Background: Peptidoglycan hydrolases, including bacterial autolysins and bacteriophage endolysins, contain generally a cell wall-binding domain (CWBD), responsible for their high affinity and specificity toward target cell walls.
Results: Two Lactobacillus casei endolysins lyse only bacterial cells with a d-Asn cross-bridge in their peptidoglycan.
Conclusion: The CWBD of these two endolysins recognizes specifically peptidoglycan with a d-Asn cross-bridge.
Significance: This CWBD is a novel type of peptidoglycan-binding domain.
Keywords: Bacteria, Bacteriophage, Cell Wall, Hydrolases, Peptidoglycan, Lactobacillus, Cell Wall-binding Domain, Endolysin, Lysis, Peptidoglycan Cross-bridge
Abstract
Peptidoglycan hydrolases (PGHs) are responsible for bacterial cell lysis. Most PGHs have a modular structure comprising a catalytic domain and a cell wall-binding domain (CWBD). PGHs of bacteriophage origin, called endolysins, are involved in bacterial lysis at the end of the infection cycle. We have characterized two endolysins, Lc-Lys and Lc-Lys-2, identified in prophages present in the genome of Lactobacillus casei BL23. These two enzymes have different catalytic domains but similar putative C-terminal CWBDs. By analyzing purified peptidoglycan (PG) degradation products, we showed that Lc-Lys is an N-acetylmuramoyl-l-alanine amidase, whereas Lc-Lys-2 is a γ-d-glutamyl-l-lysyl endopeptidase. Remarkably, both lysins were able to lyse only Gram-positive bacterial strains that possess PG with d-Ala4→d-Asx-l-Lys3 in their cross-bridge, such as Lactococcus casei, Lactococcus lactis, and Enterococcus faecium. By testing a panel of L. lactis cell wall mutants, we observed that Lc-Lys and Lc-Lys-2 were not able to lyse mutants with a modified PG cross-bridge, constituting d-Ala4→l-Ala-(l-Ala/l-Ser)-l-Lys3; moreover, they do not lyse the L. lactis mutant containing only the nonamidated d-Asp cross-bridge, i.e. d-Ala4→d-Asp-l-Lys3. In contrast, Lc-Lys could lyse the ampicillin-resistant E. faecium mutant with 3→3 l-Lys3-d-Asn-l-Lys3 bridges replacing the wild-type 4→3 d-Ala4-d-Asn-l-Lys3 bridges. We showed that the C-terminal CWBD of Lc-Lys binds PG containing mainly d-Asn but not PG with only the nonamidated d-Asp-containing cross-bridge, indicating that the CWBD confers to Lc-Lys its narrow specificity. In conclusion, the CWBD characterized in this study is a novel type of PG-binding domain targeting specifically the d-Asn interpeptide bridge of PG.
Introduction
Peptidoglycan hydrolases (PGHs)3 synthesized by Gram-positive bacteria and their bacteriophages are able to degrade the protective cell wall peptidoglycan (PG) that surrounds the bacterial cytoplasmic membrane. PG is a macromolecule consisting of glycan chains made of alternating β-1,4-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) cross-linked by peptide chains, the composition of which varies between bacterial species. Bacterial PGHs or autolysins are required for cell wall remodeling during bacterial cell growth and division (1, 2). Bacteriophage PGHs, named endolysins, are synthesized in phage-infected cells at the end of the multiplication cycle leading to bacterial lysis and release of phage progeny (3, 4). Most PGHs have a modular structure with multiple domains, including a catalytic domain with PG hydrolyzing activity and a cell wall-binding domain (CWBD) that targets the enzyme specifically to the cell wall. The ligands of CWBDs may be structural motifs present either in PG or in secondary cell wall polymers, including polysaccharides or teichoic acids that decorate PG (3).
Endolysins usually lack a signal peptide for their export and therefore rely on the synthesis of holins, which are able to insert into the cytoplasmic membrane and make pores (5). Most often, their catalytic domain is located at the N terminus and their CWBD at the C terminus (6). Generally, the catalytic domains found in endolysins belong to the same families as those encountered in bacterial PGHs (2). In contrast, certain endolysin CWBDs do not display any sequence similarity with well characterized CWBDs found in bacterial PGHs such as LysM or SH3b domains. CWBDs are considered to confer to endolysins high affinity and high specificity for their target bacteria (7).
Endolysins from bacteriophages infecting Gram-positive bacteria can lyse bacteria from the outside and thus have been proposed as alternatives for preservatives and antibiotics used to destroy pathogens in food and medical applications (3, 4, 6, 8). Because of their high affinity for the cell wall, endolysin CWBDs may also have biotechnological applications, for example, identification of bacteria by specific staining (9). Also, after fusion with a protein of interest, they could allow this protein to be displayed at the bacterial surface with potential applications such as vaccine or biocatalyst development (10).
Lactobacillus casei is a Gram-positive lactic acid bacterium (LAB) used in dairy fermentations, and certain strains have probiotic properties (i.e. show health-promoting activities for humans and animals). This species has also been proposed as a mucosal delivery vehicle for therapeutic molecules or antigens (11). For such applications, the display of proteins, including antigens by noncovalent anchoring at the surface of LAB, considered as generally recognized as safe organisms, constitutes an attractive alternative to construction of genetically modified organisms (12). Because of their high affinity for the cell wall, endolysin CWBDs constitute potential anchoring domains (13).
By searching in silico the PGH complement of L. casei BL23 in its genome sequence (14), we identified two prophage-encoded endolysins named Lc-Lys and Lc-Lys-2. Both endolysins comprise two domains. Although their C-terminal domains share high amino acid sequence identity (68%), their N-terminal catalytic domains belong to different families of amidases according to Pfam classification (15).
In this study, we first determined the hydrolytic specificity of both endolysins by identifying degradation products of purified L. casei PG. We found that Lc-Lys is actually an N-acetylmuramoyl-l-alanine amidase, and Lc-Lys-2 is a γ-d-glutamyl-l-lysyl endopeptidase. Both lysins were able to lyse specifically bacterial cells containing PG with an amidated d-Asn cross-bridge, including food LAB such as Lactococcus lactis and also the pathogen Enterococcus faecium. Moreover, we showed that their C-terminal CWBD exhibits an original and not previously described binding specificity toward amidated d-Asp (i.e. d-Asn) of PG cross-bridge.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Culture Conditions
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown in Luria-Bertani (LB) medium (Difco) with aeration at 37 °C. L. casei BL23 was grown in MRS broth (Difco) at 37 °C. L. lactis strains were grown in M17 medium (Difco) supplemented with 0.5% (w/v) glucose (M17Glc) at 30 °C. Antibiotic concentrations used were 100 μg/ml ampicillin for E. coli and 2.5 μg/ml erythromycin or 5 μg/ml chloramphenicol for L. lactis. Growth was monitored by optical density measurement at 600 nm (OD600) with a spectrophotometer (Spectronic 20 Genesys). Streptococcus thermophilus was grown in M17Glc; Lactobacillus fermentum was grown in MRS, enterococci, and other streptococci, and Listeria and Staphylococcus strains were grown in BHI (BD Biosciences) and bacilli in LB medium under shaking. E. faecium mutant M512 was grown in BHI broth in the presence of 100 μg/ml ampicillin.
TABLE 1.
Bacterial strains and plasmids
| Strains and plasmids | Characteristic(s) | Source or Ref. |
|---|---|---|
| L. casei | ||
| BL23 | Wild type strain | 44 |
| L. lactis | ||
| MG1363 | Plasmid-free and prophage-cured derivative of L. lactis NCDO712 | 45 |
| NZ9000 | MG1363 carrying pepN::nisRK | 46 |
| VES4254 | MG1363 asnH mutant, Emr | 22 |
| VES4240 | MG1363 aslA mutant carrying S. pneumoniae murMN genes on a plasmid; VES4240 (aslA/pmurMN+), Emr Cmr | 23 |
| VES4289 | MG1363 oatA mutant, Emr | 47 |
| VES4534 | MG1363 pgdA mutant, Emr | 47 |
| VES2065 | MG1363 dacB mutant, Emr | 20 |
| ΔdacA mutant | NZ9000 dacA mutant | 48 |
| E. faecium | ||
| D344S | Ampicillin-sensitive strain | 24 |
| M512 | Spontaneous mutant of D344S strain highly resistant to ampicillin | 24 |
| E. coli | ||
| TOP 10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 λ[συπι][[ι]− | Invitrogen |
| TIL 1279 | TOP 10 strain containing pBAD-Lc-Lys plasmid, Ampr | This work |
| PAR 040 | TOP 10 strain containing pBAD-Lc-LysCD plasmid, Ampr | This work |
| PAR 041 | TOP 10 strain containing pBAD-Lc-LysBD plasmid, Ampr | This work |
| PAR 042 | TOP 10 strain containing pBAD-Lc-Lys2 plasmid, Ampr | This work |
| Plasmids | ||
| pBAD/myc-His B | pBR322-derived expression vector carrying the araBAD promoter (PBAD) and allowing C-terminal myc-His6 tag fusion, Ampr | Invitrogen |
| pBAD/His B | pBR322-derived expression vector carrying the araBAD promoter (PBAD) and allowing N-terminal His6 tag fusion, Ampr | Invitrogen |
| pBAD-Lc-Lys | pBAD/myc-His B derivative carrying lcabl_11280 gene | This work |
| pBAD-Lc-LysCD | pBad/His B derivative carrying truncated lcabl_1280 gene encoding catalytic domain | This work |
| pBAD-Lc-LysBD | pBad/His B derivative carrying truncated lcabl_11280 gene encoding cell wall-binding domain | This work |
| pBAD-Lc-Lys2 | pBad/HisB derivative carrying lcabl_10020 gene | This work |
DNA Techniques and Electrotransformation
General molecular biology techniques were used as described previously (16). Electrotransformation of E. coli was performed as described previously (17) with a Gene-Pulser apparatus (Bio-Rad). PCRs were performed with the Phusion high fidelity DNA polymerase (Finnzymes) using a thermocycler (Mastercycler gradient, Eppendorf). The primers used in this study were purchased from Eurogentec and are listed in Table 2. Constructed plasmids were extracted from E. coli TOP10 using QIAprep spin miniprep kit (Qiagen).
