Abstract
Cells have evolved to regulate the asymmetric distribution of specific mRNA targets to institute spatial and temporal control over gene expression. Over the last few decades, evidence has mounted as to the importance of localization elements in the mRNA sequence and their respective RNA-binding proteins. Live imaging methodologies have shown mechanistic details of this phenomenon. In this minireview, we focus on the advanced biochemical and cell imaging techniques used to tweeze out the finer aspects of mechanisms of mRNA movement.
Keywords: Fluorescence, Gene Expression, In Vivo Imaging, RNA Transport, RNA-binding Proteins, RNA-Protein Interaction, 3′-UTR, β-Actin mRNA, Localization Element/Zipcode, mRNA
Introduction
mRNA transport and cytoplasmic compartmentalization of protein synthesis allow precise control over spatial and temporal gene expression and are essential for survival and response to extracellular cues. It has been shown that mRNA localization is essential for cell fate determination (1), directed cell movement (2), and tissue functionality (3). About 3 decades ago, it was discovered that mRNA was asymmetrically distributed in eggs and embryos (4) and in oocytes (5–7) and that this localization was necessary to convey proper embryonic patterning during development. Concurrently, it was found that mRNA could also localize in differentiated somatic cells such as migrating fibroblasts (8), oligodendrocytes (9), and neurons (Table 1) (10). It is now understood that the localization of specific mRNA targets, facilitated by RNA-binding proteins (RBPs),3 is a highly conserved mechanism to spatially restrict protein production, amplify local protein concentration, or even direct integration into macromolecular complexes, sometimes co-translationally (reviewed in Refs. 11–14). RBPs are multifunctional regulators, as they are responsible for processing, localizing, and controlling the translation of a host of mRNA targets. Sometimes, a unique RBP can carry an mRNA from the nucleus to its final destination in a translationally repressed state. Local cues altering its association with the mRNA may ensure compartmentalized translation (15, 16). Many RBPs that govern mRNA localization are preferentially expressed during critical developmental stages, where differentiation, survival, or cell migration is necessary to establish tissue patterning (3). Modifying the expression of trans-acting RBPs (i.e. knockdown/out or overexpression) or mutating cis-acting regulatory elements present in mRNA targets leads to developmental or cognitive deficiencies (17, 18) and a host of disease states (19). Advances in new imaging technologies have allowed the visualization and quantitation of mRNA localization in fixed and living cells, facilitating a more detailed analysis of the molecular mechanisms involved in the process.
TABLE 1.
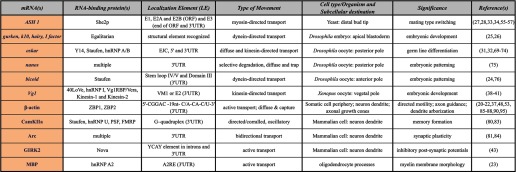
Some mRNAs, RBPs, and types of mRNA movement discussed in text
CamKIIa, Ca2+/calmodulin-dependent protein kinase II; EJC, exon-exon junction complex.
In this minireview, we discuss the current knowledge of mRNA movement from its birth to its final destination within the eukaryotic cell and the impact that biochemical approaches complemented by single molecule imaging techniques have made on the field.
How Does mRNA Localize?
mRNA localization is directed by cis-acting localization elements (LEs), also known as zipcodes, typically present in the 3′-UTR of the transcript (Fig. 1, panel IV). These cis-acting regulatory sequences can range from a few nucleotides to >1 kb in length and are recognized by diverse families of RBPs. Some mRNAs have all the information required for successful localization in a simple element. One of the earliest LE studied was the chicken zipcode in β-actin mRNA. In these early experiments, reporter plasmids expressing different elements of the 3′-UTR of β-actin mRNA narrowed the cis-acting elements to a 54-nucleotide zipcode region that mediated localization of the transcript to the leading edge of chicken fibroblasts (20). More recently, biochemical and structural characterization of this β-actin zipcode led to the identification of a bipartite LE within 28 nucleotides that is specifically recognized by ZBP1 (zipcode-binding protein 1; see “β-Actin mRNA: The Targeted mRNA”) (21, 22). Another example is the myelin basic protein mRNA, which requires an 11-nucleotide element (called A2RE) in its 3′-UTR that is recognized by heterogeneous nuclear ribonucleoprotein (hnRNP) A2 to be properly transported in oligodendrocytes (23).
FIGURE 1.
