Abstract
Constitutive activation of the Wnt/β-catenin signaling pathway is a notable feature of a large minority of cases of malignant melanoma, an aggressive and increasingly common cancer. The identification of target genes downstream from this pathway is therefore crucial to our understanding of the disease. The POU domain transcription factor Brn-2 has been implicated in control of proliferation and melanoma survival, and its expression is strongly upregulated in melanoma. We show here that in vivo Brn-2 is expressed in melanocytes but not in embryonic day 11.5 melanoblasts and that its expression is directly controlled by the Wnt/β-catenin signaling pathway in melanoma cell lines and in transgenic mice. Moreover, silent interfering RNA-mediated inhibition of Brn-2 expression in melanoma cells overexpressing β-catenin results in significantly decreased proliferation. These results, together with the observation that BRAF signaling also induces Brn-2 expression, reveal that Brn-2 is a focus for the convergence of two key melanoma-associated signaling pathways that are linked to cell proliferation.
Melanocytes originate in the neural crest as undifferentiated nonpigmented melanoblasts that migrate to the epidermis and hair follicles, where they differentiate and are responsible for skin and hair color. As melanocytes are not essential for survival and as mutations affecting the survival or differentiation of the melanocyte lineage are reflected in an obvious pigmentation phenotype, the melanocyte system represents an excellent model for understanding how signal transduction pathways coordinate the program of gene regulation underlying the genesis of a specific cell type. Importantly, constitutive activation of signaling pathways normally operating during melanocyte development is linked to the transformation of a melanocyte to a malignant melanoma (5, 16), a highly aggressive cancer, the incidence of which is increasing at an alarming rate (33).
The Wnt signaling pathway (for reviews of Wnt signaling, see references 3 and 12) is critically required for development of the melanocyte lineage; in both zebrafish and mice, overexpression of components of the Wnt signaling pathway result in an increase in the number of melanocytes at the expense of neurons and glia (9, 11), and disruption of the Wnt-1 and Wnt-3a genes leads to complete loss of melanoblasts (24). Wnt proteins interact with frizzled receptors and lead to the inhibition of serine-threonine kinase glycogen synthase kinase 3β. Phosphorylation of β-catenin by glycogen synthase kinase 3β is associated with the destabilization of β-catenin. Thus, increased Wnt signaling leads to stabilization of β-catenin and its translocation from the cytoplasm to the nucleus, where it can activate transcription via association with the Lef1 and Tcf transcription factors (1, 22, 29). A key role for Wnt signaling in melanocyte development is the activation of the promoter for the gene encoding the microphthalmia-associated transcription factor Mitf (10, 43). Mitf (19, 23) is essential for the development of the melanocyte lineage and has two key functions: in control of cell proliferation and survival and in differentiation (17).
The link between melanocyte development and melanoma is underscored by the fact that many melanomas exhibit constitutively elevated levels of nuclear β-catenin (34, 36). Given the key role played by β-catenin in development, it is likely that the elevated level of this protein observed in melanomas makes a substantial contribution to their transformed phenotype, and indeed, activation of the Wnt pathway has been linked to a metastatic or migratory phenotype (30). The identification of β-catenin target genes therefore represents an important goal if the link between melanocyte development and malignant melanoma is to be understood.
Using both cell lines and transgenic mice, we show here that the promoter for the gene encoding the POU domain transcription factor Brn-2 (also called N-Oct3 and POU3F2) that has been implicated in control of proliferation and survival of melanoma cells is a direct target for β-catenin/Lef1 and that reduction of Brn-2 expression by silent interfering RNA (siRNA) results in a decrease in proliferation. The results provide a novel insight into the link between melanocyte development and melanoma and β-catenin and regulation of proliferation.
MATERIALS AND METHODS
Cell lines and transfection assays.
The mouse melanocyte cell line melan-a was maintained in RPMI 1640 medium supplemented with 10% fetal calf serum (Gibco-BRL) and 200 nM tetradecanoyl phorbol acetate (Sigma), as was the melan-c melanocyte cell line. Mouse melanoma cell line B16 and human melanoma cell lines 501 mel and VUP were grown in RPMI 1640 medium supplemented with 10% fetal calf serum. COS7, 3T3, and XB2 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. All cells were grown at 37°C with 10% CO2, and lysates for Western blotting or band shift assays were prepared from subconfluent cultures.
Brn-2-luciferase assays were performed as described previously (15). Cells were seeded at a density of 1.5 × 103 cells/2 cm2 in a 24-well plate the day before transfection; 25 ng of promoter-reporter construct was transfected with increasing amounts of plasmid expressing activators with Fugene (Boehringer-Mannheim) according to the manufacturer's instructions. For cotransfections, an equal total amount of DNA was maintained by compensation with empty expression vector DNA. Cells were harvested 48 h after transfection and assayed for firefly luciferase activity, and the results were normalized to those with a cotransfected simian virus 40-lacZ reporter.
Brn-2-lacZ assays were performed as described previously (27). Cell lines were seeded into 2.5-cm dishes and transfected when 40% confluent with 1 μg of plasmid DNA with Fugene. A cotransfected simian virus 40-luciferase reporter was used to control for transfection efficiency. All transfection assays were repeated multiple times.
siRNA-mediated downregulation of Brn-2 and β-catenin.
A 21-base Brn-2-specific siRNA was synthesized by Dharmacon. The sequence used was 5′-GCGCAGAGCCUGGUGCAGGUU-3′ and its complement, leaving a 3′ UU overhang on both strands.
For β-catenin we used the siRNA described previously (47), 5′-AAAGCUGAUAUUGAUGGACAGdTdT-3′ and its complement, leaving a dTdT 3′ overhang on both strands. The siRNA control was 5′-UUCUCCGAACGUGUCACGUdTdT-3′ and its complement, leaving a dTdT 3′ overhang on both strands. siRNA was transfected into cells with Oligofectamine (Invitrogen) as per the manufacturer's instructions, and cells were harvested after 3 days, by which time Brn-2 or β-catenin was effectively downregulated.
Immunofluorescence and bromodeoxyuridine incorporation.
Cells treated with siRNA specific for Brn-2 or a control siRNA were incubated with medium containing bromodeoxyuridine labeling reagent (Roche) for 1 h before fixation with 4% paraformaldehyde, washed three times with phosphate-buffered saline (PBS), and permeabilized with 0.2% Triton X-100 in PBS. After washing four times with PBS, treatment for 30 min with 2 M HCl, washing again four times with PBS, and blocking for 1 h with 10% fetal bovine serum, bromodeoxyuridine incorporation was detected with a mouse monoclonal antibromodeoxyuridine primary antibody (Becton Dickinson) and a Texas Red-conjugated anti-mouse immunoglobulin secondary antibody. After being washed four times in PBS, cells were mounted in Vectashield (Vector Laboratories) containing 4′,6′-diamidino-2-phenylindole (DAPI) to visualize DNA.