TABLE 2.
Primers used in this study for cloning and validation
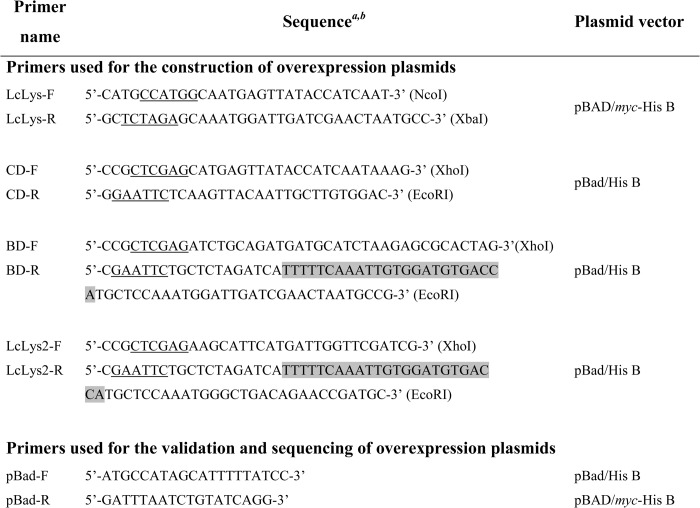
a Restriction sites are underlined in the sequences and indicated in brackets.
b Strep-tagII-encoding sequence is overlaid in gray.
Expression in E. coli and Purification of Tagged Proteins
Lc-Lys with a C-terminal His6 tag, Lc-Lys N-terminal catalytic domain (Lc-LysCD, amino acids 1–198) with an N-terminal His6 tag, Lc-Lys C-terminal domain (Lc-LysBD, amino acids 180–350) and Lc-Lys-2 with both N-terminal His6 tag and C-terminal Strep-tagII (Fig. 1A) were expressed in E. coli TOP10 with pBAD/His B or pBAD/myc-HisB expression vector as indicated in Table 1. DNA fragments were amplified by PCR from L. casei BL23 DNA using primers listed in Table 2. Strep-tagII was oligonucleotide-encoded and fused to the selected gene by PCR. The PCR fragments were then cloned into NcoI and XbaI restriction sites of pBAD/myc-HisB vector or into XhoI and EcoRI sites of pBAD/HisB. Production of tagged proteins (Lc-Lys, Lc-Lys-2, and truncated forms Lc-LysCD and Lc-LysBD) was induced in E. coli TOP10 carrying the recombinant pBAD plasmids with 0.2% l-arabinose. Bacteria were harvested 4 h after addition of l-arabinose by centrifugation and disrupted using a BAZIC Z cell disruptor (Constant Systems Ltd.) at a pressure of 1600 bars. The recombinant Lc-Lys-2, Lc-LysCD, and Lc-LysBD were purified from soluble fractions, and Lc-Lys was purified from inclusion bodies with addition of 8 m urea to all buffers. Cell extracts were loaded onto a HisTrap HP 1-ml column (GE Healthcare) connected to an FPLC AKTA chromatography system (GE Healthcare), and proteins were eluted using a linear gradient of imidazole from 30 to 400 mm.
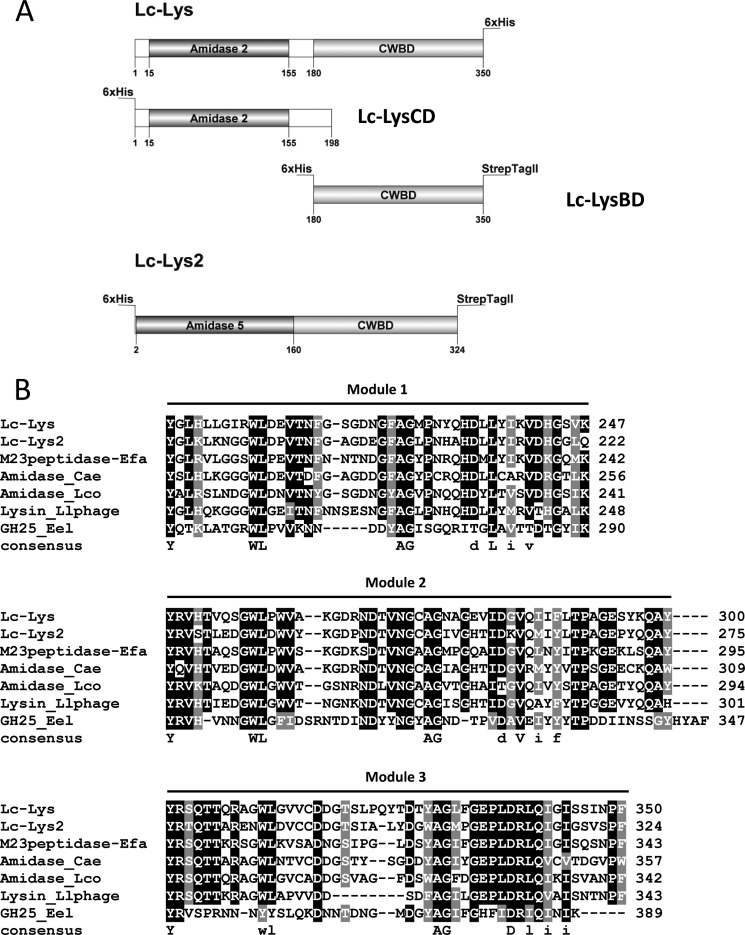
FIGURE 1.
A, schematic representation of endolysins. Lc-Lys, Lc-Lys-2, and two truncated derivatives of Lc-Lys corresponding to its catalytic domain (Lc-LysCD) and its cell wall-binding domain (Lc-LysBD) are shown. Numbers refer to amino acid positions in the complete sequences. Amidase, catalytic domain according to Pfam designations; CD, catalytic domain; BD, binding domain; CWBD, cell wall-binding domain; His6 tag and Strep-tagII, tags added to recombinant proteins. B, sequence alignments of the C-terminal CWBD of Lc-Lys and Lc-Lys-2 endolysins with the C-terminal region of other proteins. Enterolysin A was from E. faecalis (M23 peptidase_Efa) (AF249740); amidase was from C. aerofaciens (Amidase_Cae, COLAER_00250); amidase was from L. coryniformis (Amidase_Lco, LcortK3_010100004007); endolysin was from L. lactis bacteriophage 949 (LaPh949_gp055); and glycosylhydrolase family-like lysozyme was from E. eligens (EUBELI_01316). Accession numbers correspond to those of the img database (//img.jgi.doe.gov/). Multiple alignments were done with ClustalW. Black boxes, identity in at least 6 of the 7 sequences. Consensus sequence found in each of the three detected modules is indicated.
HisTrap fractions containing Lc-Lys and Lc-LysCD were desalted by Fast Desalting HR 10/10 column (GE Healthcare). Anion exchange chromatography was performed as a polishing step. Desalted samples were loaded onto a Mono Q 5/50 GL column (GE Healthcare) and eluted using a 0–250 mm linear gradient of NaCl in 20 mm Tris-HCl, pH 8.0.
HisTrap fractions containing Lc-Lys-2 and Lc-LysBD, which carry the Strep-tag II epitope, were loaded onto a 1-ml Strep-Tactin Superflow Plus Cartridge (Qiagen) and were eluted by applying a step gradient of 2.5 mm desthiobiotin.