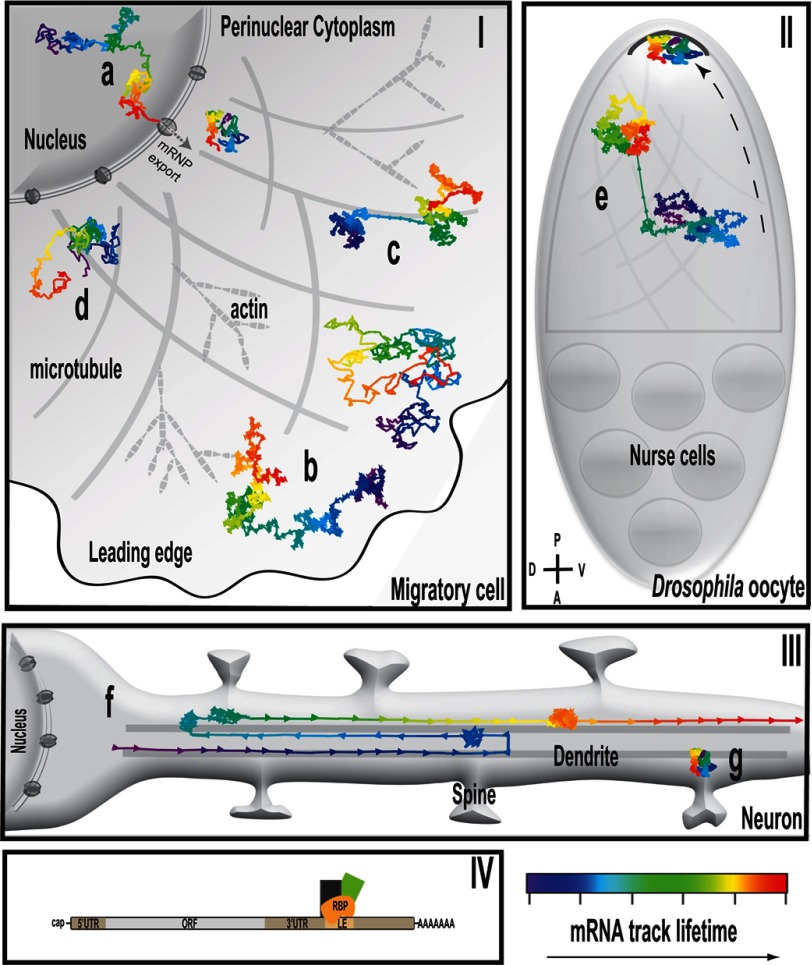
The many roads of mRNA movement. Shown is a simulation of mRNA motility within cells. mRNA tracks represent mRNA movements as a function of time, coded from purple/blue to red. Panel I, fibroblast Cell. a, nuclear mRNAs are subjected to corralled diffusion during much of the tortuous path through the nucleus due to chromatin confinement. b, in motile cells, mRNA is mainly diffusive, although it occasionally travels along the cytoskeleton. mRNAs localized to the leading edge have larger diffusion coefficients on average (c), whereas the movement of perinuclear mRNAs is more confined (d). Panel II, Drosophila oocyte. e, although oskar mRNA is largely diffusive, localization of the mRNA in oocyte stage 9 is accomplished through a slight bias in active transport of mRNA on microtubules toward the posterior pole (P; arrow) (74). A, anterior; D, dorsal; V, ventral. Panel III, neuron. f, neuronal mRNAs depend largely on microtubule-based transport for localization into dendrites. g, Arc mRNAs are seen to be docked beneath dendritic spines, indicating putative domains that maintain mRNAs in specific locations. Panel IV, schematic representation of an mRNA. The cap structure in the 5′-end, the poly(A) tail in the 3′-end, the 5′-UTR, the ORF, the 3′-UTR containing an LE, and the RBPs associated are depicted.
Zipcodes present in mRNAs may also be recognized by RBPs on the basis of secondary structures or stem-loops. For example, Drosophila bicoid mRNA contains a helical region where only secondary structure, not primary sequence, is important for transport (24). The primary sequences of LEs present in gurken, K10, hairy, and the I factor retrotransposon mRNAs are all distinct but may have similar three-dimensional structures. These LEs are recognized by Egalitarian, an RNA adaptor protein involved in dynein-directed transport, illustrating that sequence diversity can enable the recruitment of similar localization machinery (25, 26). A comparable phenomenon has been shown in yeast. The ASH1 (asymmetric synthesis of HO 1) mRNA contains four different zipcodes (E1, E2A, E2B, and E3) spread through the transcript that do not show homology in primary sequence, but each one is able to mediate mRNA localization to the bud tip using the same RBP, She2p (SWI5-dependent HO expression 2 protein) (27, 28).
A large number of LEs have been identified and characterized in different systems. However, a clear pattern in zipcode primary sequence or structure has not yet emerged, indicating the heterogeneity of these motifs (extensively reviewed in Ref. 29).
Role of Nuclear Encoding in mRNA Localization
Processing of the pre-mRNA in the nucleus influences the cytoplasmic fate of mRNA, illustrating the elegant integration of spatiotemporal events that can occur within an individual cell. Pre-mRNA modifications include the addition of the cap structure to the 5′-end, the addition of poly(A) to the 3′-end, and the deposition of the exon-exon junction complex proteins with the removal of introns during splicing (30). Deposition of RBPs onto the transcript during these events in the nucleus determines its final destination. For instance, splicing at the first intron of Drosophila oskar mRNA supports posterior pole cytoplasmic localization of the transcript with Y14, a component of the exon-exon junction complex and essential factor of the transport machinery (31, 32). In yeast, She2p is the RNA adaptor protein involved in myosin-based transport required for ASH1 mRNA to be exported to the cytoplasm and transported to the daughter cell by binding the mRNA and linking it to the cytoskeleton (33, 34). Once in the cytosol, the messenger ribonucleoprotein particle (mRNP) may gain or lose additional factors that determine whether the mRNA is freely diffusive, actively transported upon the cytoskeleton by molecular motors (kinesins, dyneins, and myosins), protected from degradation, able to translate, or anchored to a compartmentalized domain (Fig. 1 and supplemental figure and movie) (reviewed in Refs. 11 and 13). mRNAs are transported in large and diverse multiprotein complexes. In addition to RBPs, it has been suggested that noncoding RNAs and microRNAs might be components of these large complexes as well (35).
Ripping, Clipping, and Chipping Away at mRNP Composition
Defining the RBPs that associate with specific mRNAs can yield critical information about how mRNA trafficking and localization are regulated in different cellular compartments. Biochemical methodologies to study mRNA interaction with specific RBPs usually rely on standard immunoprecipitation and/or affinity purification techniques, followed by the identification and characterization of the molecules that are part of the complexes. For example, RNA affinity purification methods led to the identification of ZBP1, the founding member of the VICKZ (Vg1 RBP/Vera, IMP-1,2,3, CRD-BP, KOC, ZBP1) family of RBPs (36), in the localization of β-actin mRNA in fibroblasts (37). RNA affinity chromatography also led to the identification of the novel protein 40LoVe, a member of the hnRNP D family of proteins, in the vegetal localization of Vg1 mRNA in Xenopus oocytes. 40LoVe was found to bind specifically to two different late LEs of the Vg1 mRNA and to be a necessary component of the Vg1 LE RNP (38). hnRNP I and Vg1 RBP/Vera had previously been described as factors associating with Vg1 mRNA in the nucleus and being exported into the cytoplasm with the transcript (39). In addition, in vivo and in vitro studies showed that transport of Vg1 mRNA to the vegetal cortex of the oocyte implicates different microtubule plus-end-directed motors (40, 41). Kinesin-1 associates with Vg1 LE RNA; blocking its function by using specific antibodies prevents RNA localization, suggesting a direct role for kinesin-1 in vegetal localization of the mRNA. Kinesin-1 also interacts with kinesin-2, and both motor proteins seem to facilitate Vg1 mRNA movement to its final destination, providing new mechanistic insight into this motor-driven RNA transport process in frog oocytes (41).
