Abstract
The Ras-mitogen-activated protein (Ras-MAP) kinase pathway regulates various cellular processes, including gene expression, cell proliferation, and survival. Ribosomal S6 kinase (RSK), a key player in this pathway, modulates the activities of several cytoplasmic and nuclear proteins via phosphorylation. Here we report the characterization of the cytoskeletal protein filamin A (FLNa) as a membrane-associated RSK target. We show that the N-terminal kinase domain of RSK phosphorylates FLNa on Ser2152 in response to mitogens. Inhibition of MAP kinase signaling with UO126 or mutation of Ser2152 to Ala on FLNa prevents epidermal growth factor (EGF)-stimulated phosphorylation of FLNa in vivo. Furthermore, phosphorylation of FLNa on Ser2152 is significantly enhanced by the expression of wild-type RSK and antagonized by kinase-inactive RSK or specific reduction of endogenous RSK. Strikingly, EGF-induced, FLNa-dependent migration of human melanoma cells is significantly reduced by UO126 treatment. Together, these data provide substantial evidence that RSK phosphorylates FLNa on Ser2152 in vivo. Given that phosphorylation of FLNa on Ser2152 is required for Pak1-mediated membrane ruffling, our results suggest a novel role for RSK in the regulation of the actin cytoskeleton.
Extracellular signals that activate the Ras-mitogen-activated protein (Ras-MAP) kinase pathway have been shown to regulate many cellular processes, such as cell proliferation, differentiation, and survival. The Ras-MAP kinase pathway transduces signals from the plasma membrane to the nucleus through the recruitment and activation of various signaling molecules, including Ras, c-Raf, MEK1/2, and ERK-MAP kinase 1/2 (ERK1/2). One important ERK target is ribosomal S6 kinase (RSK) (reviewed by Frodin and Gammeltoft [14]), a unique kinase containing two structurally distinct but functional kinase domains (11). Humans express four RSK proteins (RSK1 to RSK4), and kinase-inactivating mutations in RSK2 have been associated with Coffin-Lowry syndrome, a condition characterized by mental retardation, facial and digital dysmorphisms, and skeletal defects (35).
Activation of RSK requires multiple phosphorylation events. Inputs from the upstream activators, including ERK1/2 and PDK1, as well as autophosphorylation, are necessary for RSK activation (8, 17, 23, 24, 29, 38, 40). Like phosphorylation, subcellular localization of RSK may play a role in its activation. We have recently shown that after epidermal growth factor (EGF) stimulation, RSK transiently localizes to the plasma membrane prior to nuclear translocation (6, 23). Constitutive membrane targeting of RSK leads to the phosphorylation of key regulatory sites and bypasses the requirement for ERK, indicating that membrane localization is important for activation (23, 26). Changes in subcellular localization thereby contribute to its activation and generate the potential for RSK to regulate a variety of proteins throughout the cell.
We have previously shown that filamin A (FLNa), a membrane-associated, cytoskeletal protein, is a RSK substrate (21). FLNa (also known as actin-binding protein 280, filamin 1, and nonmuscle filamin) is the most widely distributed isoform of a family of high-molecular-weight dimeric proteins that cross-link actin filaments (reviewed by Stossel et al. [28]). FLNa is a very efficient actin gelation factor, capable of inducing a liquid-to-gel transition of actin filaments at a ratio of one FLNa dimer per actin filament (15). In addition to actin, FLNa has more than 20 other binding partners, including membrane receptors, small GTPases, and stress-activated protein kinase (reviewed by Stossel et al. [28]).
FLNa is essential for mammalian cell locomotion. Human melanoma cell lines that do not express detectable levels of FLNa do not migrate (2), and restoring expression of wild-type levels of FLNa in these cells rescues their locomotion (7, 12). Mutations of the human filamin A gene on the X chromosome leading to failure of FLNa protein expression are responsible for a disease called periventricular heterotopia (PH). Males or females homozygous for PH die in utero, presumably due to severe defects in embryonic cell migration. Female PH heterozygotes present with epileptic seizures caused by the periventricular heterotopias, collections of embryonic neurons that fail to migrate from the lateral ventricles to the cerebral cortex. Heterozygous PH females often exhibit other neuronal and extraneuronal anomalies as well as premature vascular problems (13).
FLNa is known to be a phosphoprotein but how phosphorylation regulates its function is not understood. In addition to RSK (21), many kinases, including cyclic AMP (cAMP)-dependent protein kinase (PKA) (5, 16, 18, 39), CaM kinase II (20), Pak1 (36), and PKC (34), can phosphorylate FLNa in vitro. Purified human RSK2 phosphorylates the C-terminal half of FLNa in vitro at a serine residue(s) that is phosphorylated in vivo in response to lysophosphatidic acid (21). However, the RSK phosphorylation site(s) on FLNa has not been identified. Pak1 can also phosphorylate FLNa, and Ser2152 was identified as the phosphorylation site. Interestingly, mutation of Ser2152 to Ala prevents Pak1-mediated membrane ruffle formation (36). These data suggest that FLNa Ser2152 phosphorylation is required for FLNa to effect cell shape changes.
In this report, we provide clear evidence that FLNa is a membrane-associated RSK substrate. We show that RSK phosphorylates FLNa on Ser2152 in vitro and that FLNa Ser2152 phosphorylation is stimulated by a variety of agonists in cultured cells. In addition, we demonstrate that FLNa Ser2152 phosphorylation is dramatically reduced by pharmacological inhibition of ERK signaling using the MEK inhibitor UO126. In contrast, FLNa phosphorylation is not diminished by the phosphatidylinositol 3-kinase (PI3K) inhibitor wortmannin or stimulated by insulin, which potently activates PI3K and Akt. Moreover, expression of wild-type RSK potentiates EGF-stimulated FLNa phosphorylation, whereas expression of kinase-inactive RSK or RSK RNA interference (RNAi) antagonizes this phosphorylation. Supporting our hypothesis of RSK-dependent regulation of FLNa function, we show that FLNa-dependent, EGF-induced migration of human melanoma cells is significantly reduced by UO126 treatment. Since FLNa Ser2152 phosphorylation is necessary for membrane ruffling, our data suggest that RSK plays a role in control of actin cytoskeletal dynamics.
MATERIALS AND METHODS
Plasmids and antibodies.
Hemagglutinin (HA)-tagged avian RSK-WT, RSK-K112R, RSK-K464R, and RSK-K112/464R constructs have been previously described (26). HA-tagged murine Akt1 was a gift from P. Tsichlis. pGEX-4T-1-FLNa-C and pGEX-4T-1-FLNa-D consist of FLNa fragments 1 and 3 (22), respectively, cloned into pGEX-4T-1 (Pharmacia Biotech). pGEX-4T-1-FLNa-A consists of the BsmI-HincII fragment of human filamin A, and pGEX-4T-1-FLNa-B consists of the ScaI-BsaMI fragment. Full-length human filamin A was cloned into the pREP4 vector (Invitrogen) using HindIII and BamHI sites. pCMV5-FLNa (Δ20N) was previously referred to as pCMV-myc-filamin 1 (22).
Monoclonal antibodies, phospho-ERK1/2 (Sigma), and FLNa MAb1678 (Chemicon) were purchased from commercial suppliers. Rabbit polyclonal phospho-specific RSK antibody that specifically recognizes Ser380 was obtained from R&D Systems. myc antibody was produced from 9E10 hybridoma (American Type Culture Collection). HA antibody (affinity purified from 12CA5 hybridoma) was a gift from M. Chou. C-terminal peptide-specific rabbit polyclonal RSK-C3 antibody (11) and rabbit polyclonal ERK1/2 antibody (6) have been previously described. RSK1-specific antibody was generated by injecting rabbits with PIESSILAQRRVKKLPSTTL peptide, derived from amino acid residues 733 to 752 in avian RSK and corresponding to residues 716 to 735 in human RSK1 (Invitrogen). RSK2-specific antibody was made by injecting rabbits with the peptide RNQSPVLEPVGRSTL, derived from amino acids 712 to 726 in human RSK2 (Invitrogen). To make phospho-FLNa antibody, rabbits were injected with RRAPpSVANVGS peptide (containing phosphorylated Ser2152 [pS]), derived from amino acids 2148 to 2158 in human FLNa (Invitrogen).
