Background: LMAN1 is an important mammalian cargo receptor for endoplasmic reticulum (ER)-to-Golgi trafficking.
Results: Crystal structures pinpoint critical residues on LMAN1 for mannose binding, which is sensitive to Ca2+ concentration.
Conclusion: Changes in Ca2+ concentration can cause ligand release without disrupting the LMAN1·MCFD2 receptor complex.
Significance: The results shed new light on how LMAN1 binds and releases its cargo in the ER-to-Golgi transport pathway.
Keywords: Calcium-binding Proteins, Carbohydrate-binding Protein, Coagulation Factors, Endoplasmic Reticulum (ER), Golgi, Hemostasis, Membrane Trafficking, Protein Secretion, ERGIC, Hemorrhagic Disorders
Abstract
LMAN1 (ERGIC-53) is a key mammalian cargo receptor responsible for the export of a subset of glycoproteins from the endoplasmic reticulum. Together with its soluble coreceptor MCFD2, LMAN1 transports coagulation factors V (FV) and VIII (FVIII). Mutations in LMAN1 or MCFD2 cause the genetic bleeding disorder combined deficiency of FV and FVIII (F5F8D). The LMAN1 carbohydrate recognition domain (CRD) binds to both glycoprotein cargo and MCFD2 in a Ca2+-dependent manner. To understand the biochemical basis and regulation of LMAN1 binding to glycoprotein cargo, we solved crystal structures of the LMAN1-CRD bound to Man-α-1,2-Man, the terminal carbohydrate moiety of high mannose glycans. Our structural data, combined with mutagenesis and in vitro binding assays, define the central mannose-binding site on LMAN1 and pinpoint histidine 178 and glycines 251/252 as critical residues for FV/FVIII binding. We also show that mannobiose binding is relatively independent of pH in the range relevant for endoplasmic reticulum-to-Golgi traffic, but is sensitive to lowered Ca2+ concentrations. The distinct LMAN1/MCFD2 interaction is maintained at these lowered Ca2+ concentrations. Our results suggest that compartmental changes in Ca2+ concentration regulate glycoprotein cargo binding and release from the LMAN1·MCFD2 complex in the early secretory pathway.
Introduction
After undergoing folding and quality control in the endoplasmic reticulum (ER),3 many secreted proteins display distinct sorting signals that interact with ER export receptors (1–3). Such receptors are packaged into COPII-coated vesicles for anterograde transport to the Golgi. At their destinations, the receptors release their cargo proteins and recycle to the ER, potentially via COPI vesicles. The mammalian lectin LMAN1 (also called ERGIC-53) is an ER export receptor for a subset of glycoproteins, including coagulation factor V (FV) and factor VIII (FVIII) (4, 5), cathepsin C, cathepsin Z, and α1-antitrypsin (6–8). LMAN1 cycles between the ER and the ER-Golgi intermediate compartment (ERGIC). Crucially, mutations in LMAN1 cause the combined deficiency of FV and FVIII (F5F8D), an autosomal recessive bleeding disorder in which the plasma levels of FV and FVIII fall to 5–30% of normal (9).
LMAN1 is a type-I membrane protein whose ER luminal portion contains a carbohydrate recognition domain (CRD) and a putative α-helical domain that has been implicated in LMAN1 oligomerization (10). The single transmembrane helix connects the luminal domains of LMAN1 to a short cytosolic tail that contains a diphenylalanine ER exit motif, which interacts with COPII, and a dilysine ER retrieval motif, which interacts with COPI (11). After trimming of terminal glucose moieties by ER glucosidases-I and -II, properly folded glycoproteins contain the N-linked glycan Man9(GlcNAc)2, which is recognized by the LMAN1-CRD. Deletion of the CRD abolishes interactions between LMAN1 and FV/FVIII (12). In turn, inhibition of glycosylation of cathepsin Z or α1-antitrypsin markedly reduces the secretion of these proteins (13). Although glycan binding by LMAN1 is Ca2+-dependent (14), the details of the glycan-binding site and mode of binding have not been experimentally characterized. Moreover, LMAN1 interacts with a soluble luminal coreceptor, MCFD2, that is specifically required for ER export of FV/FVIII (5, 15), although not for export of cathepsins C/Z (16). Mutations in MCFD2 also give rise to combined deficiency of FV and FVIII (15). MCFD2, a double-EF-hand protein, depends on Ca2+ for its folding and interaction with LMAN1 (17, 18). MCFD2 binds to the LMAN1-CRD and also interacts directly with FV/FVIII (5).
Although the interaction between LMAN1-CRD and MCFD2 has been characterized in detail (12, 19, 20), the molecular mechanisms underlying cargo capture and release by LMAN1 are not as well understood. It is well established that receptor·cargo complexes travel through a pH gradient along secretory and endocytic pathways (21). Both the low density lipoprotein receptor and the transferrin receptor bind their cargo at neutral pH and release them in the mildly acidic environment of the early endosomes (22–24). The mannose-6 phosphate receptor binds cargo at pH 6.5 in the trans-Golgi network and releases it at pH 6 in the late endosome (25). Similarly, decreasing pH during trafficking from the ER to the ERGIC/Golgi has been proposed to trigger Ca2+ release from LMAN1, thus inducing release of glycoprotein cargo (26). However, there is no direct evidence for the acidification of pre-Golgi compartments such as the ERGIC. Moreover, a histidine residue in LMAN1 that was proposed to be a key pH sensor may not be directly involved in Ca2+ binding (14, 26).
In this study, we determined how the LMAN1-CRD binds to the terminal mannose groups of N-linked oligosaccharides on cargo glycoproteins. Furthermore, we characterized the pH and Ca2+ dependence of the interactions of the LMAN1-CRD with dimannose and with MCFD2. Our results suggest that, in the early secretory pathway, loading and unloading of FV and FVIII from LMAN1 may be directly modulated by compartmental changes in local Ca2+ concentration, whereas LMAN1 and MCFD2 remain associated as a complex.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Mutagenesis
The bacterial expression plasmid pET15-CRD was constructed by subcloning of a human LMAN1 cDNA fragment encoding the CRD (31–270) into the pET15b vector using NdeI and BamHI sites. Mammalian expression constructs of LMAN1 mannose-binding site mutants were made using the pED-FLAG-LMAN1 plasmid described previously (12). Mutagenesis was performed using the QuikChange mutagenesis kit (Agilent) and confirmed by DNA sequencing.
Expression and Purification of the LMAN1-CRD andMCFD2
pET15b-CRD was expressed in Escherichia coli BL21(DE3) cells and induced by 1 mm isopropyl-1-thio-β-d-galactopyranoside for a 5-h period. Harvested cells were lysed by sonication, and purification of LMAN1-CRD from inclusion bodies was performed by a denaturation-refolding method essentially as described previously (19). Briefly, inclusion bodies were extensively washed and solubilized in 6 m guanidinium hydrochloride. Refolding was carried out in refolding buffer (50 mm Tris-HCl, pH 8.0, 0.4 m l-arginine, 5 mm reduced glutathione, and 0.5 mm oxidized glutathione) by rapid pulse dilution. Refolded protein was concentrated by ultrafiltration and further purified using a Superdex-75 gel filtration column (GE Healthcare). A previously described pET15b-MCFD2 construct was used to express His-tagged MCFD2 in E. coli BL21(DE3) cells (15). MCFD2 was affinity-purified by nickel affinity chromatography, digested with thrombin to remove the His tag, and further purified using a Superdex-75 gel filtration column.
Crystallographic Structure Determination of LMAN1-CRD·Man-α-1,2-Man Complex
Crystals of the LMAN1-CRD with Ca2+ and Man-α-1,2-Man were grown using the hanging drop vapor diffusion method over reservoirs containing 50 mm sodium cacodylate, pH 5.8–6.4, and 16.5–18.5% PEG 8000. Crystal diffraction data were collected at beamline 4.2.2, Advanced Light Source, Lawrence Berkeley National Laboratory (space group P1) and at beamline 24-ID-E, Advanced Photon Source, Argonne National Laboratory (space group P6). Diffraction data were reduced and integrated using d*TREK (27) or XDS/SCALA (28, 29). Structures of the P1 and P6 crystal forms, respectively, contained six and one LMAN1-CRD·Man-α-1,2-Man complexes and were solved to 2.7 and 2.42 Å resolution. Structure solution was carried out by molecular replacement using the Ca2+-bound structure of human LMAN1 (Protein Data Bank (PDB) ID 3A4U, chain A) (19) using PHASER (30). Molecular replacement solutions were subjected to automated rebuilding in PHENIX (31) with RESOLVE (32) and manual model rebuilding in COOT (33). PHENIX was used to refine both structures. Each chain within the P1 structure contains one Man-α-1,2-Man with the same orientation and binding mode for all chains. Molecular structure figures were generated with PyMOL. Stereochemical analyses of the P1 and P6 structures were completed with MolProbity (34). All residues are within the favored regions of the Ramachandran plot and no poor rotamers or Cβ deviations were found in either structure. Refinement and stereochemical statistics are shown in Table 1.
