Background: Phosphatidic acid is a key lipid second messenger often generated after receptor stimulation.
Results: After P2Y6 receptor stimulation both PLD and DGK enzymes are responsible for producing PA species.
Conclusion: DGK facilitates a negative regulation of PLD and tightly controls PA levels.
Significance: Further understanding the PA signaling network is critical to developing next generation therapeutics for human diseases.
Keywords: Diacylglycerol, Phosphatidic Acid, Phospholipase D, Protein Kinase C (PKC), Purinergic Receptor, DAG Kinase, Lipidomics
Abstract
Phosphatidic acid (PA) is a lipid second messenger located at the intersection of several lipid metabolism and cell signaling events including membrane trafficking, survival, and proliferation. Generation of signaling PA has long been primarily attributed to the activation of phospholipase D (PLD). PLD catalyzes the hydrolysis of phosphatidylcholine into PA. A variety of both receptor-tyrosine kinase and G-protein-coupled receptor stimulations have been shown to lead to PLD activation and PA generation. This study focuses on profiling the PA pool upon P2Y6 receptor signaling manipulation to determine the major PA producing enzymes. Here we show that PLD, although highly active, is not responsible for the majority of stable PA being produced upon UDP stimulation of the P2Y6 receptor and that PA levels are tightly regulated. By following PA flux in the cell we show that PLD is involved in an initial increase in PA upon receptor stimulation; however, when PLD is blocked, the cell compensates by increasing PA production from other sources. We further delineate the P2Y6 signaling pathway showing that phospholipase Cβ3 (PLCβ3), PLCδ1, DGKζ and PLD are all downstream of receptor activation. We also show that DGKζ is a novel negative regulator of PLD activity in this system that occurs through an inhibitory mechanism with PKCα. These results further define the downstream events resulting in PA production in the P2Y6 receptor signaling pathway.
Introduction
G-protein-coupled receptor (GPCR)2 activation initiates signal transduction modulating several essential physiological processes in the cell. The GPCR superfamily is the largest group of cell surface receptors with more than 800 members (1). Dysfunction of GPCRs leads to many prevalent human diseases, and these proteins represent a common therapeutic potential with 50–60% of all drugs targeting these receptors (2). Downstream signaling targets of GPCR activation are also key enzymes for drug development; one of those is phospholipase D (PLD).
P2Y receptors are a diverse group of GPCRs of which there are eight subtypes (3); at least four are activated by uracil nucleotides, UDP or UTP (4). Nucleotide di- and triphosphates are important extracellular signaling components that mediate a plethora of intracellular signaling events, many of them through stimulation of P2Y receptors, and the P2Y6 receptor is the only subtype that is specifically activated by UDP (5). The P2Y6 receptor has a wide tissue distribution being found in lung, heart, spleen, placenta, intestine, brain, and thymus (6).
Previous reports on P2Y receptor activation have suggested that phospholipase D is an important downstream effector (7, 8). Specifically, the P2Y6 receptor has been shown to activate signaling pathways in the cell that turn on phospholipase C (PLC) proteins and increase diacylglycerol (DAG) production and protein kinase C (PKC) activity, which is a known activator of PLD (6). PLD, which is responsible for the conversion of phosphatidylcholine (PC) into the important signaling lipid phosphatidic acid (PA), has long been considered the essential PA producing enzyme. However, signaling PA can be generated by several different enzymes, and the location, molecular species, and timing of that production is tightly regulated (9). In addition to PLD, other biosynthetic pathways for PA production include: de novo synthesis from glycerol 3-phosphate, acylation of lysophosphatidic acid by lysophosphatidic acid acyltransferase, and phosphorylation of DAG by diacylglycerol kinase (DGK). Few studies exist that attempt to assess the compensatory roles played by different PA-producing enzymes under inhibition of key pathways. This report addresses such signaling after modulation of PLD in particular.
Two major isoforms of PLD exist, PLD1 and PLD2, each of which is uniquely regulated (10–15). Each possesses isoform-specific functions due to differential subcellular localization and modes of activation. In light of the importance of PA in cellular signaling events, PLD has become an important target for the development of small molecule inhibitors (16–20). Our laboratories have developed dual PLD1/2 inhibitors (VU0155056), a 1700-fold PLD1-specific inhibitor (VU0359595), and a 75-fold PLD2 preferring inhibitor (VU0364739) to better understand the therapeutic potential of these enzymes. Currently, PLD inhibitors are being evaluated for their potential use in brain disorders such as Alzheimer disease and stroke (21), chronic inflammation (22), and cancer (10, 23, 24).
Herein we explore how the stimulation of the P2Y6 receptor in 1321N1 astrocytoma cells affects PLD activity and PA production. We show that upon UDP stimulation of the receptor, PLD catalytic activity is enhanced, and PA production is elevated. Moreover, in this system PLD is not the major source of steady-state bulk PA under these conditions, and instead DGKζ is a significant contributor to PA production. As a result of our investigation we implicate DGKζ as a novel negative regulator of PLD catalytic activity in this signaling pathway.
EXPERIMENTAL PROCEDURES
Materials and Reagents
1-Butanol-d10 was purchased from CDN Isotopes (Quebec, Canada), and 1-butanol was purchased from EM Science (OmniSolv, Gibbstown, NJ). All solvents used for extraction or mass spectrometry were of HPLC grade or better, purchased from EMD Chemicals. R59949 and Ro32-0432 were purchased from Sigma. UDP was also purchased from Sigma and diluted in a H10D10 buffer (10 mm Hepes, 10 mm DTT, pH 7). [9,10-3H]Oleic acid (45.5 Ci/mmol) was purchased from PerkinElmer Life Sciences; Silica gel 60 Å TLC plates, 20 × 20 cm, were purchased from Whatman (Clifton, NJ); lipid standards, 32:0 phosphatidyl methanol (PtdMeOH), and 24:0 DAG as well as odd carbon glycerophospholipid standards were purchased from Avanti Polar Lipids (Alabaster, AL). Phorbol 12-myristate 13-acetate was purchased from Fisher. PLD inhibitors VU0155056, VU0359595, and VU0364739 were synthesized in house (16–19). 1321N1 and 1321N1 P2Y6 cells were a gift from Ken Harden and Rob Nicholas (Dept. of Pharmacology, University of North Carolina at Chapel Hill, NC).
Cell Culture
1321N1 astrocytoma cells expressing P2Y6 nucleotide receptors were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotic-antimycotic using G418 as a selection agent in a 37 °C humidified atmosphere with 5% CO2.
Glycerophospholipid Extraction for Mass Spectrometry
Glycerophospholipids were extracted using a modified Bligh and Dyer procedure as previously described (25). Briefly, cells were plated in 6-well tissue culture plates at 300,000 cells/well 48 h before the experiment in complete growth medium. Cell seeding was kept consistent to make experiments and conditions directly comparable. Cells were serum-starved 18 h before experiment. Each well was treated with DMEM + relevant conditions (UDP (or H10D10 buffer control), inhibitors, etc.) for 30 min at 37 °C. After treatment, the medium was removed, and cells were scraped in cold 0.1 n HCl:MeOH (1:1); suspension was then transferred to cold 1.5-ml microcentrifuge tubes and vortexed with 400 μl of cold CHCl3 for 1 min. The extraction proceeded with centrifugation (5 min, 4 °C, 18,000 × g) to separate the two phases. The lower organic layer was collected, phospholipid internal standards were added, and solvent was evaporated. The resulting lipid film was dissolved in 100 μl of isopropanol:hexane:100 mm NH4COOH (aqueous) 58:40:2 for LC-MS analysis.
Diacylglycerol Isolation and Detection
DAG isolation from total phospholipids extracts was achieved as previously described (26). Briefly, after phospholipid extraction by modified Bligh and Dyer procedure, each sample was applied to a glass Pasteur pipette column plugged with glass wool and packed with a 6-cm bed of silica gel 60 Å equilibrated with 10 ml of eluent (65:35:0.7 CHCl3:CH3OH:H2O). DAG molecular species were recovered in the first 3 ml of eluent, and solvents were evaporated in a vacuum centrifuge. Samples were dissolved in 70 μl of 9:1 CH3OH:CHCl3 containing 5 μl of 100 mm CH3COONa and analyzed by mass spectrometry as sodium adducts. For quantitation, 100 ng of 24:0 DAG was used as an internal standard.