Site-directed mutagenesis.
pGEX-4T-1-FLNa-Ala2152 was made using the QuikChange site-directed mutagenesis kit (Stratagene). pGEX-4T-1-FLNa-C was amplified with Pfu DNA polymerase, 5′ FLNa-Ala2152 primer (GCAGGCGTCGGGCTCCTGCAGTGGCCAACGTTG) and 3′ FLNa-Ala2152 primer (CAACGTTGGCCACTGCAGGAGCCCGACGCCTGC). The PCR was performed as suggested by the manufacturer's manual except that extension of template was performed at 66°C. pBluescript II SK+-FLNa (ClaI) was made by excision of a ClaI-ClaI fragment from human filamin A in pCMV5-FLNa (Δ20N) and ligation into ClaI-cut pBluescript II SK+ (Stratagene). pBluescript II SK+-FLNa-Ala2152 (ClaI) was created by amplification of pBluescript II SK+-FLNa (ClaI) with Pfu DNA polymerase and the above primers. The Ser2152-to-Ala mutation was confirmed by sequencing.
Construction of pcDNA3-myc-FLNa (human filamin A).
pcDNA3-myc-FLNa was created using the following strategy. (i) pREP4-FLNa was amplified with Ex Taq polymerase (Takara), 5′ FLNa (EcoRI) primer containing an EcoRI site (TTAGAATTCAGTAGCTCCCACTCTCGG), and 3′ FLNa (SalI/XbaI) primer containing SalI and XbaI sites (TGTTCTAGAGTCCCGCGTGTCGACG). The 70-bp PCR product was gel purified, digested with EcoRI and XbaI (New England Biolabs), and ligated into EcoRI-XbaI-digested pCMV5 vector to make pCMV5-5′FLNa. (ii) pCMV5-full-length FLNa was made by excision of an 8.6-kb SalI-SalI filamin A fragment from pCMV5-FLNa (Δ20N) and ligation into SalI-cut pCMV5-5′FLNa. (iii) pcDNA3-myc-FLNa (EcoRI-XbaI) was made by excision of a 1.9-kb EcoRI-XbaI fragment of FLNa from pCMV5-FLNa (Δ20N) and ligation into EcoRI-XbaI-digested pcDNA3 vector containing an N-terminal myc tag (27). (iv) pcDNA3-myc-FLNa was made by excision of a 6.7-kb EcoRI-EcoRI fragment from pCMV5-full-length FLNa and ligation into EcoRI-cut pcDNA3-myc-FLNa (EcoRI-XbaI).
pcDNA3-myc-FLNa-Ala2152 was created by replacement of the 2.4-kb ClaI-ClaI fragment of FLNa in pcDNA3-myc-FLNa with a ClaI-ClaI fragment containing the Ala mutation that was excised from pBluescript II SK+-FLNa-Ala2152 (ClaI).
Cell culture conditions, transfection, and lysate preparation.
Human embryonic kidney (HEK) 293E, rat 3Y1, and HeLa cells were cultured at 37°C in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum (FBS) and antibiotics. HEK 293E cells were transfected by calcium phosphate precipitation. For lysate preparation, HEK 293E or rat 3Y1 cells were washed once in 1× phosphate-buffered saline (PBS) and then harvested on ice in cold lysis buffer (10 mM KPO4, 1 mM EDTA [pH 7.05], 10 mM MgCl2, 5 mM EGTA [pH 7.2], 50 mM β-glycerophosphate [pH 7.2], 0.5% Nonidet P-40 [NP-40], 0.1% Brij-35 plus 1 mM Na3VO4, 5 μg of pepstatin ml−1, 10 μg of leupeptin ml−1, 1 mM phenylmethylsulfonyl fluoride). Lysates were cleared by centrifugation at 16,000 × g for 10 min at 4°C.
In vitro kinase assay.
Lysates were made from HEK 293E cells that had been starved for 24 h or cells that were starved and then stimulated for 10 min with 10% FBS and incubated with RSK-C3 antibody or anti-HA antibody (for overexpressed RSK or Akt) for 1 h at 4°C with rocking. Complexes were precipitated by incubation with protein A Sepharose beads for 1 h. Beads were collected by centrifugation and washed once with each of the following buffers: Hermann's buffer (pH 7.2) (10 mM Tris [pH 7.2], 100 mM NaCl, 1 mM EDTA [pH 8.0], 1% NP-40, 0.5% deoxycholate), buffer B (pH 7.2) (10 mM Tris [pH 7.2], 1 M NaCl, 0.1% NP-40), ST buffer (pH 7.2) (50 mM Tris-HCl, 5 mM Tris base, 150 mM NaCl), and kinase buffer (20 mM HEPES [pH 7.2], 10 mM MgCl2, 3 mM β-mercaptoethanol, 100 μg of bovine serum albumin ml−1). In vitro kinase assay was performed as previously described (26) using glutathione S-transferase (GST) fusion proteins purified from bacteria or proteins expressed by a baculovirus (19) as substrates. Kinase reactions were stopped by the addition of 2× sample buffer (100 mM Tris [pH 6.8], 2% sodium dodecyl sulfate [SDS], 0.1% bromophenol blue, 20% glycerol, 286 mM β-mercaptoethanol). Kinase assays were performed at least two times with comparable results.
In vivo labeling.
HEK 293E cells were transfected with plasmid DNA and then starved for about 20 h. Cells were subsequently labeled for 3 h with 32Pi (1 to 2 mCi per 10-cm-diameter dish of cells). Cells were pretreated with 5 or 10 μM UO126 (BIOMOL Research Laboratories, Inc.) for 30 min and then stimulated with 10% FBS for 10 min or with 50 ng of EGF ml−1 for 15 min. Lysates were prepared. Overexpressed FLNa was immunoprecipitated with anti-myc antibody, and endogenous RSK was immunoprecipitated from the supernatant of the myc immunoprecipitates with RSK-C3 antibody. Immunoprecipitates were washed twice with each of the following buffers: Hermann's buffer (pH 7.2), buffer B (pH 7.2), and ST buffer (pH 7.2). Samples were denatured in 2× sample buffer, boiled, and analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. Phosphate incorporation was quantitated using a phosphorimager and reported as fold activation normalized to the serum-starved samples. Each in vivo labeling experiment was performed a minimum of two times, and representative data are shown.
RNAi.
High-performance liquid chromatography-purified (>97% pure) short interfering RNA (siRNA) oligonucleotides were purchased from Qiagen. The sequences of the sense strands of the siRNA oligonucleotides follow: RSK1, 5′-GGCAACGCUGAAAGUACGUdTdT-3′, RSK2, 5′-GGCCACACUGAAAGUUCGAdTdT-3′, and nonsilencing control, 5′-UUCUCCGAACGUGUCACGUdTdT-3′. HeLa cells (2 × 105) were seeded in 35-mm-diameter wells. On the following day, cells were transfected with 200 pmol of RSK1, RSK2, or nonsilencing control siRNA oligonucleotides using Oligofectamine as described by the manufacturer (Invitrogen). Approximately 24 h later, cells were trypsinized and seeded in 60-mm-diameter plates. Cells were serum starved for 1 day and/or stimulated with EGF (with and without UO126 treatment). Cells were lysed 72 h posttransfection, since optimal silencing of RSK1 and RSK2 was observed at this time point.
Isolation of stable human melanoma cell lines.
Human melanoma cells were cultured in minimum essential medium Eagle (MEM) containing Earle's salts and l-glutamine (Mediatech, Inc.), 10% iron-enriched calf serum, 10 mM HEPES (pH 7.2), 500 μg of G418 (Invitrogen) ml−1, and antibiotics. pcDNA3 vector or pcDNA3-myc-FLNa (FLNa-WT) plasmids were each transfected into FLNa-deficient M2 human melanoma cells (7) by calcium phosphate coprecipitation. After 3 days, the transfected cells were replated, and the medium was replaced with medium containing G418 (500 μg ml−1). Cells were cultured for 17 additional days under G418 selection, and the medium was changed every few days. Colonies were picked, and resistant clonal cell lines were cultured in medium containing G418 (500 μg ml−1).
