence images from a single optical section. Left panels show transmitted light images merged with fluorescence images from a single optical section. Bars = 10 μm. H and I, BiFC analysis in tobacco epidermal cells. KEG-YN (H) or KEGAA-YN (I) was coexpressed with ABI5-YC. Bars = 100 μm. J, levels of ABI5-YC following coexpression with KEG-YN or KEGAA-YN. Analysis was done using protein extracts derived from the same samples assayed for BiFC shown in H and I. K, levels of ABI5-YC in tobacco epidermal cells coexpressing KEG-YN in the absence (−) and presence (+) of MG132. Note: ABI5-YC also contains a FLAG tag. Coomassie staining was used to confirm equal loading.
To determine whether KEG interacts directly with ABI5, BiFC assays were performed using Agrobacterium-mediated transient expression in tobacco leaf epidermal cells. Following co-expression of KEGAA fused with the amino (N)-terminal eYFP fragment (KEGAA-YN) with ABI5 fused with the carboxyl (C)-terminal eYFP fragment (ABI5-YC), BiFC fluorescence signals were observed in the cytoplasm of tobacco epidermal cells indicating that the eYFP fluorophore was reconstituted due to interaction between KEGAA and ABI5 proteins (Fig. 1I). However, co-expression of KEG fused to the N-terminal eYFP fragment (KEG-YN) with ABI5-YC did not produce an observable fluorescence signal (Fig. 1H). The lack of BiFC signal could be due to the fact that upon interaction, KEG ubiquitinates ABI5 targeting the transcription factor for degradation. To confirm this hypothesis, proteins were extracted from the same leaf sample used for BiFC analysis (Fig. 1, H and I) and used to determine ABI5 protein levels via immunoblot analysis. As shown in Fig. 1J, the ABI5-YC protein was barely detected when co-expressed with KEG-YN. In contrast, ABI5-YC accumulated when co-expressed with KEGAA-YN. To determine whether the low levels of ABI5 observed following co-expression with KEG is due to proteasomal-dependent turnover, tobacco leaves co-expressing ABI5-YC and KEG-YN were incubated with MG132, a specific inhibitor of the 26 S proteasome. As shown in Fig. 1K, ABI5 accumulated to greater levels in MG132-treated samples (Fig. 1K).
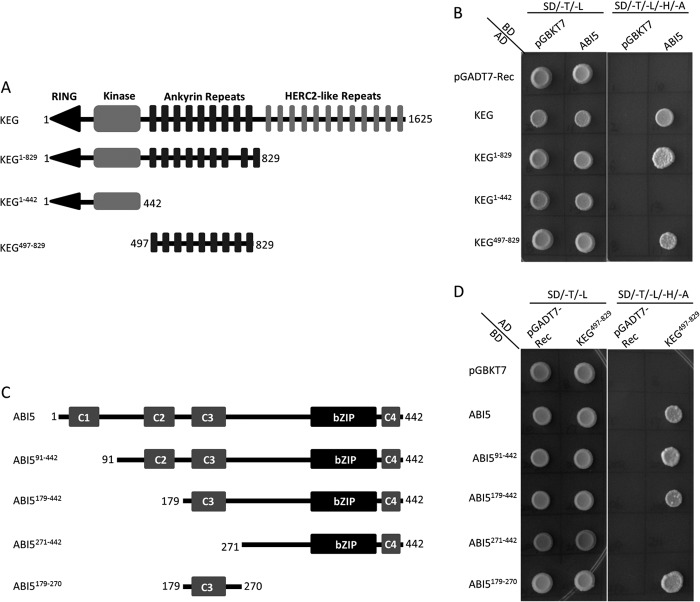
Interactions between KEG and ABI5 are facilitated by the ankyrin repeats of KEG (11, 24), however, the region of ABI5 required for this interaction is not known. Yeast two-hybrid assays were performed to identify the region of ABI5 that mediates interaction with the RING-type E3 ligase. To first demonstrate interactions between KEG and ABI5 in yeast, full-length KEG and a series of KEG truncations (KEG1–442, KEG1–829, and KEG497–829) fused with the GAL4 transcriptional activation domain (AD; prey) were co-expressed with full-length ABI5 fused with the GAL4 DNA-binding domain (BD; bait) (Fig. 2A). As shown in Fig. 2B, interaction is observed between ABI5 and KEG, KEG1–829, or KEG497–829. No interaction was observed with KEG1–442, which lacks the ankyrin repeats. ABI5 contains a conserved bZIP DNA-binding domain and four highly conserved regions, C1–C4 (25, 26) (Fig. 2C). To determine the region of ABI5 that mediates interaction with KEG, fusions between the GAL4-BD and various regions of ABI5 were tested for interaction with KEG ankyrin repeats (KEG497–829) GAL4-AD fusion in yeast (Fig. 2, C and D). As shown in Fig. 2D, a short region of ABI5 that contains the conserved C3 region (ABI5179–270) was able to interact with KEG497–829. Deletion of the conserved C3 region (ABI5271–442) prohibited ABI5 interaction with KEG497–829 (Fig. 2D).
FIGURE 2.
ABI5 conserved C3 domain facilitates interactions with KEG ankyrin repeats. A and C, schematic representations of the KEG (A) and ABI5 (C) used in the yeast two-hybrid assays. Numbers indicate amino acids. B and D, yeast two-hybrid experiments showing interactions between ABI5 and KEG. Full-length ABI5 cDNA was fused with the GAL4-binding domain (BD) and KEG constructs (as illustrated in A) were fused with the Gal4 activation domain (AD) (B). Alternatively, ABI5 cDNA constructs (as illustrated in C) were fused to the GAL4-BD and KEG cDNA (ankyrin repeats, KEG497–829) was fused to the GAL4-AD (D). Interaction between ABI5 and KEG was analyzed by growth on selection medium without Trp, Leu, His, and Ade (SD/-T/-L/-H/-A). The pGADT7 and pGBKT7-Rec empty vectors were used as negative controls.