Cross-linking and immunoprecipitation (CLIP) (42) is an effective and powerful technique for identifying LEs and has the advantage of isolating RNA-protein complexes under physiological conditions. CLIP coupled with high-throughput sequencing has recently identified an LE in the 3′-UTR of GIRK2 (G-protein-activated inwardly rectifying potassium (K) channel 2) mRNA that mediates Nova-dependent localization of its transcript in primary neurons (43). This finding suggests an attractive hypothesis in which the loading of the splicing factor Nova onto intronic sequences in the nucleus could be coupled to the mechanism of localization directed by the 3′-UTR of the same mRNA.
Characterization of zipcode sequences that are required for localizing mRNA involves visualizing the subcellular distribution of reporter RNAs carrying putative zipcode fragments. Fusing an essential LE to a reporter RNA should result in a localization pattern within the cell similar to that observed for the endogenous mRNA. Conversely, deletion of regions or point mutations that disrupt localization can be used to see how certain sequences affect mRNA localization. When the RBP is known, putative LEs can be easily characterized on the basis of their binding ability in vitro using EMSA (21, 44). Information regarding the binding specificity and affinity is valuable; however, spatial (and temporal) evidence of RNA-protein association within cells is imperative and better determined through imaging methods.
Seeing Is Believing
Originally, intracellular localization of mRNAs was only observed using in situ hybridization techniques in fixed samples (8, 45). In situ detection remains the standard tool for examining the distribution of mRNAs in fixed cells, tissues, and larger samples like Drosophila or zebrafish embryos. Lécuyer et al. (46) characterized the distribution patterns of >3000 transcripts during early developmental stages of Drosophila embryogenesis by using high-resolution fluorescence in situ hybridization procedures. Surprisingly, 70% of the mRNAs analyzed showed different subcellular localization rather than uniform distribution, suggesting that mRNA localization mechanisms are involved in the control of the majority of mRNAs.
Although most of the analysis methods to study mRNA localization in fixed cells have been qualitative (8, 47), recent advances have resulted in more objective and quantitative measurements for mRNA distribution (48, 49). Additionally, due to improvements in sensitivity and resolution of mRNA detection using live microscopy, it is now feasible to visualize single molecules of mRNA to quantify their movements in real time (50–53). Direct observation and quantitation of ensemble mRNA distributions enable further exploration of mRNA dynamics and movement mechanisms.
Prior to the widespread use of high-resolution single mRNA live imaging, mRNA localization was proposed to operate through the following mechanisms: (a) directional transport along cytoskeletal elements, (b) random diffusion and local trapping of mRNAs, (c) vectorial export from the nucleus and trapping, or (d) local protection from degradation (13). Thus far, live imaging techniques (reviewed extensively in Ref. 54) in a variety of cell lineages have provided examples of all of these behaviors. Not surprisingly, the method utilized by a cell to produce an asymmetric distribution of mRNA is finely tuned to be appropriate for the particular morphology and time constraints that the cell must overcome. For instance, during yeast cell division, ASH1 mRNA is preferentially localized into the bud tip of the daughter cell, ensuring asymmetry of HO gene expression, which is essential for mating type switching (55, 56). Pioneering the MS2 system to visualize mRNA for the first time in live cells, Bertrand et al. (57) demonstrated that ASH1 mRNA localization was due to movements consistent with myosin-directed motility. Since then, the MS2 technique has been expanded to mammalian cells and even whole organisms (50, 58–60).
RNA Movement within the Nucleus
Shav-Tal et al. (61) imaged single reporter mRNAs in the nucleus, and they found that movements of mRNA were governed by the laws of diffusion and not active transport (Fig. 1, panel I, a, and supplemental figure). Furthermore, they did not observe mRNAs docking in particular nuclear domains, indicating that mRNP assembly likely occurs co-transcriptionally. Additional studies showed that although mRNA length does not affect overall mRNA diffusion kinetics in the nucleus (∼0.005–0.02 μm2/s), larger transcripts take longer to reach the cytoplasm due to increased frequency of corralled diffusion (62), a consequence of an increased tortuous path through the chromatin (63). Mor et al. (62) also calculated that, on average, mRNA takes ∼20 min to travel from the transcription site to the nuclear pore for export (Fig. 1, panel I, a, and supplemental figure), and interestingly, inclusion of introns expedited export rates but not nucleoplasmic diffusion rates (64). Single molecule tracking analysis of mRNA through labeled nuclear pores utilizing live cell imaging determined that nuclear mRNA export takes less than one-fifth of 1 s (51), an order of magnitude slower than the transport of proteins (65). Remarkably, in 10% of the export events, mRNAs remained docked at the nuclear pores for longer periods of time (approximately seconds), indicating a rate-limiting step, possibly related to quality control (51, 66).
RNA Movement within the Cytoplasm
Similar to the nucleus, movement of mRNAs in the cytoplasm of cells is also largely governed by diffusion, although it can be up to two times faster due to a less restrictive environment (Fig. 1, panel I, and supplemental figure and movie) (62). Fusco et al. (58) were the first to observe that more than half of lacZ reporter mRNAs are freely diffusing with a diffusion coefficient of 0.45 × 10−9 cm2/s. The remainder of the mRNAs are either static or corralled, corresponding to ∼20% associated with the cytoskeleton and the remaining confined by microtubule-based domains. Five percent of the mRNAs move along microtubules in a directional manner at rates of 1–1.5 μm/s for lengths of up to 3 μm (Fig. 1, panel I, b, and supplemental figure and movie) (58). Interestingly, addition of the β-actin mRNA zipcode element to the reporter mRNA increased the frequency of active transport to 20% as well as the length of transport, implying that cis-acting elements may influence localization. A comparable mechanism is exploited during Drosophila oogenesis, where localization of oskar mRNA to the posterior pole must be accomplished at the appropriate developmental stage to allow germ line differentiation (reviewed in Ref. 67). Fixed cell imaging revealed that intact microtubules (68), kinesin (69), and several trans-acting factors such as Staufen and hnRNP A/B (70–73) are necessary for oskar mRNA localization, implicating active transport as the localization mechanism. Contrary to expectations of concerted movement to the pole, live cell imaging of oskar mRNA revealed that the majority of mRNA diffuses randomly, with only 13% being actively transported (Fig. 1, panel II, e) (74). Examination of actively transported mRNAs revealed a 7% bias in transport in the direction of the posterior pole due to a subtle bias in microtubule orientation. Likewise, nanos mRNA localizes to the posterior pole of Drosophila embryos during a critical developmental window, albeit via an alternative mechanism. nanos localization relies on diffusion and actin-dependent entrapment of mRNA at the pole, receiving help from the forces of cytoplasmic streaming toward the pole (75). bicoid mRNA localization to the anterior Drosophila oocyte in the later stages of oogenesis depends on yet another mechanism. Dynein continuously transports bicoid mRNA toward the anterior part of the oocyte, as docking there is insufficient to maintain localization (76). It is logical to speculate that the most energetically efficient and thus preferred method of mRNA localization is diffusion and docking. In the absence of these options, or alternatively, if the timing of mRNA localization is a limiting factor, cells such as neurons may rely primarily on active transport to maintain an asymmetric mRNA distribution.