Lipid Mass Spectrometry
Glycerophospholipids (including phosphatidylbutanol) were analyzed on an Applied Biosystems/MDS SCIEX 4000 Q TRAP hybrid triple quadrupole/linear ion trap mass spectrometer (Applied Biosystems, Foster City, CA) equipped with a Shimadzu high pressure liquid chromatography system with a Phenomenex Luna Silica column (2 × 250 mm, 5-μm particle size) using a gradient elution as previously described (25, 27). The identification of the individual species, accomplished by LC/MS/MS, was based on their chromatographic and mass spectral characteristics. This analysis allows identification of the two fatty acid moieties but does not determine their position on the glycerol backbone (sn-1 versus sn-2). Quantification of glycerophospholipids was achieved by the use of an LC-MS technique employing synthetic odd-carbon diacyl and lysophospholipid standards (Avanti Polar Lipids) covering a wide range of acyl chain lengths and unsaturation for which standard curves have been established.
For diacylglycerol species, mass spectral analysis was performed on a Finnigan TSQ Quantum triple quadrupole mass spectrometer (Thermo Finnigan, San Jose, CA) equipped with a Harvard Apparatus syringe pump and an electrospray source. Samples were analyzed at an infusion rate of 10 μl/min in positive ionization mode with the scan range from m/z 400 to 900. Data were collected with the Xcalibur software package (Thermo Finnigan) and analyzed with software developed in our laboratory as previously described (26).
PLD Endogenous Assays
PLD Endogenous Assay Using Radioisotopes
PLD activity was assessed by measuring accumulation of the PLD activity marker phosphatidylbutanol (PtdBuOH) that is generated in the presence of 1-butanol by a transphosphatidylation reaction as previously described (28, 29). Briefly, cells were labeled with [3H]oleic acid, 10 μCi/ml, and incubated overnight. Cells were stimulated with UDP (at various concentrations) in the presence of 0.3% 1-butanol for 30 min. Lipids were extracted and separated by TLC. Corresponding lipid spots were then imaged using a phosphorimaging tritium screen for 72 h and stained with iodine to visualize standards. PtdBuOH and PA were quantitated using Quantity One software (Bio-Rad).
PLD Endogenous Assay Using Deuterated 1-Butanol and Mass Spectrometry
MS-based PLD endogenous assay was performed essentially as described previously (29). Briefly, cells were seeded at 300,000 cells/well into 6-well tissue culture plates, and designed experiments were carried out in the presence or absence of 0.3% 1-BuOH-d10 for the desired times. At the end of the stimulation, plates were placed on ice, and media were aspirated. Glycerophospholipids were extracted by the modified Bligh and Dyer procedure as described above and analyzed in the same manner. For these samples 100 ng of 32:0 PtdMeOH was used as an internal standard to quantitate the PtdBuOH-d9 species based on offline calibration using standard curves generated from commercially available PtdBuOH standards (Avanti Polar Lipids). PtdBuOH is the unique product of PLD activity.
siRNA Protein Knockdown
1321N1-P2Y6 cells were plated at 2.2 × 105 cells/well on 6-well plates with growth medium (DMEM, 10% FBS) 24 h before transfection. Cells were ∼50% confluent at time of transfection. On-Target Plus SMART pools (O-TPSp) of siRNA (Dharmacon) of each target gene were transfected (100 nm siRNA/well) using Dharmafect 1 (Dharmacon) according to the manufacturer's protocol. After 18 h, the transfection medium was replaced with growth medium. Assays were carried out 72 h post siRNA transfection. Lipid production was then measured using either TLC or MS.
Western Blotting
Western analysis was performed to confirm the knockdown efficiency of each target gene by siRNA. Antibodies for each protein used: Gα11 and Gαq (sc-394 and sc-393), PKCα (sc-8393), RhoA (sc-418), DGKζ (sc-8721), PLCδ1 (sc-30062), and PLD1 (sc-25512) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Cdc42 (610928) was purchased from BD Biosciences, Arf1 and Arf6 (NB 100-1962 and NB 200-635) were purchased from Novus Biologicals (Littleton, CO); GAPDH (2118) was purchased from Cell Signaling Technology (Boston, MA). PLCβ3 (B521) antibody was a gift from Paul Sternweis (Dept. of Pharmacology, UT Southwestern at Dallas, TX) (30). PLD2 antibody was a gift from Sylvain Bourgoin (Laval University, Quebec, Canada) (31).
Lipid Flux Measurements
Glycerophospholipid flux was followed using incorporation of synthetic alkyne phosphatidylcholine into the cellular lipid pools and subsequent isolation and identification of both endogenous and exogenous lipid species (32, 33). Briefly, cells were plated in 60-cm tissue culture plates at 600,000 cells/plate. After 24 h cells were serum-starved. Five hours before the experiment cells were labeled with alkyne containing 34:3PC (1-(palmit-15-ynoyl)-2-oleoyl-sn-glycero-3-phosphocholine) at 2 mg/plate in DMEM contain 0.25 mg/ml fatty acid free BSA as a lipid carrier. Cells were stimulated with 50 μm UDP either with or without 0.3% 1-butanol-d10 at various time points. After stimulation, cells were extracted, and lipids were analyzed by MS as described above. Lipid identity was confirmed by fragmentation analysis.
Statistical Analysis
Mass spectrometry and siRNA results were analyzed by Student's t test to assess changes between conditions in replicate experiments (27).
RESULTS
UDP Stimulation of the P2Y6 Receptor Leads to Increased PA Formation and PLD Activity
To determine the effect of UDP stimulation of the P2Y6 receptor on lipid signaling molecules, a 1321N1 astrocytoma cell line previously developed to specifically overexpress the P2Y6 receptor was used. 1321N1 cells lack endogenous purinergic receptors and thus do not respond to extracellular nucleotides, making them an ideal model system to interrogate P2Y signaling pathways (34–37). Here UDP was used to stimulate the P2Y6 receptor in a dose-dependent manner. After ligand stimulation, PA formation was measured using quantitative lipidomic analysis developed in our laboratory (25, 27). PLD activation was also determined using a PLD-specific transphosphatidylation reaction with MS detection (29).
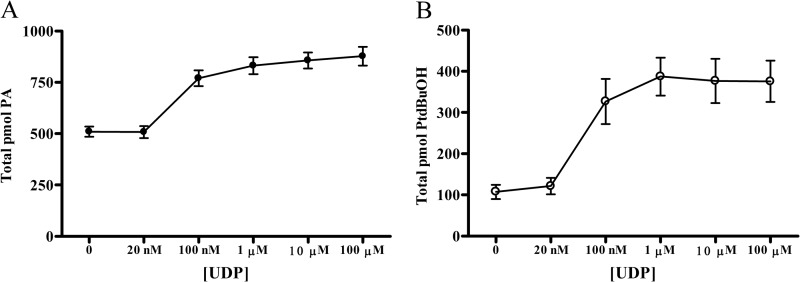
Upon UDP stimulation of the receptor there was a robust 70% increase in total PA (Fig. 1A). This is a near doubling of the total PA pool observed with doses greater than 1 μm ligand (>300 pmol absolute increase of PA). Every detectable species of PA showed increases with UDP stimulation (supplemental Fig. S1). To determine the ability of the receptor to activate PLD, product formation was measured as a function of the transphosphatidylation reaction unique to this enzyme. When cells were stimulated with UDP in the presence of 0.3% 1-butanol-d10, a marked 3.5-fold increase in total phosphatidylbutanol (PtdBuOH-d9) production was observed compared with vehicle control cells (Fig. 1B). Total PtdBuOH levels increased by nearly 300 pmol. The increase in PtdBuOH was dose-dependent with saturating stimulation seen at UDP levels above 1 μm consistent with trends for total PA production. UDP stimulation (50 μm) of 1321N1 cells lacking P2Y6 overexpression resulted in no PtdBuOH production, demonstrating that this result is a receptor-dependent response (supplemental Fig. S2).
FIGURE 1.