TABLE 1.
Data collection and refinement statistics
APS, Advanced Photon Source; ALS, Advanced Light Source. — indicates not detectable.
| Data collection | ||
| Beam line | APS 24-ID-E | ALS 4.2.2 |
| Wavelength (Å) | 0.9792 | 0.9790 |
| Space group | P6 | P1 |
| Cell dimensions | ||
| a, b, c (Å) | 110.3, 110.3, 38.3 | 39.3, 111.5, 111.4 |
| α, β, γ (°) | 90, 90, 120 | 60.1, 89.2, 86.3 |
| Resolution (Å)a | 47.75-2.42 (2.55-2.42) | 39.19-2.70 (2.80-2.70) |
| Rmergea,b | 0.082 (0.437) | 0.067 (0.299) |
| 〈I/σI〉a | 23.0 (7.7) | 6.4 (2.0) |
| Completeness (%)a | 100.0 (100.0) | 96.9 (97.8) |
| Redundancya | 10.7 (10.9) | 1.91 (1.94) |
| No. of reflections | 111,907 | 83,229 |
| No. of unique reflections | 10,429 | 43,492 |
| Refinement | ||
| Resolution (Å) | 38.33-2.42 | 37.19-2.70 |
| No. of reflections for refinement | 10,427 | 43,446 |
| Rwork/Rfree | 0.192/0.212 | 0.246/0.284 |
| No. of atoms (Average B Factors, Å2) | 1,925 | 11,508 |
| Protein | 1,799 (52.0) | 10,851 (46.8) |
| Water | 88 (58.5) | 333 (46.3) |
| Calcium | 2 (63.3) | 12 (60.0) |
| 2α-Mannobiose | — | 270 (63.8) |
| α-d-Mannose | 24 (63.7) | — |
| Glycerol | 12 (71.1) | 42 (62.4) |
| Wilson estimated B factor (Å2) | 37.5 | 62.2 |
| r.m.s.c deviations | ||
| Bond lengths (Å) | 0.008 | 0.012 |
| Bond angles (°) | 0.979 | 1.339 |
| Ramachandran plot statistics | ||
| Favored regions % | 96.9% (220/227) | 96.3% (1,320/1,371) |
| Allowed regions % | 3.1% (7/227) | 3.7% (51/1,371) |
| MolProbity validation statistics | ||
| MolProbity clash score, percentile | 11.24, 92% | 13.40, 95% |
| MolProbity score, percentile | 1.75, 98% | 1.89, 99% |
| PDB ID | 4GKY | 4GKX |
a Values in parentheses are for the highest resolution shell.
b The merging R factor is defined as Rmerge = ΣhklΣi|Ii(hkl) − I(hkl)|/ΣhklΣiIi(hkl).
c r.m.s., root mean square.
Isothermal Titration Calorimetry
Isothermal titration calorimetry (ITC) experiments were performed using an ITC200 calorimeter (GE Healthcare). To measure the interaction of the LMAN1-CRD with Ca2+ or Man-α-1,2-Man, the LMAN1-CRD (150–200 μm) in the sample cell (200 μl) was titrated with CaCl2 (20 injections of 2 μl) or Man-α-1,2-Man (14 injections of 3 μl) in the injection syringe at room temperature with stirring at 1000 rpm. To measure the interaction of the LMAN1-CRD with MCFD2, MCFD2 (∼10 μm) in the sample cell was titrated (20 injections of 2 μl) with the LMAN1-CRD (∼100 μm) in the presence of Ca2+ with indicated concentration. Each experiment was performed at least twice. Data from control experiments (using only buffer in the sample cell) were subtracted from the integrated heat data prior to curve fitting and analysis. All data were analyzed using Origin software.
Mannose Binding Assay
FLAG-tagged wild-type or mutant LMAN1 construct was transfected into COS-1 cells. Thirty-six hours after transfection, the membrane fraction of the cell lysate was incubated with d-mannose-agarose beads as described previously (12). Bound LMAN1 was detected by Western blot analysis.
Cell Culture, Transfection, and Metabolic Labeling
COS-1 cell culture, transfection of LMAN1 and FVIII constructs and, metabolic labeling with [35S]methionine/cysteine were performed as described previously (5).
Cross-linking and Immunoprecipitation
Cross-linking using dithio-bis(succinimidyl propionate) (DSP) (Thermo Scientific) and immunoprecipitation were carried out as described previously (15, 18). Immunoprecipitates were separated on a denaturing 4–12% Criterion Bis-Tris gel (Bio-Rad) and visualized by exposing to a Kodak BioMax film using a BioMax TranScreen LE.
RESULTS
Structural Basis of Mannose Recognition by the LMAN1-CRD
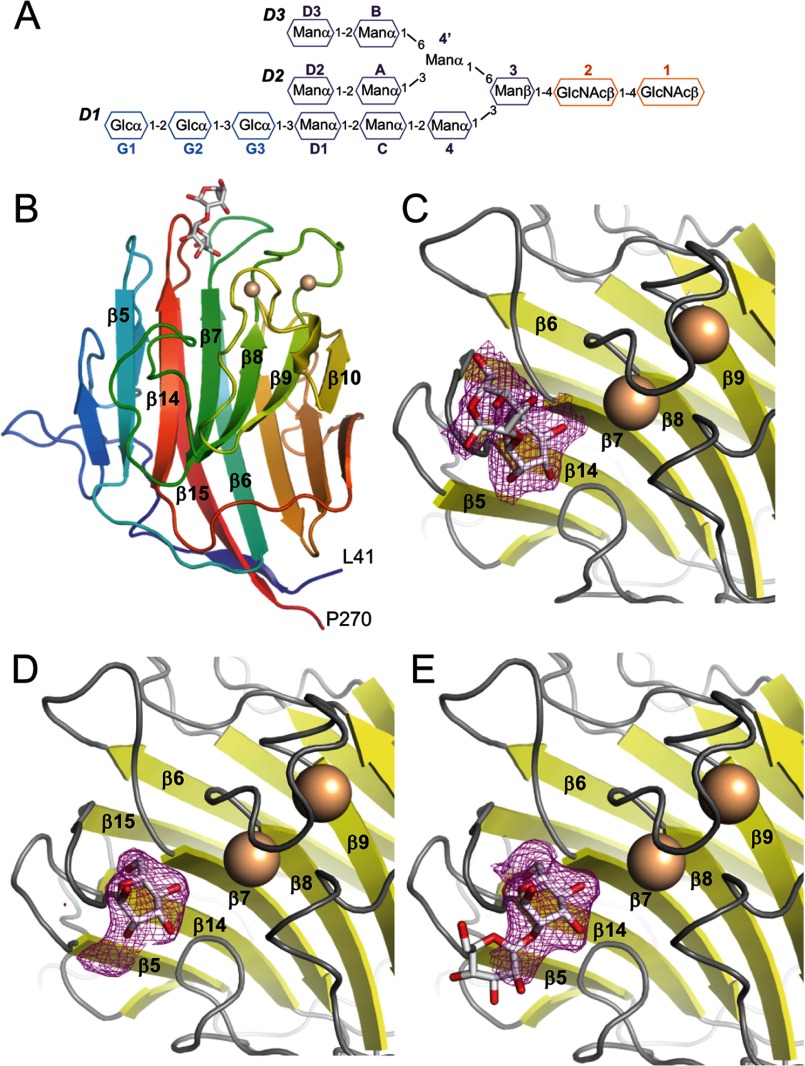
The termini of high mannose glycans contain Man-α-1,2-Man, linked via α-1,2, α-1,3 and α-1,6 linkages to successive mannose groups in the D1, D2, and D3 arms, respectively (Fig. 1A). To obtain greater insight into how LMAN1 binds to glycosylated cargo, we carried out crystallization trials of the purified LMAN1-CRD with Ca2+ and Man-α-1,2-Man, Man-α-1,3-Man, or Man-α-1,6-Man. We obtained crystals only in the presence of Man-α-1,2-Man; these crystals grew in space groups P1 and P6 (Table 1). The resulting LMAN1-CRD structures were solved at 2.7 and 2.42 Å resolution for the P1 and P6 forms, respectively. The structures closely resemble previously solved structures of Ca2+-bound, mannose-free rat and human LMAN1-CRD (14, 19, 20) with backbone root mean square deviation values of less than 0.35 Å (data not shown), but differ from Ca2+-free LMAN1-CRD (35). Ca2+ binding restructures the loop and α-helix between β-strands 9 and 10, and also reorients and restructures the flexible β8-β9 loop. Both loops contribute side chains that contact the carbohydrate and form one side of the principal Man-α-1,2-Man-binding region (Figs. 1 and 2). The opposite side of the pocket is formed by adjacent residues from the β4-β5, β6-β7, and β14-β15 loops, which undergo little change upon Ca2+ binding and mannose binding.