Purity of the recombinant proteins was monitored by SDS-PAGE. Purified proteins were then concentrated by ultrafiltration with Amicon Ultra-4 Centrifugal Filter Units (Millipore). Protein concentration was measured using the BCA method (BCA protein assay kit, Thermo Scientific). Pure proteins were stored at −80 °C.
SDS-PAGE and Zymogram
SDS-PAGE was performed with 15% (w/v) polyacrylamide separating gels. Gels were stained with GelCode Blue Stain Reagent (Thermo Scientific). Zymogram was performed to detect lytic activity as described previously (14, 18). Briefly, autoclaved bacteria (10 min at 110 °C) were used as enzyme substrates and were included at 0.4% (w/v) into polyacrylamide gels. After sample migration, the gels were washed in deionized water at room temperature and then incubated in 50 mm sodium phosphate buffer, pH 6.0, containing 0.1% (v/v) Triton X-100 overnight at 37 °C under gentle rocking. The gels were subsequently washed with deionized H2O and then stained with methylene blue. Stained SDS-polyacrylamide gels and zymogram gels were digitized with a DuoScan T1200 scanner (Agfa).
PG Extraction
PG was extracted from L. casei BL23 and from L. lactis strains according to protocols described previously (14, 19).
Determination of the Hydrolytic Bond Specificity of Lc-Lys and Lc-Lys-2
Purified PG from L. casei BL23 (2 mg dry weight) was digested separately with 50 μg of recombinant Lc-Lys or Lc-Lys-2 for 18 h in 50 mm sodium phosphate buffer, pH 6.0, at 37 °C under rotation. Soluble fractions were reduced with sodium borohydride, and PG fragments were separated by reverse phase-HPLC (RP-HPLC) with an Agilent Infinity 1290 ultra-HPLC system on a Nucleodur C18 Pyramid column (150 × 2 mm, particle size, 1.8 μm) (Macherey-Nagel) at 50 °C. Muropeptides were eluted by a 1-min isocratic step of 10 mm ammonium phosphate buffer, pH 4.5 (buffer A), and then a 59-min linear gradient (0–20%) of methanol in buffer A. Peaks were analyzed without desalting by MALDI-TOF mass spectrometry (MS) using a Voyager-DE STR mass spectrometer (Applied Biosystems) as reported previously (20).
Assay of Lc-Lys and Lc-Lys-2 Activity on Autoclaved Bacterial Cells
Autoclaved cells were used as a substrate for measuring Lc-Lys and Lc-Lys-2 activity and were prepared as follows. Bacterial strains were grown in dedicated medium up to stationary phase. Bacteria were collected by centrifugation and autoclaved 10 min at 110 °C. They were then centrifuged at 5000 × g for 15 min at 4 °C, washed in sterile conditions with ice-cold PBS followed by distilled H2O, and finally resuspended in distilled H2O. Autoclaved cells were diluted in 25 mm sodium phosphate buffer, pH 6.0, to obtain an OD600 of 0.6. Purified recombinant Lc-Lys or Lc-Lys-2 (3 μg/ml final concentration) was added in a final volume of 200 μl, and the OD600 of the cell suspension was monitored in 96-well plates with a microplate reader (Tecan). The extent of bacterial lysis was expressed as the percentage decrease in OD600.
Fluorescence Microscopy
L. lactis and L. casei cells were harvested from stationary phase cultures, washed twice with PBS, and treated for 10 min at 95 °C in 7.5% TCA solution. After TCA treatment, cells were washed twice with PBS and then with 50 mm sodium phosphate buffer at pH 6.0. TCA-treated cells (1 ml at OD 1.0) were subsequently used for Lc-LysBD binding assays as described below. They were incubated with 30 μg of purified Strep-tagged Lc-LysBD recombinant protein for 60 min at 37 °C, washed three times with PBS, and incubated with a monoclonal antibody directed against Strep-tagII (IBA GmbH) (10 μg/ml in PBS containing 2% BSA) for 1 h at room temperature. After three washes with PBS, goat anti-mouse antibody conjugated to Alexa Fluor® 555 fluorescent dye (Cell Signaling Technology) diluted 1:400 in PBS containing 2% BSA was added for 1 h at room temperature in the dark. Bacteria were washed three times with PBS. Cells were immobilized on microscope slides covered with a thin film of 1.5% agarose in H2O and then examined by epifluorescence microscopy with a Leica DMRA2 microscope. Images were acquired by using a charge-coupled device camera. Images were analyzed with the ImageJ software to make overlays of bright field and fluorescent images.
Binding of Lc-LysBD to Purified PG
Peptidoglycan samples (200 μg) extracted from L. casei BL23, L. lactis MG1363, L. lactis asnH, and L. lactis VES4240 (aslA/pmurMN+) were resuspended in 500 μl of 50 mm sodium phosphate buffer, pH 6.0. Purified recombinant Lc-LysBD (15, 30, or 45 μg) was added to the PG suspensions in a final volume of 550 μl and incubated in a ThermoMixer (Eppendorf) at 37 °C for 90 min under 600 rpm shaking speed. After incubation, the samples were centrifuged 10 min at 20,000 × g, and the pellets were washed three times with cold PBS. The pellets were then resuspended in 40 μl of denaturing buffer (50 mm Tris-HCl, pH 6.8, 2% SDS, 10% v/v, glycerol, 0.1% bromphenol blue, and 100 mm DTT), boiled for 5 min, and centrifuged for 10 min at 20,000 × g. Supernatants containing solubilized material (9 μl) were analyzed by SDS-PAGE with 15% polyacrylamide gels.
RESULTS
Sequence Analysis of L. casei BL23 Prophage Endolysins
By sequence similarity search with well characterized bacterial PGHs, we identified previously in the genome sequence of L. casei BL23 two prophage-encoded endolysins, Lc-Lys (encoded by lcabl_11280) and Lc-Lys-2 (encoded by lcabl_10020), each with a neighbor holin (14). Both of them are devoid of signal peptide and contain two domains (Fig. 1A).
The putative N-terminal catalytic domain of Lc-Lys contains Pfam PF01510 domain (Amidase_2), and the Lc-Lys-2 one contains Pfam PF05382 domain (Amidase_5), both of them predicting MurNAc-l-Ala-amidase specificity on the PG substrate. Lc-Lys exhibits high amino acid sequence identity (94%) with two previously described L. casei endolysins, PL-1 (21) and LysA2 (13). However, contradictory results were previously published regarding the hydrolytic specificity of these two endolysins with MurNAc-l-Ala-amidase specificity assigned to PL-1 (21) and d-Ala-d-Asp-endopeptidase specificity assigned to LysA2 (13), although both endolysins contain the same PF01510 (Amidase_2) catalytic domain. The C-terminal domain of LysA2 was shown to bind the L. casei cell wall (13), but the recognized motif was not identified.
The C-terminal domains of Lc-Lys and Lc-Lys-2 exhibit high sequence identity to each other (68%) and to the C-terminal domain of PL-1 and LysA2. In addition, sequence identity was found with the C-terminal part of numerous proteins found in sequence databases (Fig. 1B). These include the following: (i) enterolysin A from Enterococcus faecalis, a bacteriocin with lytic activity (67% identity); (ii) endolysins from bacteriophages infecting L. lactis (60% identity); (iii) several putative endolysins found in the genomes of LAB such as Lactobacillus rhamnosus or Lactobacillus coryniformis (90 and 65% identity, respectively) and of other Gram-positive species of the human intestinal microbiota such as Collinsella aerofaciens (63% identity); and (iv) PGH from Eubacterium eligens (35% identity), which is not prophage-encoded according to the genetic environment. From sequence alignment (Fig. 1B), it appears that the C-terminal domain of these proteins is composed of three repeated modules. Conservation of the sequences allowed us to deduce the following consensus sequence: YX(7,9)-WLX(9,17)AGX(5,6)(D/T)X(L/V/I)X(V/I/L/M/A)X(I/V/F/Y/T) present in the three related modules.
Hydrolytic Specificity of Lc-Lys and Lc-Lys-2 Endolysins toward L. casei PG
To determine the hydrolytic specificity of Lc-Lys and Lc-Lys-2 on purified PG, both proteins were tagged (Fig. 1A) and produced in E. coli. His6-tagged Lc-Lys was purified to homogeneity by nickel affinity and anion exchange chromatographies, whereas Lc-Lys-2, which bears both His6 and Strep-tagII tags, was purified by two-step affinity chromatography. In SDS-PAGE (Fig. 2, A and B, lane 1), purified recombinant Lc-Lys and Lc-Lys-2 migrated as 40- and 38-kDa proteins, respectively, as expected from the calculated mass of the tagged enzymes. Both of them displayed lytic activity in zymogram assay (Fig. 2, A and B, lane 2) on autoclaved L. casei cells used as substrate.
FIGURE 2.
