Background: Pantothenate transport is essential for Plasmodium development. The transporter that mediates entry of pantothenate is unknown.
Results: PfPAT encodes the primary pantothenate transporter of P. falciparum.
Conclusion: PfPAT plays an essential function in parasite development and thus is a valid target for antimalarial therapy.
Significance: PfPAT is the first pantothenate transporter identified and characterized in protozoan parasites and a valid target for therapy.
Keywords: Drug Delivery, Malaria, Plasmodium, Transport, Yeast
Abstract
The human malaria parasite Plasmodium falciparum is absolutely dependent on the acquisition of host pantothenate for its development within human erythrocytes. Although the biochemical properties of this transport have been characterized, the molecular identity of the parasite-encoded pantothenate transporter remains unknown. Here we report the identification and functional characterization of the first protozoan pantothenate transporter, PfPAT, from P. falciparum. We show using cell biological, biochemical, and genetic analyses that this transporter is localized to the parasite plasma membrane and plays an essential role in parasite intraerythrocytic development. We have targeted PfPAT to the yeast plasma membrane and showed that the transporter complements the growth defect of the yeast fen2Δ pantothenate transporter-deficient mutant and mediates the entry of the fungicide drug, fenpropimorph. Our studies in P. falciparum revealed that fenpropimorph inhibits the intraerythrocytic development of both chloroquine- and pyrimethamine-resistant P. falciparum strains with potency equal or better than that of currently available pantothenate analogs. The essential function of PfPAT and its ability to deliver both pantothenate and fenpropimorph makes it an attractive target for the development and delivery of new classes of antimalarial drugs.
Introduction
Nearly half of the world population is at risk of contracting malaria, a parasitic disease caused by protozoan parasites of the genus Plasmodium (1). Five species of Plasmodium infect humans, and cases of infection caused by Plasmodium knowlesi are on the rise (2). These species cause ∼250 million annual cases of clinical malaria and over 1 million deaths (3). Most deaths occur in Africa and can be ascribed to infection by Plasmodium falciparum. The lack of an effective vaccine and the emergence of drug-resistant P. falciparum strains emphasize the need for better strategies that target new pathways that are unique to the parasite and that have not yet been targeted for antimalarial therapy.
Plasmodium intraerythrocytic proliferation is fueled by nutrients, such as purine nucleosides and nucleobases, amino acids, sugars, fatty acids, and vitamins, scavenged from the host (4). The uptake of these essential nutrients involves endogenous transporters at the erythrocyte membrane as well as parasite-encoded permeases targeted to red blood cell membrane, parasite plasma membrane, or intracellular organelles (5–14). The water-soluble vitamin pantothenic acid (vitamin B5) is a precursor of the important enzyme cofactor CoA, a universal carrier of activated acyl groups involved in 9% of biochemical reactions identified in all living organisms (15). Because P. falciparum cannot synthesize pantothenate de novo, the acquisition of this vitamin from the host is an indispensable nutritional function for the parasite (Fig. 1).
FIGURE 1.
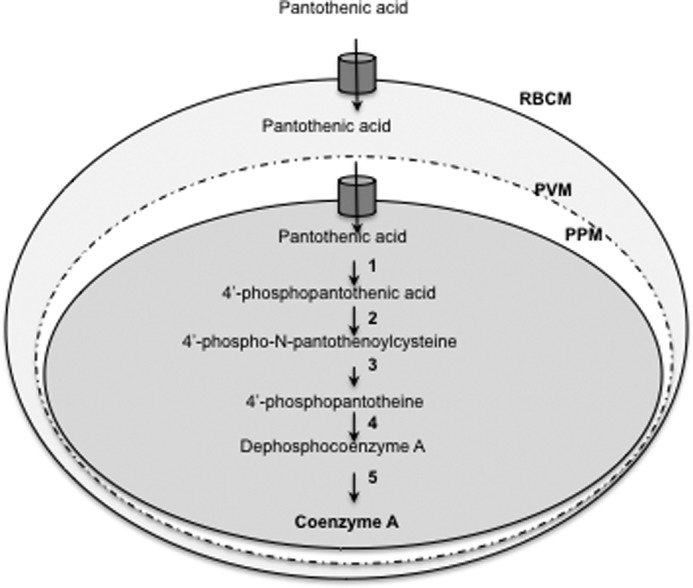
Schematic representation of pantothenate transport and CoA biosynthesis in P. falciparum-infected erythrocytes. Pantothenate is transported across the plasma membrane by the parasite pantothenate transporter and converted into CoA by the enzymes pantothenate kinase (1), phosphopantothenoylcysteine synthetase (2), phosphopantothenoylcysteine decarboxylase (3), phosphopantotheine adenyltransferase (4), and dephospho-CoA kinase (5). PPM, parasite plasma membrane; PVM, parasitophorous vacuolar membrane; RBCM, red blood cells membrane.
The transporters involved in the uptake of pantothenic acid across the red blood cell membrane or the parasite plasma membrane remain unknown, and no pantothenate transporters have yet been identified in any other protozoa. Biochemical studies have demonstrated that in contrast to the negligible pantothenate uptake in uninfected erythrocytes, pantothenate transport across infected red blood cells is rapid (16). This transport activity detected following parasite infection is mediated by the new permeation pathway. Once inside the red blood cell cytoplasm, pantothenate is transported into the parasite via a parasite-specific low affinity permease (Km = ∼23 mm) and converted into CoA via parasite-encoded enzymes (17). Unlike mammalian cells, where the transport of pantothenate is entirely dependent on the presence of Na+, the uptake of pantothenate in P. falciparum isolated trophozoites is Na+-independent, markedly dependent on pH (17), and electroneutral with pantothenate and H+ entering the parasite with a stoichiometry of 1:1 (17).
In lower eukaryotes, pantothenate transport has been characterized in Saccharomyces cerevisiae and Schizosaccharomyces pombe (18, 19). In S. cerevisiae, pantothenate is transported via a high affinity plasma membrane H+-pantothenate symporter Fen2, which is also responsible for the uptake of the antifungal drug fenpropimorph (19). Accordingly, fen2Δ knock-out mutants do not transport pantothenate, are unable to grow on media containing physiological concentrations of pantothenate (1–10 μm), and are resistant to fenpropimorph (19, 20).
Here we report the identification and functional characterization of the first protozoan pantothenate transporter, PfPAT, from P. falciparum. PfPAT plays an essential function in parasite intraerythrocytic development, and its expression in fen2Δ yeast mutant complements the growth deficiency on low pantothenate and mediates the entry of fenpropimorph into these cells.
EXPERIMENTAL PROCEDURES
Parasite Cell Culture
Plasmodium falciparum clones 3D7, NF54, and Dd2 were propagated in human RBCs at 2% hematocrit using standard growth conditions and in the presence of 0.5% AlbuMAX (21). RPMI medium was either purchased from Invitrogen or made by adding all components with the exception of pantothenic acid to make pantothenic acid-free medium.
Plasmodium Plasmid Constructs
The transfection vectors pHC1-ACP-GFP and pRZ-TK-BSD2 used in this study to create a PfPAT-GFP fusion or to knock out the PfPAT chromosomal locus were previously described (8, 22–24). To generate the Pfpat-gfp fusion construct, the open reading frame of PfPAT was amplified by PCR using genomic DNA as a template and primers 5′-GACTCTCGAGATGGCTAAAAACCAGTATATGGAGG-3′ and 5′-GACTCCTAGGTGTTAACATTTTTTTTTCTGGAATGGAATGG-3′. The PCR fragment was then cloned in the pHC1-ACP-GFP vector. The resulting expression vector, pYAN029, contains PfPAT-GFP fusion under the regulatory control of the P. falciparum CAM1 promoter and HSP86 terminator and harbors the Toxoplasma gondii DHFR-TS marker that confers resistance to pyrimethamine. To construct the targeting vector pYAN022 for PfPAT knock-out, a 585-bp fragment of PfPAT (nucleotides 35–620 of the ORF) was amplified and subcloned into the HindIII/BlpI in the pRZ-TK-BSD2 vector, yielding pYAN021. This plasmid contains a positive selectable marker BSD for selection on blasticidin (25) and a negative marker TK conferring sensitivity to ganciclovir. A second 495-bp fragment of PfPAT (nucleotides 1035–1530) was cloned into the EcoRI/KasI site of pYAN021 to generate pYAN022.