FIGURE 1.
Crystal structures of the LMAN1-CRD bound to Ca2+ and Man-α-1,2-Man. A, schematic representation of the glucosylated form of high mannose N-linked glycans, Glc3Man9(GlcNAc)2, added to secreted and membrane glycoproteins in the endoplasmic reticulum. B, overall structure of the LMAN1-CRD in the P1 crystal form; the P6 form of the protein is very similar. The two bound Ca2+ ions are shown as beige spheres, and Man-α-1,2-Man is shown in stick representation. Key secondary structure elements involved in glycan and calcium binding are labeled. C, Fo − Fc omit map density corresponding to the bound Man-α-1,2-Man in the P1 crystal, contoured at 2σ. D, Fo − Fc omit density corresponding to the bound Man-α-1,2-Man in the P6 crystal, contoured at 2σ. The density allows only one mannose group to be built. E, the “extra” density in the P6 crystal form may correspond to part of a mobile second mannose.
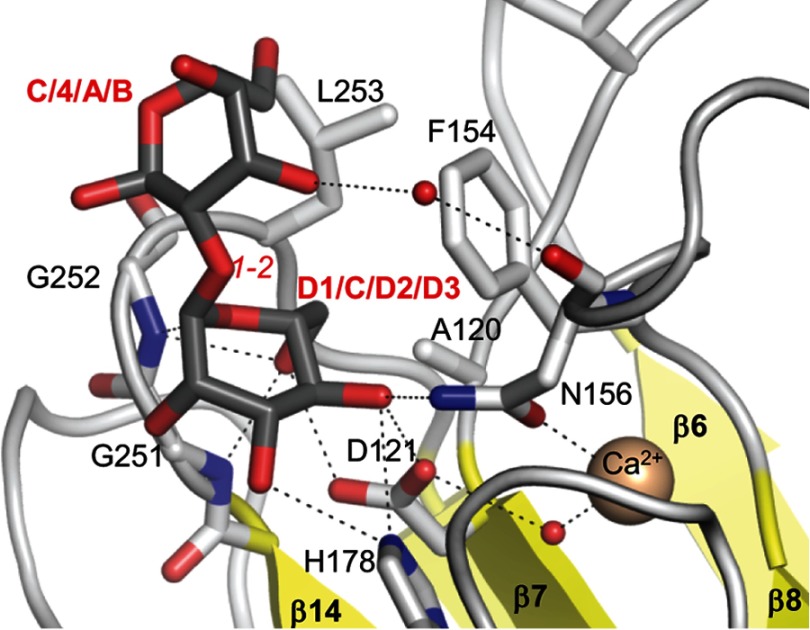
FIGURE 2.
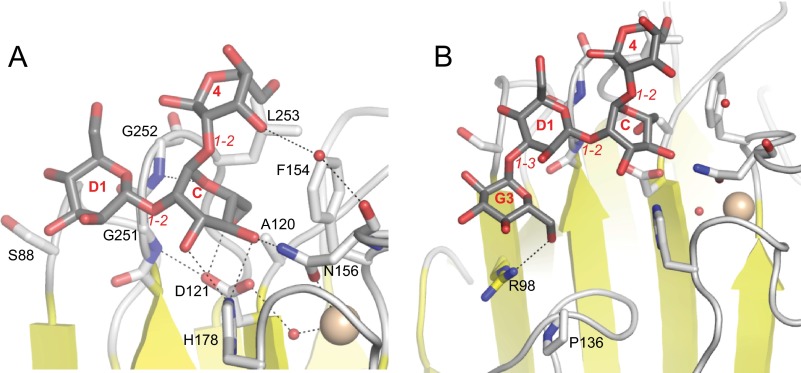
Details of Man-α-1,2-Man binding to the LMAN1-CRD. A close-up of the Man-α-1,2-Man in the P1 form and its interacting residues is shown. The mannoses are colored with gray carbons, and LMAN1 residues are shown with white carbons. Dotted lines represent potential hydrogen bonds/polar interactions. Small red spheres correspond to water molecules, and the larger beige sphere represents one of the Ca2+ ions. Italicized red text designates the inter-mannose linkage. Bold text designates the moieties of Man9(GlcNAc)2 to which the respective mannoses may correspond.
In the P1 crystal form, the nonreducing mannose of Man-α-1,2-Man occupies a characteristic pocket flanked by the β6-β7, β8-β9, and β14-β15 loops, which we refer to as the “central” mannose-binding pocket. The reducing-end mannose extends along the β6-β7 and β14-β15 loops (Fig. 1, B and C, and Fig. 2). In the P6 crystal form, however, there is strong electron density at the central position, whereas additional, weaker density extends toward β5, away from the central pocket and the Ca2+-binding sites. We therefore built a single mannose into the strong density at the central pocket (Fig. 1D), which likely corresponds to the reducing-end mannose of Man-α-1,2-Man. The additional density is not of sufficient quality to accurately define a second mannose; however, it likely corresponds to the nonreducing mannose (Fig. 1E), which appears to be highly mobile in the P6 crystal form.
The central mannose found in both structures engages in numerous polar interactions between its 3-, 4-, and 6-hydroxyl groups and both side-chain and main-chain atoms of LMAN1 (Fig. 2). The 3-OH and 4-OH groups interact with the side chains of His-178 and Asn-156, which are repositioned upon Ca2+ binding. The 4-OH and 6-OH groups interact with the side chain of Asp-121. The main-chain amides of Gly-251 and Gly-252 interact with the 6-OH and the ring oxygen, respectively. Finally, the 5- and 6-carbons pack against a “wall” of aliphatic side chains formed by Ala-120, Phe-154 (which also moves into position upon Ca2+ binding), and Leu-253 (Fig. 2). The central mannose is restricted between the β8-β9 and β14-β15 loops, leaving only the 1-OH and 2-OH positions available for linking to other sugars. The reducing-end mannose of the P1 crystal form interacts via a water molecule with the main-chain carbonyl of Asn-156. However, this mannose is primarily oriented by van der Waals interactions with LMAN1 Ala-120, Phe-154, Gly-252, and Leu-253 (Fig. 2).