Western blot analysis.
Whole-cell extracts were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, 10% polyacrylamide). Nitrocellulose membranes were used for transfer, and after being blocked with 10% skim milk (99% fat free)-0.1% Tween 20-phosphate buffer, the membranes were probed with appropriate primary antibodies (for 1 h to overnight at 4°C). Proteins were detected with peroxidase-conjugated anti-rabbit or anti-mouse immunoglobulin secondary antibody and visualized with the ECL detection kit (Amersham).
The primary antibodies used were the mouse monoclonal anti-Brn-2 antibody that we raised against bacterially expressed full-length mouse Brn-2 protein; anti-β-catenin antibody from Santa Cruz (C-18); an anti-α-tubulin antibody (Amersham); and mouse anti-bromodeoxyuridine monoclonal antibody from Becton Dickinson Biosciences. Rabbit anti-Oct1 antibody was a gift from Peter O'Hare.
Chromatin immunoprecipitation assays.
Chromatin immunoprecipitation assays were performed as described previously (15) with goat polyclonal anti-Lef1 antibody (Santa Cruz C-19), goat polyclonal anti-β-catenin antibody (Santa Cruz C-18), or 10 μl of nonspecific immunoglobulin G (IgG) (Bio-Rad). After immunoprecipitation, samples were analyzed by quantitative PCR for 25 cycles, taking care that the PCR was in the log phase of amplification. For the HSP70 high-cycle control, the PCRs were allowed to proceed for a further five cycles.
The primers used for the PCR were 5′-GAGGAGGGCTAGGAGGACTCC-3′ and 5′-CGCGTAACTGTCAATGAAAAA-3′ for the Brn-2 promoter; 5′-TCGGAAGTGGCAGTTATTCGG-3′ and 5′-TTTTAGGTGGCACCAATCC-3′ for the MITF promoter; and 5′-CCTCCAGTGAATCCCAGAAGACTCT-3′ and 5′-TGGGACAACGGGAGTCACTCTC-3′ for the HSP70 promoter.
Plasmids.
The Brn-2 promoter-luciferase reporter has been described (18). The point mutations in the Lef1 binding site were introduced by PCR with the appropriate primers.
The β-catenin expression vector has been described previously (21) and was provided by Rudi Grosschedl.
Whole-mount, histological in situ hybridizations and X-Gal staining of mouse embryos.
Riboprobes were prepared by in vitro transcription with T7 and T3 RNA polymerases from pBS-Brn2 and with digoxigenin-11-UTP (Roche) according to the manufacturer's instructions. A 403-bp PCR fragment, corresponding to nucleotides 314 to 717 starting from the ATG of mouse Brn-2, was subcloned into pBluescript II and used as a template for the Brn-2 riboprobes. Timed matings were set up to obtain staged mouse embryos, designating the noon of plug formation as embryonic day 0.5 (E0.5). E11.5 embryos and postnatal day 1 (P1) newborn mice were fixed for 4 h room temperature and overnight in 4% paraformaldehyde in PBS at 4°C, respectively. Thereafter they were either dehydrated and stored in 100% methanol at −20°C or equilibrated in 15% sucrose for 2 days at 4°C, embedded in 15% (wt/vol) sucrose-7.5% (wt/vol) gelatine in PBS and frozen at −80°C and cryosectioned (16 μm). Whole-mount and histological in situ hybridizations were performed according to a procedure described by Wehrle-Haller et al. (48). The hybridization of digoxigenin-labeled riboprobes was detected with antidigoxigenin antibodies coupled to alkaline phosphatase. Details of the protocol will be provided upon request. For detection of β-galactosidase activity in whole mounts, DCT-lacZ transgenic embryos were fixed in 0.25% glutaraldehyde in PBS for 30 min at 4°C and stained with 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) as previously described (20, 28).
Band shift assays.
The band shift assays for Lef1 were performed in a final volume of 20 μl containing 20 mM HEPES (pH 7.9), 10% glycerol, and 112 mM KCl; nuclear extracts were prepared as described previously (51). Lef1 protein was expressed as a glutathione S-transferase (GST) fusion, purified, and then removed from the GST by thrombin cleavage.
RESULTS
Expression of Brn-2 is β-catenin responsive.
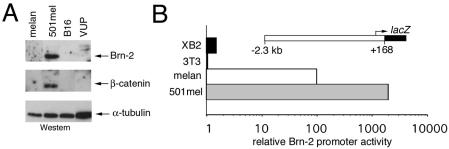
The POU domain transcription factor Brn-2 is overexpressed in a number of cancers, including Merkel cell carcinoma (26), small cell lung carcinoma (39), neuroblastoma (38, 40), and T-cell lymphoma (2), and we and others have also shown that Brn-2 mRNA expression is upregulated in many melanomas compared to melanocyte cell lines (14, 45). Since the majority of cutaneous melanomas (8), though not uveal melanomas (6, 7, 13, 35), bear activating mutations in BRAF, the elevated expression of Brn-2 observed in many melanomas may be accounted for by the observation that Brn-2 expression is strongly upregulated by BRAF-mediated activation of the mitogen-activated protein kinase signaling cascade (18). In addition to BRAF mutations, a subset of melanomas, typified by the 501 mel human melanoma cell line, are characterized by elevated levels of β-catenin (34, 36). Since Brn-2 expression is high in the 501 mel melanoma line that expresses high levels of β-catenin (Fig. 1A) but low in a human uveal melanoma line, VUP, and in the mouse B16 melanoma and melan-a melanocyte cell lines, which exhibit low β-catenin levels, we wished to determine whether Brn-2 expression was controlled by the β-catenin signaling pathway. Note that a low level of β-catenin can be detected in the other cell lines on longer exposure (data not shown).
FIG. 1.
Brn-2 and β-catenin expression in melanocyte and melanoma cell lines. (A) Extracts from the indicated melanocyte (melan-a) and melanoma (B16, 501 mel, and VUP) cell lines were subjected to Western blotting with the indicated antibodies. Note that a low level of β-catenin could be detected in all cell lines on longer exposure. (B) The Brn-2 promoter is highly active in the 501 mel cell line. The melanocyte cell line melan-c, the human melanoma cell line 501 mel, NIH 3T3 cells, and the keratinocyte cell line XB2 were transfected with the indicated promoter construct driving expression of a lacZ reporter, and β-galactosidase activity was assayed 48 h posttransfection. All results are presented relative to those obtained with a cotransfected simian virus 40 promoter-luciferase reporter.