Generation of Transgenic Parasites
pYAN022 and pYAN029 vectors were used to transfect early ring stage parasites (3D7 clone) by electroporation as previously described (8). Transfected parasites were selected on media supplemented with blasticidin (2.5 μg/ml) or pyrimethamine (100 nm), respectively. Transgenic parasites transfected with pYAN022 were subjected to several cycles of additions and removal of blasticidin and ganciclovir and tested at different times during the selection process for integration of the targeting cassette into the PfPAT locus by PCR using specific sets of oligonucleotides. Individual clones were isolated by limited dilution and further characterized by PCR.
Yeast Strains and Growth Conditions
S. cerevisiae strains BY4741 (Mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and fen2Δ (Mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 fen2Δ::Kanr) were grown in YPD medium or dextrose-based synthetic defined medium made from analytical grade chemicals and containing appropriate supplements to support cell growth but lacking pantothenate (SV medium). The cells were grown at 30 °C. To enhance membrane stability of PfPAT, a genetic deletion of the END3 gene was created in the BY4741 and fen2Δ genetic background and confirmed by PCR. Cell density was determined at A600 nm.
Yeast Plasmid Constructs
A codon-optimized version of PfPAT (PfPATco) was synthesized to improve the codon usage and decrease the A + T content (Genscript). PfPAT was inserted into the yeast expression vector pYES2.1-/V5-His-TOPO (Invitrogen) generating pYES-PfPATco-V5. The yeast FEN2 gene encoding the S. cerevisiae pantothenate transporter was also cloned into the pYES2.1-/V5-His-TOPO (Invitrogen) after amplification of the ORF using genomic DNA as a template. A chimeric form, PfPATf, consisting of the fusion of nucleotides 1–222 of FEN2 and nucleotides 250–1695 of PfPATco, including the V5 tag present in pYES-PfPATco-V5, was generated by PCR. The amplicon was cloned into the BamHI/PstI sites of the pBEVY-U (carrying the URA3 marker) expression vector (26), allowing expression of the fusion under the regulatory control of the GPD promoter. All sequences were verified by DNA sequencing. Yeast strains were transformed using a high efficiency protocol as previously described (27).
Yeast Growth Assays
To monitor yeast growth on pantothenate, yeast strains were grown overnight in 5 ml of SV medium lacking uracil and supplemented with 100 μm of pantothenate. The cells were harvested by centrifugation, washed twice in SV medium, and inoculated at a cell density of 2 × 105 cells/ml in SV medium lacking uracil supplemented with 1, 10, and 100 μm of pantothenate. Growth of fen2Δ pPfPATf was normalized based on the difference of fitness observed in BY4741 transformed with pBEVY or pPfPATf. For fenpropimorph inhibition assays, wild type and mutant yeast strains were precultured overnight in YPD medium, and this preculture was then used to inoculate new cultures at a starting cell density of 2 × 105 cells/ml in YPD lacking or supplemented with fenpropimorph (1 μg/ml).
Production and Affinity Purification of Anti-PfPAT Antibodies
The N-terminal fragment of PfPAT encoding the first 89 amino acids was cloned into pMALC2X (NEB) vector in fusion with MBP protein. PfPAT was expressed and purified according to the manufacturer recommendations. Purified recombinant protein was injected into rabbits and antibody-purified from the serum by affinity purification as previously described (28).
Localization of PfPAT in Yeast and P. falciparum
fen2Δ cells expressing Fen2, PfPATco, or PfPATf were grown to A600 0.5–1.0 in SV medium supplemented with 100 μm pantothenate. The cells were fixed, washed, depleted of their cell wall, and permeabilized, and immunofluoresence analyses were performed as previously described (5). Localization of PfPAT-GFP and native PfPAT (using PfPAT antibody) in P. falciparum-infected erythrocytes by immunofluorescence and immunoelectron microscopy were carried out as previously described (5, 29).
Targeting PfPAT for Degradation Using a Cell-penetrating Peptide-Morpholino Oligomer Conjugate
A cell-penetrating peptide-morpholino oligomer (PMO)3 conjugate was designed to target and mediate the cleavage of PfPAT mRNA and tested against P. falciparum as previously described (30). The sequence of the PMO used in this study was GUAUACGAGGUUCGAAUCCUCGGUUCUU and targeted the PfPAT mRNA from nucleotides 39 to 50 and was validated in vitro as previously described (30).
PfPAT Expression in Mammalian Cells and Transport Assays
For expression, localization and transport analyses of PfPAT in mammalian cells, the codon-optimized version of PfPAT was amplified using the following primers: 5′-GCCACCATGGCTAAAAATCAATACATG-3′ and 5′-TGTTAACATTTTCTTTTCTGGAATAGTTTC-3′, cloned into the EcoRI site of mammalian expression pCMV-3Tag3A vector, and transfected into HEK-293T and ARPE19 cells. Human retinal pigment epithelial ARPE19 cells were used because they are easy to transfect and have been successfully used to express a variety of genes from different species including the hSMVT system. Transfection was done by Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. After 48 h of transfection, [3H]pantothenic acid uptake (0.3 μCi; 6 nm) was examined in Krebs-Ringer buffer (pH 7.4) for 5 min. The cells were washed with ice-cold buffer (twice), lysed with 1 n NaOH, and neutralized with 10 n HCl, and radioactivity was counted in a liquid scintillation counter. Uptake was expressed in term of fmol/mg protein/5 min (protein was determined in cell digest by Bradford method; Bio-Rad). Mock refers to endogenous transport (where ARPE19 cells were transfected with vector alone) and used as reference to induce uptake in cells transfected with the PfPAT construct in the same vector. The data are means ± S.E. of five separate experiments (each containing three or four separate points).
RESULTS
Identification of a Candidate Malarial Pantothentate Transporter PfPAT
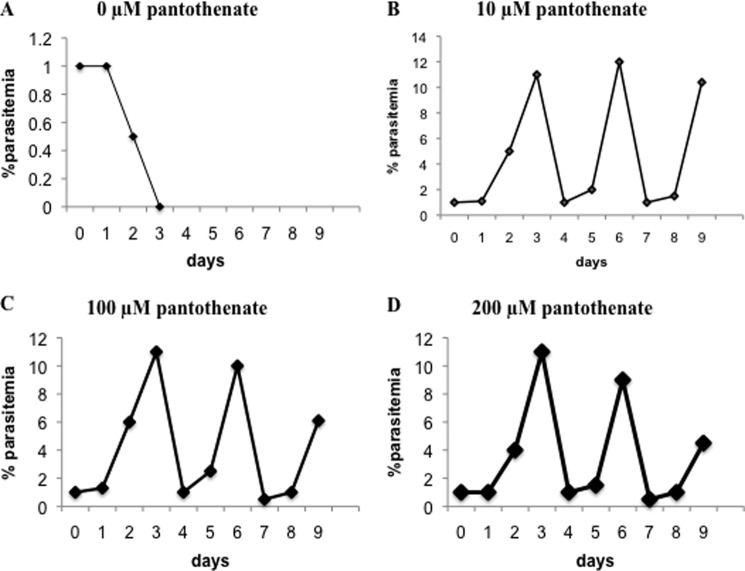
Highly synchronized P. falciparum parasites maintained in culture medium lacking pantothenate are unable to undergo normal development within human red blood cells (Fig. 2), consistent with previous reports (31, 32). Supplementation of the culture medium with increasing concentrations of pantothenate allows the parasite to undergo its normal ∼48 h cycle of development and division within red blood cells (Fig. 2). These findings are consistent with an essential role of pantothenate in parasite development within human erythrocytes.