Single-cell tracking migration assay.
Human melanoma cells (5 × 104 cells) were seeded in 25-mm-diameter flasks. On the following day, cells were washed once with PBS and starved for 16 h in MEM containing 20 mM HEPES. After the starvation period, the medium was changed to medium containing 10% serum for 1 h. EGF (5 ng ml−1) is added to the medium to induce migration. Cells were viewed using a 4× objective on a Nikon Diaphot 300 inverted microscope equipped with a heated stage. Images were captured with a charge-coupled device camera (Dage-MTI) every 15 min for 1 h beginning 15 min after EGF stimulation. Centroid displacement was determined using IP Lab Spectrum software.
RESULTS
The N-terminal kinase domain of RSK phosphorylates FLNa on Ser2152 in vitro.
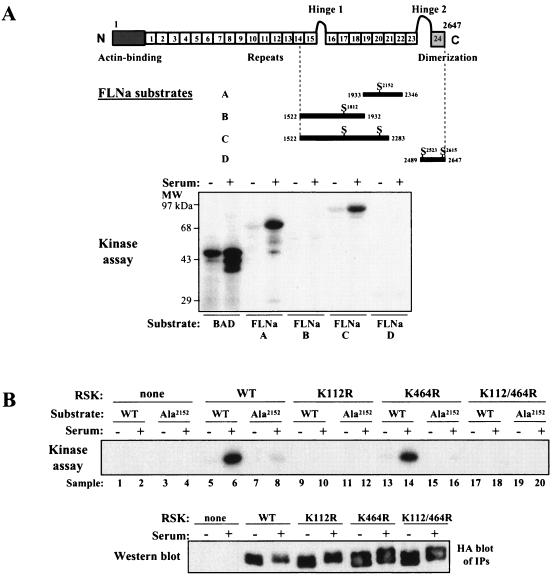
In order to identify the RSK phosphorylation site on FLNa, C-terminal pieces of FLNa were fused to GST and tested as substrates in an in vitro kinase assay. We focused on the C terminus of FLNa, because previous data showed that the mitogen-regulated phosphorylation site was located in this region (21). Endogenous RSK was immunoprecipitated from serum-starved HEK 293E cells or HEK 293E cells that had been serum starved and then stimulated with serum. The cells were incubated with substrate proteins and [γ-32P]ATP. BAD was included as a positive control, because it is a known RSK substrate (1, 26, 32). Activated RSK from serum-stimulated cells phosphorylated FLNa proteins A and C, both of which contain Ser2152 of FLNa (Fig. 1A). In addition, the sequence N-terminal to this Ser residue (R-R-R-A-P-S) fits the known consensus phosphorylation sequence for the RSK N-terminal (D1) kinase domain (R-X-R-X-X-S). To prove that Ser2152 is phosphorylated by RSK, this residue was mutated to Ala in FLNa-C (FLNa-Ala2152) and used as a substrate in a kinase reaction. Mutation of Ser2152 to Ala abolished RSK-mediated, serum-stimulated phosphorylation of FLNa, confirming that Ser2152 is the major RSK phosphorylation site (Fig. 1B, compare lanes 8 and 6). Furthermore, whereas wild-type RSK and RSK-K464R (C-terminal kinase domain mutant) phosphorylated FLNa, RSK-K112R (N-terminal kinase domain mutant) did not (Fig. 1B, compare lanes 6 and 14 to lane 10). These results demonstrate that kinase activity of the D1 kinase domain is required for FLNa phosphorylation but that activity of the D2 domain is dispensable. As expected, kinase-inactive RSK-K112/464R, which contains ATP-binding site mutations in both the N- and C-terminal kinase domains (11), did not phosphorylate FLNa, indicating that phosphorylation was not due to an associated kinase in the immunoprecipitate (Fig. 1B, lane 18). Our data suggest that the N-terminal kinase domain of RSK is responsible for FLNa phosphorylation on Ser2152.
FIG. 1.
N-terminal kinase domain of RSK phosphorylates FLNa on Ser2152 in vitro. (A) Schematic diagram of FLNa protein showing its 24 repeats, functional domains, and RSK interaction region from two-hybrid analysis. Four potential phosphorylation sites that fit the RSK consensus phosphorylation sequence R-X-X-S are located in the C-terminal half of FLNa. Endogenous RSK was immunoprecipitated from serum-starved HEK 293E cells (−) or HEK 293E cells that had been serum starved and then stimulated with serum (+). Immunoprecipitation was performed with RSK-C3 antibody. RSK kinase assays were performed using recombinant GST-tagged BAD or FLNa proteins A, B, C, and D as substrates. The positions of molecular mass markers (MW) are shown to the left of the gel. (B) HEK 293E cells were transfected with the following plasmids: RSK-WT, RSK-K112R, RSK-K464R, RSK-K112/464R, or pKH3 vector. Overexpressed RSK was immunoprecipitated from extracts made from serum-starved cells (−) or cells that had been serum starved and then stimulated with serum (+). Immunoprecipitation was performed with HA antibody. GST-FLNa-C (WT) or GST-FLNa-Ala2152-C (Ala) was used as the substrate in the kinase reaction. (C) HA-tagged Akt or RSK was transfected into HEK 293 cells. Cells were starved (−) or starved first and then stimulated with insulin (100 nM) for 20 min or with serum (10%) for 10 min (+) as indicated above the gels. Pretreatment with 100 nM wortmannin or 10 μM UO126 was for 30 min prior to stimulation. Transfected Akt or RSK was immunoprecipitated (IP) from cell extracts using HA antibody.
The sequence surrounding Ser2152 in FLNa is also consistent with the Akt consensus phosphorylation motif (R-X-R-X-X-S); therefore, we examined Akt's ability to phosphorylate FLNa. HEK 293E cells were transfected with HA-tagged Akt. After transfection and serum stimulation, cells were stimulated with insulin. We treated cells with insulin, because it potently activates Akt but only weakly activates RSK in HEK 293E cells. Cell extracts were prepared, and Akt was immunoprecipitated using HA antibody. Kinase reactions were performed as described in Materials and Methods. We found that Akt did not phosphorylate FLNa-WT or FLNa-Ala protein, despite the fact that Akt was strongly activated by insulin stimulation and phosphorylated BAD in this assay (Fig. 1C).
RSK does not phosphorylate FLNb in vitro.
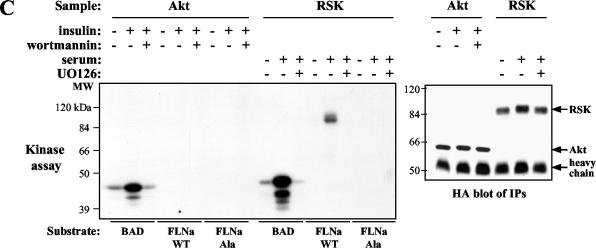
We wanted to determine whether RSK phosphorylates the other members of the filamin family, FLNb and FLNc. The Ser2152 residue of FLNa is conserved in human FLNb (Ser2107) and FLNc (Ser2126). However, FLNb and FLNc do not have arginine residues at both positions −3 and −5 of the RSK consensus sequence (Fig. 2A), suggesting that they are not likely to be good RSK substrates. FLNc is expressed only in muscle cells and has been shown to interact with sarcoglycans (33) and other muscle-specific proteins, such as myotilin (37), FATZ (10), and myozenin (30). Since a clone containing FLNc Ser2126 is not available, we were unable to test the ability of RSK to phosphorylate FLNc. To our knowledge, phosphorylation of FLNc has not been reported. Excluding a divergent hinge I region, FLNb is about 70% identical to FLNa on the amino acid level. Similar to FLNa, FLNb is expressed in many human tissues (31). In addition, FLNb has been shown to interact with FLNa in human cells (25). We tested FLNb as a substrate for RSK in an in vitro kinase assay. Overexpressed RSK was immunoprecipitated from HEK 293E cells that had been serum starved or serum starved and then stimulated with serum. Kinase reactions were performed using full-length, baculovirus-expressed human FLNa and FLNb proteins as substrates. In response to serum stimulation, RSK phosphorylated FLNa but not FLNb (Fig. 2B, compare lanes 5 and 10). These data suggest that FLNa, not FLNb, is a substrate of RSK.