The Carboxyl-terminal Region of ABI5 Is Required for Protein Turnover
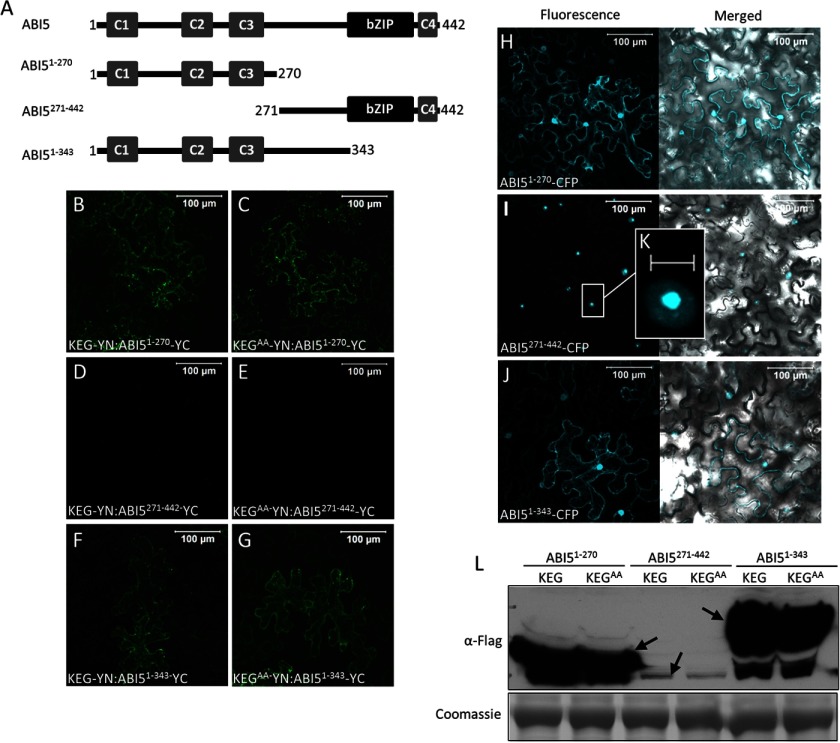
The results described above suggest that KEG modulates ABI5 abundance by interacting with the conserved C3 domain of ABI5 in cytoplasm. To further understand the mechanism underlying KEG regulation of ABI5 turnover, we aimed to determine the region of ABI5 that regulates stability. ABI5-YC (ABI51–270-YC and ABI51–343-YC) lacking the C-terminal region produced a BiFC fluorescence signal when coexpressed with both KEG-YN (Fig. 3, A, B, and F) and KEGAA-YN (Fig. 3, C and G). This is in contrast to full-length ABI5-YC, which produced a BiFC fluorescence signal only when co-expressed with KEGAA-YN (Fig. 1, H and I). ABI5-YC lacking the KEG-interacting C3 region (ABI5271–442-YC) did not produce a BiFC fluorescence signal when co-expressed with KEG-YN or KEGAA-YN (Fig. 3, A, D, and E). Loss of the C-terminal region allows for interaction with the functional KEG E3 ligase, which suggests stabilization of ABI5.
FIGURE 3.
C-terminal region of ABI5 regulates protein stability. A, schematic representation of ABI5 and ABI5 deletion mutants. Numbers indicate amino acids. B–G, BiFC analysis in tobacco epidermal cells. KEG-YN or KEGAA-YN was coexpressed with ABI51–270-YC (B and C), ABI5271–442-YC (D and E), or ABI51–343-YC (F and G). Bars = 100 μm. H–K, subcellular localizations of ABI51–270-CFP (H), ABI5271–442-CFP (I), and ABI51–343-CFP (J) in transiently transformed tobacco epidermal cells. Left panels show fluorescent images from a combined series of Z-stack. Right panels show transmitted light images overlaid with fluorescence images from a single optical section. Bars = 100 μm. K displays an enlargement of the section delimited by a white square in I. Bar = 10 μm. L, levels of ABI51–270-YC, ABI5271–442-YC, and ABI51–343-YC were determined following coexpression with KEG-YN or KEGAA-YN. Western blot analysis was done using protein extracts derived from the same tobacco leaf samples assayed for BiFC fluorescence shown in B–G. Note: ABI5-YC also contains a FLAG tag. Arrows indicate ABI5. Coomassie staining was used to confirm equal loading.
If deletion of the C-terminal region of ABI5 stabilizes the transcription factor then the deletion mutants should be observed in the cytoplasm. Localization studies show that ABI5-CFP lacking the C-terminal region was able to accumulate in the cytoplasm. Both ABI51–270-CFP and ABI51–343-CFP were observed in the cytoplasm and nucleus of transiently transformed tobacco epidermal cells (Fig. 3, H and J). Although the C-terminal region of ABI5 (ABI5271–442-CFP) remained exclusively nuclear localized (Fig. 3I). In contrast to ABI5-CFP, which is predominantly found in the nucleoplasm (Fig. 1D), ABI5271–442-CFP was observed mainly in the nucleolus (Fig. 3, I and K). These results suggests that although KEG is capable of interacting with both ABI51–270 and ABI51–343, the E3 ligase may not be able to target ABI5 lacking the C-terminal region for degradation. Therefore, the ABI5 fusion proteins are observed in the cytoplasm.
To confirm that elimination of the C-terminal region increased protein stability, the levels of ABI5 deletion mutants following co-expression with KEG and KEGAA were determined. The same leaf sample used for BiFC analysis (Fig. 3, B–G) was collected, total protein was extracted, and ABI5 protein levels were determined via immunoblot analysis. Unlike ABI5-YC (Fig. 1J), equally high levels of ABI51–270-YC and ABI51–343-YC accumulated when co-expressed with both KEG-YN (Fig. 3L, fifth and first lanes) and KEGAA-YN (Fig. 3L, second and sixth lanes). These results are consistent with localization and BiFC studies, which suggested that KEG is unable to target ABI5 lacking the C-terminal region for degradation. Unexpectedly, low levels of the ABI5271–442-YC, which lacks the KEG-interacting C3 region, were observed following co-expression with both KEG-YN and KEGAA-YC (Fig. 3L, third and fourth lanes). Reasons for this are outlined under “Discussion.”
Cell-free degradation assays were used to compare the stability of ABI5 and ABI51–343, which lack the C-terminal region. Recombinant FLAG-His-ABI5 or FLAG-His-ABI51–343 were incubated with protein extracts prepared from wild type or keg-1 seedlings in the presence or absence of MG132. Using wild type protein extracts, the levels of FLAG-His-ABI5 gradually decreased over time (Fig. 4A). The decrease in FLAG-His-ABI5 levels was considerably slower in keg-1 protein extracts demonstrating the involvement of KEG activity in the turnover of ABI5 in the experimental system (Fig. 4A). The turnover of ABI5 was also greatly reduced in the presence of proteasome inhibitor, MG132 (Fig. 4B). In contrast to FLAG-His-ABI5, decreases in FLAG-His-ABI51–343 levels were significantly slowed in the absence of MG132 (Fig. 4B), further demonstrating the involvement of the ABI5 C-terminal region in regulating protein stability.
FIGURE 4.