Validation of LMAN1 Residues That Are Critical for Mannose Binding
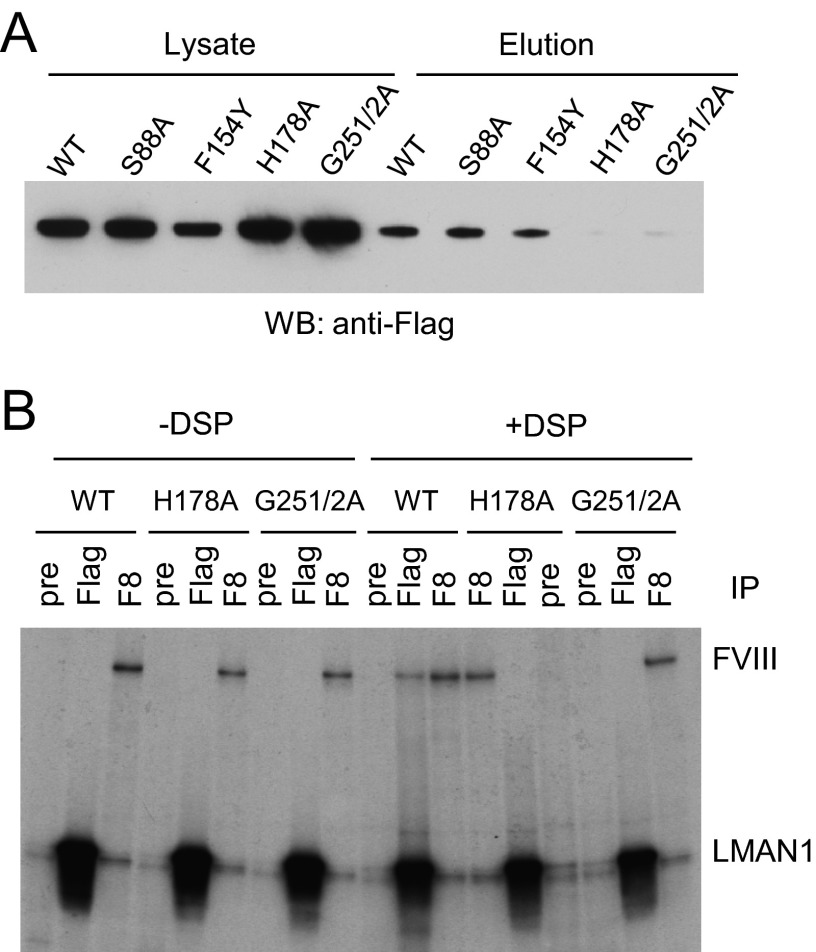
To further investigate the importance of amino acids near the central mannose-binding site, we introduced the mutations F154Y, H178A, G251A/G252A, and S88A into full-length LMAN1. The first three mutations potentially alter carbohydrate binding without affecting the ability of LMAN1 to bind Ca2+. The fourth mutant, S88A, corresponds to a residue in the structurally related lectin VIP36 that interacts with the D1 mannose of Man9(GlcNAc)2 (36). We analyzed the mutants using a cell-based mannose binding assay. COS-1 cells transfected with the mutant constructs were lysed, and the solubilized LMAN1-containing membrane fraction was incubated with mannose-agarose beads, eluted, and detected by Western blotting. As shown in Fig. 3A, H178A and G251A/G252A mutants did not bind mannose beads, whereas S88A and F154Y bound the mannose beads in a similar manner as wild-type LMAN1. These results indicate that His-178, Gly-251, and Gly-252 are critical amino acids for mannose binding, in agreement with our structural data.
FIGURE 3.
Effects of select amino acid mutations in LMAN1 on the mannose binding and the interaction with FVIII. A, mannose binding capabilities of the wild type and the indicated LMAN1 mutants. LMAN1 in cell lysate and eluted from mannose beads was detected by Western blot analysis. B, interactions of LMAN1 mutants with FVIII. COS-1 cells were co-transfected with FVIII and FLAG-tagged wild-type LMAN1 or the indicated mutant LMAN1. Cells were metabolically labeled and incubated with or without the cross-linking agent DSP before lysis. Cell lysates were immunoprecipitated with preimmune serum (pre) as a negative control, anti-FLAG (Flag) for LMAN1, and anti-FVIII (F8) for FVIII.
LMAN1 Mutants That Alter Mannose Binding Also Disrupt FVIII Binding
We previously showed that the CRD of LMAN1 interacts with FV and FVIII (12). Two point mutations in the CRD of LMAN1, N156A and D181A, abolish these interactions (12). However, these two amino acids interact directly with Ca2+, and the mutations disrupt carbohydrate binding indirectly as a consequence of impaired Ca2+ binding. We therefore used a previously validated cross-linking/co-immunoprecipitation (IP) assay (15, 18) to characterize the interactions of specific mannose-binding mutants with FVIII overexpressed in COS-1 cells. Metabolically labeled cells were treated on ice with the membrane-permeable, thiol-cleavable cross-linker DSP before lysis and IP. Cross-linking of FVIII with the H178A or G251A/G252A mutants was significantly reduced as compared with wild-type LMAN1 (Fig. 3B, compare the amount of FVIII that co-immunoprecipitates with LMAN1 in the presence and absence of DSP). These results show that specific mannose recognition at the crystallographically defined binding site is required for proper LMAN1/cargo interactions in cells.
LMAN1 Interactions with High Mannose Glycans
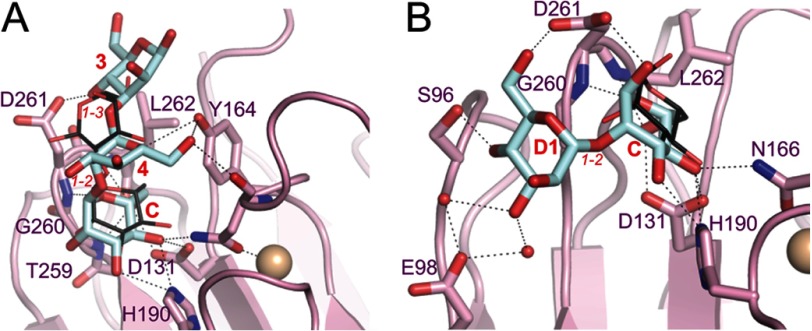
Kato and co-workers (37) previously demonstrated that the LMAN1-CRD exhibits only limited specificity for different Man9(GlcNAc)2 substructures in vitro. The putative Golgi-to-ER trafficking receptor VIP36 (38) contains a carbohydrate recognition domain that is structurally similar to the LMAN1-CRD. However, VIP36 exhibits a strong selectivity for the complete D1 arm over the D2 and D3 arms of Man9(GlcNAc)2. In a crystal structure of VIP36-CRD with Man-α-1,2-Man-α-1,3-Man (36), the reducing α-1,2-linked mannose is in a different orientation than its equivalent in our P1 structure (Fig. 4A). Both the α-1,2-linked and the α-1,3-linked mannoses make polar interactions with VIP36 residues (Asp-261 and Tyr-164) that are not conserved in LMAN1 (Gly-252 and Phe-154). A different crystal structure of VIP36-CRD with Man-α-1,2-Man is comparable with our P6 structure (Fig. 4B). However, the nonreducing mannose in the former is well resolved and interacts with VIP36 Ser-96 and VIP36 Asp-261, the latter of which is not conserved in LMAN1. Furthermore, mutation of LMAN1 Ser-88, the equivalent of VIP36 Ser-96, has no effect on LMAN1 interaction with mannose (Fig. 3A). The overall higher numbers of polar contacts in the VIP36 structures correlate with the greater specificity of VIP36 (37).
FIGURE 4.
Comparison of the carbohydrate-binding sites of VIP36 and LMAN1. In both panels, dotted lines designate putative polar interactions, small red spheres correspond to water molecules, and the beige spheres represent Ca2+ ions. Italicized red text designates inter-mannose linkages. Bold text designates the moieties of Man9(GlcNAc)2 to which the respective mannoses correspond (for the VIP36 structures). A, close-up of the carbohydrate-binding site of VIP36 with Man-α-1,2-Man-α-1,3-Man (PDB ID 2E6V). VIP36 residues are shown with pink carbons, and the carbons of the mannotriose are in blue. For comparison, the Man-α-1,2-Man from our P1 structure is superimposed (thin sticks; dark gray carbons). B, close-up of VIP36 bound to Man-α-1,2-Man (PDB ID 2DUR) colored as in panel A. For comparison, the mannose from our P6 structure is superimposed (thin sticks; dark gray carbons).
The LMAN1-CRD did not cocrystallize with Man-α-1,3-Man or Man-α-1,6-Man. We therefore used the ITC assay to measure the binding of d-mannose, Man-α-1,2-Man, Man-α-1,3-Man, or Man-α-1,6-Man to Ca2+-bound LMAN1-CRD at pH 7.4. We readily measured specific binding of the LMAN1-CRD to Man-α-1,2-Man with modest affinity (Fig. 5, A and C). The affinity of monomannose was too low to measure, as were the affinities of Man-α-1,3-Man and Man-α-1,6-Man (data not shown). We verified the specificity of the Man-α-1,2-Man binding signal by titrating Man-α-1,2-Man against the LMAN1-CRD H178A mutant, which showed only a small background signal (Fig. 5A). Docking of Man-α-1,3-Man and Man-α-1,6-Man onto the LMAN1-CRD suggests that 1,3- and 1,6-linked mannose moieties do not substantially pack against the CRD, in accordance with the lack of observed binding (data not shown).
FIGURE 5.