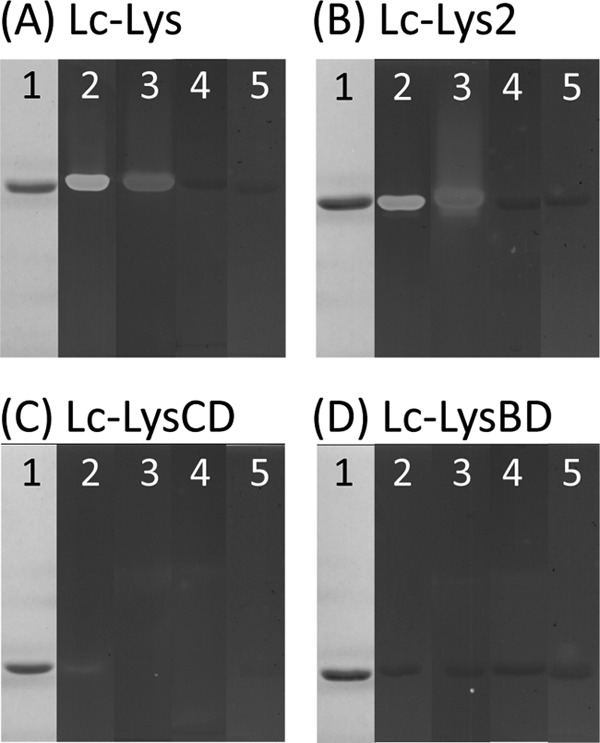
SDS-PAGE and zymogram analysis of purified recombinant endolysins and derivatives. A, Lc-Lys; B, Lc-Lys-2; C, Lc-Lys catalytic domain (Lc-LysCD); D, Lc-Lys binding domain (Lc-LysBD). After SDS-PAGE, gels were stained with Coomassie Blue (lanes 1). Zymogram assays of purified proteins were performed on autoclaved cells of L. casei BL23 (lanes 2), L. lactis MG1363 (lanes 3), L. lactis asnH (lanes 4), and L. lactis aslA(pmurMN+) (lanes 5).
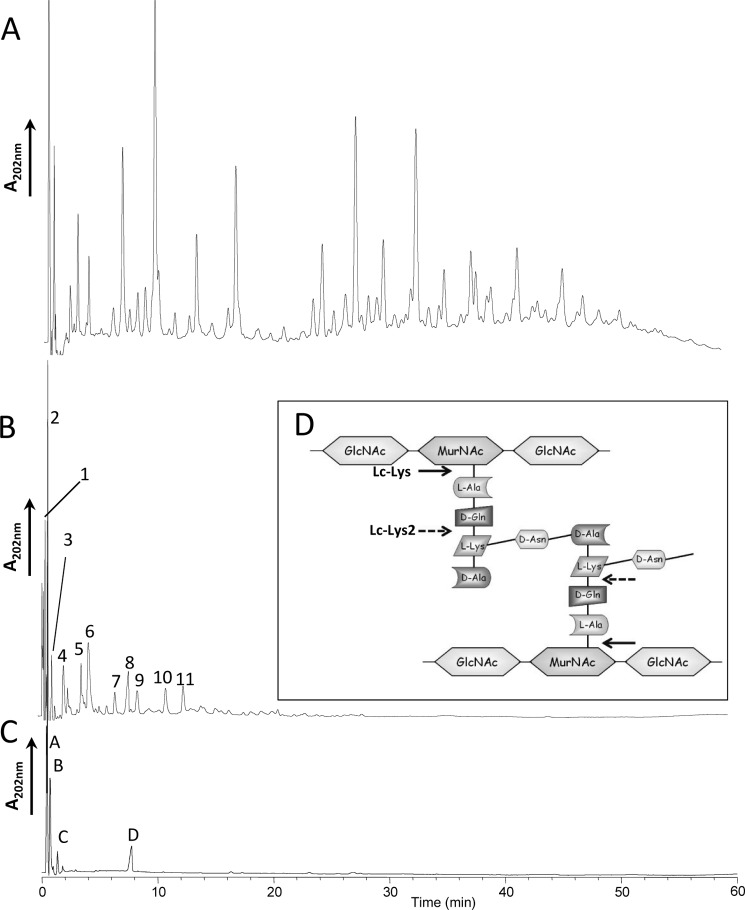
We then examined the hydrolytic specificity of recombinant pure Lc-Lys and Lc-Lys-2 on L. casei-purified PG. The PG fragments obtained by digestion with each endolysin were separated by RP-HPLC and analyzed by MALDI-TOF mass spectrometry. As shown on Fig. 3, B and C, the HPLC profiles were different, revealing distinct hydrolytic specificities for Lc-Lys and Lc-Lys-2. MS analysis of the major peaks and comparison of the obtained masses with the reference L. casei PG structure that we previously established (14) enabled us to identify peptides generated by each enzyme (Table 3) and to deduce their cleavage specificity (Fig. 3D). Lc-Lys has an N-acetylmuramoyl-l-alanine amidase specificity, whereas Lc-Lys-2 has a γ-d-glutamyl-l-lysyl endopeptidase specificity.
FIGURE 3.
Analysis of the hydrolytic specificity of Lc-Lys and Lc-Lys-2 endolysins on purified L. casei PG. A, PG digested with mutanolysin as a control for the muropeptide profile obtained with a muramidase (14); B, PG digested with recombinant endolysin Lc-Lys; C, PG digested with recombinant endolysin Lc-Lys-2. Soluble muropeptides were separated by RP-HPLC. Peaks marked with numbers and letters are identified in Table 3. D, schematic representation of L. casei PG structure showing positions of amide bonds cleaved by Lc-Lys (arrows) and Lc-Lys-2 (dashed arrows).
TABLE 3.
Molecular masses and structures of the main muropeptides obtained by digestion of L. casei PG with Lc-Lys and Lc-Lys-2
| Peaksa | Proposed structuresb |
m/zc[M + Na]+ |
|
|---|---|---|---|
| Observed | Calculated | ||
| Lc-Lys | |||
| 1 | (Asn)-Tri | 482.09 | 482.23 |
| 2 | (Asn)-Tetra | 553.08 | 553.27 |
| 3 | (Asp)-Tetra | 554.14 | 554.25 |
| 4 | (Asn)-Tetra→ (Asn)-Tri | 994.38 | 994.50 |
| 5 | (Asp)-Tetra→ (Asn)-Tri | 995.22 | 995.48 |
| 6 | (Asn)-Tetra→ (Asn)-Tetra | 1,065.45 | 1,065.54 |
| 7 | (Asp)-Tetra→ (Asn)-Tetra | 1,066.49 | 1,066.52 |
| 8 | Tetra→ (Asn)-Tetra→(Asn)-Tri | 1,392.72 | 1,392.73 |
| 9 | Tetra→ (Asn)-Tetra→(Asn)-Tetra | 1,463.58 | 1,463.77 |
| 10 | (Asn)-Tetra→ (Asn)-Tetra→(Asn)-Tri | 1,506.82 | 1,506.77 |
| 11 | (Asn)-Tetra→ (Asn)-Tetra→(Asn)-Tetra | 1,577.83 | 1,577.81 |
| Lc-Lys-2 | |||
| A | (Asn)-Lys-Ala | 354.09 | 354.17 |
| B | (Asn)-Lys-Ala→(Asn)-Lys-Ala | 667.39 | 667.35 |
| C | (Asp)-Lys-Ala→(Asn)-Lys-Ala | 668.37 | 668.33 |
| D | (Asn)-Lys-Ala→(Asn)-Lys-Ala→(Asn)-Lys-Ala | 980.51 | 980.52 |
a Peak numbers and letters refer to Fig. 3, B and C.
b Tri, tripeptide (l-Ala-d-iGln-l-Lys); Tetra, tetrapeptide (l-Ala-d-iGln-l-Lys-d-Ala); iGln, isoglutamine. See also the L. casei PG structure on Fig. 3. Assignment of d-Asp and d-Asn to either stem peptide is arbitrary. The arrows indicate the cross-links generated by dd-transpeptidation.
c m/z values correspond to monoisotopic masses.
Activity of Lc-Lys and Lc-Lys-2 toward Various Gram-positive Bacteria
The activity of Lc-Lys was tested on bacterial cells from various Gram-positive species, including LAB and pathogens with different PG chemotypes (Table 4). Pure recombinant Lc-Lys and Lc-Lys-2 were incubated with autoclaved cells of the different strains, and lysis was followed by the OD600 decrease of the suspension. As shown in Table 4, both enzymes were only active against bacterial strains exhibiting PG with d-Asx (d-Asp or d-Asn) linked to the third diamino acid l-Lys or l-Orn of the peptide stems (Fig. 4C). In addition to L. casei, these include L. lactis and E. faecium with the d-Ala4→d-Asx-l-Lys3 cross-bridge. Noteworthy, Lc-Lys lysed L. fermentum with d-Ala4→d-Asx-l-Orn3 cross-bridge; Lc-Lys-2 was also able to lyse L. fermentum cells but at a lower rate suggesting that Lc-Lys-2, which cleaves the bond between d-iGln and l-Lys, is less active when l-Lys is replaced by l-Orn.