FIGURE 2.
Essential role of exogenous pantothenate in P. falciparum intraerythrocytic development. Growth of P. falciparum parasites in the absence (pantothenic acid-free medium) (A) or presence of 10 μm (B), 100 μm (C), and 200 μm (D) pantothenate is shown. Parasite cultures were initiated at 1% parasitemia and 2% hematocrit and maintained for three developmental cycles. Cultures were diluted at days 3 and 6 to 1% when parasitemia reached 8–10%.
To identify the primary pantothenate transporter of P. falciparum, we screened a P. knowlesi cDNA library (33) made in a yeast expression vector under the constitutive ADH1 promoter for clones that can complement the growth defect of the yeast mutant fen2Δ on minimal medium containing 10 μm pantothenate. The functional complementation approach using the P. knowlesi cDNA library was recently used to isolate the malarial phosphatidylserine decarboxylase gene (33). However, attempts to complement the fen2Δ using this library did not yield transformants with a growth phenotype similar to that of the wild type or the fen2Δ+Fen2 complemented strain, harboring a wild type FEN2 gene. Several independent, partially complemented clones were identified, but sequencing of the complementing plasmids did not identify cDNAs encoding putative membrane transporters or classical enzymes of the CoA synthesis pathways. The phenotype of these partially complemented clones was likely a result of an enhanced overall fitness. These clones were not further pursued. The inability to identify the pantothenate transporter using this library could be due to either the absence of the plasmid harboring the transporter cDNA in the library or the inability of the transporter to localize to the yeast plasma membrane (5). Therefore, we searched the library of putative P. falciparum metabolite/drug transporters for possible homologs of eukaryotic and prokaryotic pantothenate and vitamin permeases. Whereas no homologs of mammalian vitamin transporters could be found in the genome of P. falciparum, our in silico analysis identified a gene PF3D7_0206200, which we named PfPAT, as a possible candidate for a vitamin transporter gene in this parasite. PfPAT is a member of the multifacilitator super family and shares significant sequence similarity (∼17% identity and ∼30% similarity; supplemental Fig. S1) with the pantothenate transporters Liz1 and Fen2 from S. pombe and S. cerevisiae (19). Phylogenetic analysis revealed the presence of PfPAT orthologs among other apicomplexa, viridiplantae, and the green algae Chlorella vulgaris (supplemental Fig. S2), suggesting that it might have been acquired following the second endosymbiotic event at the origin of alveolata (34). PfPAT encodes a 565-amino acid polypeptide with 11 predicted transmembrane domains (supplemental Fig. S3). Also, except for a 15-amino acid asparagine-rich domain, which is only present in PfPAT, orthologs in other Plasmodium species share high degree of identity and similarity throughout the length of the protein sequence (supplemental Table S1). No significant sequence similarity exists between PfPAT and the human multivitamin transporter, hSMVT (supplemental Fig. S4). Furthermore, no homologs of the human hSMVT could be found in the Plasmodium databases.
PfPAT Is Localized to the Parasite Plasma Membrane
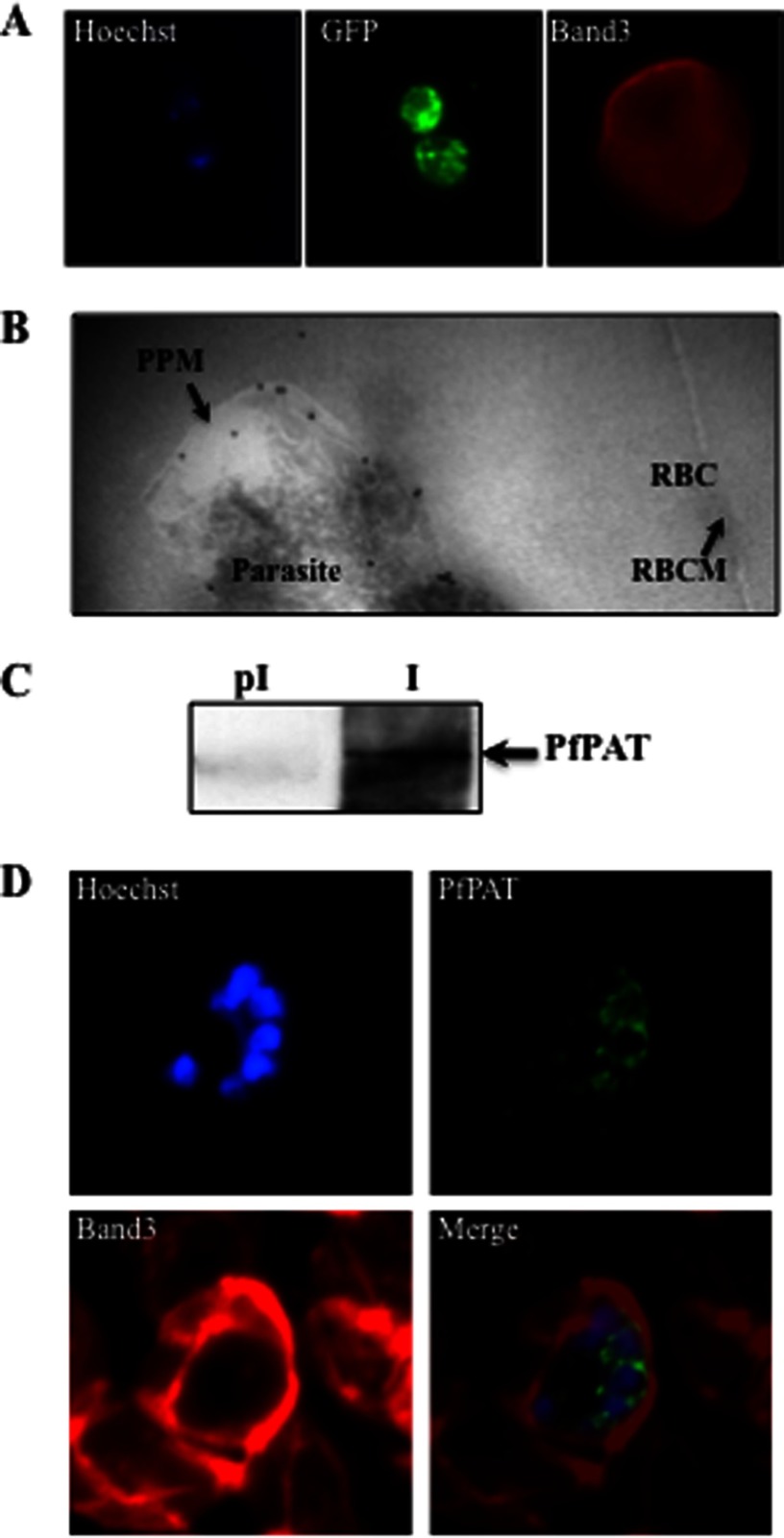
To determine the localization of PfPAT in P. falciparum, transgenic parasites expressing PfPAT fused to GFP, in a vector harboring the TgDHFR-TS positive selectable marker, were selected following transfection of the wild type P. falciparum 3D7 clone and selection on medium containing pyrimethamine. Immunofluorescence analyses showed PfPAT signal both on the parasite plasma membrane as well as associated with vesicular structures extending from the nuclear membrane in all blood stages (Fig. 3A). The latter likely resulting from overexpression of the PfPAT-GFP construct. Similar signals were previously seen with the purine transporter PfNT1 when this otherwise exclusively plasma membrane protein was overexpressed as a fusion with GFP (29). PfPAT plasma membrane localization was further confirmed by immunoelectron microscopy on ring, trophozoite, and schizont-infected erythrocytes with most PfPAT signal (∼68% of gold particles) found to be associated with the parasite plasma membrane (Fig. 3B). No PfPAT signals could be detected on the red blood cell or parasitophorous vacuolar membranes. To further demonstrate the localization of PfPAT to the parasite plasma membrane, we have raised polyclonal antibodies against the N-terminal domain of the protein. These antibodies were affinity-purified and recognized a specific band of ∼62 kDa, which was absent in preimmune sera (Fig. 3C). Immunofluorescence analyses using these antibodies showed localization of the protein to the plasma membrane of the parasite at all stages of the parasite intraerythrocytic life cycle. This localization is particularly noticeable in parasites at the schizont stage with fluorescence signal surrounding each individual merozoite (Fig. 3D). Together these results indicate that PfPAT is expressed on the parasite plasma membrane, consistent with its putative function in the uptake of pantothenate across the parasite plasma membrane.