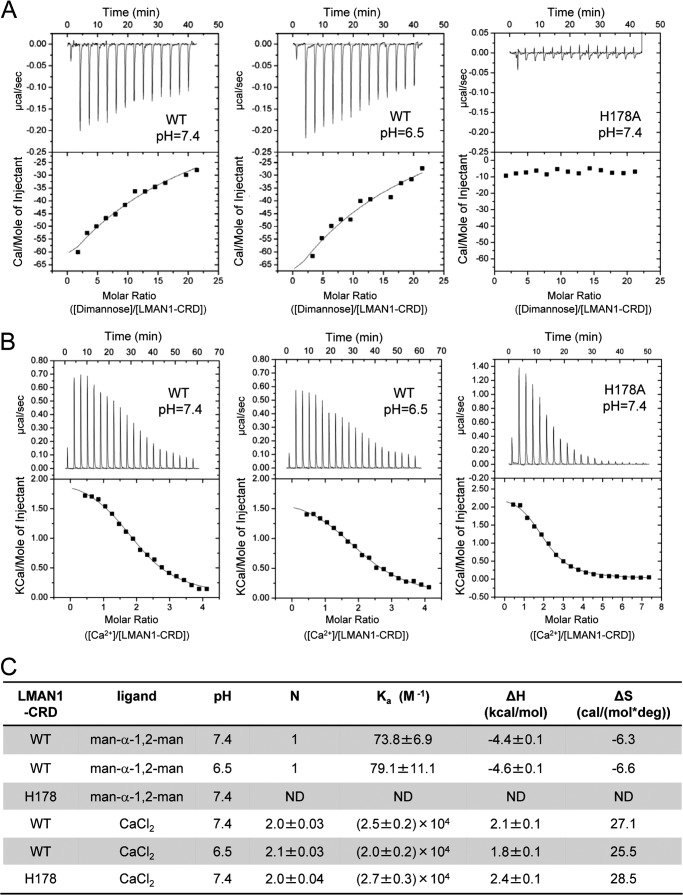
Analysis of mannose and Ca2+ binding to the LMAN1-CRD by isothermal titration calorimetry. A, the wild type (WT) (200 μm) and the H178A mutant of the LMAN1-CRD (150 μm) were titrated with 20 mm Man-α-1,2-Man in 10 mm HEPES, 150 mm NaCl, and 5 mm CaCl2 at pH 7.4 or pH 6.5 (WT only). To avoid curve-fitting errors associated with fitting the integrated ITC data from weakly binding systems (49), we fixed the stoichiometry of the complex at one Man-α-1,2-Man per LMAN1-CRD molecule, in agreement with our crystal structures. B, the WT (150 μm) and the H178A mutant (140 μm) of the LMAN1-CRD were titrated with 3 mm (WT) or 5 mm (H178A) CaCl2 in 10 mm HEPES and 150 mm NaCl at pH 7.4 or pH 6.5 (WT only). C, summary of parameters obtained from the ITC experiments for mannose and Ca2+ binding. ND, not determined; deg, degree.
We therefore hypothesize that the LMAN1-CRD binds to Man-α-1,2-Man units on any of the three arms of Man9(GlcNAc)2, in agreement with previous in vitro binding data (37). The two mannose groups in our P1 structure may therefore correspond to the D1-C, C-4, D2-A, or D3-B Man-α-1,2-Man units (Fig. 1A). Based on the weak electron density that flanks the central mannose-binding site in our P6 crystal form, we also modeled the binding of the trimannose segment Man-α-1,2-Man-α-1,2-Man from the D1 arm (Fig. 6A).
FIGURE 6.
Structure-based models of larger oligosaccharides bound to the LMAN1-CRD. Modeled oligosaccharides are colored with gray carbons, and LMAN1 residues are shown with white carbons. Italicized red text designates the inter-mannose linkage. Bold text designates moieties of Man9(GlcNAc)2 or GlcMan9(GlcNAc)2 to which the respective sugars may correspond. A, model of Man-α-1,2-Man-α-1,2-Man from the D1 arm bound to the LMAN1-CRD. B, model of Glc-α-1,3-Man-α-1,2-Man-α-1,2-Man from a monoglucosylated D1 arm bound to the LMAN1-CRD.
It was previously shown that monoglucosylation of the D1 arm is tolerated by the LMAN1-CRD (36, 37). Unlike VIP36, the LMAN1-CRD readily accommodates an additional α-1,3-linked glucose attached to the D1 mannose, near a pocket formed by Arg-98 and the long β7-β8 loop (Fig. 6B); additional glucose moieties extend past this region and appear unable to interact with the LMAN1-CRD.
Effects of pH and Ca2+ on LMAN1-CRD/Mannobiose Interactions
Because changes in local pH in the early secretory pathway have been proposed to regulate cargo glycoprotein loading and unloading from LMAN1 (26), we examined how pH regulates the binding of mannose to the LMAN1-CRD in vitro. Using ITC, we first measured the interactions of the LMAN1-CRD with Ca2+ at pH 7.4 and pH 6.5 (Fig. 5, B and C), which correspond to estimated pH values in the ER and in the cis-Golgi (21). When fit with a single-site binding model, the ITC data yielded the expected stoichiometry of two Ca2+ ions per LMAN1-CRD with an affinity of Kd = 40–50 μm. Moreover, the interaction of the LMAN1-CRD with Ca2+ is insensitive to pH in this range. It was previously suggested that His-178 acts as a pH-dependent regulator of Ca2+ binding by LMAN1 (26). We found, however, that the LMAN1-CRD H178A mutant bound to Ca2+ with a similar affinity as wild-type LMAN1-CRD (Fig. 5, B and C), suggesting that His-178 does not regulate Ca2+ binding. Similar to Ca2+ binding, Man-α-1,2-Man binding was little changed between pH 7.4 and 6.5 (Fig. 5, A and C). We also note that we were able to obtain crystals of LMAN1-CRD bound to Man-α-1,2-Man at pH values between 5.8 and 6.4.
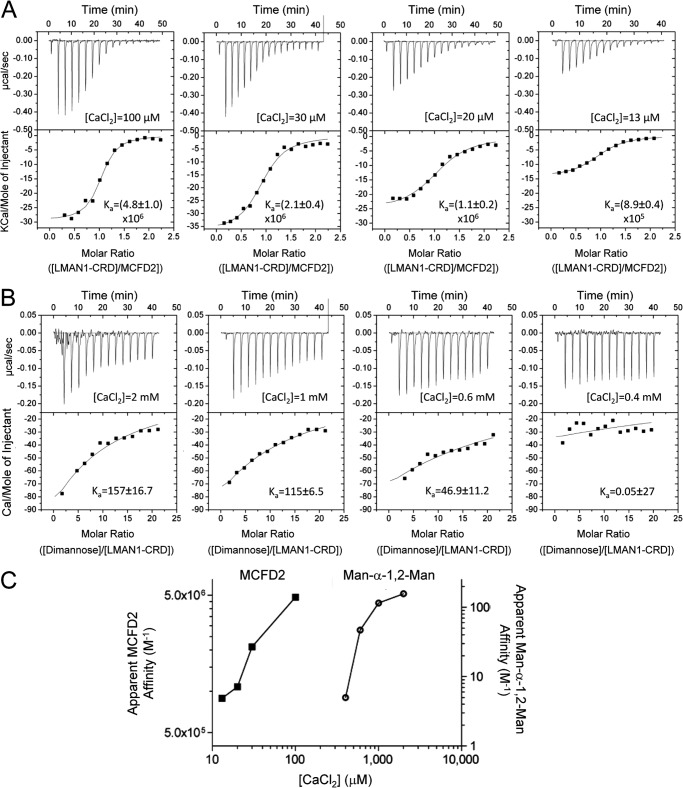
Because the Ca2+ concentration in the ERGIC may be significantly lower than that of the ER (39), we investigated the Ca2+ dependence of LMAN1/MCFD2 and LMAN1/mannobiose interactions. At 100 μm Ca2+, the purified LMAN1-CRD and MCFD2 interacted with an apparent affinity (Ka) of ∼5 × 106 m−1 (Fig. 7A), identical to values previously reported at 100 mm Ca2+ (19). A further drop in the Ca2+ concentrations decreased but did not eliminate binding; the apparent Ka values remained above 106 m−1 at 20 μm Ca2+ (Fig. 7A). In contrast, the LMAN1-CRD exhibited relatively little Man-α-1,2-Man binding even at 400–600 μm Ca2+ as compared with the binding observed at 2–5 mm Ca2+ (Fig. 7B). The difference in the dependence on Ca2+ is illustrated in Fig. 7C. Our data suggest that the LMAN1-CRD interaction with Man-α-1,2-Man is sensitive to low Ca2+ concentrations at which the LMAN1/MCFD2 interaction is maintained. A drop in compartmental Ca2+ during ER-to-ERGIC traffic may thus trigger the dissociation of glycoprotein cargo from the LMAN1-CRD in the ERGIC, without disrupting the LMAN1/MCFD2 interaction.