As a first step towards understanding whether the control of Brn-2 expression might be responsive to β-catenin, we asked whether the Brn-2 promoter was able to direct cell type-specific expression in transfected cells. To this end, a Brn-2 promoter fragment extending to about −2.5 kb with respect to the ATG initiation codon, or −2.3 kb upstream from the transcription start site, was cloned upstream from a lacZ reporter, and its activity was assayed after transfection. Similar results were obtained irrespective of whether a lacZ or luciferase (not shown) reporter was used. Several cell lines were used for transfection: melan-c, an albino melanocyte cell line, which expresses Brn-2 mRNA at a low level (18); the human 501 mel melanoma cell line, which expresses Brn-2 RNA and protein at a high level; and NIH 3T3 cells and XB2 keratinocytes, which do not express Brn-2, as assessed by Northern blotting (not shown). The results from the transfection assays (Fig. 1B), normalized for the expression of a cotransfected simian virus 40 promoter-reporter plasmid, revealed that the Brn-2 promoter is about 100-fold more efficiently expressed in the melanocyte cell line melan-c than in NIH 3T3 or XB2 cells but is also 20-fold more active in the 501 mel melanoma cell line compared to melan-c cells. Thus, 2.3 kb of the Brn-2 promoter sequence contains sufficient information to direct differential expression in cells in culture, and the expression pattern appears to reflect that of the endogenous Brn-2 gene.
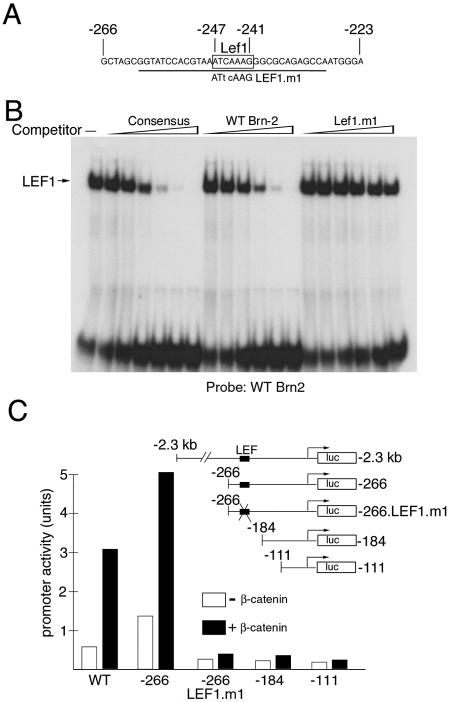
The fact that the Brn-2 promoter was so active in the 501 mel melanoma cells raised the possibility that Brn-2 expression might be regulated by β-catenin's targeting the Brn-2 promoter. Consistent with this, examination of the Brn-2 promoter revealed a consensus potential high-affinity binding site for the Lef1 and Tcf transcription factors, ATCAAAG (Fig. 2A). That this element was a potential β-catenin response element was confirmed with an in vitro DNA-binding assay (Fig. 2B). With sequences spanning the putative Lef1 binding site as a probe, this element bound Lef1 as efficiently as a control consensus binding site but did not bind Lef1 if the core sequence was mutated to ATtcAAG (Lef1.m1).
FIG. 2.
Brn-2 expression is activated by β-catenin in vitro and in vivo. (A) Sequence of a region of the Brn-2 promoter required for activity in melanocytes and melanoma, with the consensus Lef1/Tcf factor binding site indicated (−241 to −247). The sequence of the mutation in the binding site is indicated, and the underlined region was used as a probe for the DNA-binding assays presented below. (B) Lef1 binds the Brn-2 promoter in vitro. Band shift assay with a probe corresponding to the sequence underlined in panel A together with bacterially expressed Lef1. The indicated competitor oligonucleotides were used at 2, 5, 10, 20, 50, and 250 ng. The sequence of the Lef1 consensus oligonucleotide was 5′-CTAGAAGGGCACCCTTTGAAGCTCT-3′. (C) β-Catenin activates the Brn-2 promoter. The 501 mel cells were transfected with a Brn-2 promoter-luciferase reporter construct extending to either −2.3 kb (wild type) or deleted to −266, −184, or −111 or a reporter, −266/Lef.m1, in which the Lef1/Tcf site has been mutated, as illustrated in the inset. The indicated reporters were transfected alone or together with a β-catenin expression vector, and luciferase activity was determined.
To test whether this sequence was functional, we used a Brn-2 promoter-luciferase reporter together with vectors expressing β-catenin in cotransfection assays in 501 mel cells. The Brn-2 promoter used extends upstream to −2.3 kb, and the results obtained (Fig. 2C) indicate that the Brn-2 promoter can be activated by β-catenin in this assay. Deletion analysis revealed that a promoter deletion mutant lacking sequences 5′ to −266 retained its ability to respond to β-catenin but that removal of additional sequences to −184 or −111 eliminated the response, consistent with the location of the putative β-catenin-responsive element at −247 to −241. The ability of β-catenin to activate the Brn-2 promoter was also abolished if the Lef/Tcf binding site were mutated (−266.LEF.m1), confirming the critical role of this element in the response of the Brn-2 promoter to β-catenin signaling.