FIGURE 3.
PfPAT localization in P. falciparum-infected erythrocytes. A, localization of PfPAT-GFP by immunofluorescence analysis using anti-GFP (green). The red blood cell membrane marker Band3 (red) was detected using an anti-Band3 monoclonal antibody. The parasite nucleus was visualized using the Hoescht 33258 dye (blue). B, transmission electron micrograph of ultrathin cryosections of the intraerythrocytic early trophozoite stage of P. falciparum PfPAT-GFP transgenic parasites using anti-GFP antibody (18-nm gold particles; indicated with arrows). PPM, parasite plasma membrane; RBC, red blood cell; RBCM, red blood cell membrane. C, detection of native PfPAT using affinity-purified PfPAT antibodies (I). Preimmune serum (pI) is used as a control. D, localization of PfPAT in P. falciparum 3D7 parasites at the schizont stage using anti-PfPAT antibodies (green). The red blood cell membrane marker Band3 (red) was detected using an anti-Band3 monoclonal antibody. The parasite nucleus was visualized using the Hoescht 33258 dye (blue).
PfPAT Is Essential for P. falciparum Intraerythrocytic Development
To characterize the importance of PfPAT during the P. falciparum intraerythrocytic development, we attempted to knock out its chromosomal locus under physiological pantothenate concentrations (0.5 μm), as well as in the presence of supraphysiological concentrations (200 μm) of pantothenate if the gene is essential and a conditional knock-out cannot be generated. This nutritional complementation strategy was used successfully to create genetic deletions of the purine transporter gene PfNT1 in the presence of hypoxanthine and the phosphoethanolamine methyltransferase gene PfPMT in the presence of choline (8, 22). A targeting vector, pYAN022, was constructed using the pRZ-TK-BSD2 vector, as previously described (8, 22), and was used to transfect P. falciparum 3D7 parasites. Transfectants were selected for their ability to grow in the presence of blasticidin (25). After several cycles of growth of blasticidin-resistant transfectants on media lacking or containing blasticidin and/or ganciclovir, genomic DNA was isolated and analyzed by PCR for integration into the PfPAT locus. Even after several transfection attempts, no evidence for a gene replacement event following a double crossover or 5′ integration (which results in a major gene truncation) events could be detected in the cultures of parasites grown under physiological or supraphysiological concentrations of pantothenate. On the other hand, PCR analyses to examine a 3′ integration event (which does not alter the integrity of the gene) identified the expected 1.7-kb fragment (Fig. 4, A and B). Together these results indicate that deletion of the chromosomal PfPAT locus abolishes parasite growth. This was further confirmed by using a specific cell-penetrating peptide morpholino oligomer, which mediates the selective cleavage of the 5′ region of the PfPAT mRNA, following an approach recently described in P. falciparum (30). Increasing concentrations of the cell-penetrating peptide morpholino oligomer reduced the proliferation of P. falciparum with an IC50 = ∼2 μm (Fig. 4C). In addition, such a cell-penetrating peptide morpholino oligomer was equally effective on the pyrimethamine resistant strain Hb3 (Fig. 4D). Together, these results indicate that PfPAT plays an essential role during the parasite intraerythrocytic life cycle.
FIGURE 4.
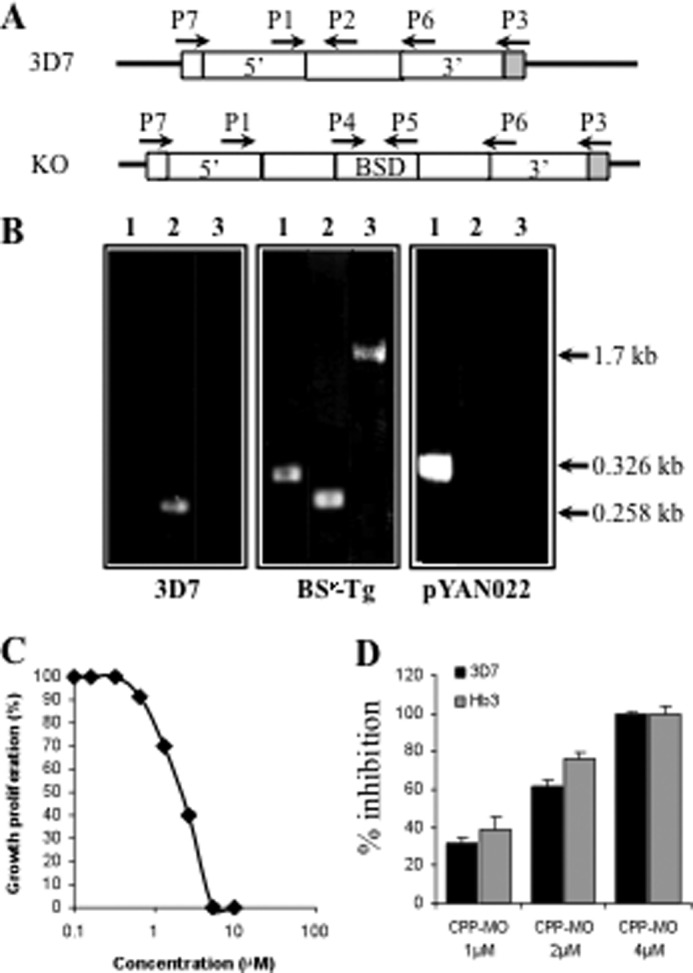
Genetic evidence for an essential role of PfPAT in P. falciparum. A, schematic representation of the PfPAT locus in 3D7 and expected knock-out parasites. B, PCR for screening knock-out parasite using primers pairs P4 and P5 (lanes 1), P1 and P2 (lanes 2), and P3 and P4 (lanes 3). Transgenic parasites harboring the pYAN022 vector were selected on blasticidin-containing medium and further subjected to several cycles of growth on media lacking or supplemented with blasticidin and/or ganciclovir. 35 clones were isolated by limited dilution and tested by PCR using the primer pairs P3 and P4. C and D, inhibition of parasite proliferation using a cell-penetrating peptide morpholino oligomer (CPP-MO), which mediates selective cleavage of the PfPAT mRNA. C, growth of 3D7 parasites in the absence of or with increasing concentrations of PfPAT PMO. D, effect of 1, 2, and 4 μm concentrations of PfPAT PMO on the growth of 3D7 and HB3 strains.