FIGURE 7.
Ca2+ concentration has a significant effect on mannose binding to the LMAN1-CRD. A, binding of the LMAN1-CRD to MCFD2 at various Ca2+ concentrations. The LMAN1-CRD (110 μm) was titrated into MCFD2 (10 μm) in buffers containing 10 mm HEPES (pH 7.4) and 150 mm NaCl with the indicated concentrations of CaCl2. B, binding of the LMAN1-CRD to Man-α-1,2-Man at various Ca2+ concentrations. The LMAN1-CRD (200 μm) was titrated with Man-α-1,2-Man (20 mm) in 10 mm HEPES (pH 7.4), 150 mm NaCl in the presence of the indicated concentrations of CaCl2. C, summary of Ca2+ dependence of the LMAN1-CRD binding to MCFD2 and Man-α-1,2-Man. The apparent affinities (Ka) of the LMAN1-CRD·MCFD2 binding (left) and the LMAN1-CRD·Man-α-1,2-Man binding (right) were plotted against the experimental CaCl2 concentrations.
Direct Comparison of Calcium Sensitivity between the LMAN1·MCFD2 Binding and the LMAN1·Mannose Binding
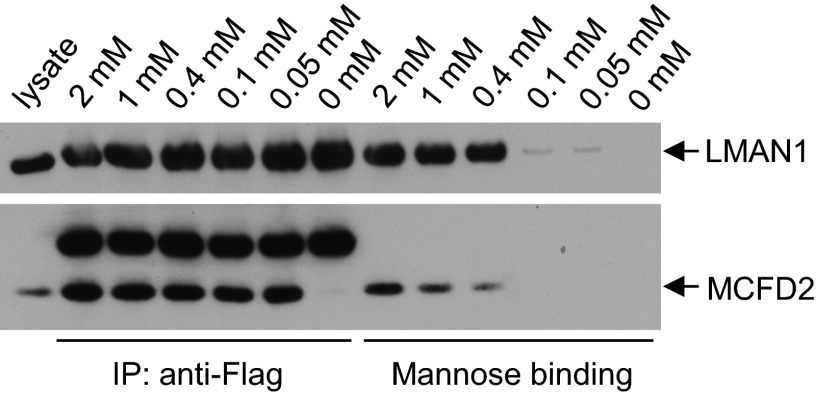
Our ITC assays have used the purified CRD of LMAN1. The full-length LMAN1 exists as a hexamer in cells. We would like to directly compare calcium sensitivities between the LMAN1·MCFD2 binding and the LMAN1·mannose binding in cells. To this end, we co-transfected COS-1 cells with constructs expressing FLAG-LMAN1 and MCFD2. FLAG-LMAN1 expressed from transfected plasmid oligomerized normally (12). We lysed the cells with buffer containing varying concentrations of CaCl2 and simultaneously performed co-IP and mannose binding assays from the same cell lysates (Fig. 8). The co-IP between LMAN1 and MCFD2 remains robust in [Ca2+] of 0.05 mm, the lowest CaCl2 concentration tested. No co-IP was detected in the absence of CaCl2, confirming the calcium dependence of the LMAN1·MCFD2 complex formation. In contrast, mannose binding by LMAN1 dropped precipitously between 0.4 and 0.1 mm of CaCl2. These results provide direct evidence suggesting that at low but physiologically relevant Ca2+ concentrations, the full-length LMAN1 can remain bound to MCFD2, whereas the LMAN1/mannose interaction is abolished.
FIGURE 8.
Direct comparison of calcium sensitivity between the LMAN1·MCFD2 binding and the LMAN1·mannose binding. COS1 cells were co-transfected with LMAN1-FLAG and MCFD2 constructs and lysed in buffers containing different concentrations of CaCl2. Cell lysates were subject to IP with an anti-FLAG antibody or the mannose binding assay. Proteins that were bound to protein G beads (left half of the gels) and to mannose beads (right half of the gels) were eluted and detected by Western blot analyses with anti-FLAG and anti-MCFD2 antibodies. The bands above MCFD2 in the anti-FLAG IP experiment represent the light chain of IgG.
DISCUSSION
Mutations in LMAN1 and MCFD2 lead to defective sorting and transport of FV and FVIII in the early secretory pathway (15, 18). LMAN1 exists primarily, possibly exclusively, as a hexamer in cells, and hexamerization is required for proper cargo export (10, 40). The LMAN1 hexamer, presumably associated with six MCFD2 coreceptor molecules, binds high mannose-glycosylated FV and FVIII molecules that have passed the ER quality control cycle. The receptor·cargo complexes traffic via COPII-coated vesicles, and cargo is released in the ERGIC, a distinct organelle that functions as a sorting station in the ER-Golgi traffic (11). Although protein epitopes in the cargo may also be important (13), high mannose glycans on the cargo are crucial for cargo interactions with LMAN1. We characterized a central mannose-binding site, part of which is constitutively present, whereas another part is formed by Ca2+-binding loops. Our structures clearly pinpoint the amino acids involved in carbohydrate recognition and rationalize how it is regulated by Ca2+ binding. Unlike previously characterized N156A, D181A, and D152S mutants (12, 26), the H178A and G251A/G252A mutants exhibit impaired carbohydrate binding (Figs. 3 and 5A), whereas still retaining Ca2+ near the mannose-binding pocket (Fig. 5B). These mutants are unable to interact with FVIII in cells (Fig. 3B), providing direct evidence that specific recognition of mannose moieties at our crystallographically defined central binding site is required.
Our crystallographic and in vitro binding data also rationalize the limited in vitro selectivity of LMAN1 for the different glycan arms (37) as the LMAN1-CRD may be able to interact with any of the four Man-α-1,2-Man moieties in the high mannose glycans, whereas selecting against Man-α-1,3-Man and Man-α-1,6-Man. This contrasts with the previously characterized preference of the related cargo receptor VIP36 for the D1 arm of Man9(GlcNAc)2 (36). In addition, LMAN1 and VIP36 exhibit different conformations at a pocket formed by β5 and the β7-β8 loop, which could accommodate a terminal glucose moiety in LMAN1 (Fig. 6B) but not in VIP36 (37). Glucosylation of the D1 arm targets glycoproteins to the ER glycoprotein quality control cycle by conferring binding to calnexin/calreticulin (41). LMAN1 may transiently bind folded glycoproteins in the ER to assist in the removal of terminal glucose by ER-resident glucosidase-II. Alternately, this binding may allow for an early escape from the ER quality control cycle before glucose trimming is complete. The functional significance of this structural difference between LMAN1 and VIP36 remains to be investigated further.
Although the measured affinity of the LMAN1-CRD/Man-α-1,2-Man interaction is modest, FV/FVIII binding to the LMAN1·MCFD2 complex could be stronger in the physiological context. It was reported that the affinity between the LMAN1-CRD and the full Man9(GlcNAc)2 was higher than the affinity of the LMAN1-CRD with Man-α-1,2-Man (37). Additional avidity may be gained from the interaction of cargo glycans and protein epitopes with both LMAN1 and MCFD2 forming ternary complexes. Finally, the LMAN1·MCFD2 hexamer may gain avidity by simultaneously engaging several glycosylation sites and epitopes on one cargo protein. Nevertheless, our mutagenesis results show that the integrity of the central mannose-binding site of LMAN1 is necessary for glycoprotein engagement within the cell (Fig. 3).
