Background: Orm proteins are key regulators of sphingolipid synthesis.
Results: Orm1 and Orm2 are phosphorylated by TORC1 and TORC2; Orm2 is transcriptionally regulated by a calcium- and calcineurin-dependent pathway.
Conclusion: The Orm proteins respond to feedback signals from the sphingolipid synthesis pathway and multiple environmental signals.
Significance: Modulation of sphingolipid synthesis is a critical component of cellular response to nutritional status and stress.
Keywords: Calcineurin, ER Stress, Phosphorylation, Signal Transduction, Sphingolipid, TOR
Abstract
Sphingolipids are structural components of membranes, and sphingolipid metabolites serve as signaling molecules. The first and rate-limiting step in sphingolipid synthesis is catalyzed by serine palmitoyltransferase (SPT). The recently discovered SPT-associated proteins, Orm1 and Orm2, are critical regulators of sphingolipids. Orm protein phosphorylation mediating feedback regulation of SPT activity occurs in response to multiple sphingolipid intermediates, including long chain base and complex sphingolipids. Both branches of the TOR signaling network, TORC1 and TORC2, participate in regulating sphingolipid synthesis via Orm phosphorylation in response to sphingolipid intermediates as well as nutritional conditions. Moreover, sphingolipid synthesis is regulated in response to endoplasmic reticulum (ER) stress by activation of a calcium- and calcineurin-dependent pathway via transcriptional induction of ORM2. Conversely, the calcium- and calcineurin-dependent pathway signals ER stress response upon lipid dysregulation in the absence of the Orm proteins to restore ER homeostasis.
Introduction
Sphingolipids are structural components of cell membranes, regulating membrane fluidity. On the outer leaflet of the membrane, sphingolipids assemble laterally with cholesterol to form membrane microdomains that provide conformational support for membrane proteins and serve as platforms for recruitment of signaling molecules (1). In addition to a structural role, sphingolipids have critical regulatory activities; sphingolipid precursors/metabolites, such as ceramide, participate as second messengers in signaling to protein kinases and phosphatases. Like phosphoinositides, which act as spatial determinants of membrane trafficking events, sphingolipid metabolites play an important role in actin cytoskeleton polarization and endocytosis (2).
Discovery of the ORMDL family of endoplasmic reticulum (ER)2 membrane proteins represents a major advance in understanding regulation of sphingolipid synthesis (3, 4). The physiologic importance of the ORMDL family is underscored by the finding that one of the human family members is an asthma susceptibility gene (5). In yeast, the ORMDL family members Orm1 and Orm2 are negative regulators of the first and rate-limiting step in sphingolipid synthesis, a reaction catalyzed by serine palmitoyltransferase (SPT). Orm1 and Orm2 proteins regulate SPT activity by physically associating with it (3, 4). Recent evidence suggests that TORC1 and TORC2 protein kinase complexes adjust sphingolipid synthesis via phosphorylation of the Orm proteins as they coordinate cell growth with environmental changes, such as nutrient availability or plasma membrane stress (6–9).
We now show that sphingolipid synthesis is regulated via Orm phosphorylation by a feedback mechanism that responds to multiple sphingolipid intermediates, including long chain base as well as complex sphingolipid(s). In response to ER stress, sphingolipid synthesis is modulated by a calcium- and calcineurin-dependent pathway that regulates Orm2 protein level. Increased Orm2 protein has been shown to increase repression of SPT activity to inhibit sphingolipid synthesis (6). Conversely, lipid dysregulation in the absence of the Orm proteins activates calcium- and calcineurin-dependent signaling to ameliorate ER stress.
EXPERIMENTAL PROCEDURES
Strains and Media
Standard yeast media and genetic manipulations were as described (10). Yeast strains are isogenic with BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and BY4742 (MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0). Orm mutants were orm1Δ::clonNATr (HXX1-7B), orm2Δ::kanr (HXX1-7A), and orm1Δ orm2Δ (HXX1-7D), as described previously (3). Strains from the deletion collection (Open Biosystems, Huntsville, AL) were confirmed by PCR. For Orm2-TAP studies, strains were made by mating ACX184-2B (MATα ORM2::TAP::HIS3 (Open Biosystems)) with MATa deletion strains as follows. ACX195-2D is ire1Δ::kanr, ACX201-5C is crz1Δ::kanr, ACX204-3B is cnb1Δ::kanr, ACX230-3B is ino2Δ::kanr, ACX231-2B is ino4Δ::kanr, ACX234-1C is cch1Δ::kanr, ACX245-1D is hac1Δ::kanr, ACX254-2B is csg2Δ::kanr, ACX261-3C is elo3Δ::kanr, ACX265-3D is isc1Δ::kanr, ACX268-4A is ire1Δ::kanr cnb1Δ::kanr, ACX275-4B is ino1Δ::kanr, and ACX233-1C is ypk1Δ::kanr. ACX264-4C is from a cross between ACX184-2B and KSY271 (kei1Δ::kei1-1::LEU2; a gift from Koji Yoda (University of Tokyo)) (11).
Plasmids
pSH14HA is a HIS3-marked centromeric plasmid bearing ORM1 tagged at the amino terminus with an HA epitope, as described (6). An MPK1-lacZ reporter construct (p1365), as described previously (12), was from David Levin (The Johns Hopkins University, Baltimore, MD). The UPRE-lacZ reporter (pJC106), as described previously (13), was from Peter Walter (University of California, San Francisco), and the INO1-lacZ reporter (pJH359), as described previously (14), was from Susan Henry (Cornell University).
Semiquantitative PCR
RNA was isolated using RNeasy (Qiagen) or Ribopure Yeast (Ambion) kits, according to the manufacturers' instructions. cDNA was generated from 2 μg of RNA using a Superscript II RT-PCR kit (Invitrogen). cDNAs were used as template for standard PCRs with 30 amplification cycles (45 s at 95 °C, 45 s at 65 °C, 1 min at 72 °C). Sequences for forward and reverse primers (569 and 571) to amplify ORM2 were AACCTGACCATGTGGGAGCAGATT and TTCCCAGCTTAGGAACGACACCAA. Sequences of primers (527 and 528) to amplify ACT1 were ACGTTCCAGCCTTCTACGTTTCCA and ACGTGAGTAACACCATCACCGGAA. ORM2 and ACT1 PCR products were analyzed on ethidium bromide-stained agarose gels and quantitated using ImageJ software.
Electrophoretic Mobility Shift Assay, Western Blot, and Enzyme Assay
To assay Orm phosphorylation, cells were harvested and frozen in liquid nitrogen and trichloroacetic acid, as described (6). Orm1 phosphorylation changes were detected as electrophoretic mobility shifts on 10% polyacrylamide gels using an N-terminal HA-tagged Orm1 construct (pSH14HA). Orm2-TAP mobility shifts were visualized after extended electrophoresis on 12% polyacrylamide gels. For phosphorylation analysis, lysate was prepared by vortexing with glass beads in the presence of trichloroacetic acid, as described previously (6). Alkaline phosphatase digestion of lysates was as described previously (6). For quantitative Western blots, lysate was prepared by vortexing with glass beads in sorbitol buffer (0.3 m sorbitol, 0.1 m NaCl, 5 mm MgCl2, 10 mm Tris, pH 7.4) with a protease inhibitor mixture and phenylmethylsulfonyl fluoride, as described previously (15). Western blots were visualized with rabbit antibody (to detect the TAP tag) and monoclonal anti-HA (Covance, Princeton, NJ) followed by horseradish peroxidase-conjugated secondary antibody and chemiluminescence detection. Western blots were scanned and quantitated using ImageJ software. For lacZ (UPRE-lacZ, MPK1-lacZ, and INO1-lacZ) assays, lysate was prepared in breaking buffer (20% glycerol, 1 mm DTT, 100 mm Tris, pH 8) or bead buffer with protease inhibitors, and β-galactosidase activity was measured as described previously (16). Samples were normalized to protein content assayed by Bradford or BCA (Pierce) assays.