FIG. 2.
RSK does not phosphorylate FLNb in vitro. (A) Amino acid sequence alignment of a region of human FLNa, FLNb, and FLNc showing the Ser2152 residue denoted with an asterisk. The residues that lie within the RSK consensus phosphorylation sequence are boxed. Identical residues (black letters on white background), nonconserved residues (white letters on black background), and similar residues (black letters on gray background) are also shown. (B) HEK 293E cells were transfected with HA-tagged RSK-WT. RSK was immunoprecipitated from serum-starved HEK 293E cells (−) or HEK 293E cells that were serum starved and then stimulated with serum (+). Immunoprecipitation was performed with HA antibody. RSK kinase assays were performed as described in the legend to Fig. 1B using recombinant, baculovirus-expressed FLNa or FLNb proteins as substrates.
In vivo phosphorylation of FLNa is sensitive to the MEK inhibitor UO126.
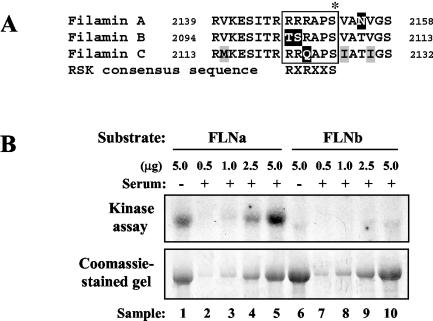
To study the role of the MAP kinase pathway in mediating FLNa phosphorylation in vivo, we examined the effect of the MEK inhibitor UO126 on phosphorylation. HEK 293E cells were transfected with a myc-tagged FLNa construct, serum starved for 1 day, and labeled with 32Pi. Extracts were prepared from labeled cells, and FLNa protein was immunoprecipitated using myc antibody. Serum stimulation resulted in a 2.5-fold increase in phosphate incorporation over the phosphorylation seen under starvation conditions. Treatment of cells with UO126 significantly reduced the serum-stimulated phosphorylation on FLNa (about 60% inhibition [Fig. 3A, left gel]). Hence, phosphorylation of FLNa is UO126 sensitive. The lack of total inhibition by UO126 may reflect the ability of other kinases, such as PKA and Pak1, to phosphorylate FLNa. As a control, endogenous RSK was immunoprecipitated from the supernatants of the myc immunoprecipitates. The 4.2-fold increase in serum-stimulated RSK phosphorylation was completely inhibited by treatment of cells with UO126 (Fig. 3A, right gel). These data strongly support a role for the MAP kinase pathway in mediating FLNa phosphorylation.
FIG. 3.
Treatment with UO126 and mutation of Ser2152 to Ala reduce in vivo FLNa phosphorylation. (A) HEK 293E cells were transfected with pCMV5-FLNa (Δ20N), serum starved, and labeled with 32Pi. Samples were treated and stimulated (+) as indicated above the gel. Cell extracts were prepared, and overexpressed FLNa was immunoprecipitated with myc antibody (left panel). Endogenous RSK was immunoprecipitated from the supernatant of the myc immunoprecipitates using RSK-C3 antibody (right panel). Immunoprecipitate samples were washed as described in Materials and Methods and analyzed by SDS-PAGE and autoradiography. Phosphorylation was quantified by phosphorimager analysis and reported as relative values to represent fold activation relative to the serum-starved samples. (B) HEK 293E cells were transfected with FLNa-WT or FLNa-Ala2152, serum starved, and metabolically labeled with 32Pi. Cells were treated with 10 μM UO126 for 30 min prior to stimulation with 10% serum for 10 min or with 50 ng of EGF ml−1 for 15 min (+) as indicated above the gels. Samples were processed as described above for panel A. The Coomassie blue-stained gel shows similar levels of FLNa-WT and FLNa-Ala2152 protein in the immunoprecipitate samples.
To determine whether FLNa is phosphorylated on Ser2152 in vivo, 32P labeling was performed in HEK 293E cells expressing myc-tagged FLNa-WT or FLNa-Ala2152 proteins. Stimulation of cells with serum or EGF led to a significant increase in phosphate incorporation by FLNa-WT (Fig. 3B, lanes 4, 5, and 7). As expected, pretreatment of cells with UO126 effectively inhibited this mitogen-induced phosphorylation (Fig. 3B, lanes 6 and 8). In contrast to FLNa-WT, FLNa-Ala2152 was not phosphorylated in response to mitogens (Fig. 3B, compare lanes 10 and 12 to lanes 5 and 7).
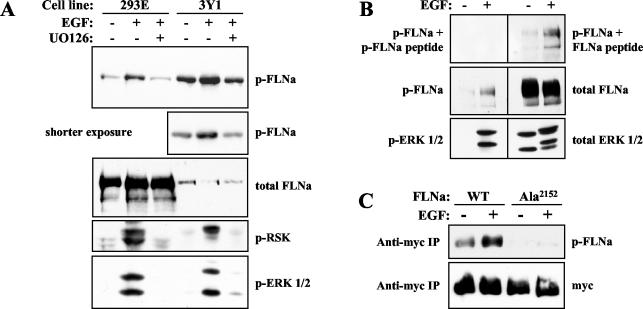
We generated a polyclonal phospho-specific FLNa (phospho-FLNa) antibody that recognizes FLNa protein phosphorylated on Ser2152 to confirm the in vivo labeling results. We tested our phospho-FLNa antibody using cell extracts prepared from HEK 293E and rat 3Y1 cells that have been serum starved or serum starved and stimulated with EGF. Phospho-FLNa antibody strongly recognized endogenous FLNa in stimulated cells, while FLNa antibody detected FLNa protein from unstimulated and stimulated cells to a similar extent (Fig. 4A). A small pool of FLNa may be basally phosphorylated, since our phospho-FLNa antibody weakly detected FLNa in unstimulated cells. We used peptide competition to validate the specificity of our phospho-specific antibody. Prior to incubation with the Western blot membrane, phospho-FLNa antibody was incubated with phosphorylated FLNa peptide or unphosphorylated FLNa peptide. Although blocking the antibody with unphosphorylated FLNa peptide had no effect (Fig. 4B, top gel, right), blocking with phosphorylated FLNa peptide abolished the signal (Fig. 4B, top gel, left). Furthermore, phospho-FLNa antibody detected overexpressed FLNa-WT protein but not FLNa-Ala2152 protein (Fig. 4C). These results suggest that our phospho-FLNa antibody specifically recognizes FLNa when it is phosphorylated on Ser2152.
FIG. 4.
Characterization of phospho-FLNa antibody that specifically recognizes FLNa phosphorylated on Ser2152. (A) HEK 293E or rat 3Y1 cells were serum starved or serum starved and then stimulated with 50 ng of EGF ml−1 (+) for 15 min. Cells were pretreated with UO126 (+) where indicated. Lysates were prepared and blotted with phospho-Ser2152-FLNa (p-FLNa), total FLNa, phospho-Ser380-RSK (p-RSK), and phospho-Thr202/Tyr204-ERK1/2 (p-ERK 1/2) antibodies. (B) Extracts were made from HEK 293E cells that were serum starved or serum starved and stimulated with EGF for 15 min. Samples were analyzed by Western blotting using p-FLNa antibody blocked with phosphorylated FLNa (p-FLNa) peptide or nonphosphorylated FLNa (FLNa) peptide, p-FLNa, total FLNa, p-ERK 1/2, and total ERK1/2 antibodies. For peptide blocking, phospho-Ser2152 FLNa antibody was incubated with 50 μg of p-FLNa peptide or FLNa peptide for 1 h at 4°C. Blocked phospho-FLNa antibody solutions were subsequently diluted 10-fold in Tris-buffered saline solution containing 0.2% Tween 20 containing 2% bovine serum albumin (final dilution of 1:5,000) and incubated with Western blot membranes. (C) HEK 293E cells were transfected with myc-tagged FLNa-WT (WT) or FLNa-Ala2152 (Ala). After serum starvation and EGF stimulation (+), FLNa was immunoprecipitated. myc immunoprecipitate (IP) samples were blotted with p-FLNa and myc antibodies.