The pH of the ER is estimated to be ∼7.2–7.4, whereas that of the cis-Golgi is ∼6.4–6.6 (21). The ERGIC likely exhibits an intermediate pH, estimated at 6.5–7.2 (42). We detected only minor differences in the interaction between the LMAN1-CRD and either Ca2+ or Man-α-1,2-Man in this pH range (Fig. 5). Moreover, a mutation of His-178, a previously suggested pH sensor for Ca2+ binding (26), had no effect on Ca2+ binding by the LMAN1-CRD (Fig. 5B). These results suggest that pH changes from the ER to the Golgi do not play a significant role in regulating cargo loading and release. We hypothesize that changes in free Ca2+ concentration in the early secretory pathway directly modulate mannose binding. Indeed, the LMAN1-CRD interaction with Man-α-1,2-Man is highly sensitive to Ca2+ concentrations in the submillimolar range (Fig. 7). The Ca2+ concentration of the ER is estimated to be in the low millimolar range, although its distribution is heterogeneous (43). In contrast, the Ca2+ concentration in the Golgi appears to be substantially lower, estimated to be ∼0.3 mm (44, 45). Although the precise luminal Ca2+ concentration of the ERGIC is unknown, several lines of evidence suggest that it is much lower than in the ER and may even be lower than that of the Golgi. The ERGIC is formed by homotypic fusion of COPII vesicles (39, 46). However, the ER Ca2+ pump SERCA (sarcoplasmic reticulum Ca2+-ATPase) is not packaged into COPII vesicles in budding reactions. Nonspecific leaks may quickly deplete Ca2+ in the ERGIC (39). Consistent with very low Ca2+ concentration in the ERGIC, high-resolution Ca2+ mapping studies did not detect any Ca2+ in the ERGIC (47).
Interaction between LMAN1 and MCFD2 is mediated by N-terminal elements from LMAN1 and by the two Ca2+-binding EF-hand domains of MCFD2 (12, 19, 20). The Ca2+-binding sites of LMAN1 are located distally from its MCFD2-interacting site (19, 20), and mutations that disrupt Ca2+ binding to LMAN1 have no effect on MCFD2 binding (12). However, Ca2+ regulates the conformations of the EF-hand domains of MCFD2 (17), and LMAN1/MCFD2 interactions are Ca2+-dependent (5). Previous studies showed that the half-lives of LMAN1 and MCFD2 are similar, suggesting that the receptor complex does not dissociate during multiple cycles of shuttling between the ER and the ERGIC/cis-Golgi (5). Although our results suggest that Ca2+ plays a major role in LMAN1 cargo release, the interaction of LMAN1 with MCFD2 remains strong (above 106 m−1) at Ca2+ levels that do not favor LMAN1-CRD/glycan interactions (Fig. 7). Therefore, a drop of Ca2+ level in the ERGIC lumen may disrupt the interaction between LMAN1 and FV/FVIII and result in the discharge of FV/FVIII, whereas the LMAN1·MCFD2 receptor complex remains intact. MCFD2 is also essential for the transport of FV/FVIII (15). We have previously shown that MCFD2 interacts with FV and FVIII and that this interaction is independent of LMAN1 binding (5, 18). We speculate that MCFD2 captures the FV/FVIII cargo in the ER lumen and that this binding is reinforced by additional interaction of LMAN1 with glycans on the cargo. Cargo interactions with both LMAN1 and MCFD2 are required to form a stable ternary complex for efficient ER exit. It is not clear whether the interaction between MCFD2 and FV/FVIII is also weakened in the ERGIC as the effects of pH and Ca2+ concentration on the interaction between MCFD2 and FV/FVIII are currently uncharacterized.
Our results do not rule out the possibility of alternative or additional mechanisms for cargo release. For example, the LMAN1·MCFD2 cargo receptor may dissociate along with the cargo in the ERGIC if the local Ca2+ concentration is sufficiently low (i.e. low or submicromolar). After moving to the cis-Golgi, LMAN1 may reassociate with MCFD2 (48). However, this model requires additional elements to restrict the movement of MCFD2 in the cis-Golgi to prevent it from diffusing away from the membrane-bound LMAN1. In addition, the α-1,2-linked mannose groups on cargo proteins may be rapidly trimmed by the Golgi mannosidases, disrupting the ability of cargo to interact with LMAN1. Further investigation of LMAN1/cargo interactions using full-length LMAN1 hexamers and full-length, glycosylated FV/FVIII promises to yield a better functional understanding of FV/FVIII trafficking in the early secretory pathway.
Acknowledgment
We thank Dr. S. Yadav for assistance with ITC data analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants HL094505 (to B. Z.) and GM080271 (to S. M.) and Postdoctoral Fellowship T32 HL007914 (to R.C.P.). This work was also supported by a postdoctoral fellowship from the American Heart Association (to C. Z.).
The atomic coordinates and structure factors (codes 4GKX and 4GKY) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ER
- endoplasmic reticulum
- ERGIC
- ER-Golgi intermediate compartment
- FV
- factor V
- FVIII
- factor VIII
- COPI
- coat protein complex I
- COPII
- coat protein complex II
- CRD
- carbohydrate recognition domain
- ITC
- isothermal titration calorimetry
- DSP
- dithio-bis(succinimidyl propionate)
- IP
- immunoprecipitation
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)-1,3-propanediol.
REFERENCES
- 1. Barlowe C. (2003) Signals for COPII-dependent export from the ER: what's the ticket out? Trends Cell Biol. 13, 295–300 [DOI] [PubMed] [Google Scholar]
- 2. Cho S., Ryoo J., Jun Y., Ahn K. (2011) Receptor-mediated ER export of human MHC class I molecules is regulated by the C-terminal single amino acid. Traffic. 12, 42–55 [DOI] [PubMed] [Google Scholar]
- 3. Mancias J. D., Goldberg J. (2007) The transport signal on Sec22 for packaging into COPII-coated vesicles is a conformational epitope. Mol. Cell 26, 403–414 [DOI] [PubMed] [Google Scholar]
- 4. Cunningham M. A., Pipe S. W., Zhang B., Hauri H. P., Ginsburg D., Kaufman R. J. (2003) LMAN1 is a molecular chaperone for the secretion of coagulation factor VIII. J. Thromb. Haemost. 1, 2360–2367 [DOI] [PubMed] [Google Scholar]
- 5. Zhang B., Kaufman R. J., Ginsburg D. (2005) LMAN1 and MCFD2 form a cargo receptor complex and interact with coagulation factor VIII in the early secretory pathway. J. Biol. Chem. 280, 25881–25886 [DOI] [PubMed] [Google Scholar]
- 6. Appenzeller C., Andersson H., Kappeler F., Hauri H. P. (1999) The lectin ERGIC-53 is a cargo transport receptor for glycoproteins. Nat. Cell Biol. 1, 330–334 [DOI] [PubMed] [Google Scholar]
- 7. Vollenweider F., Kappeler F., Itin C., Hauri H. P. (1998) Mistargeting of the lectin ERGIC-53 to the endoplasmic reticulum of HeLa cells impairs the secretion of a lysosomal enzyme. J. Cell Biol. 142, 377–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nyfeler B., Reiterer V., Wendeler M. W., Stefan E., Zhang B., Michnick S. W., Hauri H.-P. (2008) Identification of ERGIC-53 as an intracellular transport receptor of α1-antitrypsin. J. Cell Biol. 180, 705–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nichols W. C., Seligsohn U., Zivelin A., Terry V. H., Hertel C. E., Wheatley M. A., Moussalli M. J., Hauri H. P., Ciavarella N., Kaufman R. J., Ginsburg D. (1998) Mutations in the ER-Golgi intermediate compartment protein ERGIC-53 cause combined deficiency of coagulation factors V and VIII. Cell 93, 61–70 [DOI] [PubMed] [Google Scholar]
- 10. Neve E. P., Lahtinen U., Pettersson R. F. (2005) Oligomerization and interacellular localization of the glycoprotein receptor ERGIC-53 is independent of disulfide bonds. J. Mol. Biol. 354, 556–568 [DOI] [PubMed] [Google Scholar]
- 11. Hauri H. P., Kappeler F., Andersson H., Appenzeller C. (2000) ERGIC-53 and traffic in the secretory pathway. J. Cell Sci. 113, 587–596 [DOI] [PubMed] [Google Scholar]