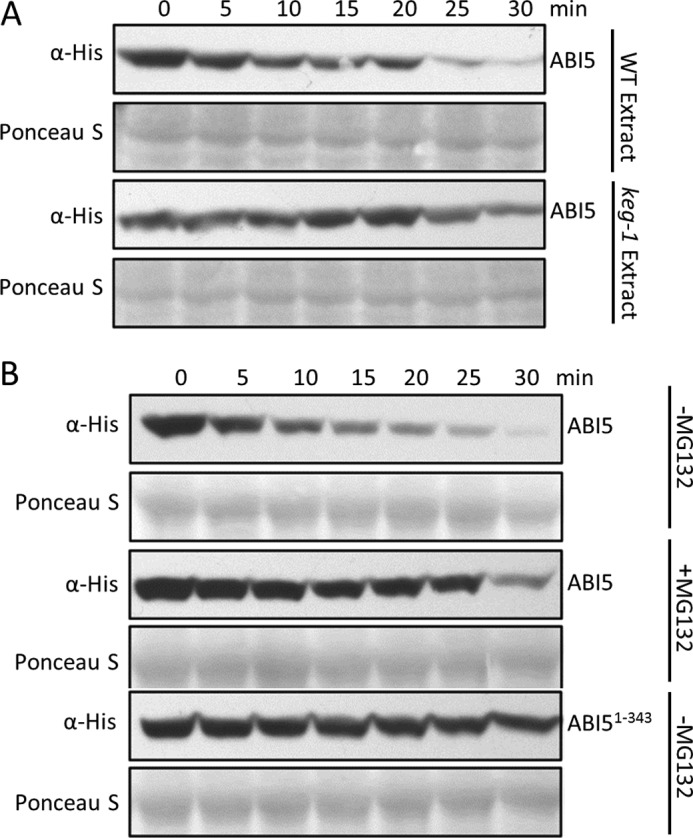
Turnover of ABI5 in cell-free degradation assays requires proteasome activity, KEG, and ABI5 C-terminal region. A, comparison of recombinant FLAG-His-ABI5 turnover in wild type (WT) Arabidopsis or keg-1 plant extracts. B, turnover of recombinant FLAG-His-ABI5 and FLAG-His-ABI51–343 in WT protein extracts in the presence (+) or absence (−) of 30 μm MG132. ABI5 protein levels were determined by Western blotting using His antibodies. Ponceau S staining was used to confirm equal loading.
The Carboxyl-terminal Region Modulates Localization as Well as Stability of ABI5
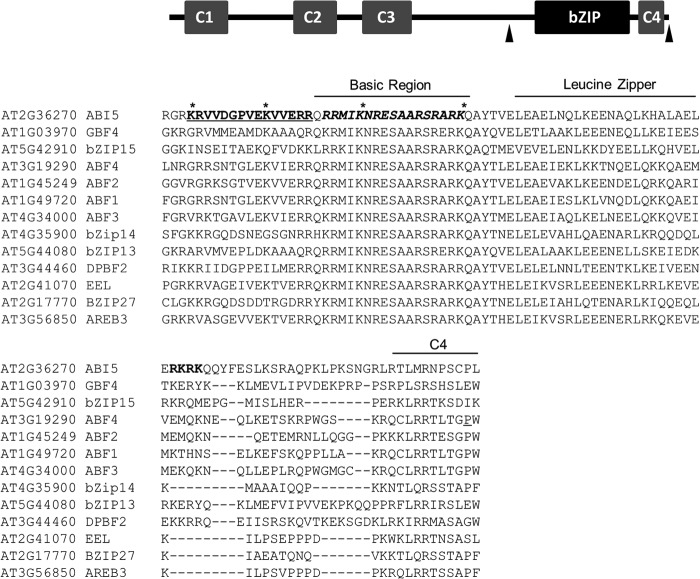
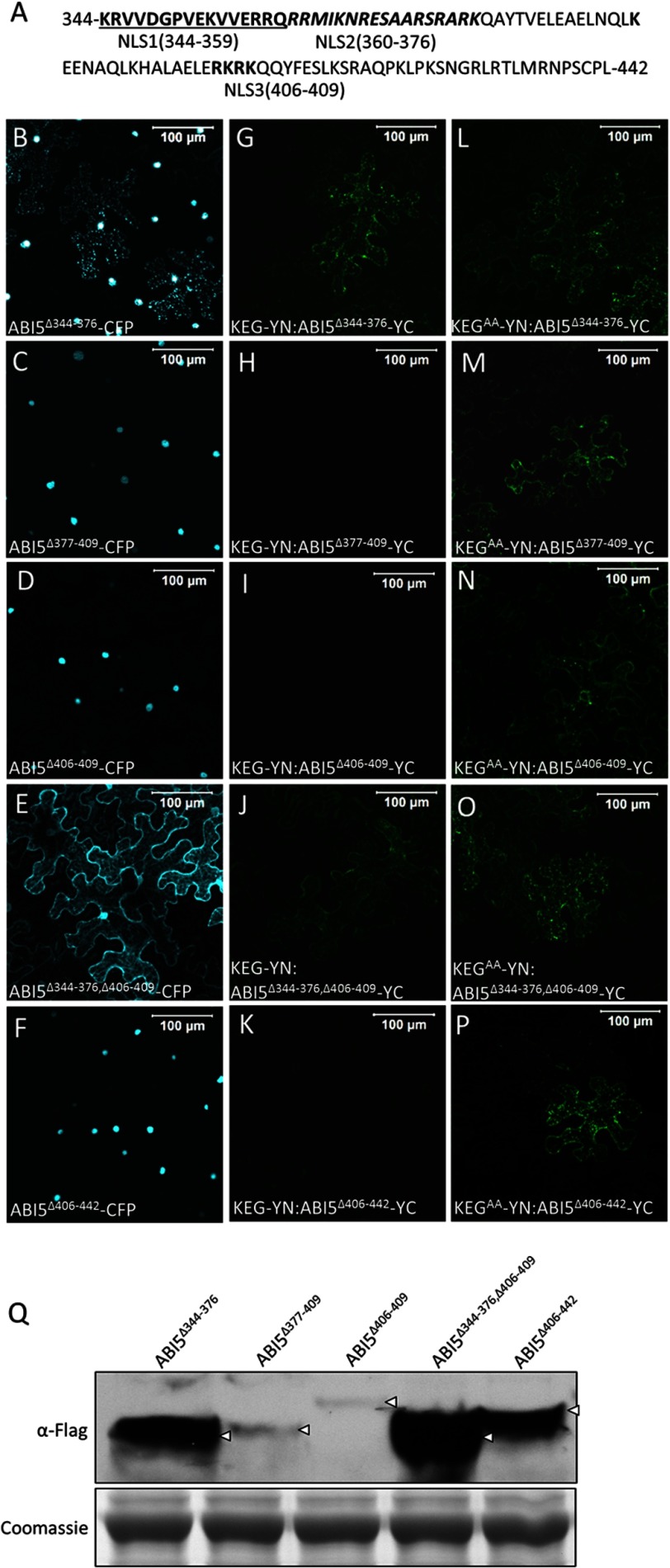
The fact that ABI5 interacts with KEG and stabilized ABI5 is observed in the cytoplasm suggests that the transcription factor is shuttled between the nucleus and the cytoplasm. Amino acid sequence analysis of ABI5 identified a potential N-terminal nuclear export signal (NES; 37LGRQSSIYSLTL48) and three potential nuclear localization signals (NLSs) within the C-terminal region, referred to as NLS1-NLS3 (Fig. 5). To confirm the presence of functional NLSs a series of CFP-FLAG-tagged ABI5 deletion mutants were generated, and subcellular localization was observed in transiently transformed tobacco epidermal cells (Fig. 6, A–F). Deletion of the NLS1/2 region (ABI5Δ344–376-CFP) resulted in a mixed localization pattern with some cells showing both nuclear and cytoplasmic localization and the remaining cells showing only a nuclear localization (Fig. 6B). Within the cytoplasm ABI5Δ344–376-CFP was observed in small punctuate structures similar to that observed for KEG-YFP (Fig. 1, A and B). The fact that ABI5Δ344–376-CFP localized predominantly to the nucleus in many cells suggests that NLS1 and NLS2 may not be necessary for nuclear localization. The observed cytoplasmic localization for ABI5Δ344–376-CFP in some cells may instead be due to stabilization of the fusion protein. To determine whether this was the case, ABI5Δ344–376-YC was co-expressed with KEG-YN or KEGAA-YN in transiently transformed tobacco cells. ABI5Δ344–376-YC interacted with both KEG-YN and KEGAA-YN, as determined by the presence of BiFC fluorescence signals (Fig. 6, G and L), indicating that KEG may not target ABI5 lacking NLS1/2 (amino acids 344–376) for degradation. Deletion of NLS3 (ABI5Δ406–409) or NLS3 plus surrounding amino acids (ABI5Δ377–409 and ABI5Δ406–442) did not alter the localization of ABI5-CFP, all fusion proteins were observed exclusively in the nucleus of tobacco epidermal cells (Fig. 6, C, D, and F). BiFC assays showed that ABI5Δ377–409-YC, ABI5Δ406–409-YC, and ABI5Δ406–442-YC produced a fluorescence signal when co-expressed with KEGAA-YN (Fig. 6, M, N, and P) but not with KEG-YN (Fig. 6, H, I, and K). These results suggest that, unlike loss of NLS1/2, deletion of NLS3 or NLS3 plus surrounding regions does not influence the stability of ABI5. Because deletion of NLS1/2 or NLS3 alone did not result in predominantly cytoplasmic localization of ABI5, the NLS1/2 and NLS3 deletions were combined (ABI5Δ344–376,Δ406–409). ABI5Δ344–376,Δ406–409-CFP was observed mainly in the cytoplasm (Fig. 6E). BiFC analysis demonstrate interaction between ABI5Δ344–376,Δ406–409-YC and both KEG-YN and KEGAA-YN (Fig. 6, J and O). Thus, loss of NLS1/2 (amino acids 344–376) in conjunction with deletion of NLS3 (amino acids 406–409) resulted in stabilization and accumulation of ABI5 in the cytoplasm.