PA formation and PLD activity following UDP stimulation. 1321N1 cells stably expressing the P2Y6 receptor were serum-starved overnight and then stimulated with varying concentrations of UDP in DMEM for 30 min. After ligand activation, cells were extracted with a modified Bligh-Dyer method to isolate cellular lipids. Lipid species were then quantified using LC-MS. A, for phosphatidic acid analysis cells were stimulated with UDP in DMEM alone for 30 min at 37 °C. B, for PLD activity assays, UDP stimulation was carried out in the presence of 0.3% 1-butanol-d10 to capture PLD lipid products as PtdBuOH-d9 species. Cellular lipids were quantified using standard methods, and values are reported as pmol of each lipid class. Data are the means ± S.E. (n = 3).
Blocking PLD-derived PA Production by 1-Butanol Has No Effect on Total PA Pools after UDP Stimulation
To determine if PLD was responsible for the robust increase in PA, we blocked PLD ability to make PA and reassessed PA levels. Primary alcohols have long been used as tools to block PLD-generated PA production (10, 38). In the presence of primary alcohols PLD carries out a preferred transphosphatidylation reaction resulting in the production of phosphatidyl alcohols rather than phosphatidic acid (39–41). The specificity constant for 1-butanol is 5700 times higher than that of water in the active site of PLD. By activating PLD in the presence of 1-butanol, PLD-derived PA production would be greatly diminished because the enzyme is mainly making PtdBuOH products (40). It is well established in the field that phosphatidyl-alcohol products are metabolically stable in intact cells with no detectible degradation observed over the course of at least 60 min (42–44). For this reason transphosphatidyl alcohols are a quantitative and reliable measure of PLD activity.
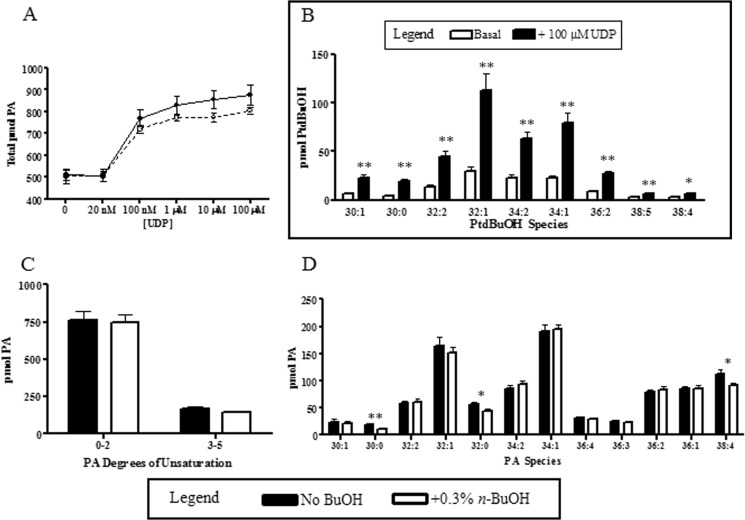
When the P2Y6 receptor was stimulated with high UDP concentrations (100 μm) in the presence or absence of 0.3% 1-butanol, there was only an ∼9% difference in the total amount of PA being produced (Fig. 2A). The small difference between the conditions was not statistically significant.
FIGURE 2.
Effects of 1-butanol addition on PA and PtdBuOH production. PA and PtdBuOH levels were measured after a 30-min stimulation of 1321N1-P2Y6 cells with 100 μm UDP either in the presence or absence of 0.3% 1-butanol-d10. After stimulation, cellular lipids were extracted and analyzed by LC-MS to quantitate lipid levels. A, total PA levels were slightly decreased (∼9%) when cells were stimulated in the presence of 1-butanol; however, those changes were not statistically significant. B, individual PtdBuOH lipid species were determined upon UDP stimulation. C, PA species were classified by the number of double bonds present, and data show that 1-butanol addition did not affect the overall lipid distribution. D, individual PA lipid species were quantitated upon UDP stimulation in the presence or absence of 1-butanol-d10. Cellular lipids were quantified using our standard methods, and values are reported as pmol of each lipid present. Data are the means ± S.E. (n = 3, *, p < 0.05; **, p < 0.01).
The lack of an effect on total PA levels upon inhibition of PLD-mediated PA production by 1-butanol was surprising due to the large increase in PLD activity observed. It is possible that individual PA species could undergo significant changes that are being diluted by total PA pool measurements. To further explore this possibility we characterized individual PLD lipid products. By using the transphosphatidylation reaction in this manner we were able to trap the PLD products as a metabolically stable compound in the cell allowing for more robust measurements. Upon stimulation of the receptor there were large increases in every PtdBuOH species detected with an average increase of >3-fold (Fig. 2B). The most abundant PtdBuOH species detected were either mono- or di-unsaturated (32:1, 34:1, 34:2 PtdBuOH) (Fig. 2B). Polyunsaturated PtdBuOH species (38:5 and 38:4) were rare, making up <3% of the total pool. These results are consistent with previously reported PLD product fatty acid specificity (45). These data suggest that PLD-generated PA is a short-lived signaling molecule that is in high flux.
After identification of the major PLD products, we further explored the specific PLD contribution to the PA pool upon receptor stimulation by looking at individual PA species. Total PA was classified by either degrees of unsaturation (Fig. 2C) or by acyl chain composition (Fig. 2D). Again there were few changes observed with 1-butanol addition. This data suggest that PLD is not a major contributor to the overall steady-state PA pool under these conditions. This could be explained in two ways; (i) the PA generated by PLD is in flux and is quickly metabolized to another lipid species in the cell, or (ii) there could be a compensatory mechanism by which other PA-producing proteins generate more PA to make up for the PA deficiency caused by blocking PLD.
Small Molecule PLD Inhibitor Treatment Does Not Affect Overall PA Levels after UDP Stimulation
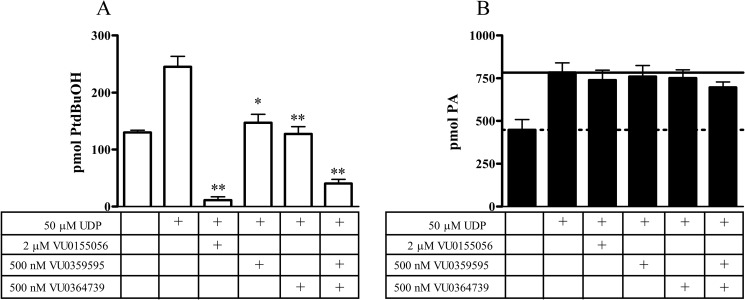
Although 1-butanol is a facile tool for blocking PLD-mediated PA production, treatment with primary alcohols could have off-target effects, and therefore, alternative methods were utilized. Primary alcohols have frequently been described incorrectly as inhibitors of PLD when in fact they mediate an alternative reaction to hydrolysis, transphosphatidylation. To further explore the role of PLD in this system, we utilized several recently developed small molecule PLD inhibitors. For this experiment the dual PLD1/2 inhibitor (VU0155056) (16) as well as the isoform preferring PLD1 (VU0359595) (18) or PLD2 (VU0364739) (19) inhibitors were used, and PLD catalytic activity was assessed. The dual PLD1/2 inhibitor, VU0155056, completely ablated PLD activity upon UDP stimulation in this system (Fig. 3A). The two isoform-selective compounds (VU0359595 and VU0364739) were used at a dose that would completely block either PLD1 or PLD2 activity, respectively, although not significantly inhibiting the other isoform (18, 19). Each compound used individually or in combination significantly decreased PLD-generated PtdBuOH levels. These data implicate both isoforms, PLD1 and PLD2, as being downstream of P2Y6 receptor activation.
FIGURE 3.
PtdBuOH and PA levels upon small molecule PLD inhibitor treatment. A, dual PLD1/2 inhibitor VU0155056 completely blocked all PLD activity as measured by the transphosphatidylation reaction and PtdBuOH production. The isoform-selective inhibitors (PLD1 VU0359595 or PLD2 VU0364739) each had a large but incomplete effect on decreasing PtdBuOH production when used individually or in combination (representative experiment shown; n = 2). B, PLD inhibitor treatment had no effect on total PA production upon UDP stimulation. There was a small (12%) decrease seen in total PA levels when VU0359595 and VU0364739 were used in combination; however, that change did not reach statistical significance. (n = 3; *, p < 0.05; **, p < 0.01).