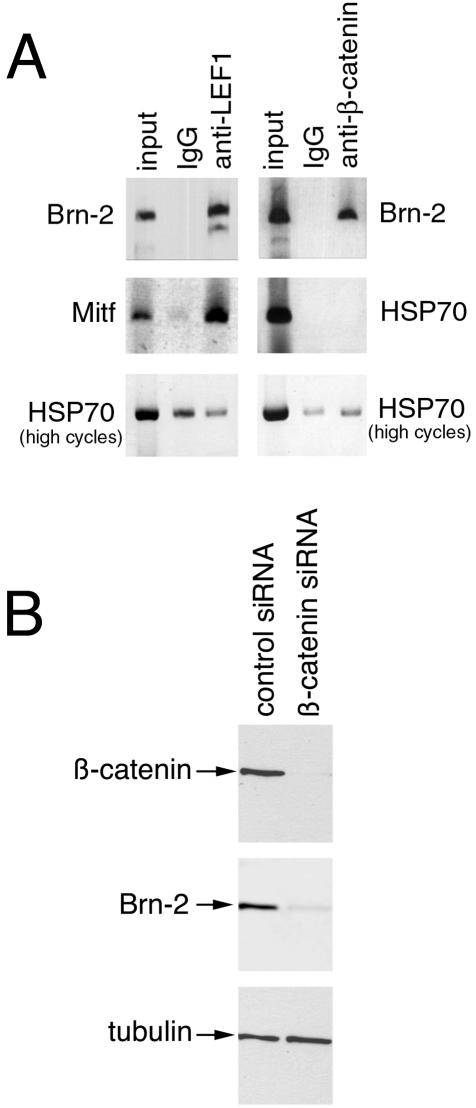
To verify that Lef1 and β-catenin could bind the Brn-2 promoter in melanoma cells, we next performed a chromatin immunoprecipitation assay. In this assay, chromatin was prepared from cross-linked 501 mel cells and sheared to an average size of about 750 bp before being subjected to immunoprecipitation with either an anti-Lef1 or an anti-β-catenin antibody. The DNA recovered after the immunoprecipitation was then used in a PCR together with primers specific for the Brn-2 promoter. As a negative control, we used primers specific for the HSP70 promoter, which is not regulated by β-catenin, and as a positive control for the Lef1 chromatin immunoprecipitation, we used primers specific for the MITF promoter, a known Lef1/β-catenin-target. The results, presented in Fig. 3A, reveal that a strong band corresponding to the Brn-2 promoter was obtained with both the anti-Lef1 and anti-β-catenin antibodies, similar to that obtained for the MITF promoter with the anti-Lef1 antibody. With nonspecific IgG in the immunoprecipitation, no PCR product above the level of the background was observed. For the HSP70 negative control, no PCR product was observed if a similar number of PCR cycles were used for the MITF and Brn-2 promoters, and if the PCR was allowed to proceed (HSP70 high cycles), an equivalent level of product was obtained irrespective of whether a specific antibody against either Lef1 or β-catenin or nonspecific IgG was used.
FIG. 3.
Brn-2 promoter is a target for Lef1 and β-catenin in vivo. (A) Chromatin immunoprecipitated from 501 mel cells with either nonspecific IgG or anti-β-catenin or anti-Lef1 antibodies was subjected to PCR with primers specific for the Brn-2 and Mitf promoters. Primers for the HSP70 promoter were used as a negative control. For the HSP70 promoter, PCR for the same number of cycles (25) used to generate a signal for the Brn-2 or Mitf promoter immunoprecipitated with anti-Lef1 or anti-β-catenin antibodies failed to reveal a signal. The PCR cycles were therefore increased to 30 so that a product was evident (high cycles). (B) siRNA-mediated downregulation of β-catenin results in decreased Brn-2 expression; 501 mel cells were transfected with either a control siRNA or an siRNA specific for β-catenin (47), harvested after 3 days, and subjected to Western blotting with the indicated antibodies.
To confirm that β-catenin was indeed regulating Brn-2 expression in melanoma cells, we transfected the 501 mel melanoma cell line with an siRNA specific to β-catenin and examined the effect of loss of β-catenin expression on Brn-2 levels. The results (Fig. 3B) indicated that the siRNA used, which has been shown previously to be both effective and specific for β-catenin (47), largely abolished β-catenin expression and moreover also resulted in a substantial downregulation of Brn-2 compared to that in cells transfected with a control nonsilencing RNA. No effect was observed on tubulin levels. These results, together with those of the chromatin immunoprecipitation assay, are consistent with the Brn-2 promoter's being bound and regulated by a Lef1/β-catenin complex in vivo.
Brn-2 is required for proliferation.
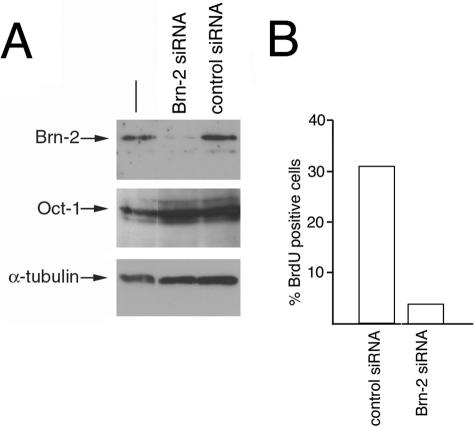
Although the results obtained so far implicate β-catenin in the regulation of Brn-2 expression, a key issue is whether Brn-2 expression contributes to proliferation. To address this question, we designed a Brn-2-specific siRNA and used it together with a control nonsilencing RNA to transfect the 501 mel cell line, in which we have established that Brn-2 expression is controlled by β-catenin. The results obtained by Western blotting (Fig. 4A) and immunofluorescence (not shown) indicate that the Brn-2 siRNA was highly specific and effective, decreasing Brn-2 expression by up to 90% while not affecting expression of the related POU domain transcription factor Oct-1 or tubulin. Significantly, transfection of the 501 mel cell line with the Brn-2 siRNA also resulted in up to an eightfold decrease in the proportion of cells incorporating bromodeoxyuridine compared to that in cells transfected with the control siRNA (Fig. 4B), although in most experiments (not shown) the bromodeoxyuridine-positive population was reduced by about fourfold. Consistent with this, treatment of the 501 mel cells with the Brn-2-specific siRNA also led to substantially reduced [3H]thymidine incorporation (not shown). However, while there was a significant decrease in the S-phase population in Brn-2-depleted 501 mel cells, we did not detect any obvious signs of increased apoptosis (not shown), suggesting that Brn-2 expression is required for proliferation but not survival of these cells.
FIG. 4.
Brn-2 expression is required for proliferation in 501 mel cells. 501 mel cells were transfected with siRNA specific for Brn-2 or a control siRNA. After 3 days, cells were grown in the presence of bromodeoxyuridine for 1 h before being subjected to either Western blotting or immunofluorescence. (A) Results of Western blotting with the indicated antibodies after Brn-2 or control siRNA treatment. (B) Quantification of the immunofluorescence results obtained by counting 300 control or Brn-2 siRNA-transfected cells stained for DNA with DAPI and for bromodeoxyuridine (BrdU) incorporation.
β-Catenin induces Brn-2 expression in transgenic mice.