RESULTS
A Calcium-dependent Signaling Pathway Regulates Orm2 Protein Levels
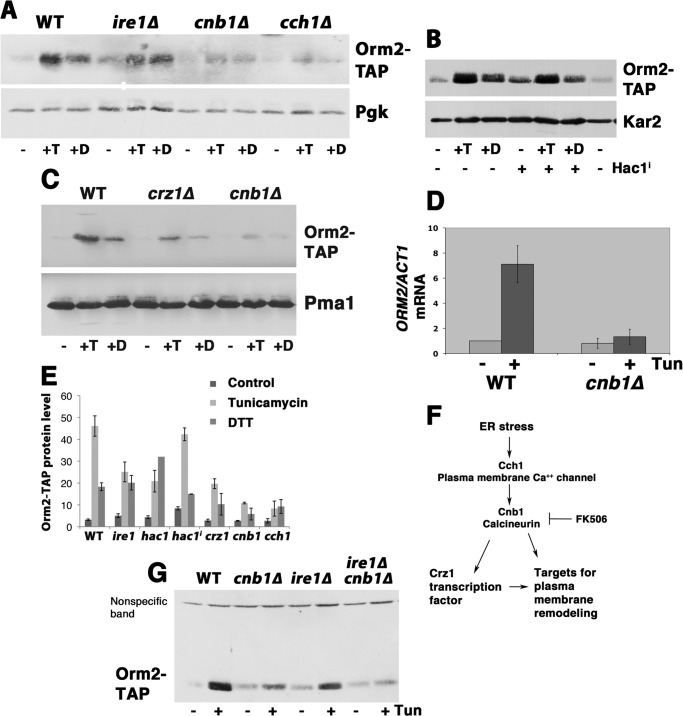
In response to protein misfolding in the ER induced by tunicamycin or DTT, Orm2 protein abundance increased, as shown by Western blot of TAP-tagged Orm2 (Fig. 1A) (3, 6). (Orm2-TAP (tagged at the chromosomal locus) is functional because it retains its association with SPT components (3).) The unfolded protein response (UPR), the major transcriptional response to protein misfolding in the ER, requires the sensor Ire1 to activate the transcription factor Hac1 (17). Surprisingly, however, increased Orm2 in response to tunicamycin or DTT was only partially abrogated in ire1Δ cells (Fig. 1A). To ask whether ORM2 is a UPR target, a constitutively active Hac1 (spliced Hac1i) was used to induce UPR in the absence of stressors (18) (Fig. 1B). As a control, the ER luminal chaperone Kar2 was induced by Hac1i or by tunicamycin or DTT addition (Fig. 1B, bottom). The Orm2 protein level was slightly increased in cells expressing Hac1i; however, the extent of the increase is far less than that induced by tunicamycin.
FIGURE 1.
Induction of Orm2 protein by ER stress is dependent on calcium-dependent signaling. Shown is a Western blot of Orm2-TAP levels in exponentially growing wild-type cells compared with cells treated with tunicamycin (T; 1 μg/ml) for 2 h or DTT (D; 1 mm) for 2 h. Increased Orm2 protein in response to ER stress is calcium-dependent because it was abrogated in cch1Δ (ACX234–1C) and reduced in cnb1Δ (ACX204-3B) and ire1Δ (ACX195-2D) cells. A, Orm2-TAP level (top) was increased after treatment with tunicamycin or DTT and partly dependent on Ire1. Bottom, phosphoglycerate kinase (Pgk) levels are shown as a loading control. B, activation of UPR by Hac1i in the absence of ER stressors had a small effect on Orm2-TAP protein level. Kar2 was blotted as a positive control for UPR activation. C, Western blot showing increased Orm2 levels in response to tunicamycin was abrogated in crz1Δ (and cnb1Δ) cells. Pma1 was blotted as a loading control (bottom). D, bar graph showing relative ORM2 mRNA levels in wild-type and cnb1Δ cells treated with tunicamycin for 2 h, as measured by semiquantitative PCR and normalized to ACT1 mRNA. Error bars, average of three independent colonies. E, summary of quantitated Western blots showing tunicamycin-stimulated increase in Orm2 protein in various deletion strains. Error bars, S.E. from n > 3 determinations. F, schematic diagram showing calcineurin-mediated signaling, activated by ER stress/lipid dysregulation and inhibited by FK506. Calcineurin signaling initiates membrane remodeling by activation of the transcription factor Crz1 and other targets. G, Western blot comparing the level of Orm2 induction in cnb1Δ and ire1Δ and double cnb1Δ ire1Δ (ACX268-4A) cells.
Although UPR contributes to increased Orm2 in response to ER stress, another signaling pathway plays an important role. As shown in Fig. 1A, Orm2 increase in response to tunicamycin and DTT requires Cch1, a plasma membrane calcium transporter. Increased Orm2 induced by tunicamycin also requires the calcium-dependent signaling molecule, calcineurin; in cnb1Δ cells, lacking the regulatory subunit of calcineurin, Orm2 response to tunicamycin is substantially diminished (Fig. 1A). Cycloheximide to inhibit protein synthesis prevents tunicamycin-stimulated Orm2 increase, suggesting that decreased Orm2 protein turnover cannot account for the net gain in Orm2 (not shown). Because increased Orm2 also depends on the calcineurin-activated transcription factor, Crz1 (Fig. 1C), semiquantitative PCR was used to assess whether ORM2 is transcriptionally activated by tunicamycin treatment. Fig. 1D shows that ORM2 mRNA transcripts, normalized to ACT1 transcripts (encoding actin), are increased in cells treated with tunicamycin; by contrast, in cnb1Δ cells, ORM2 transcript increase is abrogated. A summary of quantitated Orm2 protein levels is shown in Fig. 1E. In wild-type cells, Orm2 protein level and mRNA transcripts are increased to similar extents by tunicamycin. These results suggest that Orm2 is increased by transcriptional activation, signaled by calcineurin. Our results are consistent with identification of ORM2 as a target of calcineurin-mediated signaling in a large scale DNA microarray study (19).
As diagrammed in Fig. 1F, ER stress leads to activation of the calcium channel Cch1 and calcineurin; in response to stress, many targets of calcineurin participate in membrane remodeling, including lipid components of membranes (20, 21). The calcineurin-mediated response of Orm2 to ER stress was also observed pharmacologically; the calcineurin inhibitor FK506 abolished tunicamycin-induced Orm2 increase (see Fig. 4B). The response to ER stress was completely abrogated in ire1Δ cnb1Δ double mutants (Fig. 1G), suggesting that the Orm2 protein level is increased by calcium-dependent signaling augmented by Ire1-dependent signaling.
FIGURE 4.
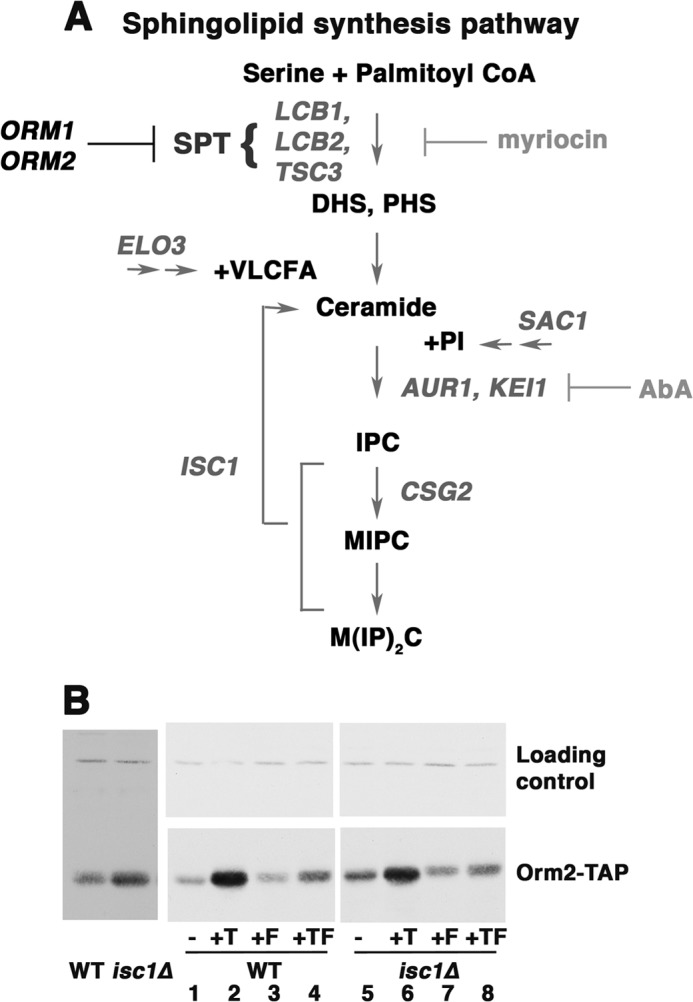
Orm2 protein level is increased in response to accumulation of complex sphingolipids in isc1Δ cells. A, diagram of sphingolipid synthesis pathway. B, Western blot showing Orm2 protein level in wild-type and isc1Δ cells. Cells were grown in SC medium with or without tunicamycin (T; 1 μg/ml for 2 h), FK506 (F; 2 μg/ml for 2 h), or tunicamycin plus FK506 (TF). Orm2 protein level was constitutively increased in isc1Δ cells in comparison with wild-type cells (left and right, compare lanes 1 and 5). The Orm2 protein level in isc1Δ cells was decreased by 2 h after FK506 addition (right, compare lanes 5 and 7).