PA levels were then measured, and there was a small but not statistically significant change (12%) in total PA levels when PLD1 and PLD2 inhibitors were used in concert (Fig. 3B). These data are consistent with results from 1-butanol treatment, again supporting the claim that PLD is not a major contributor to the stable total PA pool under these conditions. This shows PLD1 and PLD2 are active and are known sources of PA; however, the contribution to the overall stable pool is not substantial.
Characterization of P2Y6 Signaling Pathway Using siRNA Knockdown of Signaling Proteins
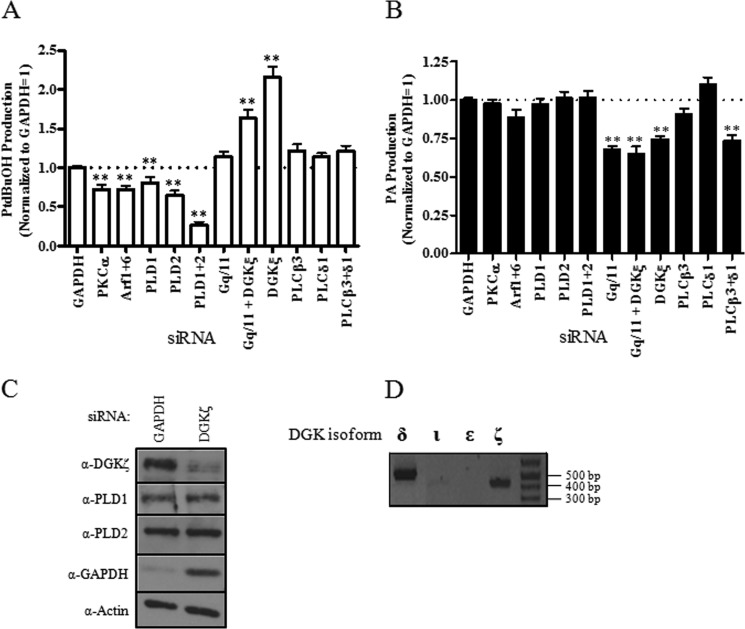
To further characterize the pathway from P2Y6 receptor to PLD activation, RNAi was used to knock down proteins previously implicated in P2Y6 receptor or PLD signaling. Cells were then labeled with [3H]oleic acid, and metabolic flux of that lipid into PtdBuOH and PA was followed by TLC. Knockdown of PKC or Arf1 and -6, known to modulate PLD signaling, had significant effects on PLD activity (Fig. 4A). Both PLD1 and PLD2 contribute to the UDP-stimulated PLD activity in this system because siRNA isoform knockdowns independently or together gave a significant decrease in overall PtdBuOH production (Fig. 4A) (also consistent with PLD inhibitor experiments). Other established PLD activating proteins including RhoA, Cdc42, and Rac1 (46–48) were knocked down with no significant effect on activity (data not shown). Additionally, an interesting and unexpected result was seen when protein levels of DGKζ were ablated; the activity of PLD increased 2-fold, suggesting a novel negative regulation of PLD activity by DGKζ (Fig. 4A).
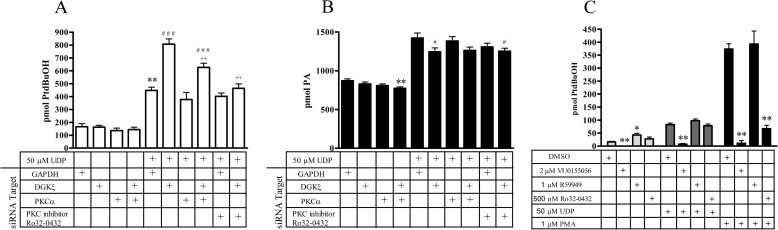
FIGURE 4.
siRNA protein knockdown of key signaling enzymes in 1321N1-P2Y6 cells. siRNA was used to knock down key proposed signaling cascade intermediates in 1321N1-P2Y6 cells labeled with [3H]oleic acid to follow lipid flux. After protein knockdown, cells were stimulated with 50 μm UDP in the presence of 1-butanol and either PLD activity (A) or PA production (B) was monitored. Data are normalized to cells with control protein GAPDH knockdown = 1. Values are the means ± S.E. (n = 3–6). **, significantly different compared with GAPDH siRNA-transfected cells (**, p < 0.01). C, Western blots of PLD1, PLD2, and DGK ζ after siRNA knockdown of DGKζ are shown. D, RT-PCR of DGK shows ζ and δ isoforms are present, and the ι and ϵ isoforms are not.
PA production was also monitored under these conditions. When known activators of PLD, such as PKC (46) and Arf (49), or PLD1 or PLD2 itself were knocked down, there was no significant effect on PA levels upon UDP stimulation (Fig. 4B). These siRNA knockdown experiments are consistent with data presented above using either 1-butanol or small molecule PLD inhibitor treatment. Significant changes in PA levels were seen with Gq/11, DGKζ or a co-knockdown of PLCβ3+ PLCδ1. The P2Y6 receptor is known to signal through Gq, activating a PLCβ pathway (6), making these conditions the ideal internal control for this experiment. PLC is responsible for the cleavage of phosphatidylinositol 4,5-bisphosphate to DAG and inositol 1,4,5-trisphosphate. DAG is then converted to PA through phosphorylation by DAG kinases. Interestingly, when PLCβ3 was knocked down on its own, there was no significant change in PA levels. PLCβ3 and PLCδ1 had to be knocked down together before any decrease was seen. Several other PLC isoforms known to be present in these cells were also knocked down either individually or in combination with no observable effect on PA production or PLD activity (supplemental Fig. S3). The lack of effect seen with PLC isoform knockdowns on PA levels suggest that PA is tightly regulated in these cells.
The 2-fold increase in PLD activity with DGKζ knockdown could possibly be explained by a subsequent induction of PLD1 or PLD2 expression. PLD1 and PLD2 protein levels were assessed by Western blot in the presence of DGKζ knockdown and no change in PLD1 or PLD2 expression was seen (Fig. 4C). All protein knockdowns were confirmed by Western blot (supplemental Fig. S4).
These data show that there is a key regulation of both PLD activity and PA production by DAG-producing (PLC) or -metabolizing (DGK) enzymes in this system. RT-PCR analysis shows the presence of both DGKζ and DGKδ isoforms (Fig. 4D), but only DGKζ protein knockdown had a significant effect on either PLD activity or PA production (DGKδ knockdown had no effect; supplemental Fig. S3).
DAG Kinase ζ Contributes to DAG-PA Conversion after P2Y6 Stimulation
Experimental results indicate that the majority of total stable PA in this system is not coming from phosphatidylcholine to phosphatidic acid conversion by PLD. Data support the hypothesis that a major contributor to the PA pool is through a phosphatidylinositol 4,5-bisphosphate-DAG-PA pathway involving PLCβ3 and PLCδ1 isoforms and DGKζ (PA decreased with protein knockdown; Fig. 4B). To further support this idea, we first show that DAG levels are elevated in response to UDP stimulation. Every species of DAG measured showed a significant increase (except 28:0 DAG) with, some increasing more than 2-fold (Fig. 5, white bars).
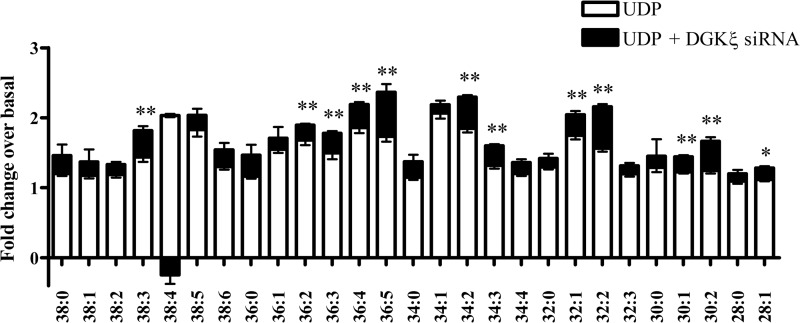
FIGURE 5.