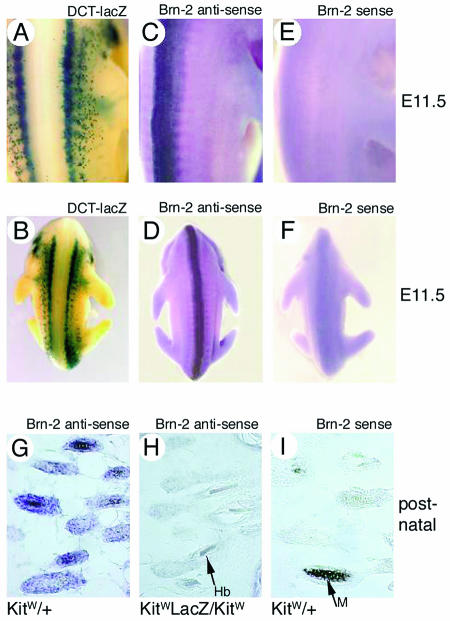
Although Brn-2 is expressed in melanoma cell lines in culture, its expression in the melanocyte lineage in vivo has not been investigated previously. To determine whether Brn-2 is expressed in melanoblasts in vivo, we performed whole-mount in situ hybridization with a sense or antisense Brn-2-specific probe and examined expression in a mouse embryo at E11.5, a time when melanoblasts are proliferating and migrating from the neural crest. As a control, we used an E11.5 embryo expressing LacZ from the DCT promoter, which directs expression in melanoblasts (28) and enables the neural crest-derived melanoblast population to be clearly visible (Fig. 5A and B). In these experiments, with an antisense probe, Brn-2 expression was clearly evident in the neural tube (Fig. 5C and D), as has been seen previously (37), and no signal was generated with a sense probe (Fig. 5E and F) as a control. However, no detectable Brn-2 expression was detected in melanoblasts or the neural crest. Although we failed to detect Brn-2 expression at this time of development, we also examined the potential for Brn-2 expression in postnatal melanocytes in the hair follicle. Here, in contrast to melanoblasts, a clear signal was obtained with the antisense Brn-2 probe in the hair follicles, consistent with the localization of wild-type melanocytes (Fig. 5G). No Brn-2 signal was present in KitW-lacZ/KitW mice, which lack the neural crest-derived melanocyte population (Fig. 5H), or if a sense probe was used on the wild-type mice (Fig. 5I).
FIG. 5.
Brn-2 is not expressed in melanoblasts in vivo but is expressed in hair follicle melanocytes. (A and B) E11.5 mouse embryo bearing a DCT-lacZ reporter transgene stained for lacZ expression as a marker of melanoblasts. (C and D) In situ hybridization of an E11.5 embryo with a Brn-2 antisense probe. (E and F) In situ hybridization of an E11.5 embryo with a Brn-2 sense probe. Note that Brn-2 expression is not detected in melanoblasts. (G to I) In situ hybridization of a cryosection of newborn mouse skin with the antisense (G and H) and sense (I) Brn-2 probes with wild-type and KitW-lacZ/KitW mice. The KitW-lacZ/KitW mice lack neural crest-derived melanocytes. The arrow in panel I indicates melanin (M), and the arrow in panel H indicates a hair bulb (hb).
To determine whether expression of Brn-2 in mice could be upregulated in vivo by β-catenin, as suggested by the results from the cell lines, we examined Brn-2 expression by reverse transcription (RT)-PCR in mouse skin from a wild-type mouse or one in which β-catenin fused to green fluorescent protein (GFP) is overexpressed in the melanocyte lineage from a tyrosinase promoter-enhancer expression cassette that specifically targets expression to the melanocyte lineage. The production of these mice and a detailed analysis of their phenotypes and the properties of their melanocytes will be described in detail elsewhere (S. Martinozzi et al., unpublished data). Consistent with the results obtained in cell lines, RT-PCR analysis of newborn skin demonstrated that Brn-2 was significantly overexpressed in the skin of the transgenic mice (GFP RNA positive) compared to their nontransgenic littermates (Fig. 6). No change was observed in the total levels of cyclin D1 or hypoxanthine phosphoribosyltransferase RNA, which were used as internal controls. Note that although the cyclin D1 gene has been reported to be a target for β-catenin signaling (44), unlike Brn-2 it is expressed in both melanocytes and keratinocytes, and as melanocytes represent only a minor population of skin cells, any elevation in cyclin D1 RNA levels in the melanocyte population is unlikely to be detected. In summary, the activation of Brn-2 expression by Wnt/β-catenin observed in cells in culture was accurately reproduced in vivo in mouse skin and provides substantial evidence to support the idea that Brn-2 expression is directly regulated by the Wnt signaling pathway.
FIG. 6.

Brn-2 is upregulated by β-catenin in vivo. β-Catenin-GFP was overexpressed in the melanocyte lineage from a tyrosinase promoter including an upstream tyrosinase locus control region (Martinozzi et al., unpublished data). RNA derived from the skin of transgenic (Tg) and nontransgenic wild-type (WT) littermates were subjected to RT-PCR with primers for the indicated genes. Primers specific for GFP were used to detect the transgene, and cyclin D1 and hypoxanthine phosphoribosyltransferase (HPRT) were used as controls.
DISCUSSION
In this paper we provide several lines of evidence to suggest that Wnt/β-catenin signaling controls Brn-2 expression. The Brn-2 promoter contains a functional Lef1 binding site, is responsive to Wnt/β-catenin signaling, and, importantly, is recognized by Lef1 and β-catenin in vivo, as determined with a chromatin immunoprecipitation assay, while siRNA-mediated downregulation of β-catenin in melanoma cells also results in decreased Brn-2 expression. Moreover, Brn-2 expression is upregulated in mice expressing activated β-catenin in melanocytes. The ability of β-catenin to upregulate Brn-2 expression will no doubt contribute to the elevated levels of Brn-2 observed in many melanomas and may also contribute to the highly specific pattern of Brn-2 expression observed during development (31, 37).
Strikingly, expression of Brn-2 is not only upregulated by β-catenin but is also elevated in response to mitogen-activated protein kinase signaling downstream from reporter tyrosine kinases (RTKs) and in particular downstream from BRAF (18), which is known to be activated by mutation in about 70% of melanomas and nevi (8, 32). Thus, the overexpression of Brn-2 is a feature of melanoma irrespective of which pathway, β-catenin or mitogen-activated protein kinase, is constitutively activated. Moreover, we demonstrate here that siRNA-mediated downregulation of Brn-2 expression dramatically reduced the S-phase population of the 501 mel cells, strongly pointing to a role for Brn-2 in proliferation. Consistent with this, we have also shown that overexpression of Brn-2 in melanocytes leads to increased proliferation, while siRNA-mediated inhibition of Brn-2 expression in melanoma cell lines expressing a constitutively activated BRAF leads to decreased [3H]thymidine incorporation (18). Previous work has also linked expression of Brn-2 in melanoma cell lines tumorigenicity and proliferation (45). Indeed, we have found that of 16 melanoma cell lines for which we have information on the status of Brn-2 expression as well as BRAF and β-catenin mutations, only three, which are apparently wild type for both BRAF and β-catenin, do not express Brn-2. Of the remaining cell lines. only one exhibits both an activating mutation on BRAF and a mutated form of β-catenin.