RNA Movement in Neurons
Neurons are highly polarized cells that rely primarily on active transport mechanisms for localizing mRNAs into dendrites (Fig. 1, panel III) (77, 78). The field of mRNA kinetics in neuronal dendrites remains largely observational, as investigations under way are still characterizing the movements as a precursor to understanding how they are achieved. One of the earlier visualizations of mRNA in neuronal dendrites revealed that a reporter mRNA (GFP-MS2-Ca2+/calmodulin-dependent protein kinase II 3′-UTR) exhibited kinesin- and microtubule-dependent oscillatory motion (79). Additional imaging studies of fluorescent RBPs have also found that mRNP granules exhibit oscillatory behavior in dendrites at speeds up to 2 μm/s (Fig. 1, panel III, f) (50, 80). Analysis of single Arc (activity-regulated cytoskeleton-associated) mRNAs in dendrites revealed that approximately half of the mRNA population was motile (81). This motile population exhibited both small bidirectional jumps and longer tracks at constant speed. The population of lengthier translocations occurred in both directions, with anterograde movements being longer than the retrograde ones.
Live cell imaging of mRNA movement in neuronal dendrites has been quantitative for studying the contribution of RBPs to localization. Currently, evidence indicates that RBPs in neurons alter the efficacy of active transport and may be an attractive target for regulation of mRNA movement. Mutation of the Drosophila FMR1 gene (fragile X mental retardation 1) was found to decrease the net distance of labeled mRNAs, implicating fragile X mental retardation protein (FMRP) as a “processivity factor” that increases mRNA interaction with motors (82). However, the total distance traveled was unaltered because the decrease in directional mRNA movement was compensated with an increase in oscillatory behavior in FMRP mutant neurons. Furthermore, the presence of wild-type FMRP in the neuron allowed increased mRNA shuttling from the soma into the dendrites as well as an increased mobile fraction of dendritic mRNA. For Chic mRNA, FMRP knockdown resulted in a reversal in the directional bias of movement from 58% anterograde (WT) to 74% retrograde (Drosophila FMR1 mutants). A subsequent study of FMRP-regulated mRNA localization in neurons confirmed that FMRP increases the fraction of motile particles as well as the displacement of dendritic Ca2+/calmodulin-dependent protein kinase II mRNA (83). An additional dimension of the regulation of mRNA localization in neurons is the synaptic activity-induced alteration of mRNA transport, which is proposed to play a role in synaptic plasticity. Rook et al. (79) found that depolarization induces an increase in anterograde mRNA motility into the dendrites. Dictenberg et al. (83) studied the increase in dendritic FMRP upon stimulation of metabotropic glutamate receptors and found that following stimulation, FMRP increases its association with its delegated motor, the kinesin KIF5, bringing it into the dendrites. Consistent with this, knock-out of FMRP reduces the steady-state localization of FMRP targets and does not exhibit metabotropic glutamate receptor stimulation-induced mRNA localization, suggesting the role of RBPs in activity-controlled transport of mRNAs essential for synaptogenesis.
The hypothesis that local translation is necessary for synapse-specific modifications suggests that mRNA should be present at the right time in a specific location to immediately contribute to the local protein pool. An example of this behavior is Arc mRNA, which is retained in domains directly beneath spines in a UTR-dependent manner (Fig. 1, panel III, g) (84). This raises intriguing questions as to how specific mRNAs are targeted and retained at the appropriate places within the cell. Dynes and Steward (81) observed that actively transported mRNAs may travel vectorially to a destination at the dendritic spine (Fig. 1, panel III). This strongly suggests that transported mRNAs may be responding to local cues to stall or be retained in certain domains.
β-Actin mRNA: The Targeted mRNA
β-Actin mRNA can be transported and targeted to subcellular compartments to undergo local translation (15, 53). Over the last 30 years, β-actin mRNA localization has provided a model system for understanding the mechanisms and purpose of mRNA localization within eukaryotic cells. Biochemical, structural analysis, and imaging approaches have provided an elegant understanding of how the zipcode and zipcode-binding proteins act together to ensure the fate of the transcript once it is synthesized in the nucleus.
β-Actin mRNA requires the presence of a zipcode in its 3′-UTR to be targeted to the leading edge in fibroblasts (20) and in dendritic filopodia and axonal growth cones (85, 86). ZBP1 has been identified by affinity purification methods (37) as the key factor that binds the β-actin mRNA zipcode in the nucleus and is involved in localization as well as translational repression of its mRNA target in the cytoplasm (15, 87–89). ZBP2, the mouse homolog of the human hnRNP protein KSRP (K-homology splicing regulator protein), also binds to the β-actin 3′-UTR in the nucleus and facilitates nuclear ZBP1 association with the transcript and further cytoplasmic localization in fibroblasts (87, 88, 90). Structural studies have recently shown that ZBP1 KH34 (third and fourth hnRNP K-homology domains) specifically binds the bipartite β-actin 3′-UTR element, with KH4 and KH3 recognizing 5′-CGGAC-3′ and 5′-(C/A)CA(C/U)-3′ sequences, respectively (21, 22). Abolition of the function of the zipcode by mutation of the element itself, treatment with specific antisense oligonucleotides, or knockdown/out of ZBP1 protein leads to the mislocalization of β-actin mRNA and subsequent alterations of cell morphology, motility, and adhesion as well as failures in synaptic growth and deficiencies in dendritic spine number, maturation, and arborization (2, 20, 22, 53, 85, 86, 91–94).