TABLE 4.
Activity of Lc-Lys and Lc-Lys-2 on various Gram-positive bacterial strains with different PG chemotypes
| Strain | PG chemotypea | Lc-Lys activityb | Lc-Lys-2 activityb |
|---|---|---|---|
| L. casei BL23 | d-Ala4→d-Asx-l-Lys3 | + | + |
| L. lactis MG1363 | d-Ala4→d-Asx-l-Lys3 | + | + |
| E. faecium DOE | d-Ala4→d-Asx-l-Lys3 | + | + |
| E. faecium D344S | d-Ala4→d-Asx-l-Lys3 | + | + |
| L. fermentum | d-Ala4→d-Asx-l-Orn3 | + | +/− |
| S. thermophilus CNRZ1358 | d-Ala4→(l-Ala)2–3-l-Lys3 | − | − |
| E. faecalis JH2–2 | d-Ala4→(l-Ala)2-l-Lys3 | − | − |
| E. faecalis OG1RF | d-Ala4→(l-Ala)2-l-Lys3 | − | − |
| S. pneumoniae 406 | d-Ala4→ l-Ala-(l-Ala/l-Ser)-l-Lys3 | − | − |
| S. agalactiae NEM316 | d-Ala4→ l-Ala-(l-Ala/l-Ser)-l-Lys3 | − | − |
| S. mutans UA159 | d-Ala4→l-Ala-l-Thr-l-Lys3 | − | − |
| S. pyogenes D471 | d-Ala4→(l-Ala)2–3-l-Lys3 | − | − |
| S. aureus RN4220 | d-Ala4→(Gly)5-l-Lys3 | − | − |
| L. monocytogenes EGD-e | d-Ala4→mDAP3 | − | − |
| B. subtilis 168 | d-Ala4→mDAP3 | − | − |
| B. thuringiensis ATCC35646 | d-Ala4→mDAP3 | − | − |
| B. cereus BGSC6A1 | d-Ala4→mDAP3 | − | − |
a Data are according to Ref. 49. d-Asx stands for d-Asp or amidated d-Asp (d-Asn).
b Data were tested on autoclaved cells from each strain by following OD600 reduction.
FIGURE 4.
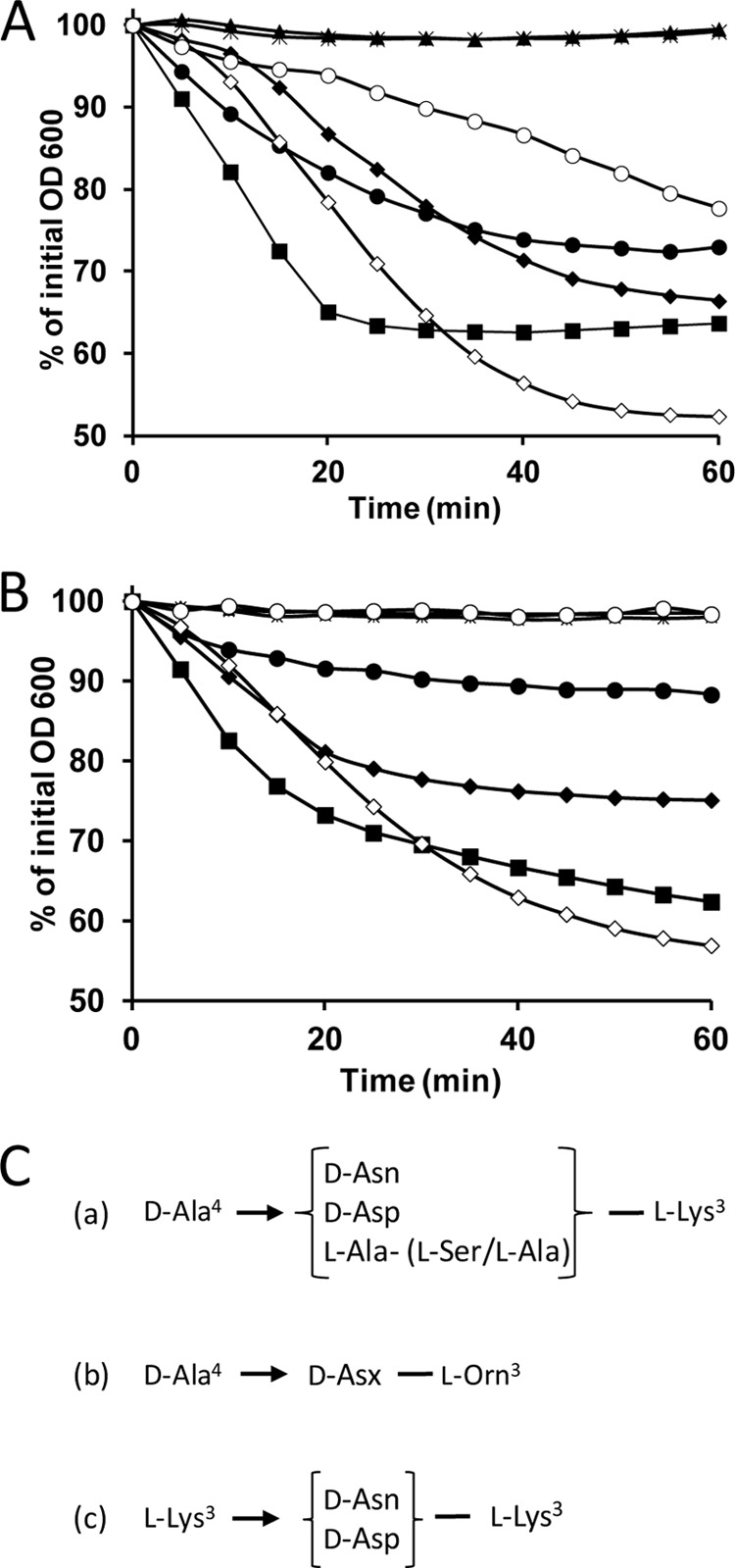
Lytic activity of purified recombinant Lc-Lys and Lc-Lys-2 toward bacteria with different PG cross-bridges and schematic representation of the cross-bridges. A, Lc-Lys; B, Lc-Lys-2 tested on L. casei BL23 (black diamonds), L. lactis MG1363 (black squares), L. lactis asnH (black triangles), L. lactis aslA(pmurMN+) (crosses), L. fermentum (black circles), E. faecium D344S (white diamonds), and E. faecium D344M512 mutant (white circles). C, schematic structure of the PG cross-bridges of the different bacterial strains tested. (a) L. lactis MG1363 wild-type (75% d-Asn and 25% d-Asp), L. lactis asnH mutant (100% d-Asp), and L. lactis VES4240 aslA(pmurMN+) (l-Ala-(l-Ser/l-Ala)), L. casei BL23 (≈100% d-Asn), and E. faecium D344S (96% d-Asn); (b) L. fermentum; (c) ampicillin-resistant E. faecium mutant M512 with 3→3 cross-links and mainly d-Asn (96%) in its cross-bridge.
Activity against L. lactis and E. faecium Cell Wall Mutants with Modified PG Structure
To examine more precisely the specificity of Lc-Lys and Lc-Lys-2 toward PG, we tested first their lytic activity on a set of L. lactis MG1363 mutants with modified PG structure, which we previously constructed (Table 1). Interestingly, the activity of Lc-Lys and Lc-Lys-2 was completely abolished on two L. lactis mutants both by the zymogram (Fig. 2, A and B, lanes 4 and 5) and spectrophotometry (Fig. 4, A and B) assays. These two L. lactis mutants have a modified interpeptide cross-bridge, which consists of 25% d-Asp and 75% d-Asn in wild-type L. lactis MG1363 (22). In the first mutant VES4240 (aslA/pmurMN+), the d-Ala4→(d-Asp/d-Asn)-l-Lys3 cross-bridge is replaced by d-Ala4→l-Ala-(l-Ala/l-Ser)-l-Lys3 (23) after inactivation of the aspartate-ligase aslA gene and expression of Streptococcus pneumoniae murMN genes on a plasmid. In the second mutant, the asnH asparagine synthase gene is disrupted, thereby preventing amidation of d-Asp (Fig. 4C) (22). In contrast, the activity of Lc-Lys and Lc-Lys-2 was only marginally affected by mutations inactivating genes encoding MurNAc-O-acetyltransferase OatA, GlcNAc deacetylase PgdA, and d-Ala-carboxypeptidases DacA and DacB (data not shown). It is worth noting that the sensitive L. casei BL23 and E. faecium D344S cells contain a high level of amidated d-Asn cross-bridge (close to 100% in L casei (14) and 96% in E. faecium (24)). Together, these results indicate that Lc-Lys and Lc-Lys-2 are specific for PG-containing d-Asn in the cross-bridge.