There is now a substantial body of evidence indicating that Brn-2 plays a key role in controlling the proliferation of many melanoma cell lines. Moreover, since Brn-2 expression lies downstream of the two genetically defined signaling pathways that are constitutively activated in melanoma, it is possible that overexpression of Brn-2 may be a necessary step in the formation of many melanomas and will confer a strong selective growth advantage. Consistent with this, expression of β-catenin in B16 melanoma cells results in increased proliferation, while expression of a dominant-negative Tcf slows melanoma cell growth (49). In this respect it is notable that in other systems, β-catenin signaling is implicated in growth control. For example, in colorectal cancer cells, expression of a dominant-negative Tcf4 leads to a G1 arrest via the induction of the p21 cyclin-dependent kinase inhibitor (46), and siRNA-mediated downregulation of β-catenin in colon cancer cells also inhibited proliferation (47). Similarly, in the pituitary, expression of the PitX1 transcription factor is induced by Wnt/β-catenin signaling and is required for proliferation (25). The fact that β-catenin can induce the expression of Brn-2 in melanoma appears to indicate that the control of proliferation by the Wnt//β-catenin signaling pathway may be a general theme.
In the melanocyte lineage, Wnt/β-catenin signaling is required to turn on expression of the Mitf transcription factor (10, 43), which plays a crucial role in melanoblast survival and is most likely a key factor in determining the identity of a melanoblast (17). Intriguingly, we found no evidence for Brn-2 expression in the neural crest in melanoblasts at E11.5, a time when these cells are both proliferating and migrating. It is possible that at this stage in development, the chromatin across the Brn-2 promoter is in a conformation that is incompatible with activation by Wnt/β-catenin signaling or that Wnt/β-catenin signaling in the melanocyte lineage operates only early in development, turns on Mitf expression, and then is no longer required. Alternatively, the levels of β-catenin expressed in melanoblasts at E11.5, while capable of activating Mitf expression, may be insufficient for activation of the Brn-2 promoter, which would only respond to the increased β-catenin expression occurring when the migrating melanoblast population arrives in the epidermis. A further possibility is that melanoblasts have the potential to express Brn-2 but that its expression can only be induced by the cooperative effect of multiple signal transduction pathways that are not simultaneously active in melanoblasts. Currently we know little about the timing of activation of signal transduction pathways in melanocyte development.
Despite the fact that Brn-2 does not appear to be expressed in melanoblasts at E11.5, it is nevertheless present in melanocytes in postnatal hair follicles, where evidently the signals required to induce Brn-2 expression are active. In these cells Brn-2 expression can be further induced by the elevated levels of β-catenin expressed in the transgenic animals, confirming the results indicating that Brn-2 expression is activated by β-catenin that were obtained by using cell lines. However, although Brn-2 may be expressed in hair follicle melanocytes, whether it plays a role in the proliferation of these cells is likely to depend on several factors, including the absolute level of Brn-2 protein expressed, which will in turn depend on the activity of the mitogen-activated protein kinase pathway downstream of RTKs and BRAF as well as the β-catenin signaling pathway. In addition, the DNA-binding activity of the Brn-2 protein in regulating gene expression will, like that of other POU domain proteins (4, 41), be controlled by phosphorylation, and it is likely that the ability of Brn-2 to cooperate or interact with other DNA-binding proteins such as Sox11, as has been seen in other cell types (50), will also be regulated. An additional level of control of Brn-2 activity may also be provided by the regulation of its ability to interact with specific transcriptional cofactors (42). Dissecting the signal transduction pathways that regulate Brn-2 and its ability to control transcription and proliferation as well as its oncogenic potential will lead to a better understanding of how mitogen-activated protein kinase and β-catenin signaling are integrated with cell cycle control.
Acknowledgments
We thank Rudi Grosschedl for the β-catenin expression vector, Kathy Jones for anti-LEF1 antibody, Marshall Nirenberg for a Brn-2 genomic clone, Ian Hart for the VUP melanoma cell line, Jean-Jacques Panthier for the newborn W/W-lacZ pups, and Hans Clevers for the Lef1 expression vector.
This work was supported by the Association for International Cancer Research (AICR), Candis and Marie Curie Cancer Care, the Fondation de France, the Ligue Nationale Contre le Cancer, and l'Association pour la Recherche contre le Cancer.