FIGURE 5.
Schematic representation of ABI5 and amino acid sequence alignment of the C-terminal region of A. thaliana ABI5-related bZIP transcription factors. Arrows delineated the region of ABI5 shown in the alignment. The basic region and leucine zipper of the bZIP DNA-binding domain and the conserved C4 region are outlined. The positions of the predicted nuclear localization signals, NLS1 (bold and underlined), NLS2 (bold and italics), and NLS3 (bold), of ABI5 are highlighted. ABI5 lysine residues (Ly344, Lys353, Lys364, and Lys376) mutated in this study are marked by asterisks (*).
FIGURE 6.
Delineating the region of ABI5 required for KEG-mediated ubiquitination. A, amino acid sequence of ABI5 C-terminal region showing the positions of the deleted regions, NLS1 (bold and underlined), NLS2 (bold and italics), and NLS3 (bold). B–F, subcellular localization of the CFP-ABI5 deletion mutants in transiently transformed tobacco leaf epidermal cells. Fluorescence images are a combined series of Z-stack. G–P, BiFC assay in tobacco leaf epidermal cells. KEG-YN or KEGAA-YN was coexpressed with the indicated ABI5-YC deletion mutant. Numbers indicate the deleted (Δ) amino acids for each ABI5 mutant. Bars = 100 μm. Q, levels of each ABI5-YC deletion mutant following coexpression with KEG-YN. Analysis was done using protein extracts derived from the same tobacco leaf samples assayed for BiFC fluorescence shown in G–K. Note: ABI5-YC also contains a FLAG tag. Arrows indicate ABI5. Coomassie staining was used to confirm equal loading.
To compare the stability of the different ABI5 deletion mutants in the presence of KEG, protein extracts were prepared from the same leaf samples used for BiFC analysis (Fig. 6, G–K) and ABI5 levels were determined via immunoblot analysis (Fig. 6Q). Consistent with the localization and BiFC results, ABI5 lacking the NLS1/2 region (ABI5Δ344–376-YC) accumulated to significantly higher levels than ABI5 with only the NLS3 region deleted (ABI5Δ337–409 and ABI5Δ406–409-YC) (Fig. 6Q, compare first lane to second and third lanes). In addition, deletion of the NLS1/2 region in conjunction with NLS3 (ABI5Δ344–376,Δ406–409-YC) resulted in even greater accumulation of ABI5 (ABI5Δ344–376-YC) (Fig. 6Q, fourth lane). These results provide further evidence for the stabilizing effects of deleting NLS1/2 (amino acids 344–376) on the ABI5 protein. Surprisingly, nuclear-localized ABI5Δ406–442-YC accumulated to much higher levels compared with other nuclear-localized ABI5 mutants (ABI5Δ337–409-YC and ABI5Δ406–409-YC) (Fig. 6Q, compare fifth lane to second and third lanes).
Lysine 344 Is Involved in ABI5 Turnover
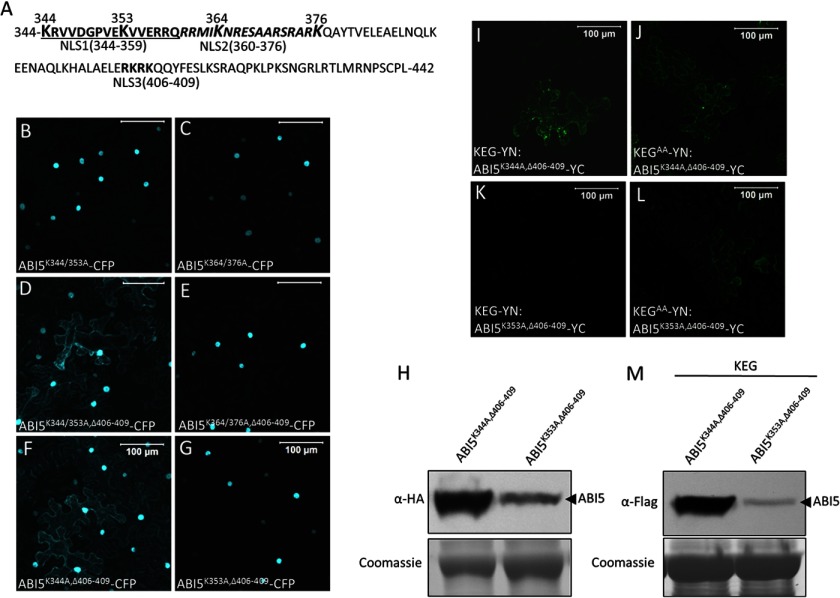
The above results suggest that residues within the 344–376 range of amino acids are involved in turnover of ABI5. Within this region there are four lysine residues (Lys344, Lys353, Lys364, and Lys376), which may serve as attachment sites of ubiquitin (Fig. 5). To determine whether one or more of these lysines are involved in degradation of ABI5, pairs of lysine residues were first changed to alanine and the ABI5 mutants were transiently expressed in tobacco cells as CFP fusion proteins (Fig. 7, A–C). Accumulation of the mutant ABI5 in the cytoplasm would indicate increased stability of the fusion protein. Replacing Lys344 and Lys353 (ABI5K344/353A-CFP) or Lys364 and Lys376 (ABI5K364/376A-CFP) with alanine did not result in any visible accumulation of ABI5-CFP in the cytoplasm (Fig. 7, B and C). The lack of cytoplasmic localization may be due to the presence of NLS3, which would direct the stabilized ABI5 to the nucleus. To address this issue, NLS3 (amino acids 406–409) deletion was combined with the lysine mutations (ABI5K344/353A,Δ406–409-CFP and ABI5K364/376A,Δ406–409-CFP). As shown in Fig. 7D, ABI5K344/353A,Δ406–409-CFP was observed in the cytoplasm as well as the nucleus. In contrast, ABI5K364/376A,Δ406–409-CFP was still only observed in the nucleus (Fig. 7E). These results indicate that Lys344 or Lys353 is utilized for targeting cytoplasmic ABI5 for degradation.