We then tested the lytic activity of Lc-Lys and Lc-Lys-2 on the ampicillin-resistant mutant M512 of E. faecium D344S (Fig. 4, A and B). Resistance of the mutant is due to bypass of the dd-transpeptidase activity of classical penicillin-binding proteins by an ld-transpeptidase. In the PG structure, bypass results in substitution of 4→3 d-Ala4-d-Asx-l-Lys3 cross-links by 3→3 l-Lys3-d-Asx-l-Lys3 cross-links. d-Asn is found in the majority, ∼ 96%, of the cross-bridges of the parental strain (344Ser) and of the mutant (M512) versus d-Asp (24). As shown in Fig. 4A, we observed that Lc-Lys is able to lyse E. faecium D344S and M512 cells indicating that the composition of the cross-bridge (d-Asn) rather than the nature of the cross-links (3→3 or 4→3) is the major specificity determinant for this enzyme. In contrast, Lc-Lys-2 was not active on the mutant cells indicating that its hydrolytic specificity of γ-d-Glu-l-Lys-endopeptidase cannot accommodate 3→3 cross-links.
Amidation of d-Asp Cross-bridge Is Crucial for Binding of the Lc-Lys C-terminal Domain
The C-terminal domain of Lc-Lys (Lc-LysBD) was produced in E. coli with an N-terminal His6 tag and a C-terminal Strep-tagII and purified by two steps of affinity chromatography (Fig. 2D, lane 1). Also, the N-terminal domain of Lc-Lys (Lc-LysCD) was expressed in E. coli with His6 tag and purified (Fig. 2C, lane 1). We first confirmed by zymogram assays that the N-terminal domain lysed L. casei cells (Fig. 2C, lane 2), although its lytic activity was reduced compared with the entire Lc-Lys (Fig. 2A, lane 2). In contrast, no activity was associated to the recombinant C-terminal domain (Fig. 2D, lane 2).
Binding of recombinant Lc-LysBD to whole bacterial cells was tested by immunofluorescence. Bacteria were first treated with TCA to remove surface polymers allowing better exposure of PG (14, 25, 26) before incubation with pure Strep-tagII-tagged Lc-LysBD. Binding was revealed after incubation with anti-Strep-tag antibody followed by secondary fluorescent antibody and observation with an epifluorescence microscope (Fig. 5). Fluorescent labeling indicated that the C-terminal domain of Lc-Lys is able to bind the L. casei cell wall (Fig. 5, A–C) as well as the L. lactis cell wall (Fig. 5, D and E). In contrast, no binding to the L. lactis asnH mutant cell wall was observed (Fig. 5, F and G).
FIGURE 5.
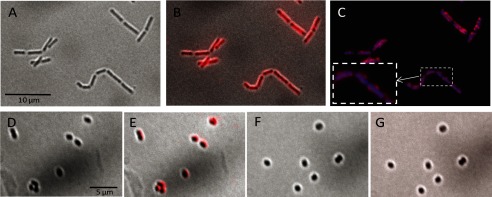
Binding of Lc-LysBD to L. casei and L. lactis cells. L. casei BL23 (A–C), L. lactis MG1363 (D and E), and L. lactis asnH mutant (F and G) cells harvested in stationary phase were treated with TCA and incubated successively with purified Strep-tagII-tagged Lc-LysBD, anti-Strep-tagII antibody, and secondary antibody coupled to Alexa Fluor 555. Bright field pictures (A, D, and F) were overlaid with the corresponding fluorescent images (B, E, and G, respectively). Additional staining of L. casei was performed with DAPI (blue) and overlaid with the corresponding fluorescent image (C).
The binding specificity of Lc-LysBD was further tested by pulldown experiments. PG extracted from L. casei BL23, L. lactis MG1363, L. lactis asnH mutant, and L. lactis VES4240 (aslA/pmurMN+) mutant was incubated with various amounts of pure recombinant Lc-LysBD, and binding of Lc-LysBD to the PG sample was assayed by SDS-PAGE (Fig. 6). Lc-LysBD bound to PG from L. casei and L. lactis in a dose-dependent manner. In the same conditions, extremely low binding and no binding could be detected on PG extracted from the mutants with the modified cross-bridge, L. lactis asnH and L. lactis VES4240 (aslA/pmurMN+), respectively (Fig. 6). Taken together, these data indicate that the C-terminal domain of Lc-Lys endolysin binds the cell wall and is highly specific for the amidated form of d-Asp (i.e. d-Asn) present in the PG cross-bridge.
FIGURE 6.
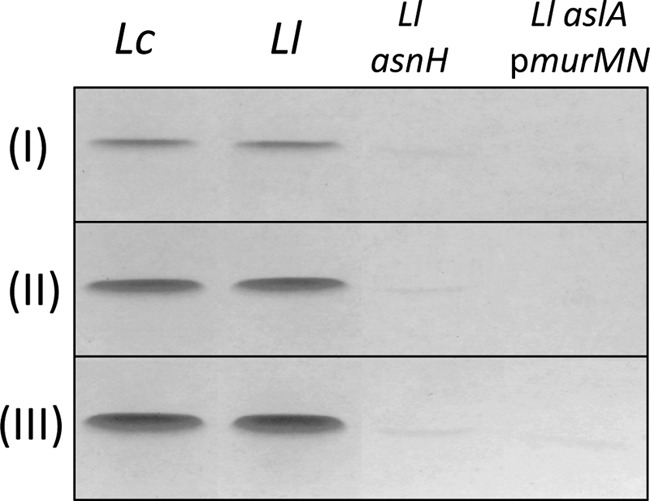
Binding of purified Lc-LysBD to purified PG containing different cross-bridge amino acids. PG samples extracted from L. casei BL23 (Lc), L. lactis MG1363 (Ll), and L. lactis asnH and L. lactis aslA(pmurMN+) were incubated with different amounts of Lc-LysBD: 15 μg (I), 30 μg (II), and 45 μg (III). Protein bound to PG was analyzed by SDS-PAGE. PG cross-bridge structure of the different strains is shown on Fig. 4C.
DISCUSSION
Most endolysins encoded by prophages or bacteriophages infecting Gram-positive bacteria have a conserved modular organization, including an N-terminal catalytic domain and a C-terminal CWBD. Until now, only a few ligands recognized by the lysin CWBDs have been identified. In this study, we characterized two endolysins from L. casei prophages that differ at the level of their catalytic domains but have identical CWBDs. Although their CWBD exhibits sequence similarity with other PGHs present in sequence databases and was shown to bind L. casei cells, the recognized structure in the cell wall was unknown up to now. We showed here that this CWBD confers to both endolysins a high specificity toward the PG cross-bridge and selectively targets PG containing an amidated d-Asp (i.e. d-Asn) cross-bridge. To our knowledge, this is the first report for such a binding specificity for a phage endolysin and more generally for a PGH.
The ligands of bacteriophage endolysin CWBDs have been characterized in a limited number of cases. Endolysin Cpl-1 from S. pneumoniae phage was shown to bind choline present in teichoic acids from this bacterial species through choline-binding domains, which are also present in bacterial PGHs (27). The CWBDs of three endolysins of phages infecting L. monocytogenes were characterized in detail. For two of them, the identified ligands were cell wall carbohydrates most likely covalently anchored to the cell wall (7), whereas the third one, PlyP35, recognizes specifically GlcNAc substituents of the polyribitol-phosphate chain constituting wall teichoic acids (28). Regarding two Bacillus anthracis bacteriophage endolysins, their CWBD was shown to be specific for cell wall polysaccharides and more precisely for their galactosylation patterns (29). Very recently, CW_7 motifs present in Cpl-7 endolysin from a S. pneumoniae bacteriophage (30) were reported to recognize most probably PG and more specifically the GlcNAc-MurNAc-l-Ala-d-iGln muropeptide (31). Finally, the CWBDs of two other L. monocytogenes endolysins were deduced to bind A1γ-type PG because wall teichoic acids restrict their access to the bacterial cell wall surface, but the exact motif recognized was not identified (32). Besides, certain endolysins contain LysM domains (33) that were shown to recognize PG glycan chains (34, 35) or SH3b domains (36) shown to bind pentaglycine interpeptide bridges in S. aureus PG (37, 38). Thus, to our knowledge, the binding specificity of the two L. casei endolysins characterized in this work is novel compared with the other studied endolysins, and this study is the first evidence of a protein domain recognizing specifically the d-Asn cross-bridge of PG. According to sequence analysis, this domain is composed of three repeated modules.