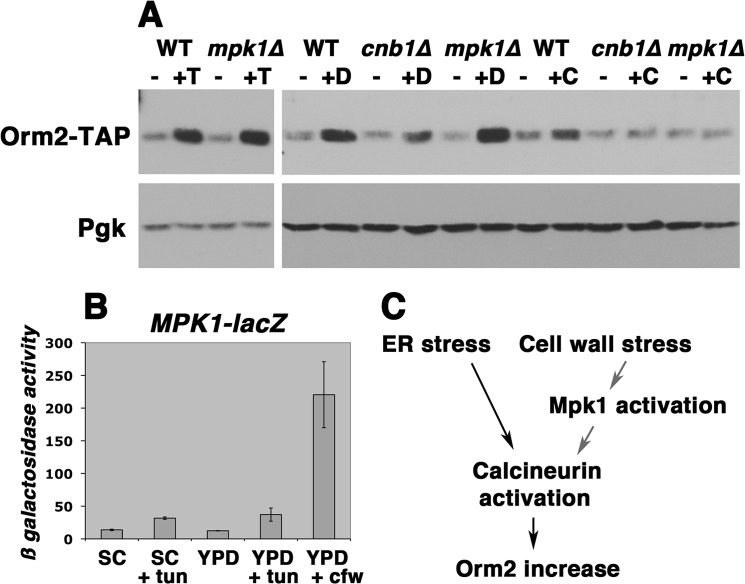
In addition to activation by protein misfolding, UPR is also induced by cell wall stress (22). To determine whether Orm2 is increased in response to cell wall stress, calcofluor white, a chitin-binding reagent, was added to cells. The Orm2 protein level was slightly increased by calcofluor white (Fig. 2A), which strongly activated cell wall stress signaling, as confirmed by induction of MPK1-lacZ, a cell wall stress reporter (Fig. 2B). By contrast, cell wall stress signaling was not significantly increased by tunicamycin, and tunicamycin-stimulated Orm2 increase occurred normally even when the cell wall integrity signaling pathway is impaired in mpk1Δ cells (23) (Fig. 2A). The Orm2 increase elicited by both tunicamycin and calcofluor white was dependent on calcineurin (Fig. 2A). As shown in the model in Fig. 2C, calcineurin mediates Orm2 increase in response to both ER stress and cell wall stress signaling pathways.
FIGURE 2.
Orm2 protein level is slightly increased by calcofluor white-induced cell wall stress. Cells were grown overnight to mid-log phase in SC-uracil medium at 30 °C and then shifted to YPD for 6 h before treatment with tunicamycin (1 μg/ml), DTT (1 mm), or calcofluor white (40 μg/ml) for an additional 2 h. Lysates were assayed for both Orm2-TAP levels and activation of cell wall stress signaling. A, top, Western blot for Orm2-TAP in cells treated with or without tunicamycin (T), DTT (D), or calcofluor white (C). Bottom, phosphoglycerate kinase (Pgk) blotted as a loading control. B, β-galactosidase activity (μmol/min/mg) of an MPK1-lacZ reporter to quantitate cell wall stress signaling. Cell wall stress was maximally induced by calcofluor white. C, diagram showing that ER stress-induced Orm2 increase is dependent on calcineurin activation, whereas cell wall stress-induced Orm2 increase is signaled through Mpk1 to calcineurin activation.
Orm2 Protein Responds to Inositol Levels
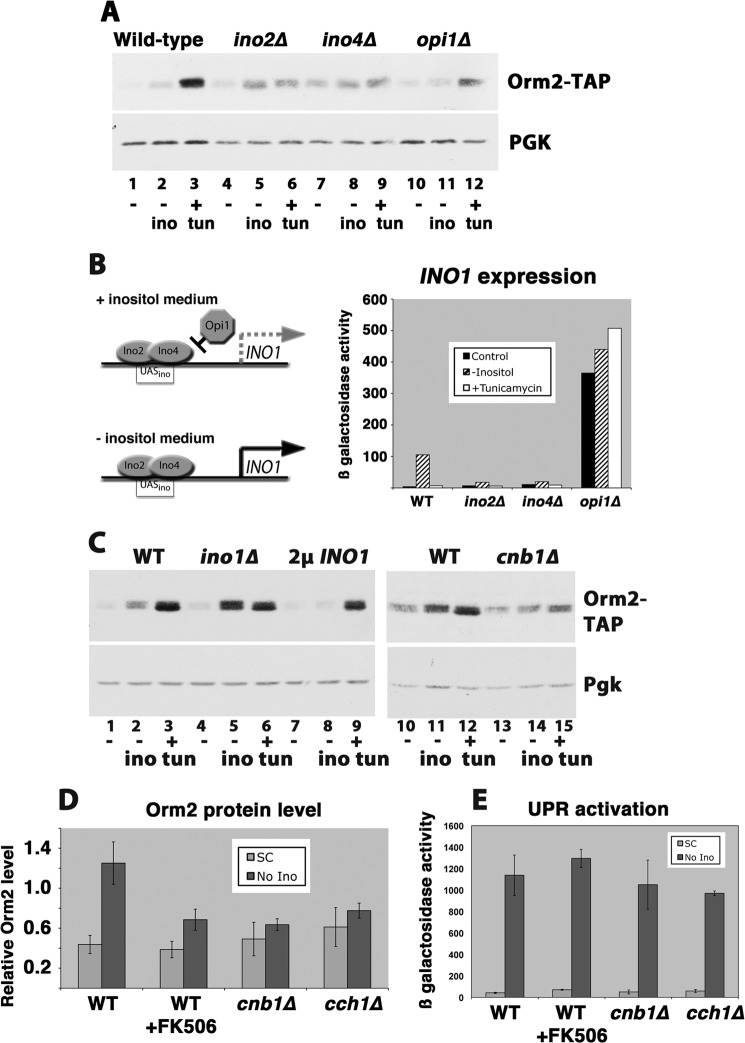
Because UPR is induced by inositol deprivation (24) and inositol is a component of the phosphatidylinositol headgroup of complex sphingolipids (25), the Orm2 protein level was examined in response to inositol removal from the medium. As shown in Fig. 3A, the Orm2 protein level was increased upon inositol removal. In wild-type cells, the removal of inositol from the medium initiated de novo synthesis of inositol by transcription of INO1, encoding the rate-limiting enzyme in inositol biosynthesis, as confirmed by INO1-lacZ measurement (Fig. 3B). In inositol-free medium, derepression of genes encoding phospholipid synthesis enzymes occurs as the repressor Opi1 dissociates from the transcriptional activators Ino2 and Ino4 (26). The Orm2 protein level was not induced in cells in which intracellular inositol did not decrease upon loss of extracellular inositol; when INO1 is constitutively transcribed (derepressed) in opi1Δ cells (Fig. 3A) or in cells overexpressing INO1 (Fig. 3C), the Orm2 protein level remained constant in inositol-free medium. By contrast, in ino2Δ and ino4Δ cells, when derepression of inositol synthesis could not occur, Orm2 was increased constitutively, and when the cells were shifted to inositol-free medium, Orm2 was increased to a greater extent (Fig. 3A). Similarly, in ino1Δ cells, Orm2 protein was increased in inositol-free medium to a greater extent than that induced in wild-type cells (Fig. 3C). It appears that Orm2 protein level is controlled by a signaling pathway that senses decreased intracellular inositol in the absence of extracellular inositol. Orm2 increased ∼3-fold in response to inositol-free medium (in comparison with 7–11-fold in response to tunicamycin (Fig. 1E)).
FIGURE 3.
Induction of Orm2 protein in response to inositol removal from the medium. A, Western blot measurement of Orm2-TAP levels in cells incubated in SC medium, inositol-free medium for 3 h, or SC medium plus tunicamycin (tun; 1 μg/ml) for 2 h at 30 °C. Increased Orm2 in response to inositol-free medium was prevented by opi1Δ; Orm2 was constitutively increased in ino2Δ and ino4Δ cells. The nitrocellulose filter was reblotted with anti-phosphoglycerate kinase (PGK) as a loading control. B, left, schematic of INO1 transcriptional regulation. Right, derepression of INO1 in cells incubated in inositol-free medium as measured by an INO1-lacZ reporter. β-Galactosidase activity in cell lysate is expressed as μmol/min/mg. C, Western blot showing response of wild-type and cnb1Δ cells to incubation in inositol-free medium for 3 h. D, quantitation of Orm2 response to inositol-free medium of wild-type cells, wild-type cells treated with FK506 (2 μg/ml for 3 h), cnb1Δ cells, and cch1Δ cells. Error bars, S.E. from n = 3 determinations. E, UPR activation in inositol-free medium. Wild-type, cnb1Δ, and cch1Δ cells were shifted to inositol-free medium for 3 h, and UPR was assayed by a UPRE-lacZ reporter; β-galactosidase activity in cell lysate is expressed as μmol/min/mg.