- 12. Zheng C., Liu H. H., Yuan S., Zhou J., Zhang B. (2010) Molecular basis of LMAN1 in coordinating LMAN1-MCFD2 cargo receptor formation and ER-to-Golgi transport of FV/FVIII. Blood 116, 5698–6706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Appenzeller-Herzog C., Nyfeler B., Burkhard P., Santamaria I., Lopez-Otin C., Hauri H. P. (2005) Carbohydrate- and conformation-dependent cargo capture for ER-exit. Mol. Biol. Cell 16, 1258–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Velloso L. M., Svensson K., Pettersson R. F., Lindqvist Y. (2003) The crystal structure of the carbohydrate-recognition domain of the glycoprotein sorting receptor p58/ERGIC-53 reveals an unpredicted metal-binding site and conformational changes associated with calcium ion binding. J. Mol. Biol. 334, 845–851 [DOI] [PubMed] [Google Scholar]
- 15. Zhang B., Cunningham M. A., Nichols W. C., Bernat J. A., Seligsohn U., Pipe S. W., Mcvey J. H., Schulte-Overberg U., de Bosch N. B., Ruiz-Saez A., White G. C., Tuddenham E. G. D., Kaufman R. J., Ginsburg D. (2003) Bleeding due to disruption of a cargo-specific ER-to-Golgi transport complex. Nat. Genet. 34, 220–225 [DOI] [PubMed] [Google Scholar]
- 16. Nyfeler B., Zhang B., Ginsburg D., Kaufman R. J., Hauri H. P. (2006) Cargo selectivity of the ERGIC-53/MCFD2 transport receptor complex. Traffic 7, 1473–1481 [DOI] [PubMed] [Google Scholar]
- 17. Guy J. E., Wigren E., Svärd M., Härd T., Lindqvist Y. (2008) New insights into multiple coagulation factor deficiency from the solution structure of human MCFD2. J. Mol. Biol. 381, 941–955 [DOI] [PubMed] [Google Scholar]
- 18. Zheng C., Liu H. H., Zhou J., Zhang B. (2010) EF hand domains of MCFD2 mediate interactions with both LMAN1 and coagulation factor V or VIII. Blood 115, 1081–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nishio M., Kamiya Y., Mizushima T., Wakatsuki S., Sasakawa H., Yamamoto K., Uchiyama S., Noda M., McKay A. R., Fukui K., Hauri H. P., Kato K. (2010) Structural basis for the cooperative interplay between the two causative gene products of combined factor V and factor VIII deficiency. Proc. Natl. Acad. Sci. U.S.A. 107, 4034–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wigren E., Bourhis J. M., Kursula I., Guy J. E., Lindqvist Y. (2010) Crystal structure of the LMAN1-CRD/MCFD2 transport receptor complex provides insight into combined deficiency of factor V and factor VIII. FEBS Lett. 584, 878–882 [DOI] [PubMed] [Google Scholar]
- 21. Paroutis P., Touret N., Grinstein S. (2004) The pH of the secretory pathway: measurement, determinants, and regulation. Physiology 19, 207–215 [DOI] [PubMed] [Google Scholar]
- 22. Arias-Moreno X., Velazquez-Campoy A., Rodríguez J. C., Pocoví M., Sancho J. (2008) Mechanism of low density lipoprotein (LDL) release in the endosome: implications of the stability and Ca2+ affinity of the fifth binding module of the LDL receptor. J. Biol. Chem. 283, 22670–22679 [DOI] [PubMed] [Google Scholar]
- 23. Byrne S. L., Chasteen N. D., Steere A. N., Mason A. B. (2010) The unique kinetics of iron release from transferrin: the role of receptor, lobe-lobe interactions, and salt at endosomal pH. J. Mol. Biol. 396, 130–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rudenko G., Henry L., Henderson K., Ichtchenko K., Brown M. S., Goldstein J. L., Deisenhofer J. (2002) Structure of the LDL receptor extracellular domain at endosomal pH. Science 298, 2353–2358 [DOI] [PubMed] [Google Scholar]
- 25. Olson L. J., Hindsgaul O., Dahms N. M., Kim J. J. (2008) Structural insights into the mechanism of pH-dependent ligand binding and release by the cation-dependent mannose 6-phosphate receptor. J. Biol. Chem. 283, 10124–10134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Appenzeller-Herzog C., Roche A. C., Nufer O., Hauri H. P. (2004) pH-induced conversion of the transport lectin ERGIC-53 triggers glycoprotein release. J. Biol. Chem. 279, 12943–12950 [DOI] [PubMed] [Google Scholar]
- 27. Pflugrath J. W. (1999) The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55, 1718–1725 [DOI] [PubMed] [Google Scholar]
- 28. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 30. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D. Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Terwilliger T. C. (2003) SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374, 22–37 [DOI] [PubMed] [Google Scholar]
- 33. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Velloso L. M., Svensson K., Schneider G., Pettersson R. F., Lindqvist Y. (2002) Crystal structure of the carbohydrate recognition domain of p58/ERGIC-53, a protein involved in glycoprotein export from the endoplasmic reticulum. J. Biol. Chem. 277, 15979–15984 [DOI] [PubMed] [Google Scholar]
- 36. Satoh T., Cowieson N. P., Hakamata W., Ideo H., Fukushima K., Kurihara M., Kato R., Yamashita K., Wakatsuki S. (2007) Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36. J. Biol. Chem. 282, 28246–28255 [DOI] [PubMed] [Google Scholar]
- 37. Kamiya Y., Kamiya D., Yamamoto K., Nyfeler B., Hauri H. P., Kato K. (2008) Molecular basis of sugar recognition by the human L-type lectins ERGIC-53, VIPL, and VIP36. J. Biol. Chem. 283, 1857–1861 [DOI] [PubMed] [Google Scholar]
- 38. Reiterer V., Nyfeler B., Hauri H. P. (2010) Role of the lectin VIP36 in post-ER quality control of human α1-antitrypsin. Traffic. 11, 1044–1055 [DOI] [PubMed] [Google Scholar]
- 39. Bentley M., Nycz D. C., Joglekar A., Fertschai I., Malli R., Graier W. F., Hay J. C. (2010) Vesicular calcium regulates coat retention, fusogenicity, and size of pre-Golgi intermediates. Mol. Biol. Cell 21, 1033–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nufer O., Kappeler F., Guldbrandsen S., Hauri H. P. (2003) ER export of ERGIC-53 is controlled by cooperation of targeting determinants in all three of its domains. J. Cell Sci. 116, 4429–4440 [DOI] [PubMed] [Google Scholar]
- 41. Spiro R. G. (2004) Role of N-linked polymannose oligosaccharides in targeting glycoproteins for endoplasmic reticulum-associated degradation. Cell Mol. Life Sci. 61, 1025–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kögler E. J. (2008) Characterization of P28, a novel ERGIC/cis-Golgi protein, required for Golgi ribbon formation; pH measurements in the early secretory pathway in vivo. Ph. D. thesis, University of Basel [Google Scholar]
- 43. Montero M., Alvarez J., Scheenen W. J., Rizzuto R., Meldolesi J., Pozzan T. (1997) Ca2+ homeostasis in the endoplasmic reticulum: coexistence of high and low [Ca2+] subcompartments in intact HeLa cells. J. Cell Biol. 139, 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pinton P., Pozzan T., Rizzuto R. (1998) The Golgi apparatus is an inositol 1,4,5-trisphosphate-sensitive Ca2+ store, with functional properties distinct from those of the endoplasmic reticulum. EMBO J. 17, 5298–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Van Baelen K., Vanoevelen J., Callewaert G., Parys J. B., De Smedt H., Raeymaekers L., Rizzuto R., Missiaen L., Wuytack F. (2003) The contribution of the SPCA1 Ca2+ pump to the Ca2+ accumulation in the Golgi apparatus of HeLa cells assessed via RNA-mediated interference. Biochem. Biophys. Res. Commun. 306, 430–436 [DOI] [PubMed] [Google Scholar]
- 46. Xu D., Hay J. C. (2004) Reconstitution of COPII vesicle fusion to generate a pre-Golgi intermediate compartment. J. Cell Biol. 167, 997–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pezzati R., Bossi M., Podini P., Meldolesi J., Grohovaz F. (1997) High-resolution calcium mapping of the endoplasmic reticulum-Golgi-exocytic membrane system. Electron energy loss imaging analysis of quick frozen-freeze dried PC12 cells. Mol. Biol. Cell 8, 1501–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kawasaki N., Ichikawa Y., Matsuo I., Totani K., Matsumoto N., Ito Y., Yamamoto K. (2008) The sugar-binding ability of ERGIC-53 is enhanced by its interaction with MCFD2. Blood 111, 1972–1979 [DOI] [PubMed] [Google Scholar]
- 49. Turnbull W. B., Daranas A. H. (2003) On the value of c: can low affinity systems be studied by isothermal titration calorimetry? J. Am. Chem. Soc. 125, 14859–14866 [DOI] [PubMed] [Google Scholar]