Regarding their hydrolytic specificity, amino acid sequence analysis predicted an N-acetylmuramoyl-l-alanine amidase specificity for both endolysins, but with different catalytic domains. Lc-Lys contains a PF01510 (Amidase_2) domain, whereas Lc-Lys-2 contains a PF05382 (Amidase_5) domain. In this study, contrary to the previous claim that LysA2 (94% similar to Lc-Lys) has d-Ala-d-Asp-endopeptidase specificity (13), we show that Lc-Lys is actually an amidase cleaving the bond between MurNAc and d-Ala inside PG (Fig. 3D). We obtained this result by digesting purified PG with Lc-Lys and analyzing the released PG fragments by HPLC and MS, which is a straightforward method compared with the indirect one used in Ref. 13. Regarding Lc-Lys-2, the same experimental approach revealed endopeptidase specificity for Lc-Lys-2 with a cleavage site between d-iGln and l-Lys inside peptide stems of the PG macromolecule (Fig. 3D). Noteworthy, another endolysin endowed with the same catalytic domain (Amidase_5) and issued from a bacteriophage infecting Streptococcus agalactiae was also reported to have γ-d-Glu-l-Lys endopeptidase activity (36).
The lytic activity of the L. casei endolysin Lc-Lys and Lc-Lys-2 was tested on a panel of bacteria with various characterized PG structures. Our results indicate that the main requirement for Lc-Lys activity is the presence of d-Asn in the PG cross-bridge. Lc-Lys was able to lyse L. casei BL23 cells, L. lactis MG1363 cells, and the pathogenic strain tested, E. faecium D344S, all with the d-Ala4→d-Asx-l-Lys3 PG cross-bridge containing a high proportion of d-Asn versus d-Asp (100% (14), 75% d-Asn (20), and 96% (24), respectively) (Fig. 4C). In contrast, Lc-Lys was not able to lyse the L. lactis asnH mutant derived from MG1363 with 100% d-Asp in its PG cross-bridge. Lc-Lys was also able to lyse L. fermentum cells with d-Ala4→d-Asx-l-Orn3 cross-bridge. Finally, it is active against the ampicillin-resistant mutant M512 derived from E. faecium D344S, which possesses 3→3 bridges mainly constituting the amidated form l-Lys3-d-Asn-l-Lys3. In addition, we showed that this specificity is associated with Lc-Lys CWBD, which binds PG with a high proportion of d-Asn but not PG with only d-Asp or other amino acids in the cross-bridges. Regarding Lc-Lys-2, the same binding specificity toward d-Asn is conferred by its CWBD as shown by comparing lytic activity on L. lactis wild-type MG1363 and asnH mutant. However, because of its cleavage specificity of γ-d-Glu-l-Lys-endopeptidase, it is less active on cells of L. fermentum with l-Orn replacing l-Lys in position 3 of PG stem peptides and not active on bacteria with the 3→3 cross-bridge.
In this study, Lc-Lys was able to lyse E. faecium DOE and D344S, whereas LysA2, the lysin of bacteriophage A2 characterized previously (13), was reported not to be active on E. faecium SC139. The apparent discrepancy could be linked to a low level of amidation of the PG cross-bridge in the SC139 strain contrary to the high level previously assessed in the D344S strain (24). Another possible reason could be the presence of hindering cell wall components at the surface of the SC139 strain.
Regarding possible applications, the L. casei endolysin Lc-Lys should be tested on a wider range of E. faecium strains to evaluate its potential as an antimicrobial agent to target E. faecium, for example in a complex microbial community. In another field of application, Lc-Lys CWBD appears already as a promising candidate to anchor noncovalent proteins at the bacterial surface as previously shown to obtain vaccines or probiotic bacteria (39).
Domains with high sequence identity (above 60%) with the CWBD domain of Lc-Lys and Lc-Lys-2 were mainly found in endolysins identified in L. lactis phages or in prophages present in complete genome sequences of firmicutes, including LAB and other species of the human gut microbiota. This domain was found associated with different catalytic domains with PG-hydrolyzing activity, including most often Amidase_2 (PF01510), but also Amidase_5 (PF05382), peptidase M23 (PF01551), and Glyco_hydro_25 (PF01183). Bacteriophages infecting LAB were intensively studied because they represent a real threat for dairy fermentations (40). Most endolysins identified in their complete sequences (41, 42) have a C-terminal domain without any sequence homology to well characterized CWBDs. To our knowledge, their ligands have not been identified so far, and our study is the first experimental identification of an endolysin ligand in the cell wall of LAB. Interestingly, the CWBD characterized in this study is also present in enterolysin A, a bacteriocin with lytic activity produced by E. faecalis and active mainly against bacteria with d-Ala4→d-Asx-l-Lys3 PG chemotype, including selected enterococci, pediococci, lactococci, and lactobacilli (43) in agreement with our results presented here.
The PG-binding domain characterized in this study is highly specific for d-Asn present in PG cross-bridges and represents a novel class of bacterial CWBD. Determination of the three-dimensional structure and the amino acid residues involved in PG recognition will help us understand the molecular basis of the high specificity of the characterized domain for its ligands.
Acknowledgments
We thank Michał Ferens, Calum MacKichan, and Marie-Françoise Noirot-Gros (Micalis, INRA, France) for their help with the epifluorescence microscopy experiments. We are very grateful to Michel Arthur (INSERM, Paris, France) for the gift of E. faecium strains and for critical reading of the manuscript.
This work was supported in part by INRA and Région Ile de France and was an associated project from the Marie Curie FP7 Initial Training Network Cross Talk (Grant Agreement 21553-2).
- PGH
- peptidoglycan hydrolase
- PG
- peptidoglycan
- MurNAc
- N-acetylmuramic acid
- RP-HPLC
- reverse-phase high performance-liquid-chromatography
- CWBD
- cell wall-binding domain
- LAB
- lactic acid bacterium
- iGln
- isoglutamine.
REFERENCES
- 1. Vollmer W., Joris B., Charlier P., Foster S. (2008) Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32, 259–286 [DOI] [PubMed] [Google Scholar]
- 2. Chapot-Chartier M.-P. (2010) in Prokaryotic Cell Wall Compounds (König H., Claus H., Varna A., eds) pp. 383–406, Springer Verlag Berlin, Heidelberg, Germany [Google Scholar]
- 3. Loessner M. J. (2005) Bacteriophage endolysins–current state of research and applications. Curr. Opin. Microbiol. 8, 480–487 [DOI] [PubMed] [Google Scholar]
- 4. Hermoso J. A., García J. L., García P. (2007) Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Curr. Opin. Microbiol. 10, 461–472 [DOI] [PubMed] [Google Scholar]
- 5. Wang I. N., Smith D. L., Young R. (2000) Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54, 799–825 [DOI] [PubMed] [Google Scholar]
- 6. Fischetti V. A. (2008) Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 11, 393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Loessner M. J., Kramer K., Ebel F., Scherer S. (2002) C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44, 335–349 [DOI] [PubMed] [Google Scholar]
- 8. O'Flaherty S., Ross R. P., Coffey A. (2009) Bacteriophage and their lysins for elimination of infectious bacteria. FEMS Microbiol. Rev. 33, 801–819 [DOI] [PubMed] [Google Scholar]
- 9. Schmelcher M., Shabarova T., Eugster M. R., Eichenseher F., Tchang V. S., Banz M., Loessner M. J. (2010) Rapid multiplex detection and differentiation of Listeria cells by use of fluorescent phage endolysin cell wall binding domains. Appl. Environ. Microbiol. 76, 5745–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee S. Y., Choi J. H., Xu Z. (2003) Microbial cell-surface display. Trends Biotechnol. 21, 45–52 [DOI] [PubMed] [Google Scholar]
- 11. Bermúdez-Humarán L. G., Kharrat P., Chatel J. M., Langella P. (2011) Lactococci and lactobacilli as mucosal delivery vectors for therapeutic proteins and DNA vaccines. Microb. Cell Fact. 10, S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leenhouts K., Buist G., Kok J. (1999) Anchoring of proteins to lactic acid bacteria. Antonie Van Leeuwenhoek 76, 367–376 [PubMed] [Google Scholar]
- 13. Ribelles P., Rodríguez I., Suárez J. E. (2012) LysA2, the Lactobacillus casei bacteriophage A2 lysin is an endopeptidase active on a wide spectrum of lactic acid bacteria. Appl. Microbiol. Biotechnol. 94, 101–110 [DOI] [PubMed] [Google Scholar]