Live imaging of β-actin mRNA in different regions of COS cells revealed that restricted mRNA is able to diffuse freely in the leading edge of the cell (Fig. 1, panel I, c, and supplemental figure), but in the perinuclear region, mRNA diffusion is restricted (Fig. 1, panel I, d, and supplemental figure). Disruption of the actin cytoskeleton by cytochalasin D delocalizes the mRNA from the leading edge and increases its mobility in the perinuclear region, indicating that the cytoskeletal actin environment strongly contributes to the location of β-actin mRNA within fibroblast cells (48, 95). Once at the leading edge, β-actin mRNA dwells around adhesions in fibroblasts to provide a novel protein source for adhesion maturation, which in turn regulates directed motility (53). In neurons, growth factor stimulation induces β-actin mRNA and ZBP1 protein transport into growth cones (86). Local translation of β-actin mRNA requires phosphorylation on ZBP1 Tyr-396 by Src kinase, a known active component of leading edge adhesions. Similarly, this regulatory mechanism was shown to be necessary for neuron growth cone turning toward a chemotactic cue (15, 94, 96–98).
Although work until now has focused largely on the β-actin mRNA-ZBP1 complex, it is worthwhile to mention that this interaction is not exclusive. β-Actin mRNA may be bound and regulated by many other proteins, and in turn, ZBP1 can bind and regulate at least 116 other mRNAs (22, 99, 100). Post-transcriptional regulation requires the proper interaction of multiple RBPs along with the mRNA. The present challenge involves solving the intricate network of associations of RBPs with motors and multiple mRNA targets. This will allow a better understanding of the molecular mechanisms that govern mRNA movement and localization in live cells.
Conclusions
In recent decades, it has become evident that localization of mRNAs within cells is a widespread and evolutionarily conserved strategy for asymmetric distribution and concentration of mRNP complexes at specific sites. A vast number of mRNAs show subcellular distribution. RNA movement and localization are cell-specific. Local environmental conditions, cytoskeletal constraints, and specific docking sites are important for diffusion-based localization. In situations where active transport is necessary, regulation is exerted on cytoskeletal orientation and the association of RBPs with motors. In all of the aforementioned examples, mRNA movement is probabilistic, with biases introduced by zipcodes and RBPs. Currently, efforts are focused on understanding similarities and differences between diverse mRNA subtypes that localize in the same manner and the molecular mechanisms that govern their targeting to specific subcompartments within the cell as well as the biological significance associated with the localization event. The field of mRNA imaging was previously hindered by technological limitations; however, it is now at a turning point, where we are able to visualize with accurate precision the methods of mRNA localization. The dynamics and stoichiometry of mRNP compositions need to be revealed. Combining high-throughput biochemical, bioinformatic, and imaging methodologies with functional analyses will provide answers to the questions of how a specific mRNA moves and localizes within cells and its phenotypic function.
Supplementary Material
Acknowledgment
We thank Dr. Brian English for generating the mRNA motility simulations for Fig. 1.
This work was supported, in whole or in part, by National Institutes of Health Grants T32 GM007491 (to Z. B. K.) and GM84364, GM57071, and GM86217 (to R. H. S.). This work was also supported by a Human Frontier Science Program (HFSP) fellowship (to C. E.).

This article contains a supplemental figure and movie.
- RBP
- RNA-binding protein
- LE
- localization element
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- mRNP
- messenger ribonucleoprotein particle
- CLIP
- cross-linking and immunoprecipitation
- FMRP
- fragile X mental retardation protein.
REFERENCES
- 1. Jedrusik A., Parfitt D. E., Guo G., Skamagki M., Grabarek J. B., Johnson M. H., Robson P., Zernicka-Goetz M. (2008) Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev. 22, 2692–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kislauskis E. H., Zhu X., Singer R. H. (1997) β-Actin messenger RNA localization and protein synthesis augment cell motility. J. Cell Biol. 136, 1263–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tolino M., Köhrmann M., Kiebler M. A. (2012) RNA-binding proteins involved in RNA localization and their implications in neuronal diseases. Eur. J. Neurosci. 35, 1818–1836 [DOI] [PubMed] [Google Scholar]
- 4. Jeffery W. R., Tomlinson C. R., Brodeur R. D. (1983) Localization of actin messenger RNA during early ascidian development. Dev. Biol. 99, 408–417 [DOI] [PubMed] [Google Scholar]
- 5. Berleth T., Burri M., Thoma G., Bopp D., Richstein S., Frigerio G., Noll M., Nüsslein-Volhard C. (1988) The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 7, 1749–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Frigerio G., Burri M., Bopp D., Baumgartner S., Noll M. (1986) Structure of the segmentation gene paired and the Drosophila PRD gene set as part of a gene network. Cell 47, 735–746 [DOI] [PubMed] [Google Scholar]
- 7. Rebagliati M. R., Weeks D. L., Harvey R. P., Melton D. A. (1985) Identification and cloning of localized maternal RNAs from Xenopus eggs. Cell 42, 769–777 [DOI] [PubMed] [Google Scholar]
- 8. Lawrence J. B., Singer R. H. (1986) Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell 45, 407–415 [DOI] [PubMed] [Google Scholar]
- 9. Trapp B. D., Moench T., Pulley M., Barbosa E., Tennekoon G., Griffin J. (1987) Spatial segregation of mRNA encoding myelin-specific proteins. Proc. Natl. Acad. Sci. U.S.A. 84, 7773–7777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garner C. C., Tucker R. P., Matus A. (1988) Selective localization of messenger RNA for cytoskeletal protein MAP2 in dendrites. Nature 336, 674–677 [DOI] [PubMed] [Google Scholar]