FLNa Ser2152 phosphorylation is induced by activators of the MAP kinase pathway and forskolin.
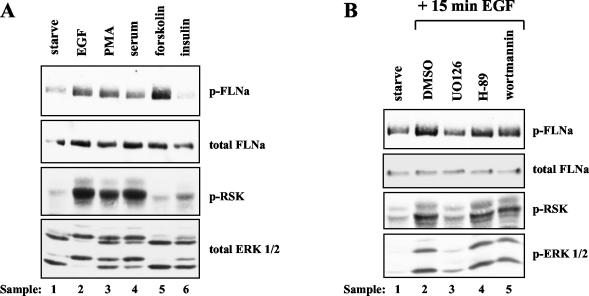
To determine which signaling pathways regulate FLNa Ser2152 phosphorylation in vivo, we stimulated 3Y1 cells with various agonists and used the phospho-FLNa antibody to detect Ser2152 phosphorylation. We found that activation of the MAP kinase pathway with agonists, such as EGF, serum, and phorbol myristate acetate, stimulated FLNa phosphorylation on Ser2152 (Fig. 5A, lanes 2, 3, and 4). In contrast, we failed to detect a significant increase in FLNa Ser2152 phosphorylation after insulin stimulation. Therefore, we conclude that the PI3K signaling pathway does not play a major role in the regulation of FLNa Ser2152 phosphorylation (Fig. 5A, lane 6). PKA also phosphorylates FLNa on Ser2152 in vitro (16). We observed an increase in Ser2152 phosphorylation after cells were treated with forskolin to activate PKA (Fig. 5A, lane 5). Thus, PKA likely phosphorylates Ser2152 in vivo. These results indicate that activation of RSK or PKA correlates with FLNa phosphorylation on Ser2152.
FIG. 5.
FLNa Ser2152 phosphorylation is stimulated by agonists of the MAP kinase pathway and specifically inhibited by UO126. (A) Rat 3Y1 cells were serum starved for 24 h. The cells were stimulated with 50 ng of EGF ml−1 or 100 ng of phorbol myristate acetate (PMA) ml−1 for 15 min, with 10% serum or 10 μM forskolin for 10 min, or with 100 nM insulin for 20 min. Lysates were blotted with phospho-Ser2152-FLNa (p-FLNa), total FLNa, phospho-Ser380-RSK (p-RSK), and total ERK1/2 antibodies. (B) Rat 3Y1 cells were serum starved and then pretreated with one of the following inhibitors for 30 min prior to EGF stimulation: DMSO control, 10 μM UO126, 10 μM H-89, or 100 nM wortmannin. Lysates were subjected to Western blot analysis using p-FLNa, total FLNa, p-RSK, and phosphorylated ERK1/2 (p-ERK 1/2) antibodies.
EGF-stimulated FLNa Ser2152 phosphorylation is specifically inhibited by UO126.
Next, we used specific inhibitors to show that the MAP kinase pathway is the major pathway responsible for EGF-stimulated FLNa Ser2152 phosphorylation. Rat 3Y1 cells were treated with inhibitors for 30 min prior to stimulation with EGF. Extracts were prepared, and FLNa Ser2152 phosphorylation was monitored using our phospho-FLNa antibody. Western blot analysis confirmed the results of the in vivo labeling experiments, as the MEK inhibitor UO126 completely blocked EGF-stimulated phosphorylation of endogenous FLNa (Fig. 5B, compare lanes 3 and 2). Treatment of cells with the PI3K inhibitor wortmannin had little or no effect on FLNa phosphorylation (Fig. 5B, lane 5), suggesting that FLNa Ser2152 phosphorylation is not regulated by the PI3K signaling pathway. Likewise, preventing PKA activation with the PKA inhibitor H-89 had little effect on EGF-stimulated FLNa Ser2152 phosphorylation, even though H-89 blocked forskolin-induced phosphorylation (Fig. 5B, lane 4, and data not shown). These data demonstrate that the FLNa Ser2152 phosphorylation induced by EGF is mediated predominantly by the MAP kinase signaling pathway.
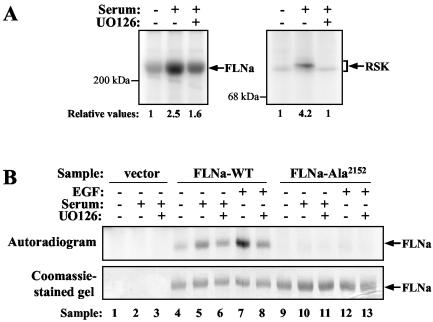
Wild-type RSK potentiates EGF-stimulated FLNa phosphorylation, while kinase-dead RSK blocks phosphorylation.
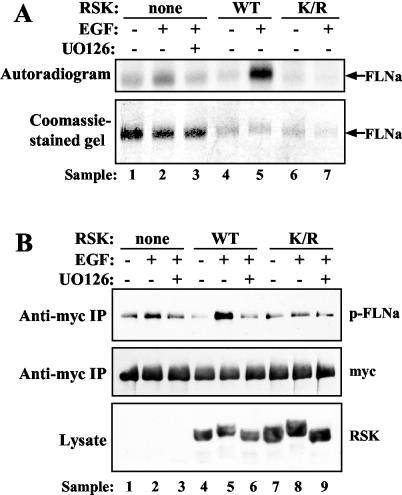
In order to provide further evidence that RSK mediates FLNa Ser2152 phosphorylation in vivo, we overexpressed RSK in HEK 293E cells and monitored FLNa phosphorylation by 32P labeling and Western blotting. In vivo labeling was performed as previously described on HEK 293E cells cotransfected with myc-tagged FLNa and HA-tagged RSK-WT, kinase-inactive RSK-K112/464R, or vector. Labeled FLNa protein was immunoprecipitated from extracts using myc antibody. Overexpression of RSK-WT led to a significant increase in phosphate incorporation by FLNa (Fig. 6A, compare lanes 5 and 2). In contrast, overexpression of RSK-K112/464R reduced EGF-induced FLNa phosphorylation to basal level (Fig. 6A, compare lanes 7 and 4). The effect of RSK on FLNa Ser2152 phosphorylation was also investigated using the phospho-FLNa antibody. HEK 293E cells were transfected with FLNa and RSK constructs, serum starved, and stimulated with EGF (with and without UO126 pretreatment). myc immunoprecipitates were analyzed by Western blotting using phospho-FLNa antibody. Similar to the results of the in vivo labeling experiment, overexpression of RSK-WT greatly enhanced EGF-stimulated FLNa Ser2152 phosphorylation (Fig. 6B, compare lanes 5 and 2). In addition, phosphorylation was completely UO126 sensitive (Fig. 6B, lane 6). Overexpression of RSK-K112/464R, on the other hand, antagonized EGF-stimulated FLNa Ser2152 phosphorylation (Fig. 6B, lane 8). These results strengthen our hypothesis that RSK is a physiological FLNa Ser2152 kinase.
FIG. 6.
EGF-stimulated FLNa Ser2152 phosphorylation is enhanced by wild-type RSK and antagonized by kinase-inactive RSK. (A) HEK 293E cells were cotransfected with myc-tagged FLNa-WT and RSK-WT (WT), RSK-K112/464R (K/R), or empty vector (none). Cells were starved, labeled with 32Pi, and stimulated with 50 ng of EGF ml−1 (+) that had been pretreated with 10 μM UO126 (+) or not pretreated with UO126 (−) as indicated. Lysates were prepared, and overexpressed FLNa was immunoprecipitated using myc antibodies. Samples were analyzed by SDS-PAGE and autoradiography. (B) HEK 293E cells were cotransfected with myc-tagged FLNa-WT and RSK-WT (WT), RSK-K112/464R (K/R), or empty vector (none). After EGF stimulation and cell lysis, samples were analyzed by Western blotting using phospho-Ser2152-FLNa (p-FLNa), total FLNa, and RSK-C3 antibodies.
Specific inhibition of RSK1 and RSK2 protein expression leads to reduction of EGF-stimulated FLNa Ser2152 phosphorylation.