Because Orm2 response to tunicamycin is mediated by a calcium- and calcineurin-dependent pathway, we tested whether the same pathway mediates response to inositol-free medium. As shown in Fig. 3C (compare lanes 13 and 14), Orm2 protein increase in the absence of environmental inositol was dependent on calcineurin because the response was abrogated in cnb1Δ cells. Consistently, the response to inositol-free medium was reduced by the calcineurin inhibitor FK506 as well as in the absence of the calcium transporter Cch1 (Fig. 3D).
UPR is activated upon loss of inositol from the medium even as INO1 is derepressed (26, 27) (Fig. 3E). Orm2 increase induced in inositol-free medium was not associated with UPR activation; FK506 or loss of Cnb1 or Cch1 inhibited Orm2 response, but calcium- and calcineurin-dependent signaling was not required for increased expression of a UPRE-lacZ construct by inositol depletion (Fig. 3E).
Orm2 Protein Level Responds to Complex Sphingolipids
To determine whether transcriptional regulation of ORM2 also acts as a feedback mechanism to control sphingolipid synthesis, mutants defective in different steps of the sphingolipid synthesis pathway were examined. Fig. 4A summarizes the key reactions in the sphingolipid synthesis pathway; SPT activity produces the long chain (sphingoid) bases dihydrosphingosine and phytosphingosine (PHS). Subsequent attachment of a very long chain fatty acid to a long chain base produces ceramide. Upon transport of ceramide from ER to Golgi, an inositol headgroup is attached, and the sphingolipid undergoes glycosylation. By contrast with mammalian cells, in which there are hundreds of complex sphingolipids differing in headgroup and/or type and number of sugar groups, there are three complex sphingolipids in yeast, each with an inositol headgroup: inositol phosphorylceramide (IPC), mannose inositol phosphorylceramide, and mannose diinositol phosphorylceramide (25).
In isc1Δ cells, defective in sphingolipid phospholipase C, complex sphingolipids are accumulated (28) (Fig. 4A). Coincident with accumulated complex sphingolipids in isc1Δ cells, there was a slight increase in Orm2 protein level (Fig. 4B). Calcineurin mediates Orm2 increase signaled by accumulated sphingolipids because the addition of FK506 reduced increased Orm2 in isc1Δ cells (Fig. 4B, compare lanes 1 and 7). As expected, FK506 also reduced Orm2 response to tunicamycin (Fig. 4B, compare lanes 2 and 4). Because sphingolipid synthesis is repressed when Orm2 protein is increased (6), it appears that increased Orm2 abundance is a feedback mechanism to down-regulate sphingolipid synthesis in isc1Δ cells. No other significant change in Orm2 protein level was observed in other sphingolipid synthesis mutants (not shown).
Orm Phosphorylation Responds to Flux through the Sphingolipid Synthesis Pathway
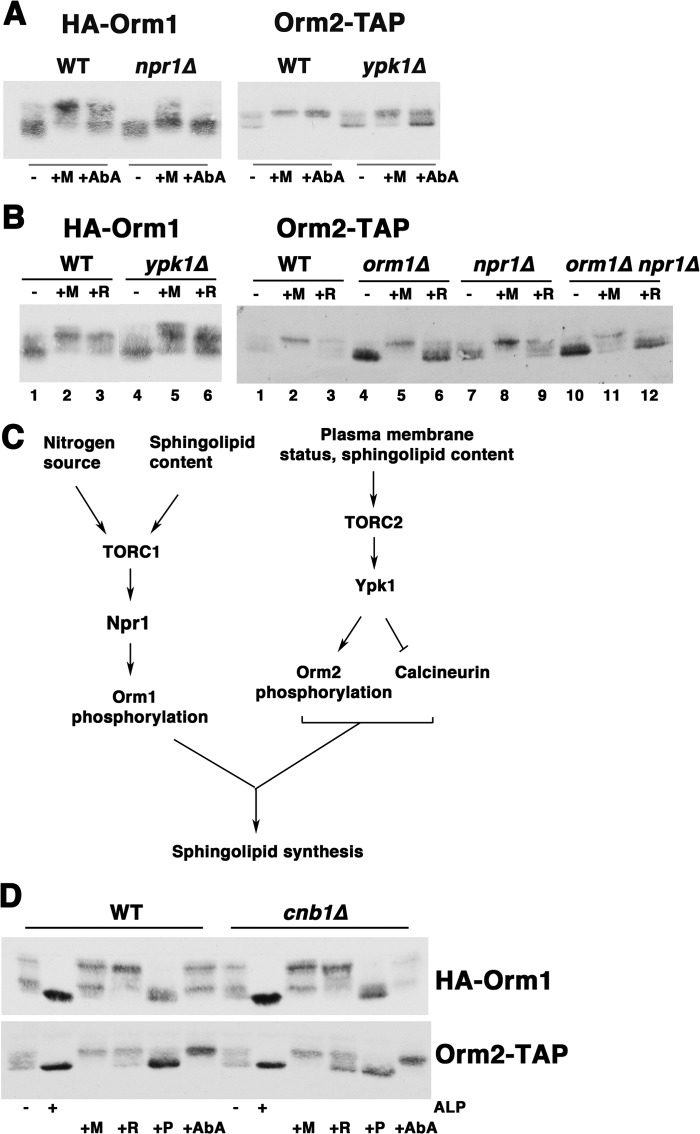
In addition to changes in Orm2 protein level, sphingolipid synthesis is regulated by phosphorylation of both Orm proteins in response to environmental conditions (6–8). Orm phosphorylation has also been suggested to respond to downstream sphingolipid biosynthetic intermediates (4). To identify the sphingolipid intermediate(s) that signal Orm response, combined genetic and chemical approaches were taken to study Orm phosphorylation when the sphingolipid synthesis pathway is perturbed. The extent of phosphorylation of HA-tagged Orm1 was assayed by changes in electrophoretic mobility, as described previously (6). Similarly, electrophoretic mobility changes of Orm2-TAP (tagged at the chromosomal locus) reflect changes in Orm2 phosphorylation. Reduced mobility of Orm2-TAP and HA-Orm1 reflects hyperphosphorylation because alkaline phosphatase treatment collapsed slower migrating bands to a single fast migrating band (Figs. 5A and 7D).
FIGURE 5.