DAG species detected in 1321N1-P2Y6 cells. White bars, DAG species were quantitated in 1321N1-P2Y6 cells and are expressed here as fold change. 1321N1-P2Y6 cells were stimulated with 50 μm UDP or vehicle for 20 min, after which DAGs were isolated and analyzed by ESI-MS. Data are normalized to basal conditions = 1 and are expressed as fold change. Every species of DAG significantly increased in response to UDP stimulation with p values <0.01; n = 12 (except 28:0 species, which did not reach significance). Black bars, 1321N1-P2Y6 cells were transfected with either siRNA targeting GAPDH or DGKζ and then stimulated with 50 μm UDP for 20 min. Additional fold increase over UDP-stimulated DAG levels is indicated by black bars plotted with respect to basal conditions. DAG species 38:4 is plotted differently to indicate a decrease (24%) with respect to the UDP-stimulated level when DGKζ was knocked down. (n = 2; black bars: *, p < 0.05; **, p < 0.01).
To determine if DGKζ is a key isoform catalyzing DAG to PA conversion, we also measured DAG levels when DGKζ was knocked down to confirm that DAG is building up and not being converted to PA via another DGK isoform. Cells were transfected with siRNA against GAPDH control protein or DGKζ and then stimulated with UDP. There was a significant increase in overall DAG levels with several species showing significant elevation with DGKζ knockdown above UDP-stimulated control levels (Fig. 5, black bars). This paired with observed decreases in PA after DGKζ knockdown confirm that DGKζ is a key enzyme contributing to DAG metabolism and PA generation upon UDP stimulation of the P2Y6 receptor.
PLD Is Regulated by a PKCα-DGKζ Mechanism
The large potentiation of PLD activity upon DGKζ protein knockdown observed in the lipid flux experiments suggests that the protein is essential for the negative regulation of PLD activity. Previous studies from other laboratories have found PKCα and DGKζ exist in a regulatory complex (50, 51). Our data suggest that this signaling pathway may be under the control of a similar PKC-DGK regulatory mechanism. To test this hypothesis, siRNA was used to knock down PKCα or DGKζ alone or in conjunction, and endogenous lipid levels were assayed with a quantitative MS based lipidomic approach. Consistent with previous experiments (involving radiolabeled oleic acid labeling; Fig. 4), DGKζ knockdown resulted in a large activation of PLD (Fig. 6A). Interestingly when DGKζ and PKCα were knocked down together, the large increase in PLD activation was attenuated significantly (>50% decrease). The activation of PLD upon DGKζ knockdown did not occur in the absence of receptor stimulation (Fig. 6A). In addition to siRNA knockdown we also used a small molecule PKC inhibitor to further explore this mechanism. When DGKζ was knocked down and PKC activity was inhibited (with Ro32-0432), the DGKζ-mediated PLD activation was ablated to near control levels (Fig. 6A). These data support the hypothesis that DGKζ regulates PLD through a PKCα-dependent mechanism, possibly through a PKCα-DGKζ protein complex previously described (50, 51). When PKCα was knocked down or inhibited chemically, there was only a small decrease in PLD activity, suggesting that there are additional non-PKC modes for activating PLD after P2Y6 receptor stimulation.
FIGURE 6.
DGKζ is negatively regulating PLD activation through PKCα in UDP-stimulated 1321N1-P2Y6 cells. siRNA was used to knock down key proposed signaling cascade intermediates DGKζ and PKCα in 1321N1-P2Y6 cells. After protein knock down, cells were stimulated with 50 μm UDP in the presence of 1-butanol, and PLD activity was monitored using quantitative LC-MS analysis of endogenous lipids. A, PtdBuOH production is shown. GAPDH protein knockdown was used as a control. PKC inhibitor Ro32-0432 was used at 500 nm. Values are the means ± S.E. (n = 4). **, p < 0.01 versus Basal GAPDH siRNA condition; ###, p < 0.001 versus UDP stimulated GAPDH siRNA; ++, p < 0.01 versus UDP-stimulated DGKζ siRNA condition. B, PA production is shown. **, p < 0.01 versus basal GAPDH siRNA condition; #, p < 0.05 versus UDP stimulated GAPDH siRNA condition; n = 4. C, 1321N1 P2Y6 cells were pretreated with small molecule inhibitors or vehicle control for 10 min. After pretreatment, cells were stimulated with either 50 μm UDP, 1 μm phorbol 12-myristate 13-acetate, or vehicle control ± each inhibitor in the presence of 0.3% 1-butanol-d10 for 30 min. PLD inhibitor, VU0155056; DGK inhibitor, R59949 PKC inhibitor, Ro32-0432. Data here show totals of 32:1, 34:1, 34:2, and 36:2 PtdBuOH species (n = 3). *, p < 0.05; **, p < 0.01, versus DMSO for each activator.
We also assessed PA production under these conditions. UDP stimulation increased PA production (>60%), and PA production was decreased when DGKζ protein was knocked down (15% decrease; p < 0.05). No other large changes in PA were observed with siRNA knockdown either under basal or UDP-stimulated conditions (Fig. 6B and supplemental Fig. S5). To determine if this was a universal PLD regulation in these cells we stimulated the endogenously expressed thrombin receptor. Thrombin stimulation led to increased PLD activity; however, DGKζ protein knockdown had no effect on either PLD levels or PA production, suggesting that this is a signaling pathway specific effect (supplement Fig. S6).
To better determine that this mechanism was DGKζ-centric and did not involve other classes of DGK isoforms, we used a small molecule DGK inhibitor (R59949) that is specific to type I DGK isoforms and is not effective against DGKζ (52, 53). Treatment with R59949 had no effect on either UDP or phorbol 12-myristate 13-acetate stimulated PLD activity (Fig. 6C). This would suggest that the mechanism by which DGK regulates PLD is through an inhibitor resistant DGK isoform. To confirm that the DGK inhibitor works properly, DAG levels were assessed and shown to be elevated under these conditions (data not shown). Unfortunately, there are no small molecule inhibitors specific to DGKζ currently available. Treatment with PKC inhibitor Ro32-0432 led to differential effects depending on stimulation. Under UDP stimulation no change in PLD activity was seen with PKC inhibition; however, when stimulated with phorbol ester (phorbol 12-myristate 13-acetate) there was a large decrease in PLD activation when PKC was blocked. This is consistent with the known mechanism of PLD activation via PKC upon phorbol ester stimulation (54) and suggests that after P2Y6 receptor stimulation PLD is activated in ways that are both dependent and independent of PKC.
PLD Contribution to the PA Pool Is Small but Immediate after P2Y6 Receptor Activation
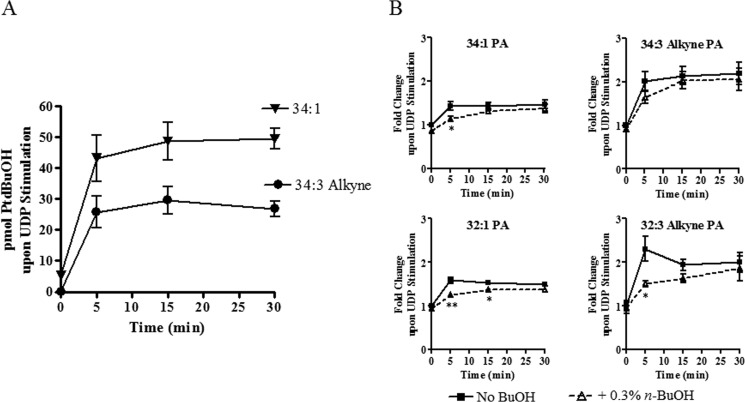
To more closely assess the time frame of signaling activation, we carried out a time course and followed specific lipid species to determine if there was a more significant temporal element to PLD activation in this system. Monitoring lipid distribution and flux in the cell is difficult due the plethora of molecular species and metabolic pathways. To simplify this monitoring process we utilized a MS-based alkyne-tagged lipid method previously developed in our laboratory (32, 33). We labeled 1321N1 P2Y6 cells with an alkyne containing PC (34:3), an exogenous version of a major PLD substrate (34:1 PC), and followed the alkynyl chain as it was incorporated into other phospholipid classes after receptor stimulation. By using this exogenous lipid we can monitor if a specific PC is being remodeled, metabolized, or converted to a different lipid class in a time-dependent manner. We did this both in the presence or absence of the alternative PLD substrate 1-butanol. After UDP stimulation of the P2Y6 receptor, there was an immediate increase (5 min) in PtdBuOH levels (Fig. 7A), suggesting PLD activation occurs quickly. This was true for both exogenous alkyne lipids (34:3) as well as endogenous lipid species (34:1), showing that PLD was able to use the alkyne lipid efficiently, consistent with previous reports (32).