FIGURE 7.
Lysine 344 of ABI5 is involved in protein stability and KEG-dependent turnover. A, amino acid sequence of the ABI5 C-terminal region showing lysine (K) residues that were changed to alanine (A). B–G, subcellular localization of the indicated ABI5-CFP mutants in transiently transformed tobacco epidermal cells. Fluorescent images represent a combined series of Z-stack. H, protein levels of ABI5K343A/Δ406–409-CFP and ABI5K343A/Δ406–409-CFP. Analysis was done using protein extracts derived from the same tobacco leaf samples assayed for localization shown in F and G. Note: ABI5-CFP also contains a HA tag. I–L, BiFC assays in tobacco epidermal cells. KEG-YN or KEGAA-YN was coexpressed with the indicated ABI5-YC mutant. Bars = 100 μm. M, levels of ABI5K343A/Δ406–409-YC and ABI5K343A/Δ406–409-YC following co-expression with KEG-YN. Analysis was done using protein extracts derived from the same tobacco leaf samples assayed for BiFC fluorescence shown in I and K. Note: ABI5-YC also contains a FLAG tag. Coomassie staining was used to confirm equal loading.
To distinguish between the requirement for Lys344 and/or Lys354, single lysine to alanine substitutions were combined with deletion of NLS3 (ABI5K344A/Δ406–409-CFP and ABI5K353A,Δ406–409-CFP) (Fig. 7, A, F, and G). Replacing Lys344 with alanine was sufficient for significant accumulation of ABI5K344A,Δ406–409-CFP in the cytoplasm (Fig. 7F), whereas loss of K353 did not affect localization of ABI5K353A,Δ406–409-CFP (Fig. 7G). Consistent with the localization studies, immunoblot analysis showed that ABI5K344A,Δ406–409-CFP (Fig. 7H, lane 1) accumulated to much higher levels compared with ABI5K353A,Δ406–409-CFP in the tobacco cells (Fig. 7H, lane 2). Using BiFC analysis, ABI5K344A,Δ406–409-YC was found to interact with both KEG-YN and KEGAA-YN (Fig. 7, I and J). In contrast, ABI5K353A,Δ406–409-YC was found to produce BiFC fluorescence signals with KEGAA-YN and not with KEG-YN (Fig. 7, K and L). These results suggest that Lys344 is involved in turnover of ABI5. The levels of ABI5K344A,Δ406–409-YC and ABI5K353A,Δ406–409-YC following co-expression with KEG-YN were also determined. The same leaf samples used for BiFC analysis (Fig. 7, I and K) were collected, and ABI5 protein levels were determined via immunoblot analysis. Compared with ABI5K353A,Δ406–409-YC, significantly higher levels of ABI5K344A,Δ406–409-YC accumulated when co-expressed with KEG-YN (Fig. 7M). These results are consistent with the BiFC analysis, which suggested that KEG does not promote the turnover of ABI5 lacking Lys344.
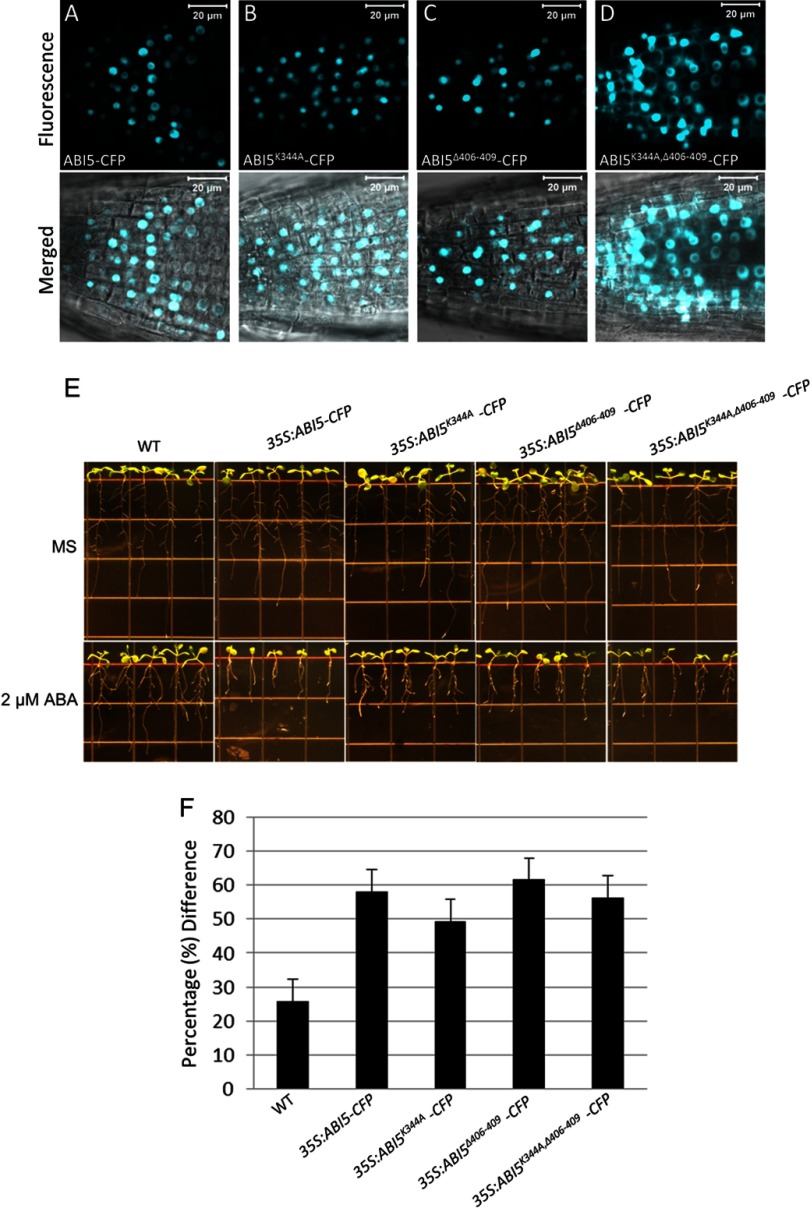
To further assess the role of lysine 344 in the turnover of ABI5, transgenic Arabidopsis plants expressing CFP-tagged wild type and mutant versions of ABI5 were generated and used for localization studies. Two independently generated transgenic lines were obtained for each transgene. Similar to ABI5-CFP, ABI5K344A-CFP and ABI5Δ406–409-CFP are observed only within the nucleus of Arabidopsis root cells (Fig. 8, A–C). However, ABI5K344A,Δ406–409-CFP was observed within the nucleus and cytoplasm (Fig. 8D). Therefore, similar to results obtained using tobacco cells (Fig. 7F), loss of lysine 344 in conjunction with deletion of NLS3 (amino acids 406–409) resulted in stabilization of ABI5 and accumulation of the transcription factor in the cytoplasm.