REFERENCES
- 1.Behrens, J., J. P. von Kries, M. Kuhl, L. Bruhn, D. Wedlich, R. Grosschedl, and W. Birchmeier. 1996. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 382:638-642. [DOI] [PubMed] [Google Scholar]
- 2.Bert, A. G., J. Burrows, A. Hawwari, M. A. Vadas, and P. N. Cockerill. 2000. Reconstitution of T cell-specific transcription directed by composite NFAT/Oct elements. J. Immunol. 165:5646-5655. [DOI] [PubMed] [Google Scholar]
- 3.Cadigan, K. M., and R. Nusse. 1997. Wnt signaling: a common theme in animal development. Genes Dev. 11:3286-3305. [DOI] [PubMed] [Google Scholar]
- 4.Caelles, C., H. Hennemann, and M. Karin. 1995. M-phase-specific phosphorylation of the POU transcription factor GHF-1 by a cell cycle-regulated protein kinase inhibits DNA binding. Mol. Cell. Biol. 15:6694-6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin, L., G. Merlino, and R. A. Depinho. 1998. Malignant melanoma: modern black plague and genetic black box. Genes Dev. 12:3467-3481. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, Y., N. Goldenberg-Cohen, P. Parrella, I. Chowers, S. L. Merbs, J. Pe'er, and D. Sidransky. 2003. Lack of BRAF mutation in primary uveal melanoma. Investig. Ophthalmol. Vis. Sci. 44:2876-2878. [DOI] [PubMed] [Google Scholar]
- 7.Cruz, F., 3rd, B. P. Rubin, D. Wilson, A. Town, A. Schroeder, A. Haley, T. Bainbridge, M. C. Heinrich, and C. L. Corless. 2003. Absence of BRAF and NRAS mutations in uveal melanoma. Cancer Res. 63:5761-5766. [PubMed] [Google Scholar]
- 8.Davies, H., G. R. Bignell, C. Cox, P. Stephens, S. Edkins, S. Clegg, J. Teague, H. Woffendin, M. J. Garnett, W. Bottomley, N. Davis, E. Dicks, R. Ewing, Y. Floyd, K. Gray, S. Hall, R. Hawes, J. Hughes, V. Kosmidou, A. Menzies, C. Mould, A. Parker, C. Stevens, S. Watt, S. Hooper, R. Wilson, H. Jayatilake, B. A. Gusterson, C. Cooper, J. Shipley, D. Hargrave, K. Pritchard-Jones, N. Maitland, G. Chenevix-Trench, G. J. Riggins, D. D. Bigner, G. Palmieri, A. Cossu, A. Flanagan, A. Nicholson, J. W. Ho, S. Y. Leung, S. T. Yuen, B. L. Weber, H. F. Seigler, T. L. Darrow, H. Paterson, R. Marais, C. J. Marshall, R. Wooster, M. R. Stratton, and P. A. Futreal. 2002. Mutations of the BRAF gene in human cancer. Nature 417:949-954. [DOI] [PubMed] [Google Scholar]
- 9.Dorsky, R. I., R. T. Moon, and D. W. Raible. 1998. Control of neural crest cell fate by the Wnt signalling pathway. Nature 396:370-373. [DOI] [PubMed] [Google Scholar]
- 10.Dorsky, R. I., D. W. Raible, and R. T. Moon. 2000. Direct regulation of nacre, a zebrafish MITF homolog required for pigment cell formation, by the Wnt pathway. Genes Dev. 14:158-162. [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn, K. J., B. O. Williams, Y. Li, and W. J. Pavan. 2000. Neural crest-directed gene transfer demonstrates wnt1 role in melanocyte expansion and differentiation during mouse development. Proc. Natl. Acad. Sci. USA 97:10050-10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eastman, Q., and R. Grosschedl. 1999. Regulation of LEF-1/TCF transcription factors by Wnt and other signals. Curr. Opin. Cell Biol. 11:233-240. [DOI] [PubMed] [Google Scholar]
- 13.Edmunds, S. C., I. A. Cree, F. Di Nicolantonio, J. L. Hungerford, J. S. Hurren, and D. P. Kelsell. 2003. Absence of BRAF gene mutations in uveal melanomas in contrast to cutaneous melanomas. Br. J. Cancer 88:1403-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisen, T., D. J. Easty, D. C. Bennett, and C. R. Goding. 1995. The POU domain transcription factor Brn-2: elevated expression in malignant melanoma and regulation of melanocyte-specific gene expression. Oncogene 11:2157-2164. [PubMed] [Google Scholar]
- 15.Galibert, M. D., S. Carreira, and C. R. Goding. 2001. The Usf-1 transcription factor is a novel target for the stress-responsive p38 kinase and mediates UV-induced tyrosinase expression. EMBO J. 20:5022-5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goding, C. R. 2000. Melanocyte development and malignant melanoma. Forum (Genoa) 10:176-187. [PubMed] [Google Scholar]
- 17.Goding, C. R. 2000. Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes Dev. 14:1712-1728. [PubMed] [Google Scholar]
- 18.Goodall, J., C. Wellbrock, T. J. Dexter, K. Roberts, R. Marais, and C. R. Goding. 2004. The Brn-2 transcription factor links activated BRAF to melanoma proliferation. Mol. Cell. Biol. 24:2924-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodgkinson, C. A., K. J. Moore, A. Nakayama, E. Steingrimsson, N. G. Copeland, N. A. Jenkins, and H. Arnheiter. 1993. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 74:395-404. [DOI] [PubMed] [Google Scholar]
- 20.Hou, L., J. J. Panthier, and H. Arnheiter. 2000. Signaling and transcriptional regulation in the neural crest-derived melanocyte lineage: interactions between KIT and MITF. Development 127:5379-5389. [DOI] [PubMed] [Google Scholar]
- 21.Hsu, S. C., J. Galceran, and R. Grosschedl. 1998. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol. Cell. Biol. 18:4807-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber, O., R. Korn, J. McLaughlin, M. Ohsugi, B. G. Herrmann, and R. Kemler. 1996. Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech. Dev. 59:3-10. [DOI] [PubMed] [Google Scholar]
- 23.Hughes, M. J., J. B. Lingrel, J. M. Krakowsky, and K. P. Anderson. 1993. A helix-loop-helix transcription factor-like gene is located at the mi locus. J. Biol. Chem. 268:20687-20690. [PubMed] [Google Scholar]
- 24.Ikeya, M., S. M. K. Lee, J. E. Johnson, A. P. McMahon, and S. Takada. 1997. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature 289:966-970. [DOI] [PubMed] [Google Scholar]
- 25.Kioussi, C., P. Briata, S. H. Baek, D. W. Rose, N. S. Hamblet, T. Herman, K. A. Ohgi, C. Lin, A. Glieberman, J. Wang, V. Brault, P. Ruiz-Lozano, H. D. Nguyen, R. kemler, C. K. Glass, Wynshaw-Boris, and M. G. Rosenfeld. 2002. Identification of a Wnt/Dvl/β-catenin>Pitx1 pathway mediating cell-type-specific proliferation during development. Cell 111:673-685. [DOI] [PubMed] [Google Scholar]
- 26.Leonard, J. H., and J. R. Bell. 1997. Insights into the Merkel cell phenotype from Merkel cell carcinoma cell lines. Aust. J. Dermatol. 38(Suppl. 1):S91-S98. [DOI] [PubMed] [Google Scholar]