FIGURE 7.
PLD activation occurs immediately upon UDP stimulation. Comprehensive lipidomic analysis was carried out as a time course of UDP stimulation (50 μm). A, PLD activation occurs immediately after UDP stimulation. B, PA production showed a time lag when PLD was blocked (with 0.3% 1-butanol). Significant changes in PA levels ± 0.3% of n-BuOH were observed at early time points after UDP stimulation. n = 2; *, p < 0.05; **, p < 0.01.
When PLD-mediated PA generation was blocked with primary alcohol, interesting results were seen. There was a statistically significant difference in PA levels at early time points after receptor activation in the presence of 1-butanol (Fig. 7B). Again this was true for endogenous (34:1 and 32:1) and exogenous (34:3) as well as remodeled (32:3) lipid species. Under these conditions all PA species showed a distinct time shift when 1-butanol was present (supplemental Fig. S7, A and B). These data suggest that the immediate increase in PA levels after P2Y6 receptor stimulation is heavily dependent on PLD activity. This also shows that when PLD generation of PA is blocked, the cell compensates over time to adjust PA levels back to those seen in the absence of a PLD blocking agent, showing the tight regulation of PA pools. Thirty minutes post-stimulation there is no difference in PA levels either with or without primary alcohol.
Other lipid classes were also analyzed. Both phosphatidylserine and phosphatidylethanolamine contained alkyne lipids showing that exogenous PC was metabolized into other glycerophospholipid classes (data not shown). Another interesting trend was seen with phosphatidylinositol lipids. Both endogenous phosphatidylinositol (PI; 34:1) and alkynyl-PI (34:3) levels decreased over time after P2Y6 stimulation, and in the presence of a primary alcohol that decrease was more pronounced (supplemental Fig. S7C).
DISCUSSION
Cellular PA is produced through multiple mechanisms, but PLD has long been represented in the literature as the main source of signaling PA. Because of the essential roles attributed to PA in the cell as well as the importance of PC breakdown (55), PLD has received much attention.
The data presented herein demonstrate that the cellular PA pool is a dynamic population that is tightly regulated. Modulation and inhibition of one key PA-producing enzyme, PLD, has little effect on total steady-state PA levels in 1321N1 cells under these conditions. When the P2Y6 receptor is stimulated, it is clear that both PLD activity and PA levels increase greatly, but siRNA knockdown or chemical inhibition of PLD indicates the enzyme is not the only source of bulk-PA production. These data do not show that PLD-generated PA is unimportant; rather, it is possible that the flux of the PLD-generated PA is so fast that we are unable to trap that population using total lipid pool measurements. Our lipid flux experiments further support the claim that PLD is responsible for the initial immediate generation of PA after UDP treatment. When this essential enzyme is blocked, the cell eventually overcomes this blockade by increased activity from other sources. This would suggest that PLD-generated PA is a critical signaling pool that is rapidly metabolized.
PA has dichotomous roles in the cell as a structural lipid important in inducing negative membrane curvature, protein recruitment, and vesicle formation as well as an important signaling second messenger (56). Due to the different roles this lipid fills, global measurements do not provide a complete picture. More research into distinguishing PA pools in the cell is currently under way, and it is likely that small subcellular PA pools are those essential for many of the important signaling properties associated with this molecule. PLD1 and PLD2 are differentially localized, with PLD1 being mostly found in intercellular membranes and PLD2 found constitutively at the plasma membrane (10). Our laboratory (16–19) as well as others (20, 57) have published several reports implicating small molecule PLD inhibitors as possible anti-cancer agents. It has been shown that PLD inhibitor treatment leads to decreased cancer cell invasion (16), cell spreading, chemotaxis (20), and possibly could be a tool to decrease cancer metastasis (58). The utility of these small molecules to inhibit such essential cellular processes while not effecting bulk steady-state PA levels is intriguing and would suggest that there is a small subpopulation of essential signaling PA that is tightly regulated and critical to these cellular processes.
Identification of other novel regulatory mechanisms of PLD activity provides additional opportunities for small molecule therapeutic development. The identification of DGKζ as a negative regulator of PLD activity is one of these novel mechanisms. Mérida and co-workers (59) have proposed that DGKζ is part of a much larger protein complex that scaffolds lipid metabolizing enzymes and effectors near cellular membranes. The possible role of DGKζ as a scaffolding protein further supports the importance of local modulation of the signaling event.
It has been established in several cell systems that DGKζ and PKCα are found associated in a regulatory complex (51). Based on literature evidence and the results reported herein, we propose that that regulation is intact in this system.
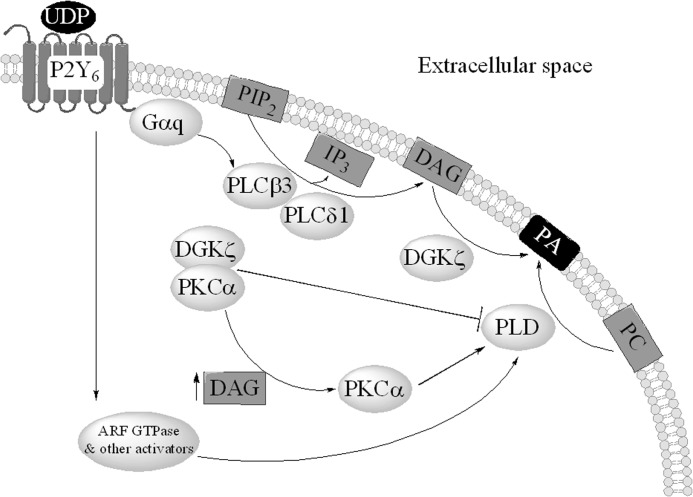
This study further delineates the mechanism of PLD activation downstream of P2Y6 receptor stimulation (Fig. 8). UDP activates the P2Y6 receptor, which is a GPCR signaling through Gαq-activating PLC enzymes; specifically β3 and δ1 isoforms are required. PLCs catalyze the conversion of phosphatidylinositol 4,5-bisphosphate into DAG and inositol 1,4,5-trisphosphate. DAG is then phosphorylated by DGKζ to generate PA. Increased DAG levels also activate PKCα, which in turn activates PLD. PLD can also be turned on in a PKC-independent manner through Arf (or other small GTPases). We have shown that both PLD1 and PLD2 isoforms are involved in this pathway. When DGKζ protein is not present, the pathway is deregulated, and PLD is hyperactive.
FIGURE 8.
Mechanism of PA regulation downstream of P2Y6 activation.
Studies presented in this work are the first direct evidence showing that DGKζ is a negative regulator of PLD activity. It is also shown that DGK and PLD are in a tightly regulated compensatory mechanism by which steady-state PA levels are controlled. PLD is activated by P2Y6 receptor stimulation, but that activity is highly dynamic and difficult to discriminate as part of the overall PA population. Activation of PLD downstream of several P2Y receptors (P2Y1, P2Y2, P2Y4, and P2Y6) is known (8, 60, 61), and exploration of these signaling pathways could provide new insight into the relevant role of this phospholipase. Despite its limitations, work in the field of the purinergic receptors has frequently used the 1321N1 cell line. It has been a useful model system in which to explore signaling pathways.
This work further illustrates that the PA signaling network is complex and highly integrated. Attempts to successfully modulate this signaling pathway for therapeutic development will require a detailed understanding of these interconnections. Determining why cells have evolved such a dynamic network designed to maintain cellular PA levels even to the point of having signaling pathways compete remains to be fully elucidated. This reinforces our understanding of the essential roles that signaling PA pools play in cellular functions and that abnormalities have roles in pathology.
Supplementary Material
Acknowledgments
We thank Dr. Ken Harden and Dr. Robert Nicholas for providing 1321N1 P2Y6 stable cell lines and Ronald Bruntz for helpful discussion and generation of reagents. We also thank Dr. Bourgoin for providing PLD2 antibody.
This work was supported, in whole or in part, by National Institutes of Health Grants/Molecular Libraries Probe Production Centers Network U54 MH084659, Grants P01-ES013125 and U54 069338, and Grant T32 MH093366 (postdoctoral training in CNS Drug Discovery Research (to T. P. M.). This work also supported in part by The McDonnell Foundation for Brain Cancer Research.