FIGURE 8.
Effects of the lysine 344 mutation on ABI5 localization and function in Arabidopsis. A–D, localization of ABI5-CFP, ABI5K344A-CFP, ABI5Δ406–409-CFP, and ABI5K344A,Δ406–409-CFP in root cells of transgenic Arabidopsis seedlings. Upper panels show fluorescence images from a single optical section. Lower panels show transmitted light images merged with fluorescence images from a single optical section. Bars = 20 μm. E, ABA root sensitivity assays. 8-Day-old wild type (WT) and transgenic seedlings expressing CFP-ABI5 or the indicated CFP-ABI5 mutant were grown for 5 days without (top panels) or with 2 μm ABA (bottom panel). F, the graph illustrates the percentage difference in root length between ABA-treated (2 μm) and untreated roots for each genotype. Error bars represent mean ± S.E.
To ensure that the cytoplasmic localization of ABI5 is due to stabilization and not to accumulation of a nonfunctional protein, the ability of transgenic seedlings to respond to ABA was assessed. There were no observable differences between wild type and transgenic seedlings grown in the absence of ABA (Fig. 8E). Similar to overexpression of ABI5-CFP, transgenic seedlings overexpressing ABI5K344A-CFP, ABI5Δ406–409-CFP, or ABI5K344A,Δ406–409-CFP were sensitive to the inhibitory effects of ABA on primary root elongation (Fig. 8, E and F). This demonstrates that the mutant versions of ABI5 are functional and capable of facilitating plant response to ABA.
DISCUSSION
Our findings demonstrate that KEG directly interacts with the transcription factor ABI5 and this interaction is critically involved in the regulation of ABI5 abundance. This conclusion is based on a number of supporting observations. First, despite the apparent differences in subcellular localization KEG interacts directly with ABI5 in the cytoplasm/TGN/EE. The interaction occurs via the conserved C3 region of ABI5 and KEG ankyrin repeats. Second, using BiFC analysis, we were only able to detect interactions between KEG and ABI5 following inactivation of the KEG RING E3 ligase domain, suggesting turnover of ABI5 by the functional KEG protein. In addition, ABI5 was found to accumulate in tobacco cells co-expressing the KEG RING mutant but not in cells co-expressing wild type KEG. Furthermore, treatment of tobacco cells co-expressing KEG and ABI5 with proteasome inhibitors resulted in accumulation of ABI5. These results support the notion that KEG binds to and ubiquitinates ABI5, which is then degraded by the 26 S proteasome. Finally, using deletion mutants, we show that the C-terminal region of ABI5 regulates stability. Specifically, we demonstrate that lysine 344 is involved in turnover of ABI5 in the cytoplasm. Loss of the C-terminal region of ABI5 or elimination of lysine 344 stabilizes ABI5 resulting in accumulation of the transcription factor in the cytoplasm and also facilitates interactions with wild type KEG. Overall, this study provides strong evidence for the functional relationship between ABI5 and KEG.
ABI5 and ABI5-related bZIP transcription factors (AREBs (ABA responsive element-binding proteins), ABFs (ABRE binding Factors), and/or DPBFs (Dc3 promoter binding factors)), all possess similar domain architecture, consisting of a bZIP DNA-binding domain and four highly conserved regions, C1–C4 (25–27). The interaction between KEG and ABI5 occurs via the conserved C3 region of ABI5. Therefore, it is possible that KEG may also interact with and regulate other family members. However, lysine 344, which is required for KEG regulation of ABI5 abundance, is less conserved among other family members (Fig. 5). For most ABI5-related proteins, a lysine residue can be found within the surrounding amino acids. However, a similarly positioned lysine residue is not the only criteria that may determine regulation by KEG. In addition to the position of the ubiquitin acceptor site, the amino acid sequence or local structure surrounding the site also serves as a determinant for degron function (4). These factors may work together to decide the substrate specificity for KEG among the ABI5-related transcription factors. Nevertheless, this work opens the way to study the regulation of the other ABI5-related bZIP transcription factors.
ABI5 is predicted to be a nucleocytoplasmic protein. Movement across the nuclear envelope requires the presence of nuclear import and export signals, which are recognized by receptor proteins that mediate nucleocytoplasmic transport (28). The position of these signals within the shuttled protein is also of importance because they must be accessible to the nuclear transport machinery (28, 29). ABI5 is predicted to contain two basic NLSs and a leucine-rich NES, which may facilitate nucleocytoplasmic transport. Although the presented results suggest nucleocytoplasmic localization further experiments are required to demonstrate that the predicted NLS and NES sequences are directly involved in the transport of ABI5 across the nuclear envelope. Additional experiments are also required to determine how nuclear import and export of ABI5 may be regulated. The presence of the NLS and NES sequences also suggests that subcellular relocalization may play a role in regulating ABI5 function. Many transcription factors are regulated in this manner including bZIP-type repression of shoot growth (RSG) and common plant regulatory factor 2 (CPRF2), which are retained in the cytoplasm and relocalized to the nucleus in response to external signals such as light or stress (30, 31). However, relocalization may not be the major regulatory step in modulating ABI5 function. Instead, in the absence of ABA, ABI5 seems to be turned over by the ubiquitin proteasome system within the cytoplasm. Evidence provided in this and previous studies demonstrates that KEG interacts with and ubiquitinates ABI5 targeting the protein for proteasomal degradation (11). The KEG-dependent turnover of ABI5 within the cytoplasm may account for the constitutive nuclear localization observed for the transcription factor (14, 15). This notion is supported by the fact that mutations in ABI5 such as deletions within the C-terminal region (e.g. amino acids 344–376) or replacing lysine 344 with alanine, which prohibit degradation and allow for interactions with KEG, facilitate accumulation of ABI5 in the cytoplasm. Based on the results, the C-terminal region of ABI5 seems to regulate both nuclear localization as well as stability. This would explain the mainly cytoplasmic localization of ABI5 when the entire C-terminal region is deleted. Also, ABI5 lacking a nuclear localization signal (NLS3) is only observed in the cytoplasm when the deletion is combined with the stabilizing lysine 344 to alanine mutation. In response to ABA or stress, ABI5 levels increase dramatically due in part to a reduction in proteasomal degradation (7, 9). ABA has been shown to promote KEG self-ubiquitination and subsequent degradation by the proteasome, which would allow for accumulation of ABI5 (12). Despite the decrease in ubiquitin-dependent turnover, ABI5 is not observed in the cytoplasm in the presence of the hormone (14). This is most likely due to the predicted nuclear localization signals, which may immediately shuttle the stabilized transcription factor into the nucleus.