- 11. Holt C. E., Bullock S. L. (2009) Subcellular mRNA localization in animal cells and why it matters. Science 326, 1212–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marchand V., Gaspar I., Ephrussi A. (2012) An intracellular transmission control protocol: assembly and transport of ribonucleoprotein complexes. Curr. Opin. Cell Biol. 24, 202–210 [DOI] [PubMed] [Google Scholar]
- 13. Medioni C., Mowry K., Besse F. (2012) Principles and roles of mRNA localization in animal development. Development 139, 3263–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Condeelis J., Singer R. H. (2005) How and why does β-actin mRNA target? Biol. Cell 97, 97–110 [DOI] [PubMed] [Google Scholar]
- 15. Hüttelmaier S., Zenklusen D., Lederer M., Dictenberg J., Lorenz M., Meng X., Bassell G. J., Condeelis J., Singer R. H. (2005) Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438, 512–515 [DOI] [PubMed] [Google Scholar]
- 16. Stöhr N., Köhn M., Lederer M., Glass M., Reinke C., Singer R. H., Hüttelmaier S. (2012) IGF2BP1 promotes cell migration by regulating MK5 and PTEN signaling. Genes Dev. 26, 176–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Darnell J. C., Van Driesche S. J., Zhang C., Hung K. Y., Mele A., Fraser C. E., Stone E. F., Chen C., Fak J. J., Chi S. W., Licatalosi D. D., Richter J. D., Darnell R. B. (2011) FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miller S., Yasuda M., Coats J. K., Jones Y., Martone M. E., Mayford M. (2002) Disruption of dendritic translation of CaMKIIα impairs stabilization of synaptic plasticity and memory consolidation. Neuron 36, 507–519 [DOI] [PubMed] [Google Scholar]
- 19. Liu-Yesucevitz L., Bassell G. J., Gitler A. D., Hart A. C., Klann E., Richter J. D., Warren S. T., Wolozin B. (2011) Local RNA translation at the synapse and in disease. J. Neurosci. 31, 16086–16093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kislauskis E. H., Zhu X., Singer R. H. (1994) Sequences responsible for intracellular localization of β-actin messenger RNA also affect cell phenotype. J. Cell Biol. 127, 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chao J. A., Patskovsky Y., Patel V., Levy M., Almo S. C., Singer R. H. (2010) ZBP1 recognition of β-actin zipcode induces RNA looping. Genes Dev. 24, 148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Patel V. L., Mitra S., Harris R., Buxbaum A. R., Lionnet T., Brenowitz M., Girvin M., Levy M., Almo S. C., Singer R. H., Chao J. A. (2012) Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev. 26, 43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Munro T. P., Magee R. J., Kidd G. J., Carson J. H., Barbarese E., Smith L. M., Smith R. (1999) Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J. Biol. Chem. 274, 34389–34395 [DOI] [PubMed] [Google Scholar]
- 24. Macdonald P. M., Kerr K. (1998) Mutational analysis of an RNA recognition element that mediates localization of bicoid mRNA. Mol. Cell. Biol. 18, 3788–3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bullock S. L., Ringel I., Ish-Horowicz D., Lukavsky P. J. (2010) A′-form RNA helices are required for cytoplasmic mRNA transport in Drosophila. Nat. Struct. Mol. Biol. 17, 703–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dienstbier M., Boehl F., Li X., Bullock S. L. (2009) Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 23, 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chartrand P., Meng X. H., Singer R. H., Long R. M. (1999) Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol. 9, 333–336 [DOI] [PubMed] [Google Scholar]
- 28. Gonzalez I., Buonomo S. B., Nasmyth K., von Ahsen U. (1999) ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr. Biol. 9, 337–340 [DOI] [PubMed] [Google Scholar]
- 29. Jambhekar A., Derisi J. L. (2007) Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA 13, 625–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hocine S., Singer R. H., Grünwald D. (2010) RNA processing and export. Cold Spring Harb. Perspect. Biol. 2, a000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hachet O., Ephrussi A. (2001) Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 11, 1666–1674 [DOI] [PubMed] [Google Scholar]
- 32. Hachet O., Ephrussi A. (2004) Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature 428, 959–963 [DOI] [PubMed] [Google Scholar]
- 33. Kruse C., Jaedicke A., Beaudouin J., Bohl F., Ferring D., Guttler T., Ellenberg J., Jansen R. P. (2002) Ribonucleoprotein-dependent localization of the yeast class V myosin Myo4p. J. Cell Biol. 159, 971–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Long R. M., Gu W., Lorimer E., Singer R. H., Chartrand P. (2000) She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J. 19, 6592–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bramham C. R., Wells D. G. (2007) Dendritic mRNA: transport, translation and function. Nat. Rev. Neurosci. 8, 776–789 [DOI] [PubMed] [Google Scholar]
- 36. Yisraeli J. K. (2005) VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biol. Cell 97, 87–96 [DOI] [PubMed] [Google Scholar]
- 37. Ross A. F., Oleynikov Y., Kislauskis E. H., Taneja K. L., Singer R. H. (1997) Characterization of a β-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 17, 2158–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Czaplinski K., Köcher T., Schelder M., Segref A., Wilm M., Mattaj I. W. (2005) Identification of 40LoVe, a Xenopus hnRNP D family protein involved in localizing a TGF-β-related mRNA during oogenesis. Dev Cell 8, 505–515 [DOI] [PubMed] [Google Scholar]
- 39. Kress T. L., Yoon Y. J., Mowry K. L. (2004) Nuclear RNP complex assembly initiates cytoplasmic RNA localization. J. Cell Biol. 165, 203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Betley J. N., Heinrich B., Vernos I., Sardet C., Prodon F., Deshler J. O. (2004) Kinesin II mediates Vg1 mRNA transport in Xenopus oocytes. Curr. Biol. 14, 219–224 [DOI] [PubMed] [Google Scholar]
- 41. Messitt T. J., Gagnon J. A., Kreiling J. A., Pratt C. A., Yoon Y. J., Mowry K. L. (2008) Multiple kinesin motors coordinate cytoplasmic RNA transport on a subpopulation of microtubules in Xenopus oocytes. Dev. Cell 15, 426–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ule J., Jensen K. B., Ruggiu M., Mele A., Ule A., Darnell R. B. (2003) CLIP identifies Nova-regulated RNA networks in the brain. Science 302, 1212–1215 [DOI] [PubMed] [Google Scholar]