- 14. Regulski K., Courtin P., Meyrand M., Claes I. J., Lebeer S., Vanderleyden J., Hols P., Guillot A., Chapot-Chartier M. P. (2012) Analysis of the peptidoglycan hydrolase complement of Lactobacillus casei and characterization of the major γ-d-glutamyl-l-lysyl endopeptidase. PLoS One 7, e32301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A. (2010) The Pfam protein families database. Nucleic Acids Res. 38, D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 17. Dower W. J., Miller J. F., Ragsdale C. W. (1988) High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16, 6127–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huard C., Miranda G., Wessner F., Bolotin A., Hansen J., Foster S. J., Chapot-Chartier M. P. (2003) Characterization of AcmB, an N-acetylglucosaminidase autolysin from Lactococcus lactis. Microbiology 149, 695–705 [DOI] [PubMed] [Google Scholar]
- 19. Meyrand M., Boughammoura A., Courtin P., Mézange C., Guillot A., Chapot-Chartier M.-P. (2007) Peptidoglycan N-acetylglucosamine deacetylation decreases autolysis in Lactococcus lactis. Microbiology 153, 3275–3285 [DOI] [PubMed] [Google Scholar]
- 20. Courtin P., Miranda G., Guillot A., Wessner F., Mézange C., Domakova E., Kulakauskas S., Chapot-Chartier M. P. (2006) Peptidoglycan structure analysis of Lactococcus lactis reveals the presence of an ld-carboxypeptidase involved in peptidoglycan maturation. J. Bacteriol. 188, 5293–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kashige N., Nakashima Y., Miake F., Watanabe K. (2000) Cloning, sequence analysis, and expression of Lactobacillus casei phage PL-1 lysis genes. Arch. Virol. 145, 1521–1534 [DOI] [PubMed] [Google Scholar]
- 22. Veiga P., Erkelenz M., Bernard E., Courtin P., Kulakauskas S., Chapot-Chartier M. P. (2009) Identification of the asparagine synthase responsible for d-Asp amidation in the Lactococcus lactis peptidoglycan interpeptide cross-bridge. J. Bacteriol. 191, 3752–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Veiga P., Piquet S., Maisons A., Furlan S., Courtin P., Chapot-Chartier M. P., Kulakauskas S. (2006) Identification of an essential gene responsible for d-Asp incorporation in the Lactococcus lactis peptidoglycan cross-bridge. Mol. Microbiol. 62, 1713–1724 [DOI] [PubMed] [Google Scholar]
- 24. Mainardi J. L., Legrand R., Arthur M., Schoot B., van Heijenoort J., Gutmann L. (2000) Novel mechanism of β-lactam resistance due to bypass of dd-transpeptidation in Enterococcus faecium. J. Biol. Chem. 275, 16490–16496 [DOI] [PubMed] [Google Scholar]
- 25. Chapot-Chartier M. P., Vinogradov E., Sadovskaya I., Andre G., Mistou M. Y., Trieu-Cuot P., Furlan S., Bidnenko E., Courtin P., Péchoux C., Hols P., Dufrêne Y. F., Kulakauskas S. (2010) The cell surface of Lactococcus lactis is covered by a protective polysaccharide pellicle. J. Biol. Chem. 285, 10464–10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bosma T., Kanninga R., Neef J., Audouy S. A., van Roosmalen M. L., Steen A., Buist G., Kok J., Kuipers O. P., Robillard G., Leenhouts K. (2006) Novel surface display system for proteins on nongenetically modified Gram-positive bacteria. Appl. Environ. Microbiol. 72, 880–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. García E., García J. L., García P., Arrarás A., Sánchez-Puelles J. M., López R. (1988) Molecular evolution of lytic enzymes of Streptococcus pneumoniae and its bacteriophages. Proc. Natl. Acad. Sci. U.S.A. 85, 914–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eugster M. R., Haug M. C., Huwiler S. G., Loessner M. J. (2011) The cell wall binding domain of Listeria bacteriophage endolysin PlyP35 recognizes terminal GlcNAc residues in cell wall teichoic acid. Mol. Microbiol. 81, 1419–1432 [DOI] [PubMed] [Google Scholar]
- 29. Mo K. F., Li X., Li H., Low L. Y., Quinn C. P., Boons G. J. (2012) Endolysins of Bacillus anthracis bacteriophages recognize unique carbohydrate epitopes of vegetative cell wall polysaccharides with high affinity and selectivity. J. Am. Chem. Soc. 134, 15556–15562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bustamante N., Campillo N. E., García E., Gallego C., Pera B., Diakun G. P., Sáiz J. L., García P., Díaz J. F., Menéndez M. (2010) Cpl-7, a lysozyme encoded by a pneumococcal bacteriophage with a novel cell wall-binding motif. J. Biol. Chem. 285, 33184–33196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bustamante N., Rico-Lastres P., García E., García P., Menéndez M. (2012) Thermal Stability of Cpl-7 Endolysin from the Streptococcus pneumoniae bacteriophage Cp-7; cell wall targeting of its CW_7 motifs. PLoS One 7, e46654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eugster M. R., Loessner M. J. (2012) Wall teichoic acids restrict access of bacteriophage endolysin Ply118, Ply511, and PlyP40 cell wall binding domains to the Listeria monocytogenes peptidoglycan. J. Bacteriol. 194, 6498–6506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hu S., Kong J., Kong W., Guo T., Ji M. (2010) Characterization of a novel LysM domain from Lactobacillus fermentum bacteriophage endolysin and its use as an anchor to display heterologous proteins on the surfaces of lactic acid bacteria. Appl. Environ. Microbiol. 76, 2410–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Steen A., Buist G., Leenhouts K. J., El Khattabi M., Grijpstra F., Zomer A. L., Venema G., Kuipers O. P., Kok J. (2003) Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J. Biol. Chem. 278, 23874–23881 [DOI] [PubMed] [Google Scholar]
- 35. Frankel M. B., Schneewind O. (2012) Determinants of murein hydrolase targeting to cross-wall of Staphylococcus aureus peptidoglycan. J. Biol. Chem. 287, 10460–10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pritchard D. G., Dong S., Kirk M. C., Cartee R. T., Baker J. R. (2007) LambdaSa1 and LambdaSa2 prophage lysins of Streptococcus agalactiae. Appl. Environ. Microbiol. 73, 7150–7154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gründling A., Schneewind O. (2006) Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus. J. Bacteriol. 188, 2463–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lu J. Z., Fujiwara T., Komatsuzawa H., Sugai M., Sakon J. (2006) Cell wall-targeting domain of glycylglycine endopeptidase distinguishes among peptidoglycan cross-bridges. J. Biol. Chem. 281, 549–558 [DOI] [PubMed] [Google Scholar]
- 39. Ribelles P., Benbouziane B., Langella P., Suárez J. E., Bermúdez-Humarán L. G. (2013) Protection against human papillomavirus type 16-induced tumors in mice using nongenetically modified lactic acid bacteria displaying E7 antigen at its surface. Appl. Microbiol. Biotechnol. 97, 1231–1239 [DOI] [PubMed] [Google Scholar]
- 40. Garneau J. E., Moineau S. (2011) Bacteriophages of lactic acid bacteria and their impact on milk fermentations. Microb. Cell Fact. 10, S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lortal S., Chapot-Chartier M. P. (2005) Role, mechanisms and control of lactic acid bacteria lysis in cheese. Int. Dairy J. 15, 857–871 [Google Scholar]
- 42. Gasson M. J. (1996) Lytic systems in lactic acid bacteria and their bacteriophages. Antonie Van Leeuwenhoek 70, 147–159 [DOI] [PubMed] [Google Scholar]
- 43. Nilsen T., Nes I. F., Holo H. (2003) Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl. Environ. Microbiol. 69, 2975–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Acedo-Félix E., Pérez-Martínez G. (2003) Significant differences between Lactobacillus casei subsp. casei ATCC 393T and a commonly used plasmid-cured derivative revealed by a polyphasic study. Int. J. Syst. Evol. Microbiol. 53, 67–75 [DOI] [PubMed] [Google Scholar]
- 45. Gasson M. J. (1983) Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kuipers O. P., de Ruyter P. G., Kleerebezem M., de Vos W. M. (1998) Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64, 15–21 [Google Scholar]
- 47. Veiga P., Bulbarela-Sampieri C., Furlan S., Maisons A., Chapot-Chartier M.-P., Erkelenz M., Mervelet P., Noirot P., Frees D., Kuipers O. P., Kok J., Gruss A., Buist G., Kulakauskas S. (2007) SpxB regulates O-acetylation-dependent resistance of Lactococcus lactis peptidoglycan to hydrolysis. J. Biol. Chem. 282, 19342–19354 [DOI] [PubMed] [Google Scholar]
- 48. Roces C., Courtin P., Kulakauskas S., Rodríguez A., Chapot-Chartier M. P., Martínez B. (2012) Isolation of Lactococcus lactis mutants simultaneously resistant to the cell wall-active bacteriocin Lcn972, lysozyme, nisin, and bacteriophage c2. Appl. Environ. Microbiol. 78, 4157–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schleifer K. H., Kandler O. (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477 [DOI] [PMC free article] [PubMed] [Google Scholar]