Mutant analysis identified two regions within the C-terminal end of ABI5 that modulate protein stability. As discussed above, the first region containing lysine 344 (amino acids 344–376) is involved in KEG-mediated turnover of ABI5 in the cytoplasm. A second region may reside within amino acids 410–442. ABI5 deletion mutants, ABI5Δ377–409, ABI5Δ406–409, and ABI5Δ406–442 are only observed in the nucleus and all are potential targets for KEG E3 ligase activity (Fig. 6). However, when co-expressed with KEG, ABI5 mutants lacking amino acids 410–442 (ABI5Δ406–442) accumulate to much higher levels compared with other deletion mutants (ABI5Δ377–409 and ABI5Δ406–409) that contain these amino acids (Fig. 6Q). This suggests the presence of another E3 ligase, possibly nuclear localized, which contributes to the turnover of ABI5. The actions of a nuclear-localized E3 ligase may account for the significantly higher accumulation of ABI5Δ344–376,Δ406–409, which combines deletion of amino acids 344–376 and a nuclear localization signal (NSL3), compared with ABI5Δ344–376 that only lacks amino acids 344–376 (Fig. 6Q). Impaired nuclear localization would prohibit degradation by the unknown nuclear localized E3, further reducing the turnover of ABI5. Alternatively, the difference in protein levels may be due to variations in expression, however, this is unlikely as the results shown are consistently observed over independently repeated trials. The presence of another E3 ligase that targets ABI5 for degradation may also account for the increase in ABI5 levels observed following treatment of keg-1 seedlings with ABA (11). Additionally, the rise in ABI5 levels seen in ABA-treated keg-1 seedlings can be attributed to an increase in transcript levels.
The abundance of ABI5 is regulated by ABA at both the transcriptional and post-translational levels. ABI5 transcript is abundant in developing and mature/dried seeds (6, 7). Post-germination, the ABI5 transcript is undetectable but increases in the presence of ABA via the action of multiple ABI loci including ABI5, ABI3, and ABI4 (6, 7). Similar to ABI5, ABI3 and ABI4 encode for transcription factors that may directly regulate ABI5 expression (7, 8). Post-translation regulation of ABI5 abundance requires the function of multiple E3 ligases (10). Each E3 ligase seems to regulate ABI5 abundance under certain circumstances and within specific subcellular compartments. A Cullin-based RING E3 ligase complex, which utilize DWD hypersensitive to ABA 1 (DWA1) and DWA2 as substrate receptors has also been identified as a negative regulator of ABI5 protein abundance (32). Evidence for DWA1/2 involvement in regulating ABI5 abundance comes from the observation that dwa1 or dwa2 single or double mutant plants accumulate higher levels of ABI5 than wild type plants following treatment with ABA (32). Although KEG and DWA1/2 regulate ABI5 stability they do so under different circumstances. KEG functions to keep ABI5 low in the absence of ABA, whereas DWA1 and DWA2 ubiquitinate ABI5 in the nucleus to possibly attenuate the ABA response. Evidence supporting these assumptions is as follows. First, KEG is localized in the cytoplasm/TGN/EE, whereas DWA1 and DWA2 heterodimer is found in the nucleus. Second, keg seedlings exhibit strong post-germinative growth arrest without the need for ABA treatment. By contrast, dwa1 and dwa2 mutants do not show any phenotypic differences compared with the wild type in the absence of ABA. Third, ABI5 is strikingly up-regulated in keg seedlings regardless of the presence or absence of the hormone, whereas ABI5 is not detectable in dwa1 and dwa2 and only accumulates after ABA treatment. Thus KEG and DWA1/2 function cooperatively to modulate ABI5 stability to ensure that protein levels remain low in the absence of ABA or stress stimuli and then kept in check so that the plant can recover following removal of the stress stimulus.
Based on this and previous reports, a model is suggested for modulation of ABI5 abundance (Fig. 9). The presence of both potential nuclear localization and export signals suggests that ABI5 protein is continually shuttled between the nucleus and cytoplasm. Post-germination, in the absence of ABA, the ABI5 transcript and protein levels are low. Turnover of the ABI5 protein involves KEG-mediated ubiquitination and subsequent degradation by the proteasome in the cytoplasm. The KEG-interacting ABI5 may be a nuclear-exported transcription factor that accumulated in developing/mature seeds. In seeds/seedlings lacking KEG function, this pool of ABI5 would be stable and arrest growth post-germination. Additionally, another E3 may regulate nuclear ABI5 through amino acid residues 410–442 to further ensure that ABI5 function is fully suppressed. In response to ABA, ABI5 protein accumulates via increased transcription and decrease in proteasomal degradation. The increase in ABI5 abundance is due in part to the impediment of KEG E3 activity by ABA-induced KEG self-ubiquitination and subsequent degradation. Nuclear ABI5 is activated by phosphorylation and modulates the expression of ABA-responsive genes. When the function of ABI5 is no longer required or needs to be reduced, the transcription factor is degraded by nuclear-localized E3s, such as DWA1 and DWA2 containing Cullin-based RING E3 ligases.
FIGURE 9.
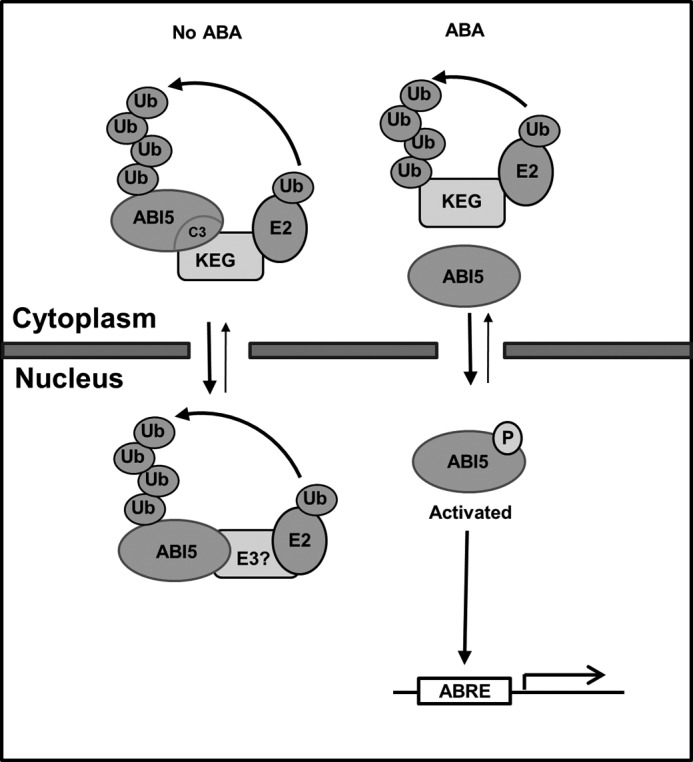
Model for regulating ABI5 protein abundance in the absence and presence of ABA. In the absence of ABA, KEG interacts with the C3 domain of ABI5, ubiquitinates and targets ABI5 for degradation by the proteasome in the cytoplasm. Another E3 may regulate the abundance of nuclear ABI5 utilizing amino acids 410–442. In response to ABA, ABI5 accumulates due to increase transcription and impediment of KEG E3 activity by ABA-mediated KEG self-ubiquitination and degradation. ABI5 is activated by phosphorylation, which leads to the expression of ABA-responsive genes. When the activity of ABI5 is no longer required or needs to be reduced, the transcription factor is degraded by other E3s such as DWA1/2 containing Cullin-based RING E3 ligase.