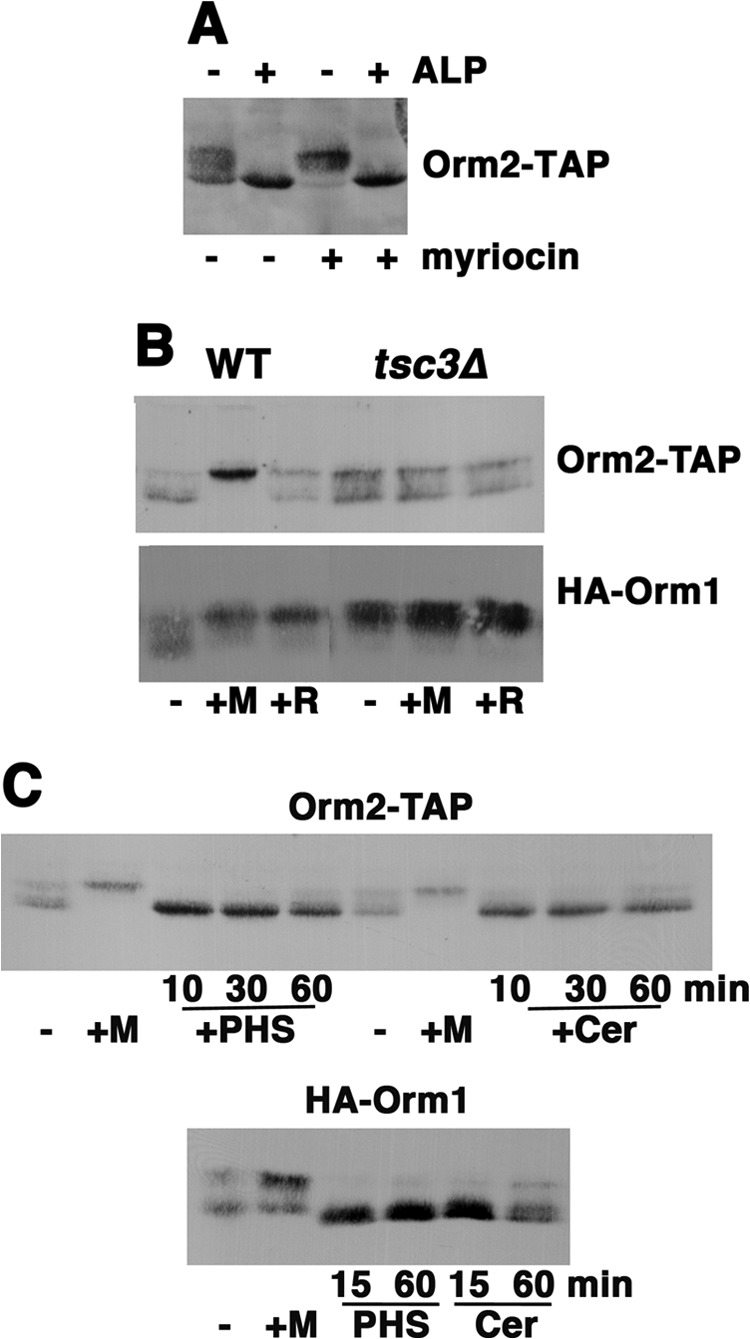
Phosphorylation of the Orm proteins responds to early intermediates in the sphingolipid synthesis pathway. A, Western blot of Orm2-TAP; electrophoretic mobility reflects Orm2 phosphorylation. Cells were treated with or without myriocin (0.15 μg/ml) for 1 h. Lysate was prepared in the presence of TCA and treated with or without alkaline phosphatase (ALP) for 1 h at 37 °C, as described previously (6). The fastest migrating band appears unphosphorylated because its mobility remained the same after alkaline phosphatase treatment. B, Western blot showing electrophoretic mobilities of HA-Orm1 and Orm2-TAP from cells with or without 0.15 μg/ml myriocin (M) or 20 μm rapamycin (R) for 1 h at 30 °C. In tsc3Δ cells, phosphorylation of both Orm proteins was constitutively increased. C, HA-Orm1 and Orm2-TAP proteins were dephosphorylated in response to the addition of myriocin (1 h) or PHS (20 μm) or C2-ceramide (Cer; 10 μm) for various times.
FIGURE 7.
Regulation of Orm protein phosphorylation of the Orm protein by TORC1 and TORC2 signaling pathways. A, electrophoretic mobility assay of HA-Orm1 and Orm2-TAP phosphorylation. Cells were treated with or without myriocin (0.15 μg/ml) or AbA (2 μg/ml) for 1 h. Left, orm1Δ (HXX1-7B) and orm1Δ npr1Δ (ACX251-10B) bearing pSH14HA. Right, wild-type (HXX1-7C) and ypk1Δ (ACX233-1C) cells with chromosomal ORM2 tagged with a TAP tag. B, Western blot of HA-Orm1 and Orm2-TAP phosphorylation in response to rapamycin treatment. Cells were treated with myriocin (+M; 0.15 ng/ml) or rapamycin (+R; 20 μm) for 1 h. Left, wild-type (HXX1-7C) and ypk1Δ cells bearing pSH14HA. Right, strains with chromosomal ORM2 tagged with TAP: wild type (ACX251-9B), orm1Δ (ACX190-3A), npr1Δ (251-6D), and orm1Δ npr1Δ (ACX251-9A). C, schematic diagram showing signaling pathways involved in regulating sphingolipid synthesis. TORC1 plays a major role in regulating Orm1 phosphorylation via Npr1. Regulation of Orm2 activity by phosphorylation is primarily under the control of TORC2 via Ypk1. Calcineurin signaling regulates Orm2 protein levels; increased Orm2 represses SPT activity (6). D, Western blot of Orm phosphorylation in wild-type and cnb1Δ cells. Cells were incubated with 0.15 μg/ml myriocin (+M), 20 μm rapamycin (+R), 20 μm phytosphingosine (+P), or 2 μg/ml aureobasidin A (+AbA) for 1 h at 30 °C. Lysates were treated with or without alkaline phosphatase (ALP) for 1 h at 37 °C as indicated.
In cells deleted of TSC3, encoding the small stimulatory subunit of SPT, there is a severe reduction in SPT activity (29). In tsc3Δ cells, mobility shifts indicate increased phosphorylation of both HA-Orm1 and Orm2-TAP (Fig. 5B). Constitutive phosphorylation of Orm1 appeared maximal in tsc3Δ cells because its mobility was similar to that stimulated by the addition of myriocin for 1 h. These results support a model in which Orm1 phosphorylation is a compensatory response to SPT activity acutely inhibited by myriocin or chronically depressed in tsc3Δ cells. Phosphorylation of Orm2 was also constitutively increased in tsc3Δ cells; however, Orm2 phosphorylation was not further increased to the maximal extent by myriocin. One possible explanation for an apparent diminished response to myriocin is that Orm2 phosphorylation may respond to different extents to multiple downstream sphingolipid intermediates (discussed further below).
A time course of Orm phosphorylation upon the acute addition of downstream SPT products, long chain base (PHS) and ceramide (a C-2 ceramide analog), is shown in Fig. 5C. Both Orm1 and Orm2 underwent rapid hypophosphorylation, suggesting a direct effect of the downstream intermediates on the Orm-SPT complex. To examine whether Orm phosphorylation is also adjusted in response to the level of complex sphingolipids, aureobasidin A (AbA) was added to inhibit attachment of the inositol headgroup to ceramide to form IPC (Fig. 4A) (30). Strikingly, AbA induced increased phosphorylation of both Orm1 and Orm2 (Fig. 6A); these results suggest that decreased IPC levels also feed back to the Orm proteins to increase SPT activity. Feedback control of SPT by IPC was studied further in cells bearing a temperature-sensitive allele of KEI1, encoding a subunit of IPC synthase (11). The kei1-1 mutant was analyzed at the semipermissive temperature (30 °C) to avoid effects on sphingolipid synthesis that occur at 37 °C (31). As shown in Fig. 6B, phosphorylation of Orm1 and Orm2 was constitutively increased in kei1-1 cells, supporting the idea that both Orm proteins respond to IPC levels.
FIGURE 6.
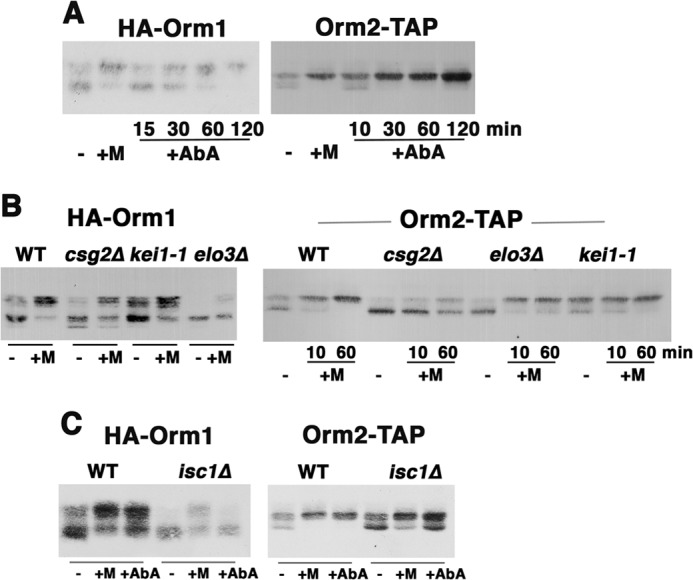
Phosphorylation of the Orm proteins in response to sphingolipid metabolites in the distal sphingolipid synthesis pathway. A, Western blot showing electrophoretic mobility changes of HA-Orm1 and Orm2-TAP in exponentially growing wild-type control cells in comparison with cells incubated with myriocin (0.15 μg/ml) for 1 h or for the indicated times with AbA (2 μg/ml). B, electrophoretic mobility assay of Orm proteins in wild-type, elo3Δ, csg2Δ, and kei1-1 cells with or without myriocin for 10 min and/or 1 h. C, electrophoretic mobility assay of Orm proteins in wild-type and isc1Δ cells. Cells were treated with or without myriocin or AbA for 1 h.