- 27.Lowings, P., U. Yavuzer, and C. R. Goding. 1992. Positive and negative elements regulate a melanocyte-specific promoter. Mol. Cell. Biol. 12:3653-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackenzie, M. A., S. A. Jordan, P. S. Budd, and I. J. Jackson. 1997. Activation of the receptor tyrosine kinase Kit is required for the proliferation of melanoblasts in the mouse embryo. Dev. Biol. 192:99-107. [DOI] [PubMed] [Google Scholar]
- 29.Molenaar, M., M. van de Wetering, M. Oosterwegel, J. Peterson-Maduro, S. Godsave, V. Korinek, J. Roose, O. Destree, and H. Clevers. 1996. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell 86:391-399. [DOI] [PubMed] [Google Scholar]
- 30.Murakami, T., S. Toda, M. Fujimoto, M. Ohtsuki, H. R. Byers, T. Etoh, and H. Nakagawa. 2001. Constitutive activation of Wnt/beta-catenin signaling pathway in migration-active melanoma cells: role of LEF-1 in melanoma with increased metastatic potential. Biochem. Biophys. Res. Commun. 288:8-15. [DOI] [PubMed] [Google Scholar]
- 31.Nakai, S., H. Kawano, T. Yudate, M. Nishi, J. Kuno, A. Nagate, K.-i. Jishage, H. Hamada, H. Fujii, K. Kawamura, K. Shiba, and T. Noda. 1995. The POU domain trnscription factor Brn-2 is required for the determination of specific neuronal linages in the hypothalamus of the mouse. Genes Dev. 9:3109-3121. [DOI] [PubMed] [Google Scholar]
- 32.Pollock, P. M., U. L. Harper, K. S. Hansen, L. M. Yudt, M. Stark, C. M. Robbins, T. Y. Moses, G. Hostetter, U. Wagner, J. Kakareka, G. Salem, T. Pohida, P. Heenan, P. Duray, O. Kallioniemi, N. K. Hayward, J. M. Trent, and P. S. Meltzer. 2003. High frequency of BRAF mutations in nevi. Nat. Genet. 33:19-20. [DOI] [PubMed] [Google Scholar]
- 33.Rigel, D. S., R. J. Friedman, and A. W. Kopf. 1996. The incidence of malignant melanoma in the United States: issues as we approach the 21st century. J. Am. Acad. Dermatol. 34:839-847. [DOI] [PubMed] [Google Scholar]
- 34.Rimm, D. L., K. Caca, G. Hu, F. B. Harrison, and E. R. Fearon. 1999. Frequent nuclear/cytoplasmic localization of beta-catenin without exon 3 mutations in malignant melanoma. Am. J. Pathol. 154:325-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rimoldi, D., S. Salvi, D. Lienard, F. J. Lejeune, D. Speiser, L. Zografos, and J. C. Cerottini. 2003. Lack of BRAF mutations in uveal melanoma. Cancer Res. 63:5712-5715. [PubMed] [Google Scholar]
- 36.Rubinfeld, B., P. Robbins, M. El-Gamil, I. Albert, E. Porfiri, and P. Polakis. 1997. Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science 275:1790-1792. [DOI] [PubMed] [Google Scholar]
- 37.Schonemann, M. D., A. K. Ryan, R. J. McEvilly, S. M. O'Connell, C. A. Arias, K. A. Kalla, P. Li, P. E. Sawchenko, and M. G. Rosenfeld. 1995. Development and survival of the endocrine hypothalamus and posterior pituitary gland requires the neuronal POU domain factor Brn-2. Genes Dev. 9:3122-3155. [DOI] [PubMed] [Google Scholar]
- 38.Schreiber, E., K. Harshman, I. Kemler, U. Malipiero, W. Schaffner, and A. Fontana. 1990. Astrocytes and glioblastoma cells express novel octamer-DNA binding proteins distinct from the ubiquitous Oct-1 and B cell type Oct-2 proteins. Nucleic Acids Res. 18:5495-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schreiber, E., A. Himmelmann, U. Malpiero, A. Tobler, R. Stahel, and A. Fontana. 1992. Human small cell lung cancer expresses the octamer DNA-binding and nervous system-specific transcription factor N-Oct 3 (Brain-2). Cancer Res. 52:6121-6124. [PubMed] [Google Scholar]
- 40.Schreiber, E., R. E. Merchant, O. D. Wiestler, and A. Fontana. 1994. Primary brain tumors differ in their expression of octamer deoxyribonucleic acid-binding transcription factors from long-term cultured glioma cell lines. Neurosurgery 34:129-135. [PubMed] [Google Scholar]
- 41.Segil, N., B. Roberts, and N. Heintz. 1991. Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science 254:1814-1816. [DOI] [PubMed] [Google Scholar]
- 42.Smit, D. J., A. G. Smith, P. G. Parsons, G. E. Muscat, and R. A. Sturm. 2000. Domains of Brn-2 that mediate homodimerization and interaction with general and melanocytic transcription factors. Eur. J. Biochem. 267:6413-6422. [DOI] [PubMed] [Google Scholar]
- 43.Takeda, K., K. Yasumoto, R. Takada, S. Takada, K. Watanabe, T. Udono, H. Saito, K. Takahashi, and S. Shibahara. 2000. Induction of melanocyte-specific microphthalmia-associated transcription factor by Wnt-3a. J. Biol. Chem. 275:14013-14016. [DOI] [PubMed] [Google Scholar]
- 44.Tetsu, O., and F. McCormick. 1999. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398:422-426. [DOI] [PubMed] [Google Scholar]
- 45.Thomson, J. A., K. Murphy, E. Baker, G. R. Sutherland, P. G. Parsons, and R. A. Sturm. 1995. The brn-2 gene regulates the melanocytic phenotype and tumorigenic potential of human melanoma cells. Oncogene 11:690-700. [PubMed] [Google Scholar]
- 46.van de Weterring, M., E. Sancho, C. Verwij, W. de Lau, I. Oving, A. Hurlstone, K. van der Horn, E. Batlle, D. Coudreuse, and A.-P. Hramis. 2002. The β-catenin/Tcf-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111:241-250. [DOI] [PubMed] [Google Scholar]
- 47.Verma, U. N., R. M. Surabhi, A. Schmaltieg, C. Becerra, and R. B. Gaynor. 2003. Small interfering RNAs directed against beta-catenin inhibit the in vitro and in vivo growth of colon cancer cells. Clin. Cancer Res. 9:1291-1300. [PubMed] [Google Scholar]
- 48.Wehrle-Haller, B., M. Meller, and J. A. Weston. 2001. Analysis of melanocyte precursors in Nf1 mutants reveals that MGF/KIT signaling promotes directed cell migration independent of its function in cell survival. Dev. Biol. 232:471-483. [DOI] [PubMed] [Google Scholar]
- 49.Widlund, H. R., M. A. Horstmann, E. R. Price, J. Cui, S. L. Lessnick, M. Wu, X. He, and D. E. Fisher. 2002. β-Catenin-induced melanoma growth requires the downstream target Microphthalmia-associated transcription factor. J. Cell Biol. 158:1079-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiebe, M. S., T. K. Nowling, and A. Rizzino. 2003. Identification of novel domains within Sox-2 and Sox-11 involved in autoinhibition of DNA binding and partnership specificity. J. Biol. Chem. 278:17901-17911. [DOI] [PubMed] [Google Scholar]
- 51.Yavuzer, U., and C. R. Goding. 1994. Melanocyte-specific gene expression: role of repression and identification of a melanocyte-specific factor, MSF. Mol. Cell. Biol. 14:3494-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]