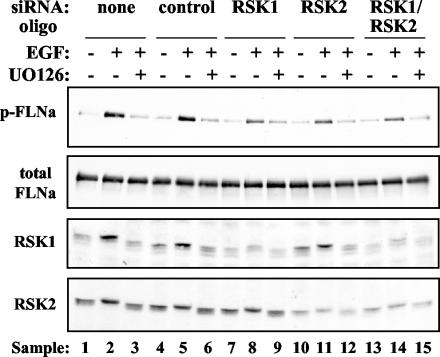
We directly examined the role of RSK in FLNa Ser2152 phosphorylation using RNAi techniques to inhibit expression of endogenous RSK. HeLa cells were transfected with control, RSK1, RSK2, or a combination of RSK1 and RSK2 siRNA oligonucleotides. Forty-eight hours posttransfection, cells were serum starved for 1 day. After starvation, cells were pretreated with UO126 prior to EGF stimulation. Lysates were prepared and analyzed by Western blotting using phospho-FLNa antibody. Reduction of RSK1 or RSK2 protein levels by RNAi caused a substantial decrease in EGF-stimulated FLNa Ser2152 phosphorylation (Fig. 7, compare lanes 8 and 11 to lane 5). Although FLNa Ser2152 phosphorylation was not completely abolished by RSK RNAi, our data clearly reveal an important role for RSK in FLNa phosphorylation. In addition, some RSK1 and RSK2 protein was still expressed in the RNAi samples and may account for the small amount of FLNa phosphorylation remaining. Alternatively, RSK3, RSK4, or additional kinases may also contribute to EGF-stimulated FLNa Ser2152 phosphorylation.
FIG. 7.
EGF-stimulated FLNa Ser2152 phosphorylation is reduced by RSK RNAi. HeLa cells were transfected with 200 pmol of each siRNA oligonucleotide (oligo) (for combined targeting of RSK1 and RSK2, 100 pmol of each oligonucleotide was used), serum starved or serum starved first and then stimulated with 50 ng of EGF ml−1 (+) pretreated with 10 μM UO126 (+) or not pretreated with UO126 (−) as indicated above the gels. Lysates were prepared 72 h posttransfection and analyzed by Western blotting using FLNa, RSK1, and RSK2 antibodies (10 μg of total protein was loaded per lane). The Western blot membrane was stripped of FLNa antibody and reprobed with phospho-Ser2152-FLNa (p-FLNa) antibody to ensure equal FLNa protein levels (top blot). The data shown are representative of five experiments.
Inhibition of MAP kinase signaling by UO126-mediated inhibition of MEK dramatically reduces FLNa-dependent, EGF-stimulated migration of human melanoma cells.
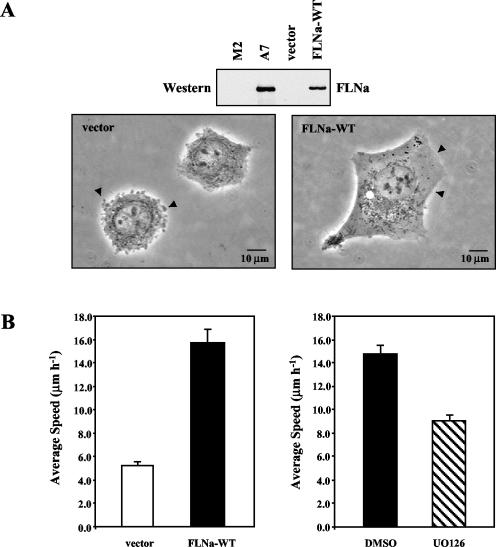
In order to determine whether inhibition of FLNa Ser2152 phosphorylation influences a biological process, we utilized a human melanoma cell line to study cell migration. We transfected FLNa-deficient human melanoma (M2) cells with empty vector or vector containing myc-tagged, full-length FLNa (FLNa-WT). Stable clones were selected with Geneticin and analyzed for FLNa expression by Western blotting using anti-myc and anti-FLNa monoclonal antibodies. Given that maximal migration of melanoma cells requires a specific amount of FLNa protein, we chose a cell line that expresses FLNa at a level similar to that found in the FLNa-replete A7 line (Fig. 8A). Resembling the parental FLNa-deficient M2 line, vector control cells are round and display extensive membrane blebbing. FLNa-WT cells, on the other hand, have smooth plasma membrane surfaces, and the morphology of FLNa-WT cells is similar to the morphology of A7 cells (Fig. 8A) (7, 12).
FIG. 8.
FLNa-dependent, EGF-induced migration of human melanoma cells is inhibited by UO126. (A) Lysates were prepared from human melanoma cells and analyzed by Western blotting using FLNa antibody (30 μg of total protein was loaded per lane). Phase-contrast images of human melanoma cells were taken on the following day with a 40× objective. (B) A total of 5 × 104 FLNa-WT or vector control melanoma cells were seeded in 25-mm-diameter flasks. On the following day, cells were washed once with PBS and serum starved overnight. The medium was then changed to medium containing 10% serum for 1 h. Migration was stimulated by the addition of EGF (5 ng ml−1). Images were captured every 15 min for 1 h beginning 15 min after EGF stimulation. Displacement of the centroid was used to calculate the distance migrated. Results represent the mean speed ± standard deviation. A minimum of 140 cells from at least two separate experiments was analyzed for each cell line (left panel). FLNa-WT cells were treated with 50 μM UO126 or DMSO for 5 h prior to EGF stimulation. The migration of at least 300 cells for each condition was analyzed (right panel).
To assess the migratory ability of vector control and FLNa-WT melanoma cells, we performed single-cell tracking assays. After EGF stimulation, vector control cells migrated with an average speed of only 5.3 ± 0.3 μm per hour. In contrast, the average speed of FLNa-WT cells was 15.7 ± 1.2 μm per hour under the same conditions (Fig. 8B, left panel). Thus, EGF-induced migration of these melanoma cells is dependent on the expression of FLNa. To determine whether MAP kinase signaling is required for this FLNa-dependent migration, we treated cells with the MEK inhibitor UO126. We found that EGF-induced migration of FLNa-WT cells was greatly impaired after UO126 treatment. UO126-treated FLNa-WT cells migrated with an average speed of 9.1 ± 0.45 μm per hour, significantly slower than control (dimethyl sulfoxide [DMSO]-treated) cells, which migrated with a speed of 14.8 ± 0.7 μm per hour (Fig. 8B, right panel). Thus, inhibition of MAP kinase signaling and RSK caused a 60% decrease in FLNa-dependent migration of human melanoma cells.
DISCUSSION
The molecular basis for how the Ras-MAP kinase pathway contributes to the regulation of plasma membrane dynamics is not well defined. Our data suggest that the Ras-MAP kinase pathway may control actin cytoskeletal changes via RSK phosphorylation of the FLNa protein. We found that RSK phosphorylated FLNa on Ser2152 and mutation of Ser2152 to Ala dramatically reduced serum- and EGF-induced FLNa phosphorylation in vivo. Moreover, EGF-stimulated FLNa Ser2152 phosphorylation was specifically inhibited by UO126, indicating that the MAP kinase pathway plays a major role in regulating FLNa phosphorylation. In contrast, the PI3K pathway did not appear to contribute to FLNa phosphorylation, since Ser2152 phosphorylation was neither increased by insulin nor inhibited by wortmannin. In addition, we demonstrated that expression of wild-type RSK markedly augments FLNa phosphorylation and that kinase-inactive RSK prevents phosphorylation.
Other kinases, such as PKA and Pak1, have also been shown to phosphorylate FLNa protein. Several groups have demonstrated that treatment of cells with cAMP or agonists that increase intracellular cAMP levels activates PKA and stimulates FLNa phosphorylation in platelets, smooth muscle cells, and fibroblasts (4, 5, 9, 39, 41). The in vivo PKA phosphorylation site(s) was not identified in these studies. The effect of PKA-mediated FLNa phosphorylation on FLNa function is somewhat controversial. One group reported that PKA phosphorylation of FLNa in vitro confers resistance to cleavage by calpain (5). Another study concluded that phosphorylation of FLNa is required for the ability of FLNa to cross-link F-actin in vitro (42). Ser2152 on FLNa has been identified as a PKA phosphorylation site in vitro (16); however, we present the first evidence that Ser2152 phosphorylation is stimulated in vivo after PKA activation (Fig. 5A, lane 5).