In the sphingolipid synthesis pathway, Elo3 catalyzes the last of a series of reactions to synthesize very long chain fatty acids required for the formation of ceramide, causing a backlog in the pathway at ceramide synthesis (Fig. 4A) (25, 32). In elo3Δ cells, high levels of PHS are accumulated (32). Fig. 6B shows that in the absence of Elo3, the Orm proteins were dephosphorylated. The response to myriocin was impaired (Fig. 6B) because constitutively elevated PHS in elo3Δ cells may interfere with the ability of myriocin to signal increased Orm phosphorylation. A similar phenotype was seen in csg2Δ cells lacking a calcium-binding protein required in catalyzing mannosylation of IPC; IPC is accumulated in csg2Δ cells (25, 33). In csg2Δ cells, both Orm proteins were constitutively dephosphorylated in comparison with wild-type cells (Fig. 6B). The response to myriocin by hyperphosphorylation was impaired in csg2Δ cells so that neither Orm protein can achieve maximal phosphorylation (Fig. 6B). It seems possible that the effect of myriocin to decrease downstream products is quelled by elevated levels of IPC in csg2Δ cells (34).
Further evidence for Orm response to IPC is shown in Fig. 6C. Without complex sphingolipase activity in isc1Δ cells (28), Orm1 was constitutively dephosphorylated, and to a lesser extent, Orm2 was also constitutively dephosphorylated. These responses to accumulated complex sphingolipids are consistent with feedback repression of sphingolipid synthesis (Fig. 6C). In isc1Δ cells, hyperphosphorylation in response to AbA or myriocin addition for 1 h was impaired to a greater and lesser extent for Orm1 and Orm2, respectively (Fig. 6C); the effect of the inhibitors to decrease IPC production was probably blunted by built-up complex sphingolipids. Together, these data suggest that feedback regulation of SPT activity is mediated via Orm phosphorylation by multiple downstream products in the sphingolipid synthesis pathway, including long chain base, ceramide, and the complex sphingolipid IPC.
TOR Signaling Regulates Orm Phosphorylation
Phosphorylation of Orm1 and Orm2 is regulated by the TOR signaling pathway via Npr1 and Ypk1, respectively, representing TORC1 and TORC2 branches (6, 8, 35). Hyperphosphorylation of Orm1 in response to inhibition of sphingolipid synthesis by myriocin or AbA treatment was abrogated in npr1Δ cells (Fig. 7A, left). Orm2 hyperphosphorylation in response to myriocin or AbA was diminished in ypk1Δ cells (Fig. 7A, right). Thus, feedback response of Orm phosphorylation to flux through the sphingolipid synthesis pathway occurs via TOR pathway kinases.
In addition to feedback regulation, Orm1 phosphorylation is also regulated by Npr1 in response to nutritional status (6, 9). Orm1 phosphorylation is increased upon rapamycin treatment to inhibit TORC1, mimicking amino acid starvation (6, 9). Although Orm1 phosphorylation responded maximally to rapamycin, Orm2 response was reduced (Fig. 7B, left and right, compare lanes 3). Orm2 response to rapamycin was minimal in comparison with its feedback response (as assayed by its response to myriocin treatment) (Fig. 7B, right, compare lanes 2 and 3). Indeed, Orm2 response to rapamycin was further diminished in orm1Δ cells by dephosphorylation (Fig. 7B, right, compare lane 6 with lanes 3 and 9), suggesting the possibility that Orm2 response to rapamycin occurs indirectly via Orm1.
Fig. 7C presents a model in which Orm1 is a major focus of TORC1 regulation via Npr1. Orm2 phosphorylation remained unaffected in npr1Δ cells (Fig. 7, compare A (left) with B (right)). Orm2 is a major focus of TORC2 via Ypk1, and Orm1 phosphorylation was minimally affected in ypk1Δ cells (Fig. 7, compare A (right) with B (left)).
Because TOR and calcineurin have a mutually antagonistic relationship (36), we tested whether Orm phosphorylation is regulated by calcineurin's phosphatase activity. As shown in Fig. 7D, phosphorylation of HA-Orm1 and Orm2-TAP were examined in cnb1Δ cells. Phosphorylation of both Orm proteins appeared identical in wild-type and cnb1Δ cells; both Orm proteins displayed increased phosphorylation in response to myriocin and AbA addition to inhibit SPT and IPC synthase, respectively. In wild-type and cnb1Δ cells, both Orm proteins were dephosphorylated upon the addition of PHS (Fig. 7D). Thus, calcineurin does not play a significant role in regulating Orm phosphorylation, although it does modulate Orm2 protein level (Fig. 1).
UPR Activation in Response to Lipid Dysregulation Is Dependent on Calcium and Calcineurin
In the absence of regulation via the Orm proteins, perturbation of lipid homeostasis causes ER stress, and orm1Δ orm2Δ cells have constitutive activation of UPR and impaired growth in the presence of tunicamycin (3) (Fig. 8). Constitutive UPR was prevented in ire1Δ orm1Δ orm2Δ cells because Ire1 is required for UPR activation, as measured by a UPRE-lacZ reporter (Fig. 8) (3). Because calcineurin regulates Orm2 abundance and lipid synthesis (20), a role for calcineurin was tested in UPR activation in orm1Δ orm2Δ cells. Fig. 8 shows that constitutive UPR requires both calcium and calcineurin because its induction was prevented in cch1Δ orm1Δorm2Δ and cnb1Δ orm1Δ orm2Δ cells. Moreover, calcium- and calcineurin-mediated signaling to activate the Crz1 transcription factor is required for constitutive UPR because it was abolished in crz1 orm1Δ orm2Δ cells (Fig. 8). UPR was also constitutively activated when complex sphingolipids were accumulated in isc1Δ cells as well as when sphingolipid synthesis was constitutively decreased in tsc3Δ cells (Fig. 8). Strikingly, calcium and calcineurin were required to mediate UPR signaling in isc1Δ and tsc3Δ cells as well (Fig. 8), consistent with a role for this signaling pathway to maintain sphingolipid homeostasis. UPR induction by tunicamycin treatment (which increases both protein misfolding and calcineurin signaling) was not significantly impaired in any of the tested mutants except for ire1Δ cells (Fig. 8). Thus, calcium- and calcineurin-mediated signaling appear to elicit the UPR specifically in response to sphingolipid dysregulation.
FIGURE 8.
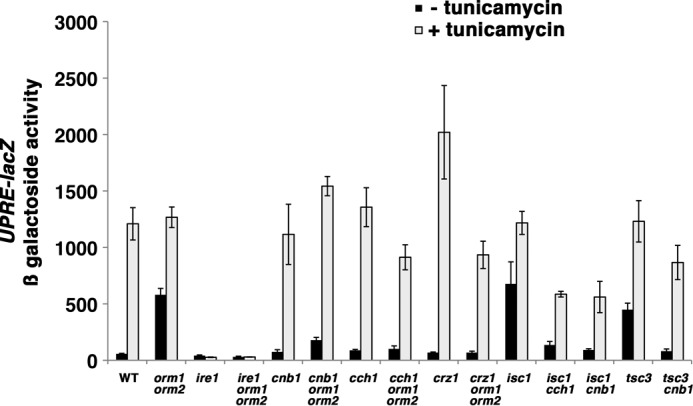
ER stress in orm1Δ orm2Δ cells is signaled via calcium and calcineurin. Constitutive UPR activation in orm1Δ orm2Δ cells is dependent on calcium- and calcineurin-dependent signaling. Exponentially growing cells bearing a UPRE-lacZ reporter (pJC104) (52) were grown in the presence or absence of tunicamycin (1 μg/ml) for 2 h. β-Galactosidase activity was measured in cell lysate, as described (6). Error bars, S.E.
DISCUSSION
Regulation of sphingolipid synthesis is mediated by phosphorylation of Orm1 and Orm2 (6, 8) as well as by up-regulation of Orm2 protein levels (6) (Fig. 1). A major finding of this paper is that ER stress signaling to increase Orm2 abundance occurs via a calcium- and calcineurin-dependent pathway. Although UPR activation is the major response to ER stress, activated Hac1i in the absence of tunicamycin or DTT stressor induces only a small increase in Orm2; UPR makes only a partial contribution to inducing Orm2. Previous work has shown that ER stress triggers calcium influx through the Cch1-Mid1 calcium channel and calcineurin activation; this calcium-calcineurin signaling pathway acts coordinately with the UPR and ensures cell survival during ER stress (21, 37–39). In addition to increasing Orm2 to repress SPT, calcium/calcineurin-mediated signaling regulates late steps in sphingolipid synthesis: the mannosylation of IPC to mannose inositol phosphorylceramide via Csg1/Sur1 and Csg2 (19) and complex sphingolipid breakdown via Isc1 activity (40). Together, sphingolipids and calcineurin regulate Slm1 and Slm2 proteins, which are proposed to act as integrators of different inputs in the TORC2 protein kinase network (40–42).