Supplementary Material
Acknowledgments
We thank Dr. Yuhai Cui for kindly providing the pEarleygate201-YN, pEarleygate202-YC, pGBKT7-DEST, and pGADT7-DEST vectors and Dr. Judy Callis for providing keg seeds. We thank Wendy Lyzenga and Daryl McNeilly for reading of the manuscript.
This work was supported in part by a Natural Science and Engineering Research Council of Canada grant and Human Frontier Science Program Center Developmental Award (to S. L. S.).

This article contains supplemental Table 1.
- ABI5
- abscisic acid insensitive 5
- bZIP
- basic leucine zipper
- ABA
- abscisic acid
- KEG
- KEEP ON GOING
- NLS
- nuclear localization signal
- RING
- really interesting new gene
- TGN/EE
- trans-Golgi network/early endosome
- YC
- carboxyl-terminal fragment of enhanced yellow fluorescence protein
- YN
- amino-terminal fragment of enhanced yellow fluorescence protein
- AD
- activation domain
- BD
- binding domain
- nt
- nucleotide
- BiFC
- bimolecular fluorescence complementation.
REFERENCES
- 1. Vierstra R. D. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10, 385–397 [DOI] [PubMed] [Google Scholar]
- 2. Hammond-Martel I., Yu H., Affar el B. (2012) Roles of ubiquitin signaling in transcription regulation. Cell. Signal. 24, 410–421 [DOI] [PubMed] [Google Scholar]
- 3. Strieter E. R., Korasick D. A. (2012) Unraveling the complexity of ubiquitin signaling. ACS Chem. Biol. 7, 52–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ravid T., Hochstrasser M. (2008) Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 9, 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Radivojac P., Vacic V., Haynes C., Cocklin R. R., Mohan A., Heyen J. W., Goebl M. G., Iakoucheva L. M. (2010) Identification, analysis, and prediction of protein ubiquitination sites. Proteins 78, 365–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Finkelstein R. R., Lynch T. J. (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lopez-Molina L., Mongrand S., Chua N. H. (2001) A post-germination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 98, 4782–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brocard I. M., Lynch T. J., Finkelstein R. R. (2002) Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 129, 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smalle J., Kurepa J., Yang P., Emborg T. J., Babiychuk E., Kushnir S., Vierstra R. D. (2003) Pleiotropic role of the 26 S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15, 965–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu H., Stone S. L. (2011) E3 ubiquitin ligases and abscisic acid signaling. Plant Signal. Behav. 6, 344–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stone S. L., Williams L. A., Farmer L. M., Vierstra R. D., Callis J. (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18, 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu H., Stone S. L. (2010) Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22, 2630–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gu Y., Innes R. W. (2011) The KEEP ON GOING protein of Arabidopsis recruits the ENHANCED DISEASE RESISTANCE1 protein to trans-Golgi network/early endosome vesicles. Plant Physiol. 155, 1827–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lopez-Molina L., Mongrand S., McLachlin D. T., Chait B. T., Chua N. H. (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32, 317–328 [DOI] [PubMed] [Google Scholar]
- 15. Lopez-Molina L., Mongrand S., Kinoshita N., Chua N. H. (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 17, 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nakai K., Horton P. (1999) PSORT, a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34–36 [DOI] [PubMed] [Google Scholar]
- 17. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) The CLUSTAL_X windows interface, flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu Q., Tang X., Tian G., Wang F., Liu K., Nguyen V., Kohalmi S. E., Keller W. A., Tsang E. W., Harada J. J., Rothstein S. J., Cui Y. (2010) Arabidopsis homolog of the yeast TREX-2 mRNA export complex, components and anchoring nucleoporin. Plant J. 61, 259–270 [DOI] [PubMed] [Google Scholar]
- 19. Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., Stevenson D. K., Zimmerman J., Barajas P., Cheuk R., Gadrinab C., Heller C., Jeske A., Koesema E., Meyers C. C., Parker H., Prednis L., Ansari Y., Choy N., Deen H., Geralt M., Hazari N., Hom E., Karnes M., Mulholland C., Ndubaku R., Schmidt I., Guzman P., Aguilar-Henonin L., Schmid M., Weigel D., Carter D. E., Marchand T., Risseeuw E., Brogden D., Zeko A., Crosby W. L., Berry C. C., Ecker J. R. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 [DOI] [PubMed] [Google Scholar]
- 20. Sparkes I. A., Runions J., Kearns A., Hawes C. (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025 [DOI] [PubMed] [Google Scholar]
- 21. Earley K. W., Haag J. R., Pontes O., Opper K., Juehne T., Song K., Pikaard C. S. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629 [DOI] [PubMed] [Google Scholar]
- 22. Clough S. J., Bent A. F. (1998) Floral dip, a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 23. Wang F., Zhu D., Huang X., Li S., Gong Y., Yao Q., Fu X., Fan L. M., Deng X. W. (2009) Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21, 2378–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sedgwick S. G., Smerdon S. J. (1999) The ankyrin repeat, a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24, 311–316 [DOI] [PubMed] [Google Scholar]
- 25. Finkelstein R., Gampala S. S., Lynch T. J., Thomas T. L., Rock C. D. (2005) Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol. Biol. 59, 253–267 [DOI] [PubMed] [Google Scholar]
- 26. Kim S. Y., Ma J., Perret P., Li Z., Thomas T. L. (2002) Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiol. 130, 688–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bensmihen S., Rippa S., Lambert G., Jublot D., Pautot V., Granier F., Giraudat J., Parcy F. (2002) The homologous ABI5 and EEL transcription factors function antagonistically to fine-tune gene expression during late embryogenesis. Plant Cell 14, 1391–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meier I., Somers D. E. (2011) Regulation of nucleocytoplasmic trafficking in plants. Curr. Opin. Plant Biol. 14, 538–546 [DOI] [PubMed] [Google Scholar]
- 29. Suo Y., Miernyk J. A. (2004) Regulation of nucleocytoplasmic localizatioin of the atDjC6 chaperone protein. Protoplasma 224, 79–89 [DOI] [PubMed] [Google Scholar]
- 30. Igarashi D., Ishida S., Fukazawa J., Takahashi Y. (2001) 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13, 2483–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kircher S., Wellmer F., Nick P., Rügner A., Schäfer E., Harter K. (1999) Nuclear import of the parsley bZIP transcription factor CPRF2 is regulated by phytochrome photoreceptors. J. Cell Biol. 144, 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee J. H., Yoon H. J., Terzaghi W., Martinez C., Dai M., Li J., Byun M. O., Deng X. W. (2010) DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 22, 1716–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.