PfPAT Expression on the Yeast Plasma Membrane Complements Pantothenate Transport Deficiency
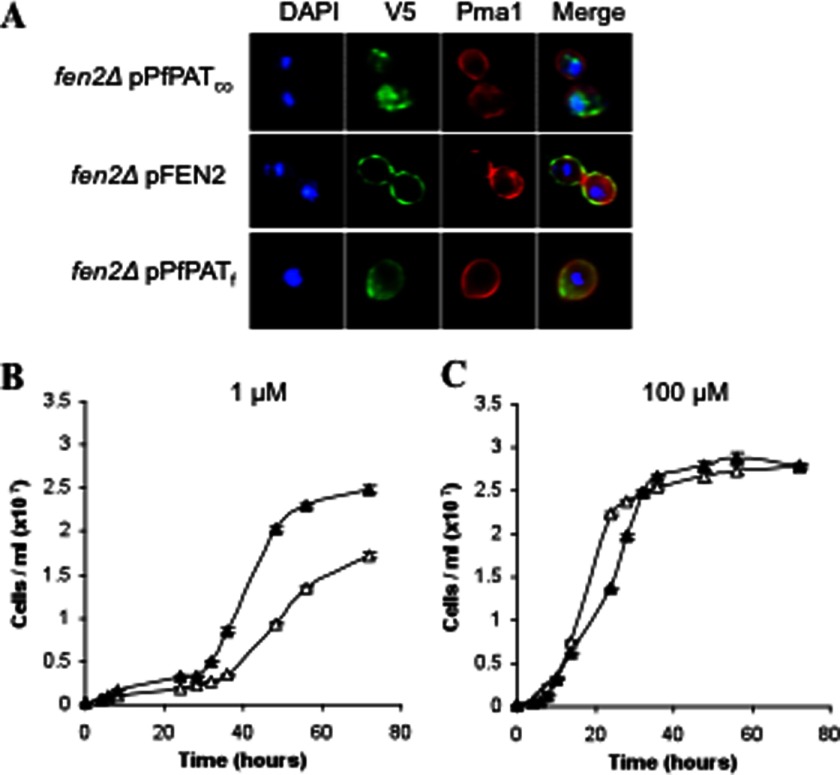
Previous studies have shown that, more often than not, expression and characterization of P. falciparum transporter genes in the yeast S. cerevisiae requires codon optimization of the gene of interest, reduction of the overall A + T base content, and amelioration of the distribution of codon usage frequency along the length of the gene sequence (5, 28, 35, 36). Therefore, for successful expression of PfPAT, the codon usage and GC content of this gene were adjusted to produce PfPATco. The gene PfPATco was then subcloned into the pYES2.1 vector for expression in yeast under the regulatory control of the strong and inducible GAL1 promoter. The pYES-PfPATco was used to transform the yeast fen2Δ, which lacks the yeast's only pantothenate transporter and shows severe growth delays on low pantothenate concentrations. The full-length PfPATco, however, did not complement the growth defect of the fen2Δ yeast knock-out on low pantothenate concentrations. Consistent with these findings, immunofluorescence assays revealed that the full-length protein was not targeted to the yeast plasma membrane (Fig. 5A). As a control, the yeast Fen2 expressed under similar conditions was successfully localized to the plasma membrane (Fig. 5A). Therefore, to target PfPATco to the yeast plasma membrane, we employed a series of molecular and genetic modifications that we have previously successfully employed to target the P. falciparum purine transporter PfNT2 to the yeast the plasma membrane (5). First, we constructed a chimeric form of PfPATco (PfPATf) in which the N-terminal domain of PfPAT, preceding the first predicted transmembrane domain, was replaced with that of the yeast Fen2. Second, we replaced the expression vector pYES2.1 with the pBEVY-U vector to express the chimeric form under the regulatory control of the constitutive and weaker GPD promoter (26). As shown in Fig. 5A, addition of the N-terminal domain of the Fen2 protein allowed targeting of PfPATf to the yeast plasma membrane. PfPAT localization pattern was similar to that of the yeast plasma membrane marker Pma1p (37) (Fig. 5A).
FIGURE 5.
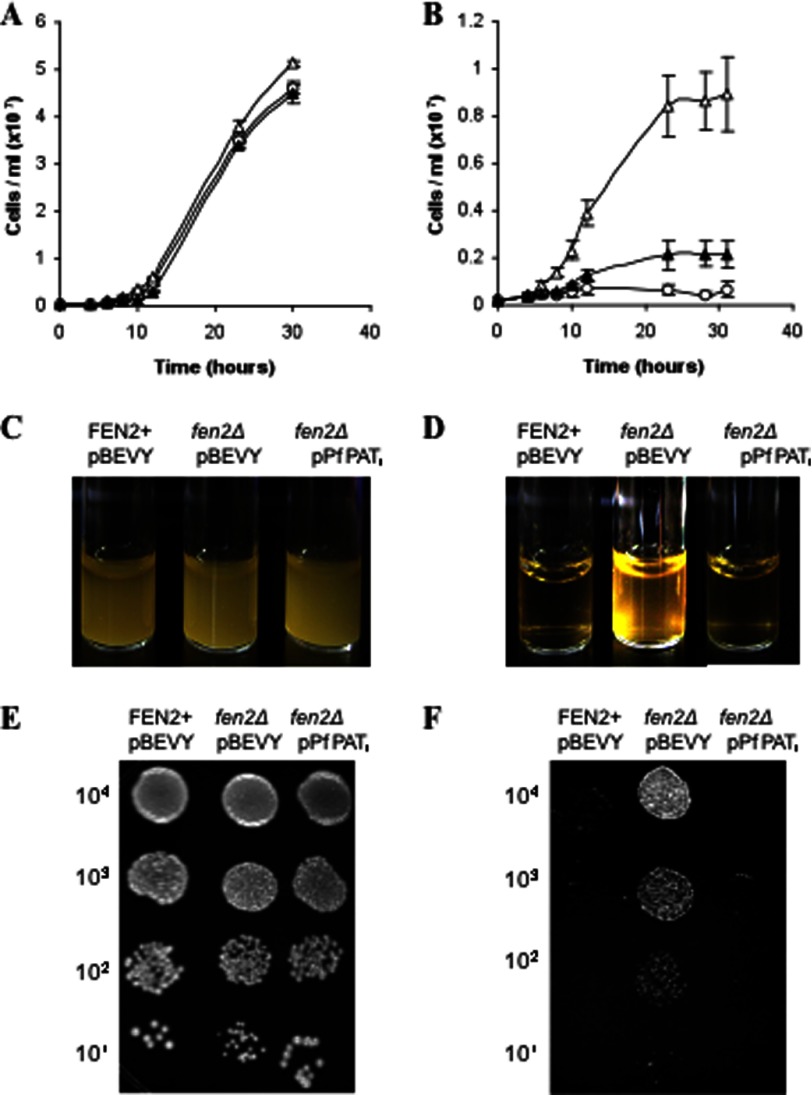
Expression and functional characterization of PfPAT in yeast. A, targeting PfPAT to the yeast plasma membrane for functional analysis. fen2Δ cells were transformed with plasmids harboring the codon optimized version of PfPAT (PfPATco), the yeast pantothenate transporter Fen2 (pFEN2) or a chimeric form (PfPATf). All constructs result in a V5 epitope in the C-terminal region of the fusion proteins. Plasma membrane localization was confirmed using anti-Pma1 antibodies. Nuclear staining was achieved using DAPI. B and C, complementation of fen2Δ pantothenate uptake defect by PfPAT. fen2Δ cells harboring the empty pBEVY-U vector (open triangles) or the pPFPATf (closed triangles) expression vector were precultured in SV medium supplemented with 100 μm of pantothenate, washed twice, and inoculated to an initial cell density of 2 × 105 cells/ml in medium supplemented with 1 μm (B) or 100 μm (C) of pantothenate. All cultures were initiated at pH 5.7.