Another environmental stimulus that induces increased Orm2 is loss of inositol from the medium (Fig. 3). Induction of Orm2 protein by inositol-free medium is independent of INO1 derepression and inositol biosynthesis because Orm2 is increased in ino1Δ, ino2Δ, and ino4Δ cells. Instead, increased Orm2 in response to loss of medium inositol is triggered by calcineurin-mediated signaling (Fig. 3C); our results are in agreement with a previous report showing activation of calcineurin signaling in response to inositol starvation (43). Several genes have been identified that are under negative control by inositol (44) and the transcription factors Ino2 and Ino4 (45); like these, ORM2 appears transcriptionally activated in ino2Δ or ino4Δ cells (Fig. 3A). Signaling via calcineurin affects plasma membrane remodeling in response to a variety of stress conditions (20). Control of sphingolipid synthesis upon depletion of intracellular inositol and perturbation of phospholipid homeostasis underscores the link to the lipid component of calcineurin-mediated membrane restructuring.
The Orm proteins participate in a feedback mechanism to modulate SPT activity in response to flux through the sphingolipid synthesis pathway (4). We now show that multiple intermediates in the sphingolipid synthesis pathway serve as signals to regulate Orm phosphorylation, including long chain base, ceramide, and complex sphingolipid. Our results show that both Orm proteins become hypophosphorylated upon the addition of the long chain base PHS (in agreement with a previous report (8)) as well as a C2-ceramide (Fig. 5). Hyperphosphorylation of the Orm proteins is triggered by the addition of myriocin to produce an acute drop in long chain base and ceramide levels. Complex sphingolipid is also a feedback signal to Orm phosphorylation because the addition of AbA to decrease IPC acutely elicits hyperphosphorylation of both Orm proteins (Fig. 6). Conversely, accumulation of IPC and other complex sphingolipids in isc1Δ cells generates constitutively dephosphorylated Orm phosphorylation (Fig. 6).
In several mutants impaired in different steps in the sphingolipid synthesis pathway, there is a blunted response of Orm phosphorylation to myriocin and AbA. These results are consistent with contradictory signals; for instance, in csg2Δ cells, constitutively high IPC levels conflict with an AbA-mediated IPC decrease, resulting in impaired response to AbA (Fig. 6). Strikingly, in addition to regulating phosphorylation, IPC accumulation modulates Orm2 protein level (Fig. 4). Thus, build-up of the end product down-regulates the first committed step of the pathway via two different regulatory mechanisms to decrease Orm phosphorylation and increase Orm2 protein.
Feedback regulation of Orm phosphorylation is mediated by both branches of the TOR signaling pathway (Fig. 7). A recent paper reported that distinct sites on Orm1 are phosphorylated in response to sphingolipid status (after myriocin or AbA treatment) or nutritional status (after rapamycin treatment) (9). Previously, we showed that the Npr1 protein kinase, downstream of TORC1, regulates Orm1 phosphorylation (6). Abrogation of HA-Orm1 phosphorylation in npr1Δ cells supports the idea that Npr1 mediates, in large part, response to both nutritional status (6) and feedback response (Fig. 7A) (6). By contrast with its effect on Orm1, Npr1 appears to play a less significant role in Orm2 regulation; in npr1Δ cells, feedback phosphorylation of Orm2-TAP is essentially unaffected (Fig. 7, A and B). Orm2 appears largely regulated by TORC2. There is a slight Orm2 response to the TORC1 inhibitor rapamycin (Fig. 7B), but feedback phosphorylation of Orm2-TAP is substantially diminished in the absence of the AGC family kinase, Ypk1, acting in the TORC2 pathway (Fig. 7A) (7, 8). Conversely, feedback Orm1 phosphorylation and Orm1 response to rapamycin is not detectably affected in ypk1Δ cells (Fig. 7B). Thus, Orm1 and Orm2 are overlapping but distinct targets of TORC1 and TORC2, respectively. A model is shown in Fig. 7C depicting Orm regulation via branches of the TOR signaling network.
In the absence of the Orm proteins, dysregulation of SPT and sphingolipid homeostasis results in pleiotropic phenotypes (3), including constitutive activation of the UPR (Fig. 8) (3). UPR activation in ormΔ cells requires calcium influx via Cch1 and calcineurin. By contrast, UPR activation by tunicamycin is calcineurin-independent, suggesting that calcineurin-mediated signaling in orm1Δ orm2Δ cells is not activated by protein misfolding per se. Instead, UPR induction through calcium and calcineurin signaling appears to reflect a reaction to lipid perturbation, and also requires Ire1. Our result is consistent with distinct domains of Ire1 responding to different ER stresses (46). More importantly, our results support accruing evidence that ER homeostasis is maintained by cross-talk between UPR and other signaling pathways (47, 48).
Calcium and calcineurin are at the hub of a signaling network that maintains sphingolipid homeostasis. TORC2 has been reported to regulate ceramide synthesis in a calcineurin-dependent manner (49). Our observations here suggest that TORC2, as a negative regulator of calcineurin (50, 51), probably regulates sphingolipid synthesis by calcineurin-mediated modulation of Orm2 levels (Fig. 1). Strikingly, calcineurin does not appear to regulate Orm phosphorylation (Fig. 7D). Calcineurin-mediated regulation of sphingolipid homeostasis contributes to our understanding of how the plasma membrane landscape is sculpted in response to cellular needs and environmental signals. The importance of sphingolipids in membrane biogenesis and cell growth is underscored by regulation by both branches of TOR signaling.
This work was supported by a Margaret and Herman Sokol Faculty Award and an Elizabeth Crosby Faculty grant from the University of Michigan.
- ER
- endoplasmic reticulum
- SPT
- serine palmitoyltransferase
- UPR
- unfolded protein response
- PHS
- phytosphingosine
- AbA
- aureobasidin A
- TAP
- tandem affinity purification.
REFERENCES
- 1. Lingwood D., Simons K. (2010) Lipid rafts as a membrane-organizing principle. Science 327, 46–50 [DOI] [PubMed] [Google Scholar]
- 2. Souza C. M., Pichler H. (2007) Lipid requirements for endocytosis in yeast. Biochim. Biophys. Acta 1771, 442–454 [DOI] [PubMed] [Google Scholar]
- 3. Han S., Lone M. A., Schneiter R., Chang A. (2010) Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. 107, 5851–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Breslow D. K., Collins S. R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C. S., Weissman J. S. (2010) Orm family proteins mediate sphingolipid homeostasis. Nature 463, 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moffatt M. F., Kabesch M., Liang L., Dixon A. L., Strachan D., Heath S., Depner M., von Berg A., Bufe A., Rietschel E., Heinzmann A., Simma B., Frischer T., Willis-Owen S. A., Wong K. C., Illig T., Vogelberg C., Weiland S. K., von Mutius E., Abecasis G. R., Farrall M., Gut I. G., Lathrop G. M., Cookson W. O. (2007) Genetic variants regulating ORMDL expression contribute to the risk of childhood asthma. Nature 448, 470–473 [DOI] [PubMed] [Google Scholar]
- 6. Liu M., Huang C. J., Polu S. R., Schneiter R., Chang A. (2012) Regulation of sphingolipid synthesis via Orm1 and Orm2 in yeast. J. Cell Sci. 125, 2428–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roelants F. M., Breslow D. K., Muir A., Weissman J. S., Thorner J. (2011) Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 108, 19222–19227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sun Y., Miao Y., Yamane Y., Zhang C., Shokat K. M., Takematsu H., Kozutsumi Y., Drubin D. G. (2012) Orm protein phosphoregulation mediates transient sphingolipid biosynthesis response to heat stress via the Pkh-Ypk and Cdc55-PP2A pathways. Mol. Biol. Cell 23, 2388–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shimobayashi M., Oppliger W., Moes S., Jenö P., Hall M. N. (2013) TORC1-regulated protein kinase Npr1 phosphorylates Orm to stimulate complex sphingolipid synthesis. Mol. Biol. Cell 24, 870–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sherman F., Hicks J. B., Fink G. R. (1986) Methods in Yeast Genetics: A Laboratory Manual, pp. 523–585, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 11. Sato K., Noda Y., Yoda K. (2009) Kei1. A novel subunit of inositolphosphorylceramide synthase, essential for its enzyme activity and Golgi localization. Mol. Biol. Cell 20, 4444–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jung U. S., Sobering A. K., Romeo M. J., Levin D. E. (2002) Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46, 781–789 [DOI] [PubMed] [Google Scholar]