Pak1 has also been shown to phosphorylate FLNa on Ser2152 in vitro (36). Treatment of MCF-7 cells with heregulin increased FLNa phosphorylation, and overexpression of kinase-inactive Pak1 or the Pak1 autoinhibitory domain antagonized this phosphorylation. Activated Pak1 (Thr423-to-Glu mutant) stimulated ruffling in FLNa-expressing melanoma cells but not in FLNa-deficient cells, suggesting that FLNa is required for Pak1-mediated ruffle formation. In addition, ruffling was induced in FLNa-deficient melanoma cells expressing activated Pak1 and FLNa, but not in cells expressing activated Pak1 and FLNa-Ala2152 mutant (36). These data provide convincing evidence that phosphorylation of FLNa on Ser2152 regulates actin cytoskeletal changes.
Our data strongly support the conclusion that RSK phosphorylates FLNa on Ser2152 in vivo. Although Pak1 phosphorylates FLNa on the same site, we believe that RSK and Pak1 are likely activated under different conditions. First, we demonstrated that EGF-stimulated FLNa phosphorylation is sensitive to the MEK inhibitor UO126. UO126 specifically inhibits MEK1/2 and has not been reported to affect Pak1 activation. Second, whereas expression of wild-type RSK greatly enhanced EGF-stimulated FLNa Ser2152 phosphorylation, kinase-inactive RSK inhibited phosphorylation. Third, FLNa Ser2152 phosphorylation was greatly diminished by specific inhibition of endogenous RSK1 and/or RSK2 expression using siRNA oligonucleotides. Finally, inhibition of MAP kinase signaling to RSK significantly reduced FLNa-dependent migration of human melanoma cells in response to EGF. Taken together, these results strengthen our hypothesis that RSK is a physiological FLNa kinase.
What are the downstream events controlled by phosphorylation of FLNa on Ser2152? FLNa Ser2152 phosphorylation may directly stimulate binding or cross-linking of actin filaments. Preliminary data suggest, however, that these functions are not affected by phosphorylation. FLNa-Ala2152 proteins retained the ability to bind actin in sedimentation assays and to cross-link actin filaments into three-dimensional gels in vitro (F. Nakamura, personal communication).
Alternatively, FLNa Ser2152 phosphorylation may regulate binding to and/or activity of one of its many binding partners. To date, more than 20 proteins have been reported to interact with FLNa (reviewed by Stossel et al. [28]). Two FLNa binding proteins are the Rho family GTPases and β integrins. Given that Rho family GTPases play a major role in the regulation of actin cytoskeletal dynamics, their connection to FLNa is intriguing. FLNa interacted with the small GTPase RalA in a GTP-dependent manner, and induction of RalA-induced filopodia did not occur in FLNa-deficient cells, indicating that FLNa protein is necessary for the formation of membrane protrusions (22). β integrins have also been shown to interact with FLNa. Interestingly, valine-to-isoleucine substitutions in the cytoplasmic tail of β1A integrin caused β1A integrin to bind FLNa with greater affinity. Furthermore, increased binding of β integrin to FLNa inhibited cell migration (3). Whether FLNa Ser2152 phosphorylation influences the binding of FLNa to GTPases and β integrins has yet to be determined.
In conclusion, we provide the first evidence that RSK mediates phosphorylation of FLNa on Ser2152, a site that regulates the biological function of FLNa. Exploring this connection between RSK and FLNa will give us a better understanding of how activation of the Ras-MAP kinase pathway elicits actin cytoskeletal changes.
Acknowledgments
We thank F. Nakamura for generously sharing unpublished data and reagents and M. Chou, P. James, and S. Hollenberg for providing reagents. We are grateful to members of the Blenis lab for helpful discussions and critical reading of the manuscript.
This work was supported in part by grant CA09595 from NCI/NIH (M.S.W.), grants RO1 CA46595 and RO1 GM51405 from the NIH (J.B.), USPHS grant HL19429, and the American Cancer Society (T.P.S.).
REFERENCES
- 1.Bonni, A., A. Brunet, A. E. West, S. R. Datta, M. A. Takasu, and M. E. Greenberg. 1999. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science 286:1358-1362. [DOI] [PubMed] [Google Scholar]
- 2.Byers, H. R., T. Etoh, J. R. Doherty, A. J. Sober, and M. C. Mihm, Jr. 1991. Cell migration and actin organization in cultured human primary, recurrent cutaneous and metastatic melanoma. Time-lapse and image analysis. Am. J. Pathol. 139:423-435. [PMC free article] [PubMed] [Google Scholar]
- 3.Calderwood, D. A., A. Huttenlocher, W. B. Kiosses, D. M. Rose, D. G. Woodside, M. A. Schwartz, and M. H. Ginsberg. 2001. Increased filamin binding to beta-integrin cytoplasmic domains inhibits cell migration. Nat. Cell Biol. 3:1060-1068. [DOI] [PubMed] [Google Scholar]
- 4.Carroll, R. C., and J. M. Gerrard. 1982. Phosphorylation of platelet actin-binding protein during platelet activation. Blood 59:466-471. [PubMed] [Google Scholar]
- 5.Chen, M., and A. Stracher. 1989. In situ phosphorylation of platelet actin-binding protein by cAMP-dependent protein kinase stabilizes it against proteolysis by calpain. J. Biol. Chem. 264:14282-14289. [PubMed] [Google Scholar]
- 6.Chen, R. H., C. Sarnecki, and J. Blenis. 1992. Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Mol. Cell. Biol. 12:915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham, C. C., J. B. Gorlin, D. J. Kwiatkowski, J. H. Hartwig, P. A. Janmey, H. R. Byers, and T. P. Stossel. 1992. Actin-binding protein requirement for cortical stability and efficient locomotion. Science 255:325-327. [DOI] [PubMed] [Google Scholar]
- 8.Dalby, K. N., N. Morrice, F. B. Caudwell, J. Avruch, and P. Cohen. 1998. Identification of regulatory phosphorylation sites in mitogen-activated protein kinase (MAPK)-activated protein kinase-1a/p90rsk that are inducible by MAPK. J. Biol. Chem. 273:1496-1505. [DOI] [PubMed] [Google Scholar]
- 9.Davies, P., Y. Shizuta, K. Olden, M. Gallo, and I. Pastan. 1977. Phosphorylation of filamin and other proteins in cultured fibroblasts. Biochem. Biophys. Res. Commun. 74:300-307. [DOI] [PubMed] [Google Scholar]
- 10.Faulkner, G., A. Pallavicini, A. Comelli, M. Salamon, G. Bortoletto, C. Ievolella, S. Trevisan, S. Kojic, F. Dalla Vecchia, P. Laveder, G. Valle, and G. Lanfranchi. 2000. FATZ, a filamin-, actinin-, and telethonin-binding protein of the Z-disc of skeletal muscle. J. Biol. Chem. 275:41234-41242. [DOI] [PubMed] [Google Scholar]
- 11.Fisher, T. L., and J. Blenis. 1996. Evidence for two catalytically active kinase domains in pp90rsk. Mol. Cell. Biol. 16:1212-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flanagan, L. A., J. Chou, H. Falet, R. Neujahr, J. H. Hartwig, and T. P. Stossel. 2001. Filamin A, the Arp2/3 complex, and the morphology and function of cortical actin filaments in human melanoma cells. J. Cell Biol. 155:511-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox, J. W., E. D. Lamperti, Y. Z. Eksioglu, S. E. Hong, Y. Feng, D. A. Graham, I. E. Scheffer, W. B. Dobyns, B. A. Hirsch, R. A. Radtke, S. F. Berkovic, P. R. Huttenlocher, and C. A. Walsh. 1998. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron 21:1315-1325. [DOI] [PubMed] [Google Scholar]