To assess whether plasma membrane-localized PfPAT can function as a pantothenate transporter, the fen2Δ knock-out strain was transformed with an expression plasmid harboring PfPATf or the empty vector, and the transformants were inoculated at 2 × 105 cells/ml and monitored over time for their rate of growth on media containing 1 or 100 μm pantothenic acid at pH 5.7. As shown in Fig. 5B, whereas the fen2Δ strain harboring the empty vector reached a cell density of 1.5 × 107/ml in 56 h, fen2Δ cells harboring PfPATf reached 2.4 × 107/ml at that time point. These results suggest that PfPAT expression on the yeast plasma membrane allows entry of pantothenate into the cell. As expected, all strains showed similar growth on the supraphysiological concentration of 100 μm (Fig. 5B).
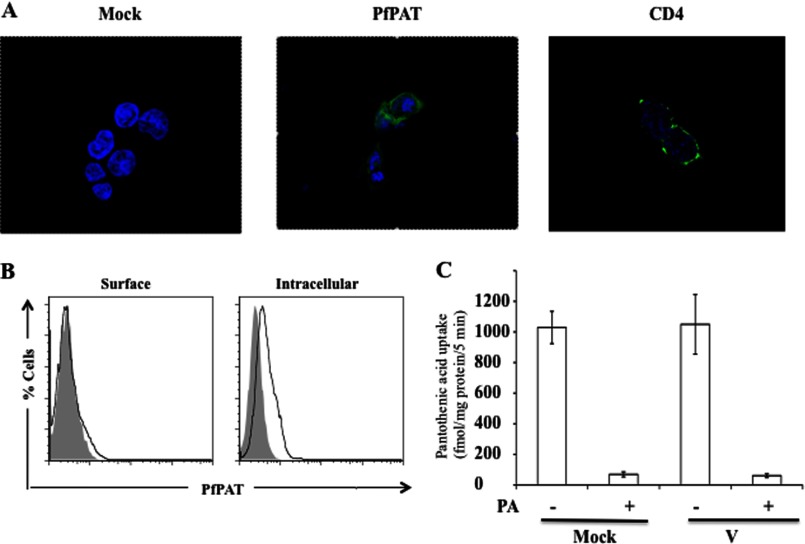
The successful expression of PfPAT on the yeast plasma membrane and its ability to complement fen2Δ growth defect on low pantothenate concentrations led us to investigate whether its expression in mammalian cells can be achieved to allow direct determination of its biochemical properties. HEK-293T cells were transfected with a vector expressing PfPAT harboring a C-terminal FLAG tag and examined for plasma membrane localization by confocal microscopy and FACS analysis. Whereas successful expression of PfPAT in mammalian cells was achieved, the protein did not localize to the plasma membrane (Fig. 6, A and B). As a control, cells transfected with CD4-GFP showed plasma membrane localization of this fusion protein by confocal microscopy (Fig. 6A). Consistent with the lack of expression of PfPAT on the plasma membrane of mammalian cells, no uptake of [3H]pantothenic acid could be detected in these cells (Fig. 6C).
FIGURE 6.
A, confocal microscopy of HEK-293T cells transfected with PfPAT-FLAG (middle) and stained with anti-FLAG FITC (green) or transfected with CD4-GFP as a control (right). Cell nucleus was visualized by staining with Hoescht 33258 dye (blue). B, flow cytometric analysis of HEK-293T cells mock transfected (gray) or transfected with PfPAT-FLAG (black) and surface stained or permeabilized for intracellular staining with anti-FLAG FITC. C, uptake of [3H]pantothenic acid (PA) by ARPE19 cells transiently transfected with pCMV-3Tag vector alone or with PfPAT cloned in the vector examined 48 h after transfection. Mock denotes the endogenous pantothenic acid transport by ARPE19 cells. The presence of carrier was confirmed by competing the [3H]pantothenic acid uptake by excess of unlabeled pantothenic acid. The data shown are means ± S.E. of five independent sets of experiments.
PfPAT Mediates Fenpropimorph Uptake into fen2Δ Cells
Because Fen2 expression is important for yeast sensitivity to the fungicidal drug (19, 20) fenpropimorph, we assessed whether PfPATf could mediate the transport of fenpropimoph into fen2Δ cells, which are resistant to this drug. Therefore, we compared the growth of fen2Δ expressing an empty vector with that of the fen2Δ strain harboring the PfPATf gene on liquid and solid media lacking or containing 1 mg/ml fenpropimorph. Whereas all strains grew at the same rate in the absence of fenpropimorph (Fig. 7, A and C), significant growth differences between the two strains were seen on fenpropimorph (Fig. 7, B and D). One day following inoculation, the fen2Δ strain harboring an empty vector reached 8.6 × 106 cells/ml in fenpropimorph-containing medium, whereas fen2Δ-PfPATf harboring PfPAT only reached 2.1 × 106 cells/ml in this medium (Fig. 7B). fen2Δ-PfPATf sensitivity was similar to that of the wild type strain harboring an endogenous Fen2 (Fig. 7B). Serial dilutions of the respective yeast culture on agar medium supplemented with fenpropimorph showed similar results (Fig. 7, E and F).
FIGURE 7.
PfPAT mediates entry of fenpropimorph into yeast cells. BY4741-derived end3Δ strain lacking the END3 gene harboring the pBEVY-U vector (open circles), end3Δfen2Δ pBEVY-U (open triangles), and end3Δfen2Δ-pPFPATf (closed triangles) were precultured in SV medium supplemented with 100 μm of pantothenate, washed twice, and inoculated to an initial cell density of 2 105 cells/ml in YPD medium in the absence (A and C) or presence (B and D) of 1 μg/ml fenpropimorph. E and F, for agar assay, cells were cultured in SV medium supplemented with 100 μm of pantothenate and washed twice and 10-fold serial dilutions of cells were plated onto YPD medium (E) or YPD medium supplemented with 1 μg/ml fenpropimorph (F) and incubated at 30 °C.
Fenpropimorph Inhibits P. falciparum Intraerythrocytic Development
Previous studies have shown that the pantothenate analog and antibiotic CJ-15 801 (IC50: 39 μm), pantothenol (IC50: 60 μm), and N-pantoyl-substituted amines (IC50 ranging between 15 and 200 μm) inhibit the growth of P. falciparum in vitro (32, 38, 39). The finding that PfPATf complements the loss of Fen2 in yeast by rendering fen2Δ cells sensitive to fenpropimorph led us to investigate the possible antimalarial activity of this compound. The sensitivity of two chloroquine-sensitive strains (3D7 and NF54) and one chloroquine-resistant strain Dd2 to fenpropimorph was assessed by monitoring parasite multiplication in the absence or presence of increasing concentrations of the compound (40). Fenpropimorph inhibited the growth of the three strains with IC50 = ∼30 μm (Fig. 8).
FIGURE 8.
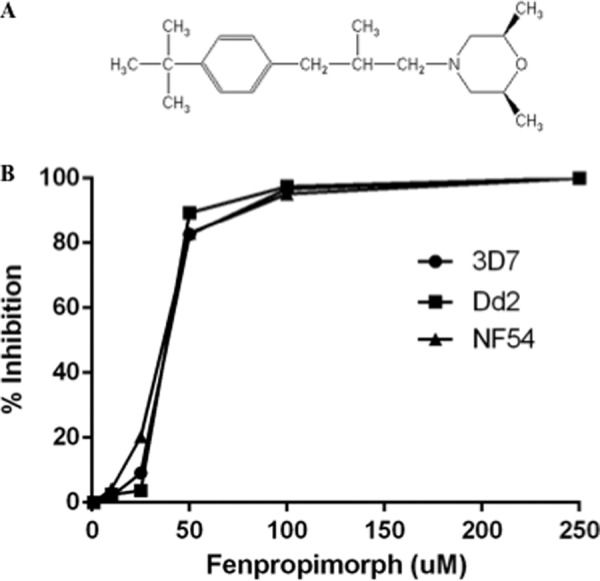
P. falciparum parasites are sensitive to fenpropimorph. A, chemical structure of fenpropimorph. B, growth of P. falciparum 3D7, NF54, and Dd2 strains in the absence or presence of increasing concentrations of fenpropimorph using the SYBR green proliferation assay (43).