This article contains supplemental Figs. S1[en]S7.
- GPCR
- G-protein-coupled receptor
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PLD
- phospholipase D
- PLC
- phospholipase C
- DAG
- diacylglycerol
- DGK
- DAG kinase
- Ptd
- phosphatidyl
- PtdBuOH
- phosphatidylbutanol.
REFERENCES
- 1. Kroeze W. K., Sheffler D. J., Roth B. L. (2003) G-protein-coupled receptors at a glance. J. Cell Sci. 116, 4867–4869 [DOI] [PubMed] [Google Scholar]
- 2. Lundstrom K. (2009) An overview on GPCRs and drug discovery. Structure-based drug design and structural biology on GPCRs. Methods Mol. Biol. 552, 51–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. von Kügelgen I., Harden T. K. (2011) Molecular pharmacology, physiology, and structure of the P2Y receptors. Adv. Pharmacol. 61, 373–415 [DOI] [PubMed] [Google Scholar]
- 4. King B. F., Townsend-Nicholson A., Burnstock G. (1998) Metabotropic receptors for ATP and UTP. Exploring the correspondence between native and recombinant nucleotide receptors. Trends Pharmacol. Sci. 19, 506–514 [DOI] [PubMed] [Google Scholar]
- 5. Nicholas R. A., Watt W. C., Lazarowski E. R., Li Q., Harden K. (1996) Uridine nucleotide selectivity of three phospholipase C-activating P2 receptors. Identification of a UDP-selective, a UTP-selective, and an ATP- and UTP-specific receptor. Mol. Pharmacol. 50, 224–229 [PubMed] [Google Scholar]
- 6. von Kügelgen I. (2006) Pharmacological profiles of cloned mammalian P2Y-receptor subtypes. Pharmacol. Ther. 110, 415–432 [DOI] [PubMed] [Google Scholar]
- 7. Purkiss J., Owen P. J., Jones J. A., Boarder M. R. (1992) Stimulation of phosphatidic acid synthesis in bovine aortic endothelial cells in response to activation of P2-purinergic receptors. Biochem. Pharmacol. 43, 1235–1242 [DOI] [PubMed] [Google Scholar]
- 8. Ralevic V., Burnstock G. (1998) Receptors for purines and pyrimidines. Pharmacol. Rev. 50, 413–492 [PubMed] [Google Scholar]
- 9. Wang X., Devaiah S. P., Zhang W., Welti R. (2006) Signaling functions of phosphatidic acid. Prog. Lipid Res. 45, 250–278 [DOI] [PubMed] [Google Scholar]
- 10. Selvy P. E., Lavieri R. R., Lindsley C. W., Brown H. A. (2011) Phospholipase D. Enzymology, functionality, and chemical modulation. Chem. Rev. 111, 6064–6119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hammond S. M., Altshuller Y. M., Sung T. C., Rudge S. A., Rose K., Engebrecht J., Morris A. J., Frohman M. A. (1995) Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J. Biol. Chem. 270, 29640–29643 [DOI] [PubMed] [Google Scholar]
- 12. Colley W. C., Sung T. C., Roll R., Jenco J., Hammond S. M., Altshuller Y., Bar-Sagi D., Morris A. J., Frohman M. A. (1997) Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization. Curr. Biol. 7, 191–201 [DOI] [PubMed] [Google Scholar]
- 13. Hammond S. M., Jenco J. M., Nakashima S., Cadwallader K., Gu Q., Cook S., Nozawa Y., Prestwich G. D., Frohman M. A., Morris A. J. (1997) Characterization of two alternately spliced forms of phospholipase D1. Activation of the purified enzymes by phosphatidylinositol 4,5-bisphosphate, ADP-ribosylation factor, and Rho family monomeric GTP-binding proteins and protein kinase Cα. J. Biol. Chem. 272, 3860–3868 [DOI] [PubMed] [Google Scholar]
- 14. Park S. K., Provost J. J., Bae C. D., Ho W. T., Exton J. H. (1997) Cloning and characterization of phospholipase D from rat brain. J. Biol. Chem. 272, 29263–29271 [DOI] [PubMed] [Google Scholar]
- 15. Exton J. H. (1997) New developments in phospholipase D. J. Biol. Chem. 272, 15579–15582 [DOI] [PubMed] [Google Scholar]
- 16. Scott S. A., Selvy P. E., Buck J. R., Cho H. P., Criswell T. L., Thomas A. L., Armstrong M. D., Arteaga C. L., Lindsley C. W., Brown H. A. (2009) Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat. Chem. Biol. 5, 108–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lavieri R., Scott S. A., Lewis J. A., Selvy P. E., Armstrong M. D., Alex Brown H., Lindsley C. W. (2009) Design and synthesis of isoform-selective phospholipase D (PLD) inhibitors. Part II. Identification of the 1,3,8-triazaspiro[4,5]decan-4-one privileged structure that engenders PLD2 selectivity. Bioorg. Med. Chem. Lett. 19, 2240–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lewis J. A., Scott S. A., Lavieri R., Buck J. R., Selvy P. E., Stoops S. L., Armstrong M. D., Brown H. A., Lindsley C. W. (2009) Design and synthesis of isoform-selective phospholipase D (PLD) inhibitors. Part I. Impact of alternative halogenated privileged structures for PLD1 specificity. Bioorg. Med. Chem. Lett. 19, 1916–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lavieri R. R., Scott S. A., Selvy P. E., Kim K., Jadhav S., Morrison R. D., Daniels J. S., Brown H. A., Lindsley C. W. (2010) Design, synthesis, and biological evaluation of halogenated N-(2-(4-oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-8-yl)ethyl)benzamides. Discovery of an isoform-selective small molecule phospholipase D2 inhibitor. J. Med. Chem. 53, 6706–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Su W., Yeku O., Olepu S., Genna A., Park J. S., Ren H., Du G., Gelb M. H., Morris A. J., Frohman M. A. (2009) 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI), a phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Mol. Pharmacol. 75, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lindsley C. W., Brown H. A. (2012) Phospholipase D as a therapeutic target in brain disorders. Neuropsychopharmacology 37, 301–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Steed P. M., Chow A. H. (2001) Intracellular signaling by phospholipase D as a therapeutic target. Curr. Pharm. Biotechnol. 2, 241–256 [DOI] [PubMed] [Google Scholar]
- 23. Huang P., Frohman M. A. (2007) The potential for phospholipase D as a new therapeutic target. Expert Opin. Ther. Targets 11, 707–716 [DOI] [PubMed] [Google Scholar]
- 24. Foster D. A., Xu L. (2003) Phospholipase D in cell proliferation and cancer. Mol. Cancer Res. 1, 789–800 [PubMed] [Google Scholar]
- 25. Ivanova P. T., Milne S. B., Byrne M. O., Xiang Y., Brown H. A. (2007) Glycerophospholipid identification and quantitation by electrospray ionization mass spectrometry. Methods Enzymol. 432, 21–57 [DOI] [PubMed] [Google Scholar]
- 26. Callender H. L., Forrester J. S., Ivanova P., Preininger A., Milne S., Brown H. A. (2007) Quantification of diacylglycerol species from cellular extracts by electrospray ionization mass spectrometry using a linear regression algorithm. Anal. Chem. 79, 263–272 [DOI] [PubMed] [Google Scholar]
- 27. Myers D. S., Ivanova P. T., Milne S. B., Brown H. A. (2011) Quantitative analysis of glycerophospholipids by LC-MS. Acquisition, data handling, and interpretation. Biochim. Biophys. Acta 1811, 748–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Walker S. J., Brown H. A. (2004) Measurement of G protein-coupled receptor-stimulated phospholipase D activity in intact cells. Methods Mol. Biol. 237, 89–97 [DOI] [PubMed] [Google Scholar]
- 29. Brown H. A., Henage L. G., Preininger A. M., Xiang Y., Exton J. H. (2007) Biochemical analysis of phospholipase D. Methods Enzymol. 434, 49–87 [DOI] [PubMed] [Google Scholar]
- 30. Smrcka A. V., Sternweis P. C. (1993) Regulation of purified subtypes of phosphatidylinositol-specific phospholipase Cβ by G protein α and βγ subunits. J. Biol. Chem. 268, 9667–9674 [PubMed] [Google Scholar]