- 43. Racca C., Gardiol A., Eom T., Ule J., Triller A., Darnell R. B. (2010) The neuronal splicing factor Nova co-localizes with target RNAs in the dendrite. Front. Neural Circuits 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ryder S. P., Recht M. I., Williamson J. R. (2008) Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol. Biol. 488, 99–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Singer R. H., Ward D. C. (1982) Actin gene expression visualized in chicken muscle tissue culture by using in situ hybridization with a biotinated nucleotide analog. Proc. Natl. Acad. Sci. U.S.A. 79, 7331–7335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lécuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., Hughes T. R., Tomancak P., Krause H. M. (2007) Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131, 174–187 [DOI] [PubMed] [Google Scholar]
- 47. Latham V. M., Jr., Kislauskis E. H., Singer R. H., Ross A. F. (1994) β-Actin mRNA localization is regulated by signal transduction mechanisms. J. Cell Biol. 126, 1211–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yamagishi M., Shirasaki Y., Funatsu T. (2009) Size-dependent accumulation of mRNA at the leading edge of chicken embryo fibroblasts. Biochem. Biophys. Res. Commun. 390, 750–754 [DOI] [PubMed] [Google Scholar]
- 49. Park H. Y., Trcek T., Wells A. L., Chao J. A., Singer R. H. (2012) An unbiased analysis method to quantify mRNA localization reveals its correlation with cell motility. Cell Rep. 1, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lionnet T., Czaplinski K., Darzacq X., Shav-Tal Y., Wells A. L., Chao J. A., Park H. Y., de Turris V., Lopez-Jones M., Singer R. H. (2011) A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat. Methods 8, 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Grünwald D., Singer R. H. (2010) In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature 467, 604–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu B., Chao J. A., Singer R. H. (2012) Fluorescence fluctuation spectroscopy enables quantitative imaging of single mRNAs in living cells. Biophys. J. 102, 2936–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Katz Z. B., Wells A. L., Park H. Y., Wu B., Shenoy S. M., Singer R. H. (2012) β-Actin mRNA compartmentalization enhances focal adhesion stability and directs cell migration. Genes Dev. 26, 1885–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Weil T. T., Parton R. M., Davis I. (2010) Making the message clear: visualizing mRNA localization. Trends Cell Biol. 20, 380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Long R. M., Singer R. H., Meng X., Gonzalez I., Nasmyth K., Jansen R. P. (1997) Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277, 383–387 [DOI] [PubMed] [Google Scholar]
- 56. Takizawa P. A., Sil A., Swedlow J. R., Herskowitz I., Vale R. D. (1997) Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature 389, 90–93 [DOI] [PubMed] [Google Scholar]
- 57. Bertrand E., Chartrand P., Schaefer M., Shenoy S. M., Singer R. H., Long R. M. (1998) Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2, 437–445 [DOI] [PubMed] [Google Scholar]
- 58. Fusco D., Accornero N., Lavoie B., Shenoy S. M., Blanchard J. M., Singer R. H., Bertrand E. (2003) Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol. 13, 161–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Belaya K., St Johnston D. (2011) Using the mRNA-MS2/MS2CP-FP system to study mRNA transport during Drosophila oogenesis. Methods Mol. Biol. 714, 265–283 [DOI] [PubMed] [Google Scholar]
- 60. Grünwald D., Singer R. H., Czaplinski K. (2008) Cell biology of mRNA decay. Methods Enzymol. 448, 553–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shav-Tal Y., Darzacq X., Shenoy S. M., Fusco D., Janicki S. M., Spector D. L., Singer R. H. (2004) Dynamics of single mRNPs in nuclei of living cells. Science 304, 1797–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mor A., Suliman S., Ben-Yishay R., Yunger S., Brody Y., Shav-Tal Y. (2010) Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat. Cell Biol. 12, 543–552 [DOI] [PubMed] [Google Scholar]
- 63. Siebrasse J. P., Veith R., Dobay A., Leonhardt H., Daneholt B., Kubitscheck U. (2008) Discontinuous movement of mRNP particles in nucleoplasmic regions devoid of chromatin. Proc. Natl. Acad. Sci. U.S.A. 105, 20291–20296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rodríguez-Navarro S., Hurt E. (2011) Linking gene regulation to mRNA production and export. Curr. Opin. Cell Biol. 23, 302–309 [DOI] [PubMed] [Google Scholar]
- 65. Grünwald D., Singer R. H., Rout M. (2011) Nuclear export dynamics of RNA-protein complexes. Nature 475, 333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Siebrasse J. P., Kaminski T., Kubitscheck U. (2012) Nuclear export of single native mRNA molecules observed by light sheet fluorescence microscopy. Proc. Natl. Acad. Sci. U.S.A. 109, 9426–9431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Becalska A. N., Gavis E. R. (2009) Lighting up mRNA localization in Drosophila oogenesis. Development 136, 2493–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Clark I., Giniger E., Ruohola-Baker H., Jan L. Y., Jan Y. N. (1994) Transient posterior localization of a kinesin fusion protein reflects anteroposterior polarity of the Drosophila oocyte. Curr. Biol. 4, 289–300 [DOI] [PubMed] [Google Scholar]
- 69. Brendza R. P., Serbus L. R., Duffy J. B., Saxton W. M. (2000) A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science 289, 2120–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Huynh J. R., Munro T. P., Smith-Litière K., Lepesant J. A., St Johnston D. (2004) The Drosophila hnRNPA/B homolog, Hrp48, is specifically required for a distinct step in osk mRNA localization. Dev. Cell 6, 625–635 [DOI] [PubMed] [Google Scholar]
- 71. St Johnston D., Beuchle D., Nüsslein-Volhard C. (1991) staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell 66, 51–63 [DOI] [PubMed] [Google Scholar]
- 72. Kim-Ha J., Smith J. L., Macdonald P. M. (1991) oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell 66, 23–35 [DOI] [PubMed] [Google Scholar]