DISCUSSION
Molecular characterization of nutrient transporters from P. falciparum has been limited to only few transporters such as the purine transporters PfNT1 and PfNT2, the hexose transporter PfHT1, and the potassium channel PfK1 (5, 8, 41). For most candidate permeases, however, this remains a difficult task because of poor expression, mislocalization, or inadequate membrane orientation of the transporter in the heterologous system used. Furthermore, because of the high A+T content of P. falciparum genes, further codon optimization or harmonization is needed to increase the expression efficiency and prevent early transcription termination (5, 28, 36, 42). In this study, we attempted to identify the primary pantothenate transporter of Plasmodium by genetic complementation in yeast using a P. knowlesi cDNA library made in a yeast expression vector under a constitutive promoter. This approach, however, failed to identify such a transporter. Further analysis of the library using specific PCR primers showed that the cDNA encoding the full-length P. knowlesi PAT ortholog is not represented in the library. PfPAT was thus found in silico based on sequence similarity with the S. cerevisiae Fen2. The best homologues of PfPAT are found in apicomplexa and Viridiplantae (green algae and land plants). Because pantothenate transport has not yet been characterized in plants, the finding of several plant orthologs likely resulting from duplication events (supplemental Fig. S2), suggests that this process might be critical in plant physiology.
So far, attempts to target PfPAT to the plasma membrane of mammalian cells to measure its transport activity have not been successful (Fig. 6). Future efforts aimed to create chimeric proteins similar to those generated in yeast could make it possible to measure pantothenate uptake in heterologous systems and determine its kinetics parameters and inhibition profile.
The successful localization of a chimeric form of PfPAT to the yeast plasma membrane following addition of the N-terminal domain of Fen2 allowed us to perform complementation assays in yeast to demonstrate the function of PfPAT in pantothenate transport. PfPAT complemented the growth defect of the yeast fen2Δ strain on media containing low pantothenate concentrations, thus providing the first evidence that PfPAT functions in pantothenate uptake. Our genetic studies in P. falciparum provided strong evidence that the PfPAT gene plays an essential function in parasite development. Unlike PfNT1, whose deletion could be complemented by the addition of high concentrations of exogenous purine nucleosides and nucleobases (8), we were unable to generate a genetically null mutant for PfPAT even in the presence of pantothenate at a concentration 400-fold higher than its physiological concentration. Furthermore, targeting PfPAT mRNA degradation using specific PMO conjugates resulted in parasite death. Together, these results suggest that PfPAT is essential for P. falciparum intraerythrocytic development, and no alternative mode of pantothenate entry exists in this parasite. PfPAT does not share homology with the human multivitamin transporter, hSMVT, suggesting that this transporter might be a good target for the development of selective antimalarial drugs.
Expression of PfPATf in yeast renders fen2Δ more susceptible to the fungicidal drug fenpropimorph, suggesting that PfPAT may transport both pantothenate and fenpropimorph. This finding led us to investigate whether the drug can inhibit P. falciparum intraerythrocytic development. Interestingly, we found that fenpropimorph inhibits the growth of chloroquine and pyrimethamine-sensitive and -resistant strains with IC50 values below 40 μm. Fenpropimorph's antimalarial potency is equal or better than most of the pantothenate analogs discovered so far (38). Although it is unclear whether fenpropimorph exerts its antimalarial activity by inhibiting pantothenate transport, CoA biosynthesis, or another metabolic pathway, the finding that it requires PfPAT for entry suggests that the transporter may also serve to selectively deliver antimalarial drugs into the parasite. Safer analogs of fenpropimorph or chimera of fenpropimorph analogs and other drugs might define new classes of antimalarials, which have not been used in malaria therapy.
Our in silico analyses identified orthologs of PfPAT in other protozoa (supplemental Fig. S2 and Table S2). Biochemical and genetic characterization of these candidate transporters might help advance our understanding of pantothenate transport during the entire life cycle of protozoan parasites in the arthropod vector and mammalian host. These studies may also help identify new classes of chemicals to treat and prevent parasitic diseases.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant AI097218 and AI077603 (to C. B. M.).

This article contains supplemental Tables S1 and S2 and Figs. S1–S4.
- PMO
- peptide-morpholino oligomer.
REFERENCES
- 1. World Malaria Report (2011) World Health Organization, Geneva [Google Scholar]
- 2. Singh B., Kim Sung L., Matusop A., Radhakrishnan A., Shamsul S. S., Cox-Singh J., Thomas A., Conway D. J. (2004) A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet 363, 1017–1024 [DOI] [PubMed] [Google Scholar]
- 3. Murray C. J., Rosenfeld L. C., Lim S. S., Andrews K. G., Foreman K. J., Haring D., Fullman N., Naghavi M., Lozano R., Lopez A. D. (2012) Global malaria mortality between 1980 and 2010. A systematic analysis. Lancet 379, 413–431 [DOI] [PubMed] [Google Scholar]
- 4. Saliba K. J., Kirk K. (2001) Nutrient acquisition by intracellular apicomplexan parasites. Staying in for dinner. Int. J. Parasitol. 31, 1321–1330 [DOI] [PubMed] [Google Scholar]
- 5. Downie M. J., El Bissati K., Bobenchik A. M., Nic Lochlainn L., Amerik A., Zufferey R., Kirk K., Ben Mamoun C. (2010) PfNT2, a permease of the equilibrative nucleoside transporter family in the endoplasmic reticulum of Plasmodium falciparum. J. Biol. Chem. 285, 20827–20833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Downie M. J., Saliba K. J., Bröer S., Howitt S. M., Kirk K. (2008) Purine nucleobase transport in the intraerythrocytic malaria parasite. Int. J. Parasitol. 38, 203–209 [DOI] [PubMed] [Google Scholar]
- 7. El Bissati K., Downie M. J., Kim S. K., Horowitz M., Carter N., Ullman B., Ben Mamoun C. (2008) Genetic evidence for the essential role of PfNT1 in the transport and utilization of xanthine, guanine, guanosine, and adenine by Plasmodium falciparum. Mol. Biochem. Parasitol. 161, 130–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. El Bissati K., Zufferey R., Witola W. H., Carter N. S., Ullman B., Ben Mamoun C. (2006) The plasma membrane permease PfNT1 is essential for purine salvage in the human malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 103, 9286–9291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kirk K., Saliba K. J. (2007) Targeting nutrient uptake mechanisms in Plasmodium. Curr. Drug Targets 8, 75–88 [DOI] [PubMed] [Google Scholar]
- 10. Martin R. E., Ginsburg H., Kirk K. (2009) Membrane transport proteins of the malaria parasite. Mol. Microbiol. 74, 519–528 [DOI] [PubMed] [Google Scholar]