- 31. Freyberg Z., Bourgoin S., Shields D. (2002) Phospholipase D2 is localized to the rims of the Golgi apparatus in mammalian cells. Mol. Biol. Cell 13, 3930–3942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Milne S. B., Tallman K. A., Serwa R., Rouzer C. A., Armstrong M. D., Marnett L. J., Lukehart C. M., Porter N. A., Brown H. A. (2010) Capture and release of alkyne-derivatized glycerophospholipids using cobalt chemistry. Nat. Chem. Biol. 6, 205–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tallman K. A., Armstrong M. D., Milne S. B., Marnett L. J., Brown H. A., Porter N. A. (2013) Cobalt carbonyl complexes as probes for alkyne-tagged lipids. J. Lipid Res. 54, 859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Communi D., Parmentier M., Boeynaems J.-M. (1996) Cloning, functional expression and tissue distribution of the human P2Y6 receptor. Biochem. Biophys. Res. Commun. 222, 303–308 [DOI] [PubMed] [Google Scholar]
- 35. Filtz T. M., Li Q., Boyer J. L., Nicholas R. A., Harden T. K. (1994) Expression of a cloned P2Y purinergic receptor that couples to phospholipase C. Mol. Pharmacol. 46, 8–14 [PubMed] [Google Scholar]
- 36. Parr C. E., Sullivan D. M., Paradiso A. M., Lazarowski E. R., Burch L. H., Olsen J. C., Erb L., Weisman G. A., Boucher R. C., Turner J. T. (1994) Cloning and expression of a human P2U nucleotide receptor, a target for cystic fibrosis pharmacotherapy. Proc. Nat. Acad. Sci. 91, 3275–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Communi D., Pirotton S., Parmentier M., Boeynaems J.-M. (1995) Cloning and functional expression of a human uridine nucleotide receptor. J. Biol. Chem. 270, 30849–30852 [DOI] [PubMed] [Google Scholar]
- 38. Hu T., Exton J. H. (2005) 1-Butanol interferes with phospholipase D1 and protein kinase Cα association and inhibits phospholipase D1 basal activity. Biochem. Biophys. Res. Commun. 327, 1047–1051 [DOI] [PubMed] [Google Scholar]
- 39. Dawson R. M. (1967) The formation of phosphatidylglycerol and other phospholipids by the transferase activity of phospholipase D. Biochem. J. 102, 205–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chalifa-Caspi V., Eli Y., Liscovitch M. (1998) Kinetic analysis in mixed micelles of partially purified rat brain phospholipase D activity and its activation by phosphatidylinositol 4,5-bisphosphate. Neurochem. Res. 23, 589–599 [DOI] [PubMed] [Google Scholar]
- 41. Yang S. F., Freer S., Benson A. A. (1967) Transphosphatidylation by phospholipase D. J. Biol. Chem. 242, 477–484 [PubMed] [Google Scholar]
- 42. Seidler L., Kaszkin M., Kinzel V. (1996) Primary alcohols and phosphatidylcholine metabolism in rat brain synaptosomal membranes via phospholipase D. Pharmacol. Toxicol. 78, 249–253 [DOI] [PubMed] [Google Scholar]
- 43. Billah M. M., Pai J. K., Mullmann T. J., Egan R. W., Siegel M. I. (1989) Regulation of phospholipase D in HL-60 granulocytes. Activation by phorbol esters, diglyceride, and calcium ionophore via protein kinase-independent mechanisms. J. Biol. Chem. 264, 9069–9076 [PubMed] [Google Scholar]
- 44. Randall R. W., Bonser R. W., Thompson N. T., Garland L. G. (1990) A novel and sensitive assay for phospholipase D in intact cells. FEBS Lett. 264, 87–90 [DOI] [PubMed] [Google Scholar]
- 45. Pettitt T. R., McDermott M., Saqib K. M., Shimwell N., Wakelam M. J. (2001) Phospholipase D1b and D2a generate structurally identical phosphatidic acid species in mammalian cells. Biochem. J. 360, 707–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Singer W. D., Brown H. A., Bokoch G. M., Sternweis P. C. (1995) Resolved phospholipase D activity is modulated by cytosolic factors other than Arf. J. Biol. Chem. 270, 14944–14950 [DOI] [PubMed] [Google Scholar]
- 47. Walker S. J., Wu W. J., Cerione R. A., Brown H. A. (2000) Activation of phospholipase D1 by Cdc42 requires the Rho insert region. J. Biol. Chem. 275, 15665–15668 [DOI] [PubMed] [Google Scholar]
- 48. Henage L. G., Exton J. H., Brown H. A. (2006) Kinetic analysis of a mammalian phospholipase D. Allosteric modulation by monomeric GTPases, protein kinase C, and polyphophoinositides. J. Biol. Chem. 281, 3408–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brown H. A., Gutowski S., Moomaw C. R., Slaughter C., Sternweis P. C. (1993) ADP-ribosylation factor, a small GTP-dependent regulatory protein, stimulates phospholipase D activity. Cell 75, 1137–1144 [DOI] [PubMed] [Google Scholar]
- 50. Luo B., Prescott S. M., Topham M. K. (2003) Protein kinase Cα phosphorylates and negatively regulates diacylglycerol kinase ζ. J. Biol. Chem. 278, 39542–39547 [DOI] [PubMed] [Google Scholar]
- 51. Luo B., Prescott S. M., Topham M. K. (2003) Association of diacylglycerol kinase ζ with protein kinase Cα. Spatial regulation of diacylglycerol signaling. J. Cell Biol. 160, 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bunting M., Tang W., Zimmerman G. A., McIntyre T. M., Prescott S. M. (1996) Molecular cloning and characterization of a novel human diacylglycerol kinase ζ. J. Biol. Chem. 271, 10230–10236 [PubMed] [Google Scholar]
- 53. Jiang Y., Sakane F., Kanoh H., Walsh J. P. (2000) Selectivity of the diacylglycerol kinase inhibitor 3-[2-(4-[bis-(4-fluorophenyl)methylene]-1-piperidinyl)ethyl]-2, 3-dihydro-2-thioxo-4(1H)quinazolinone (R59949) among diacylglycerol kinase subtypes. Biochem. Pharmacol. 59, 763–772 [DOI] [PubMed] [Google Scholar]
- 54. Exton J. H. (1999) Regulation of phospholipase D. Biochim. Biophys. Acta 1439, 121–133 [DOI] [PubMed] [Google Scholar]
- 55. Exton J. H. (1990) Signaling through phosphatidylcholine breakdown. J. Biol. Chem. 265, 1–4 [PubMed] [Google Scholar]
- 56. Yang C. Y., Frohman M. A. (2012) Mitochondria. Signaling with phosphatidic acid. Int. J. Biochem. Cell Biol. 44, 1346–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ye Q., Kantonen S., Gomez-Cambronero J. (2013) Serum deprivation confers the MDA-MB-231 breast cancer line with an EGFR/JAK3/PLD2 system that maximizes cancer cell invasion. J. Mol. Biol. 425, 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Su W., Chen Q., Frohman M. A. (2009) Targeting phospholipase D with small-molecule inhibitors as a potential therapeutic approach for cancer metastasis. Future Oncol. 5, 1477–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rincón E., Gharbi S. I., Santos-Mendoza T., Mérida I. (2012) Diacylglycerol kinase ζ. At the crossroads of lipid signaling and protein complex organization. Prog. Lipid Res. 51, 1–10 [DOI] [PubMed] [Google Scholar]
- 60. Berg K. A., Evans K. L., Cropper J. D., Clarke W. P. (2003) Temporal regulation of agonist efficacy at 5-hydroxytryptamine (5-HT)1A and 5-HT 1B receptors. J. Pharmacol. Exp. Ther. 304, 200–205 [DOI] [PubMed] [Google Scholar]
- 61. Neary J. T., Kang Y., Bu Y., Yu E., Akong K., Peters C. M. (1999) Mitogenic signaling by ATP/P2Y purinergic receptors in astrocytes. Involvement of a calcium-independent protein kinase C, extracellular signal-regulated protein kinase pathway distinct from the phosphatidylinositol-specific phospholipase C/calcium pathway. J. Neurosci. 19, 4211–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.