- 14.Frodin, M., and S. Gammeltoft. 1999. Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol. Cell. Endocrinol. 151:65-77. [DOI] [PubMed] [Google Scholar]
- 15.Ito, T., A. Suzuki, and T. P. Stossel. 1992. Regulation of water flow by actin-binding protein-induced actin gelatin. Biophys. J. 61:1301-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jay, D., E. J. Garcia, J. E. Lara, M. A. Medina, and M. de la Luz Ibarra. 2000. Determination of a cAMP-dependent protein kinase phosphorylation site in the C-terminal region of human endothelial actin-binding protein. Arch. Biochem. Biophys. 377:80-84. [DOI] [PubMed] [Google Scholar]
- 17.Jensen, C. J., M. B. Buch, T. O. Krag, B. A. Hemmings, S. Gammeltoft, and M. Frodin. 1999. 90-kDa ribosomal S6 kinase is phosphorylated and activated by 3-phosphoinositide-dependent protein kinase-1. J. Biol. Chem. 274:27168-27176. [DOI] [PubMed] [Google Scholar]
- 18.Kawamoto, S., and H. Hidaka. 1984. Ca2+-activated, phospholipid-dependent protein kinase catalyzes the phosphorylation of actin-binding proteins. Biochem. Biophys. Res. Commun. 118:736-742. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura, F., E. Osborn, P. A. Janmey, and T. P. Stossel. 2002. Comparison of filamin A-induced cross-linking and Arp2/3 complex-mediated branching on the mechanics of actin filaments. J. Biol. Chem. 277:9148-9154. [DOI] [PubMed] [Google Scholar]
- 20.Ohta, Y., and J. H. Hartwig. 1995. Actin filament cross-linking by chicken gizzard filamin is regulated by phosphorylation in vitro. Biochemistry 34:6745-6754. [DOI] [PubMed] [Google Scholar]
- 21.Ohta, Y., and J. H. Hartwig. 1996. Phosphorylation of actin-binding protein 280 by growth factors is mediated by p90 ribosomal protein S6 kinase. J. Biol. Chem. 271:11858-11864. [DOI] [PubMed] [Google Scholar]
- 22.Ohta, Y., N. Suzuki, S. Nakamura, J. H. Hartwig, and T. P. Stossel. 1999. The small GTPase RalA targets filamin to induce filopodia. Proc. Natl. Acad. Sci. USA 96:2122-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards, S. A., V. C. Dreisbach, L. O. Murphy, and J. Blenis. 2001. Characterization of regulatory events associated with membrane targeting of p90 ribosomal S6 kinase 1. Mol. Cell. Biol. 21:7470-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards, S. A., J. Fu, A. Romanelli, A. Shimamura, and J. Blenis. 1999. Ribosomal S6 kinase 1 (RSK1) activation requires signals dependent on and independent of the MAP kinase ERK. Curr. Biol. 9:810-820. [DOI] [PubMed] [Google Scholar]
- 25.Sheen, V. L., Y. Feng, D. Graham, T. Takafuta, S. S. Shapiro, and C. A. Walsh. 2002. Filamin A and filamin B are co-expressed within neurons during periods of neuronal migration and can physically interact. Hum. Mol. Genet. 11:2845-2854. [DOI] [PubMed] [Google Scholar]
- 26.Shimamura, A., B. A. Ballif, S. A. Richards, and J. Blenis. 2000. Rsk1 mediates a MEK-MAP kinase cell survival signal. Curr. Biol. 10:127-135. [DOI] [PubMed] [Google Scholar]
- 27.Stephens, L., K. Anderson, D. Stokoe, H. Erdjument-Bromage, G. F. Painter, A. B. Holmes, P. R. Gaffney, C. B. Reese, F. McCormick, P. Tempst, J. Coadwell, and P. T. Hawkins. 1998. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science 279:710-714. [DOI] [PubMed] [Google Scholar]
- 28.Stossel, T. P., J. Condeelis, L. Cooley, J. H. Hartwig, A. Noegel, M. Schleicher, and S. S. Shapiro. 2001. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2:138-145. [DOI] [PubMed] [Google Scholar]
- 29.Sutherland, C., D. G. Campbell, and P. Cohen. 1993. Identification of insulin-stimulated protein kinase-1 as the rabbit equivalent of rskmo-2: identification of two threonines phosphorylated during activation by mitogen-activated protein kinase. Eur. J. Biochem. 212:581-588. [DOI] [PubMed] [Google Scholar]
- 30.Takada, F., D. L. Vander Woude, H. Q. Tong, T. G. Thompson, S. C. Watkins, L. M. Kunkel, and A. H. Beggs. 2001. Myozenin: an alpha-actinin- and gamma-filamin-binding protein of skeletal muscle Z lines. Proc. Natl. Acad. Sci. USA 98:1595-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takafuta, T., G. Wu, G. F. Murphy, and S. S. Shapiro. 1998. Human β-filamin is a new protein that interacts with the cytoplasmic tail of glycoprotein Ibα. J. Biol. Chem. 273:17531-17538. [DOI] [PubMed] [Google Scholar]
- 32.Tan, Y., H. Ruan, M. R. Demeter, and M. J. Comb. 1999. p90RSK blocks Bad-mediated cell death via a protein kinase C-dependent pathway. J. Biol. Chem. 274:34859-34867. [DOI] [PubMed] [Google Scholar]
- 33.Thompson, T. G., Y. M. Chan, A. A. Hack, M. Brosius, M. Rajala, H. G. Lidov, E. M. McNally, S. Watkins, and L. M. Kunkel. 2000. Filamin 2 (FLN2): a muscle-specific sarcoglycan interacting protein. J. Cell Biol. 148:115-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tigges, U., B. Koch, J. Wissing, B. M. Jockusch, and W. H. Ziegler. 2003. The F-actin cross-linking and focal adhesion protein filamin A is a ligand and in vivo substrate for protein kinase C alpha. J. Biol. Chem. 278:23561-23569. [DOI] [PubMed] [Google Scholar]
- 35.Trivier, E., D. De Cesare, S. Jacquot, S. Pannetier, E. Zackai, I. Young, J. L. Mandel, P. Sassone-Corsi, and A. Hanauer. 1996. Mutations in the kinase Rsk-2 associated with Coffin-Lowry syndrome. Nature 384:567-570. [DOI] [PubMed] [Google Scholar]
- 36.Vadlamudi, R. K., F. Li, L. Adam, D. Nguyen, Y. Ohta, T. P. Stossel, and R. Kumar. 2002. Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nat. Cell Biol. 4:681-690. [DOI] [PubMed] [Google Scholar]
- 37.van der Ven, P. F., S. Wiesner, P. Salmikangas, D. Auerbach, M. Himmel, S. Kempa, K. Hayess, D. Pacholsky, A. Taivainen, R. Schroder, O. Carpen, and D. O. Furst. 2000. Indications for a novel muscular dystrophy pathway: γ-filamin, the muscle-specific filamin isoform, interacts with myotilin. J. Cell Biol. 151:235-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vik, T. A., and J. W. Ryder. 1997. Identification of serine 380 as the major site of autophosphorylation of Xenopus pp90rsk. Biochem. Biophys. Res. Commun. 235:398-402. [DOI] [PubMed] [Google Scholar]
- 39.Wallach, D., P. J. Davies, and I. Pastan. 1978. Cyclic AMP-dependent phosphorylation of filamin in mammalian smooth muscle. J. Biol. Chem. 253:4739-4745. [PubMed] [Google Scholar]
- 40.Williams, M. R., J. S. Arthur, A. Balendran, J. van der Kaay, V. Poli, P. Cohen, and D. R. Alessi. 2000. The role of 3-phosphoinositide-dependent protein kinase 1 in activating AGC kinases defined in embryonic stem cells. Curr. Biol. 10:439-448. [DOI] [PubMed] [Google Scholar]
- 41.Wu, M. P., D. Jay, and A. Stracher. 1994. Existence of multiple phosphorylated forms of human platelet actin binding protein. Cell. Mol. Biol. Res. 40:351-357. [PubMed] [Google Scholar]
- 42.Zhuang, Q. Q., S. Rosenberg, J. Lawrence, and A. Stracher. 1984. Role of actin binding protein phosphorylation in platelet cytoskeleton assembly. Biochem. Biophys. Res. Commun. 118:508-513. [DOI] [PubMed] [Google Scholar]