- 13. Cox J. S., Shamu C. E., Walter P. (1993) Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73, 1197–1206 [DOI] [PubMed] [Google Scholar]
- 14. Lopes J. M., Hirsch J. P., Chorgo P. A., Schulze K. L., Henry S. A. (1991) Analysis of sequences in the INO1 promoter that are involved in its regulation by phospholipid precursors. Nucleic Acids Res. 19, 1687–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chang A., Slayman C. W. (1991) Maturation of the yeast plasma membrane [H+]ATPase involves phosphorylation during intracellular transport. J. Cell Biol. 115, 289–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rose M. D., Winston F., Hieter P. (1990) Methods in Yeast Genetics: A Laboratory Manual, p. 198, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 17. Ron D., Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 18. Shamu C. E. (1997) Signal transduction. Splicing together the unfolded-protein response. Curr. Biol. 7, R67–R70 [DOI] [PubMed] [Google Scholar]
- 19. Yoshimoto H., Saltsman K., Gasch A. P., Li H. X., Ogawa N., Botstein D., Brown P. O., Cyert M. S. (2002) Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 277, 31079–31088 [DOI] [PubMed] [Google Scholar]
- 20. Cyert M. S. (2003) Calcineurin signaling in Saccharomyces cerevisiae. How yeast go crazy in response to stress. Biochem. Biophys. Res. Commun. 311, 1143–1150 [DOI] [PubMed] [Google Scholar]
- 21. Bonilla M., Nastase K. K., Cunningham K. W. (2002) Essential role of calcineurin in response to endoplasmic reticulum stress. EMBO J. 21, 2343–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Scrimale T., Didone L., de Mesy Bentley K. L., Krysan D. J. (2009) The unfolded protein response is induced by the cell wall integrity mitogen-activated protein kinase signaling cascade and is required for cell wall integrity in Saccharomyces cerevisiae. Mol. Biol. Cell 20, 164–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Levin D. E. (2005) Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69, 262–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cox J. S., Chapman R. E., Walter P. (1997) The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell 8, 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Funato K., Vallée B., Riezman H. (2002) Biosynthesis and trafficking of sphingolipids in the yeast Saccharomyces cerevisiae. Biochemistry 41, 15105–15114 [DOI] [PubMed] [Google Scholar]
- 26. Henry S. A., Kohlwein S. D., Carman G. M. (2012) Metabolism and regulation of glyerolipids in the yeast Saccharomyces cerevisiae. Genetics 190, 317–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang H. J., Jones E. W., Henry S. A. (2002) Role of the unfolded protein response pathway in regulation of INO1 and in the sec14 bypass mechanism in Saccharomyces cerevisiae. Genetics 162, 29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matmati N., Hannun Y. A. (2008) ISC1 (inositol phosphosphingolipid-phospholipase C), the yeast homologue of neutral sphingomyelinases. J. Lipid Res. 49, 922–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gable K., Slife H., Bacikova D., Monaghan E., Dunn T. M. (2000) Tsc3 is an 80-amino acid protein associated with serine palmitoyltransferase and required for optimal enzyme activity. J. Biol. Chem. 275, 7597–7603 [DOI] [PubMed] [Google Scholar]
- 30. Cerantola V., Guillas I., Roubaty C., Vionnet C., Uldry D., Knudsen J., Conzelmann A. (2009) Aureobasidin A arrests growth of yeast cells through both ceramide intoxication and deprivation of essential inositolphosphorylceramamides. Mol. Microbiol. 71, 1523–1537 [DOI] [PubMed] [Google Scholar]
- 31. Dickson R. C., Nagiec E. E., Skrzypek M., Tillman P., Wells G. B., Lester R. L. (1997) Sphingolipids are potential heat stress signals in Saccharomyces cerevisiae. J. Biol. Chem. 272, 30196–30200 [DOI] [PubMed] [Google Scholar]
- 32. Kobayashi S. D., Nagiec M. M. (2003) Ceramide/long-chain base phosphate rheostat in Saccharomyces cerevisiae. Regulation of ceramide synthesis by Elo3p and Cka2p. Eukaryot. Cell 2, 284–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Uemura S., Kihara A., Iwaki S., Inokuchi J., Igarashi Y. (2007) Regulation of the transport and protein levels of the inositol phosphorylceramide mannosyltransferases Csg1 and Csh1 by the Ca2+-binding protein Csg2. J. Biol. Chem. 282, 8613–8621 [DOI] [PubMed] [Google Scholar]
- 34. Zhao C., Beeler T., Dunn T. (1994) Suppressors of the Ca2+-sensitive yeast mutant (csg2) identify genes involved in sphingolipid biosynthesis. J. Biol. Chem. 269, 21480–21488 [PubMed] [Google Scholar]
- 35. MacGurn J. A., Hsu P.-C., Smolka M. B., Emr S. D. (2011) TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell 147, 1104–1117 [DOI] [PubMed] [Google Scholar]
- 36. Mulet J. M., Martin D. E., Loewith R., Hall M. N. (2006) Mutual antagonism of target of rapamycin and calcineurin signaling. J. Biol. Chem. 281, 33000–33007 [DOI] [PubMed] [Google Scholar]
- 37. Bonilla M., Cunningham K. W. (2003) Mitogen-activated protein kinase stimulation of Ca2+ signaling is required for survival of endoplasmic reticulum stress in yeast. Mol. Biol. Cell 14, 4296–4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hong M. P., Vu K., Bautos J., Gelli A. (2010) Cch1 restores intracelllar Ca2+ in fungal cells during endoplasmic reticulum stress. J. Biol. Chem. 285, 10951–10958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dudgeon D. D., Zhang N., Ositelu O. O., Kim H., Cunningham K. W. (2008) Nonapoptotic death of Saccharomyces cerevisiae cells that is stimulated by Hsp90 and inhibited by calcineurin and Cmk2 in response to endoplasmic reticulum stresses. Eukaryot. Cell 7, 2037–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tabuchi M., Audhya A., Parsons A. B., Boone C., Emr S. D. (2006) The phosphatidyl 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol. Cell. Biol. 26, 5861–5875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Daquinag A., Fadri M., Jung S. Y., Qin J., Kunz J. (2007) The yeast PH domain proteins Slm1 and Slm2 are targets of sphingolipid signaling during the response to heat. Mol. Cell. Biol. 27, 633–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bultynck G., Heath V. L., Majeed A. P., Galan J. M., Haguenauer-Tsapis R., Cyert M. S. (2006) Slm1 and Slm2 are novel substrates of the calciuneurin phosphatase required for heat stress-induced endocytosis of the yeast uracil permease. Mol. Cell. Biol. 26, 4729–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Villa-Garcia M. J., Choi M. S., Hinz F. I., Gaspar M. L., Jesch S. A., Henry S. A. (2011) Genome-wide screen for inositol auxotrophy in Saccharomyces cerevisiae implicates lipid metabolism in stress response signaling. Mol. Genet. Genomics 285, 125–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jesch S. A., Zhao X., Wells M. T., Henry S. A. (2005) Genome-wide analysis reveals inositol, not choline, as the major effector of Ino2p-Ino4p and unfolded protein response target gene expression in yeast. J. Biol. Chem. 280, 9106–9118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Santiago T. C., Mamoun C. B. (2003) Genome expression analysis in yeast reveals novel transcriptional regulation by inositol and choline and new regulatory functions for Opi1p, Ino2p, and Ino4p. J. Biol. Chem. 278, 38723–38730 [DOI] [PubMed] [Google Scholar]
- 46. Promlek T., Ishiwata-Kimata Y., Shido M., Sakuramoto M., Kohno K., Kimata Y. (2011) Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol. Biol. Cell 22, 3520–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rutkowski D. T., Hegde R. S. (2010) Regulation of basal cellular physiology by the homeostatic unfolded protein response. J. Cell Biol. 189, 783–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang S., Kaufman R. J. (2012) The impact of the unfolded protein response on human disease. J. Cell Biol. 197, 857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Aronova S., Wedaman K., Aronov P. A., Fontes K., Ramos K., Hammock B. D., Powers T. (2008) Regulation of ceramide biosynthesis by TOR complex 2. Cell Metabolism 7, 148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Niles B. J., Mogri H., Hill A., Vlahakis A., Powers T. (2012) Plasma membrane recruitment and activation of the AGC kinase Ypk1 is mediated by target of rapamycin complex 2 (TORC2) and its effector proteins Slm1 and Slm2. Proc. Natl. Acad. Sci. 109, 1536–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Berchtold D., Piccolis M., Chiaruttini N., Riezman I., Riezman H., Roux A., Walther T. C., Loewith R. (2012) Plasma membrane stress induces relocalization of Slm proteins and activation of TORC2 to promote sphingolipid synthesis. Nat. Cell Biol. 14, 542–547 [DOI] [PubMed] [Google Scholar]
- 52. Cox J. S., Walter P. (1996) A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87, 391–404 [DOI] [PubMed] [Google Scholar]