- 73. Yano T., López de Quinto S., Matsui Y., Shevchenko A., Shevchenko A., Ephrussi A. (2004) Hrp48, a Drosophila hnRNPA/B homolog, binds and regulates translation of oskar mRNA. Dev. Cell 6, 637–648 [DOI] [PubMed] [Google Scholar]
- 74. Zimyanin V. L., Belaya K., Pecreaux J., Gilchrist M. J., Clark A., Davis I., St Johnston D. (2008) In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell 134, 843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Forrest K. M., Gavis E. R. (2003) Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr. Biol. 13, 1159–1168 [DOI] [PubMed] [Google Scholar]
- 76. Weil T. T., Forrest K. M., Gavis E. R. (2006) Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Dev. Cell 11, 251–262 [DOI] [PubMed] [Google Scholar]
- 77. Wang D. O., Martin K. C., Zukin R. S. (2010) Spatially restricting gene expression by local translation at synapses. Trends Neurosci. 33, 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Doyle M., Kiebler M. A. (2011) Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J. 30, 3540–3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Rook M. S., Lu M., Kosik K. S. (2000) CaMKIIα 3′ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J. Neurosci. 20, 6385–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Knowles R. B., Sabry J. H., Martone M. E., Deerinck T. J., Ellisman M. H., Bassell G. J., Kosik K. S. (1996) Translocation of RNA granules in living neurons. J. Neurosci. 16, 7812–7820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Dynes J. L., Steward O. (2007) Dynamics of bidirectional transport of Arc mRNA in neuronal dendrites. J. Comp. Neurol. 500, 433–447 [DOI] [PubMed] [Google Scholar]
- 82. Estes P. S., O'Shea M., Clasen S., Zarnescu D. C. (2008) Fragile X protein controls the efficacy of mRNA transport in Drosophila neurons. Mol. Cell. Neurosci. 39, 170–179 [DOI] [PubMed] [Google Scholar]
- 83. Dictenberg J. B., Swanger S. A., Antar L. N., Singer R. H., Bassell G. J. (2008) A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev. Cell 14, 926–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Dynes J. L., Steward O. (2012) Arc mRNA docks precisely at the base of individual dendritic spines indicating the existence of a specialized microdomain for synapse-specific mRNA translation. J. Comp. Neurol. 520, 3105–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Eom T., Antar L. N., Singer R. H., Bassell G. J. (2003) Localization of a β-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J. Neurosci. 23, 10433–10444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhang H. L., Eom T., Oleynikov Y., Shenoy S. M., Liebelt D. A., Dictenberg J. B., Singer R. H., Bassell G. J. (2001) Neurotrophin-induced transport of a β-actin mRNP complex increases β-actin levels and stimulates growth cone motility. Neuron 31, 261–275 [DOI] [PubMed] [Google Scholar]
- 87. Farina K. L., Huttelmaier S., Musunuru K., Darnell R., Singer R. H. (2003) Two ZBP1 KH domains facilitate β-actin mRNA localization, granule formation, and cytoskeletal attachment. J. Cell Biol. 160, 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Pan F., Hüttelmaier S., Singer R. H., Gu W. (2007) ZBP2 facilitates binding of ZBP1 to β-actin mRNA during transcription. Mol. Cell. Biol. 27, 8340–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Oleynikov Y., Singer R. H. (2003) Real-time visualization of ZBP1 association with β-actin mRNA during transcription and localization. Curr. Biol. 13, 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Gu W., Pan F., Zhang H., Bassell G. J., Singer R. H. (2002) A predominantly nuclear protein affecting cytoplasmic localization of β-actin mRNA in fibroblasts and neurons. J. Cell Biol. 156, 41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Shestakova E. A., Singer R. H., Condeelis J. (2001) The physiological significance of β-actin mRNA localization in determining cell polarity and directional motility. Proc. Natl. Acad. Sci. U.S.A. 98, 7045–7050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Perycz M., Urbanska A. S., Krawczyk P. S., Parobczak K., Jaworski J. (2011) Zipcode binding protein 1 regulates the development of dendritic arbors in hippocampal neurons. J. Neurosci. 31, 5271–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gu W., Katz Z., Wu B., Park H. Y., Li D., Lin S., Wells A. L., Singer R. H. (2012) Regulation of local expression of cell adhesion and motility-related mRNAs in breast cancer cells by IMP1/ZBP1. J. Cell Sci. 125, 81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Welshhans K., Bassell G. J. (2011) Netrin-1-induced local β-actin synthesis and growth cone guidance requires zipcode binding protein 1. J. Neurosci. 31, 9800–9813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yamagishi M., Ishihama Y., Shirasaki Y., Kurama H., Funatsu T. (2009) Single-molecule imaging of β-actin mRNAs in the cytoplasm of a living cell. Exp. Cell Res. 315, 1142–1147 [DOI] [PubMed] [Google Scholar]
- 96. Leung K.-M., van Horck F. P. G., Lin A. C., Allison R., Standart N., Holt C. E. (2006) Asymmetrical β-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat. Neurosci. 9, 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yao J., Sasaki Y., Wen Z., Bassell G. J., Zheng J. Q. (2006) An essential role for β-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat. Neurosci. 9, 1265–1273 [DOI] [PubMed] [Google Scholar]
- 98. Sasaki Y., Welshhans K., Wen Z., Yao J., Xu M., Goshima Y., Zheng J. Q., Bassell G. J. (2010) Phosphorylation of zipcode binding protein 1 is required for brain-derived neurotrophic factor signaling of local β-actin synthesis and growth cone turning. J. Neurosci. 30, 9349–9358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jønson L., Vikesaa J., Krogh A., Nielsen L. K., Hansen T. vO., Borup R., Johnsen A. H., Christiansen J., Nielsen F. C. (2007) Molecular composition of IMP1 ribonucleoprotein granules. Mol. Cell. Proteomics 6, 798–811 [DOI] [PubMed] [Google Scholar]
- 100. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M., Jr., Jungkamp A. C., Munschauer M., Ulrich A., Wardle G. S., Dewell S., Zavolan M., Tuschl T. (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.