- 11. Martin R. E., Henry R. I., Abbey J. L., Clements J. D., Kirk K. (2005) The “permeome” of the malaria parasite. An overview of the membrane transport proteins of Plasmodium falciparum. Genome Biol. 6, R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Slavic K., Straschil U., Reininger L., Doerig C., Morin C., Tewari R., Krishna S. (2010) Life cycle studies of the hexose transporter of Plasmodium species and genetic validation of their essentiality. Mol. Microbiol. 75, 1402–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Woodrow C. J., Burchmore R. J., Krishna S. (2000) Hexose permeation pathways in Plasmodium falciparum-infected erythrocytes. Proc. Natl. Acad. Sci. U.S.A. 97, 9931–9936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Woodrow C. J., Penny J. I., Krishna S. (1999) Intraerythrocytic Plasmodium falciparum expresses a high affinity facilitative hexose transporter. J. Biol. Chem. 274, 7272–7277 [DOI] [PubMed] [Google Scholar]
- 15. Spry C., Kirk K., Saliba K. J. (2008) Coenzyme A biosynthesis. An antimicrobial drug target. FEMS Microbiol. Rev. 32, 56–106 [DOI] [PubMed] [Google Scholar]
- 16. Saliba K. J., Horner H. A., Kirk K. (1998) Transport and metabolism of the essential vitamin pantothenic acid in human erythrocytes infected with the malaria parasite Plasmodium falciparum. J. Biol. Chem. 273, 10190–10195 [DOI] [PubMed] [Google Scholar]
- 17. Saliba K. J., Kirk K. (2001) H+-coupled pantothenate transport in the intracellular malaria parasite. J. Biol. Chem. 276, 18115–18121 [DOI] [PubMed] [Google Scholar]
- 18. Stolz J., Caspari T., Carr A. M., Sauer N. (2004) Cell division defects of Schizosaccharomyces pombe liz1-mutants are caused by defects in pantothenate uptake. Eukaryot. Cell 3, 406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stolz J., Sauer N. (1999) The fenpropimorph resistance gene FEN2 from Saccharomyces cerevisiae encodes a plasma membrane H+-pantothenate symporter. J. Biol. Chem. 274, 18747–18752 [DOI] [PubMed] [Google Scholar]
- 20. Marcireau C., Joets J., Pousset D., Guilloton M., Karst F. (1996) FEN2. A gene implicated in the catabolite repression-mediated regulation of ergosterol biosynthesis in yeast. Yeast 12, 531–539 [DOI] [PubMed] [Google Scholar]
- 21. Trager W., Jensen J. B. (1976) Human malaria parasites in continuous culture. Science 193, 673–675 [DOI] [PubMed] [Google Scholar]
- 22. Witola W. H., El Bissati K., Pessi G., Xie C., Roepe P. D., Mamoun C. B. (2008) Disruption of the Plasmodium falciparum PfPMT gene results in a complete loss of phosphatidylcholine biosynthesis via the serine-decarboxylase-phosphoethanolamine-methyltransferase pathway and severe growth and survival defects. J. Biol. Chem. 283, 27636–27643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Witola W. H., Pessi G., El Bissati K., Reynolds J. M., Mamoun C. B. (2006) Localization of the phosphoethanolamine methyltransferase of the human malaria parasite Plasmodium falciparum to the Golgi apparatus. J. Biol. Chem. 281, 21305–21311 [DOI] [PubMed] [Google Scholar]
- 24. Foth B. J., Ralph S. A., Tonkin C. J., Struck N. S., Fraunholz M., Roos D. S., Cowman A. F., McFadden G. I. (2003) Dissecting apicoplast targeting in the malaria parasite Plasmodium falciparum. Science 299, 705–708 [DOI] [PubMed] [Google Scholar]
- 25. Mamoun C. B., Gluzman I. Y., Goyard S., Beverley S. M., Goldberg D. E. (1999) A set of independent selectable markers for transfection of the human malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 96, 8716–8720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miller C. A., 3rd, Martinat M. A., Hyman L. E. (1998) Assessment of aryl hydrocarbon receptor complex interactions using pBEVY plasmids. Expression vectors with bi-directional promoters for use in Saccharomyces cerevisiae. Nucleic Acids Res. 26, 3577–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gietz D., St Jean A., Woods R. A., Schiestl R. H. (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Santiago T. C., Zufferey R., Mehra R. S., Coleman R. A., Mamoun C. B. (2004) The Plasmodium falciparum PfGatp is an endoplasmic reticulum membrane protein important for the initial step of malarial glycerolipid synthesis. J. Biol. Chem. 279, 9222–9232 [DOI] [PubMed] [Google Scholar]
- 29. Rager N., Mamoun C. B., Carter N. S., Goldberg D. E., Ullman B. (2001) Localization of the Plasmodium falciparum PfNT1 nucleoside transporter to the parasite plasma membrane. J. Biol. Chem. 276, 41095–41099 [DOI] [PubMed] [Google Scholar]
- 30. Augagneur Y., Wesolowski D., Tae H. S., Altman S., Ben Mamoun C. (2012) Gene selective mRNA cleavage inhibits the development of Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 109, 6235–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Divo A. A., Geary T. G., Davis N. L., Jensen J. B. (1985) Nutritional requirements of Plasmodium falciparum in culture. I. Exogenously supplied dialyzable components necessary for continuous growth. J. Protozool. 32, 59–64 [DOI] [PubMed] [Google Scholar]
- 32. Saliba K. J., Ferru I., Kirk K. (2005) Provitamin B5 (pantothenol) inhibits growth of the intraerythrocytic malaria parasite. Antimicrob. Agents Chemother. 49, 632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Choi J. Y., Augagneur Y., Ben Mamoun C., Voelker D. R. (2012) Identification of gene encoding Plasmodium knowlesi phosphatidylserine decarboxylase by genetic complementation in yeast and characterization of in vitro maturation of encoded enzyme. J. Biol. Chem. 287, 222–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J. F., Guindon S., Lefort V., Lescot M., Claverie J. M., Gascuel O. (2008) Phylogeny.fr. Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pessi G., Choi J. Y., Reynolds J. M., Voelker D. R., Mamoun C. B. (2005) In vivo evidence for the specificity of Plasmodium falciparum phosphoethanolamine methyltransferase and its coupling to the Kennedy pathway. J. Biol. Chem. 280, 12461–12466 [DOI] [PubMed] [Google Scholar]
- 36. Reynolds J. M., Takebe S., Choi J. Y., El Bissati K., Witola W. H., Bobenchik A. M., Hoch J. C., Voelker D. R., Mamoun C. B. (2008) Biochemical and genetic analysis of the phosphoethanolamine methyltransferase of the human malaria parasite Plasmodium falciparum. J. Biol. Chem. 283, 7894–7900 [DOI] [PubMed] [Google Scholar]
- 37. Serrano R., Kielland-Brandt M. C., Fink G. R. (1986) Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+-ATPases. Nature 319, 689–693 [DOI] [PubMed] [Google Scholar]
- 38. Spry C., Chai C. L., Kirk K., Saliba K. J. (2005) A class of pantothenic acid analogs inhibits Plasmodium falciparum pantothenate kinase and represses the proliferation of malaria parasites. Antimicrob. Agents Chemother. 49, 4649–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saliba K. J., Kirk K. (2005) CJ-15,801, a fungal natural product, inhibits the intraerythrocytic stage of Plasmodium falciparum in vitro via an effect on pantothenic acid utilisation. Mol. Biochem. Parasitol. 141, 129–131 [DOI] [PubMed] [Google Scholar]
- 40. Johnson J. D., Dennull R. A., Gerena L., Lopez-Sanchez M., Roncal N. E., Waters N. C. (2007) Assessment and continued validation of the malaria SYBR green I-based fluorescence assay for use in malaria drug screening. Antimicrob. Agents Chemother. 51, 1926–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Waller K. L., McBride S. M., Kim K., McDonald T. V. (2008) Characterization of two putative potassium channels in Plasmodium falciparum. Malar. J. 7, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Baro N. K., Pooput C., Roepe P. D. (2011) Analysis of chloroquine resistance transporter (CRT) isoforms and orthologues in S. cerevisiae yeast. Biochemistry 50, 6701–6710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rason M. A., Randriantsoa T., Andrianantenaina H., Ratsimbasoa A., Menard D. (2008) Performance and reliability of the SYBR Green I based assay for the routine monitoring of susceptibility of Plasmodium falciparum clinical isolates. Trans. R. Soc. Trop. Med. Hyg. 102, 346–351 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.