Background: Light-driven proton pumps are utilized to control the neural activity.
Results: We have succeeded to produce a blue-shifted proton pump. The rotation of the β-ionone ring contributes to the spectral shift.
Conclusion: The designed color variant provides a tool that allows the control of neural activity by blue light.
Significance: The knowledge will help to understand the color-tuning mechanism and can be utilized for optogenetics.
Keywords: 7-Helix Receptor, Membrane Proteins, Photoreceptors, Proton Pumps, Rhodopsin, Retinal
Abstract
Ion-transporting rhodopsins are widely utilized as optogenetic tools both for light-induced neural activation and silencing. The most studied representative is Bacteriorhodopsin (BR), which absorbs green/red light (∼570 nm) and functions as a proton pump. Upon photoexcitation, BR induces a hyperpolarization across the membrane, which, if incorporated into a nerve cell, results in its neural silencing. In this study, we show that several residues around the retinal chromophore, which are completely conserved among BR homologs from the archaea, are involved in the spectral tuning in a BR homolog (HwBR) and that the combination mutation causes a large spectral blue shift (λmax = 498 nm) while preserving the robust pumping activity. Quantum mechanics/molecular mechanics calculations revealed that, compared with the wild type, the β-ionone ring of the chromophore in the mutant is rotated ∼130° because of the lack of steric hindrance between the methyl groups of the retinal and the mutated residues, resulting in the breakage of the π conjugation system on the polyene chain of the retinal. By the same mutations, similar spectral blue shifts are also observed in another BR homolog, archearhodopsin-3 (also called Arch). The color variant of archearhodopsin-3 could be successfully expressed in the neural cells of Caenorhabditis elegans, and illumination with blue light (500 nm) led to the effective locomotory paralysis of the worms. Thus, we successfully produced a blue-shifted proton pump for neural silencing.
Introduction
Light-absorbing photoactive proteins show characteristic colors originating from a species specific energy gap between their ground state and excited state, which leads to different characteristic absorption maxima (λmax). Among these proteins and their cognate chromophores, the rhodopsins are known to show a large variation in their absorption spectra depending on the interaction between the apoprotein (opsin) and the retinal chromophore, which is called the opsin shift (1). Actually, microbial (type 1) rhodopsins show a wide range of λmax ranging from 485 to 590 nm while keeping the same isomeric composition, all-trans-retinal (2). Three types of such microbial molecules, proton pumping rhodopsins such as archearhodopsin-3 (AR3),2 chloride pumping rhodopsins such as halorhodopsin (HR), and cation channel rhodopsins such as channelrhodopsin-2 (ChR2), have already been utilized as optogenetic tools for controlling the neural activity (3, 4). The ion pumps (AR3 and HR) and the ion channel (ChR2) induce a hyperpolarization used for neural silencing and a depolarization used for neural activation, respectively (3–5). As for fluorescent proteins, the color variants of ion-transporting rhodopsins make it possible to use various wavelengths of light.
The λmax of a molecule corresponds to the most probable transition from its ground to excited state (1). Empirical and theoretical studies of the spectral tuning of rhodopsins have suggested several mechanisms including the following: (i) an alteration in the strength of the electrostatic interaction between the retinal protonated Schiff base (RPSB) and its counter ion or hydrogen bond acceptor; (ii) an alteration in the polarity or polarizability of the environment of the chromophore-binding pocket, caused by the arrangement of polar or aromatic residues; and (iii) an isomerization around the 6-S bond (6,7-torsion angle) connecting the polyene chain to the β-ionone ring (1, 6–12). By combinations of these effects, rhodopsins show various absorption maxima. All microbial rhodopsins are quite similar in their primary and tertiary structures, especially in the chromophore-binding site (13), suggesting that the λmax is mainly controlled by the slight structural difference(s) of their side chain(s). For example, the λmax value of sensory rhodopsin II (SRII) (∼ 500 nm) is remarkably different from that of other rhodopsins such as bacteriorhodopsin (BR), HR, or sensory rhodopsin I (SRI) (560–590 nm) (2). This is reasonable because SRII is a negative sensor for light which contains harmful UV radiation (14). Previously, we have investigated the color-tuning mechanism in rhodopsins experimentally (15, 16). In those studies that aimed to determine the key residue(s) contributing to the color change of SRII, several SRII mutants were analyzed in which each residue was replaced by its corresponding residue in BR. In addition to this, we also performed similar experiments for SRI (17). From the results, we have found the key residues (factors) contributing to the color change from orange (∼500 nm) to purple (∼560 nm), as well as several unique factors, as the interaction between the D- and E-helices for SRII (16) or the chloride ion binding around the β-ionone ring for SRI (18), respectively.
On the basis of this knowledge combined with other findings, we aimed, in this study, to produce blue-shifted color variants of BR (Fig. 1A). As mentioned above, it can be estimated that the introduction of polarity or polarizability around the β-ionone ring, or the removal of those around the Schiff base would cause a spectral blue shift (Asp-123 → Ala, Ser-149 → Ala, and Ala-223 → Thr in Fig. 1B). Moreover, it can be predicted that a volume change around the β-ionone ring would cause a change in interaction between the retinal and the replaced residues (Met-126 → Ala and Ser-149 → Ala in Fig. 1B), resulting in a spectral shift caused by the isomerization around the 6-S bond. According to the amino acid sequence alignment of BR homologs, we identified four candidates for the color-tuning residues, which are completely conserved among BR homologs (Fig. 1B). It should be noted that, because of the property of the well studied BR from Halobacterium salinarum (HsBR) of not forming proper functional rhodopsins outside its native halobacterial membrane, the investigation of its color-tuning mechanism has been performed using refolded HsBR salvaged from denatured Escherichia coli inclusion bodies (19). This might cause unexpected effects such as improper protein folding and improper interaction with retinal in HsBR. In contrast, a new microbial BR-like rhodopsin from Haloquadratum walsbyi (HwBR), which we have found recently, functions as a robust proton pump also in E. coli cells (20). Utilizing this protein and another BR homolog (AR3), we achieved the production of large spectral blue-shifted orange pigments (λmax = 498 nm and λmax = 500 nm, respectively) by combination of the mutations while keeping their robust pumping activity. Quantum mechanics/molecular mechanics (QM/MM) calculations, which exhibit the strengths associated with both methods (QM (accuracy) and MM (speed)), revealed that the origin of the spectral shift was in the rotation of the 6-S bond. Furthermore, these color variants provided us with a tool to control the neural activity of Caenorhabditis elegans effectively upon illumination with blue light (500 nm). Thus, neural silencing in living cells is now controllable by different wavelengths of light (500–560 nm), including those also absorbed by ChR2 and its variant.
FIGURE 1.
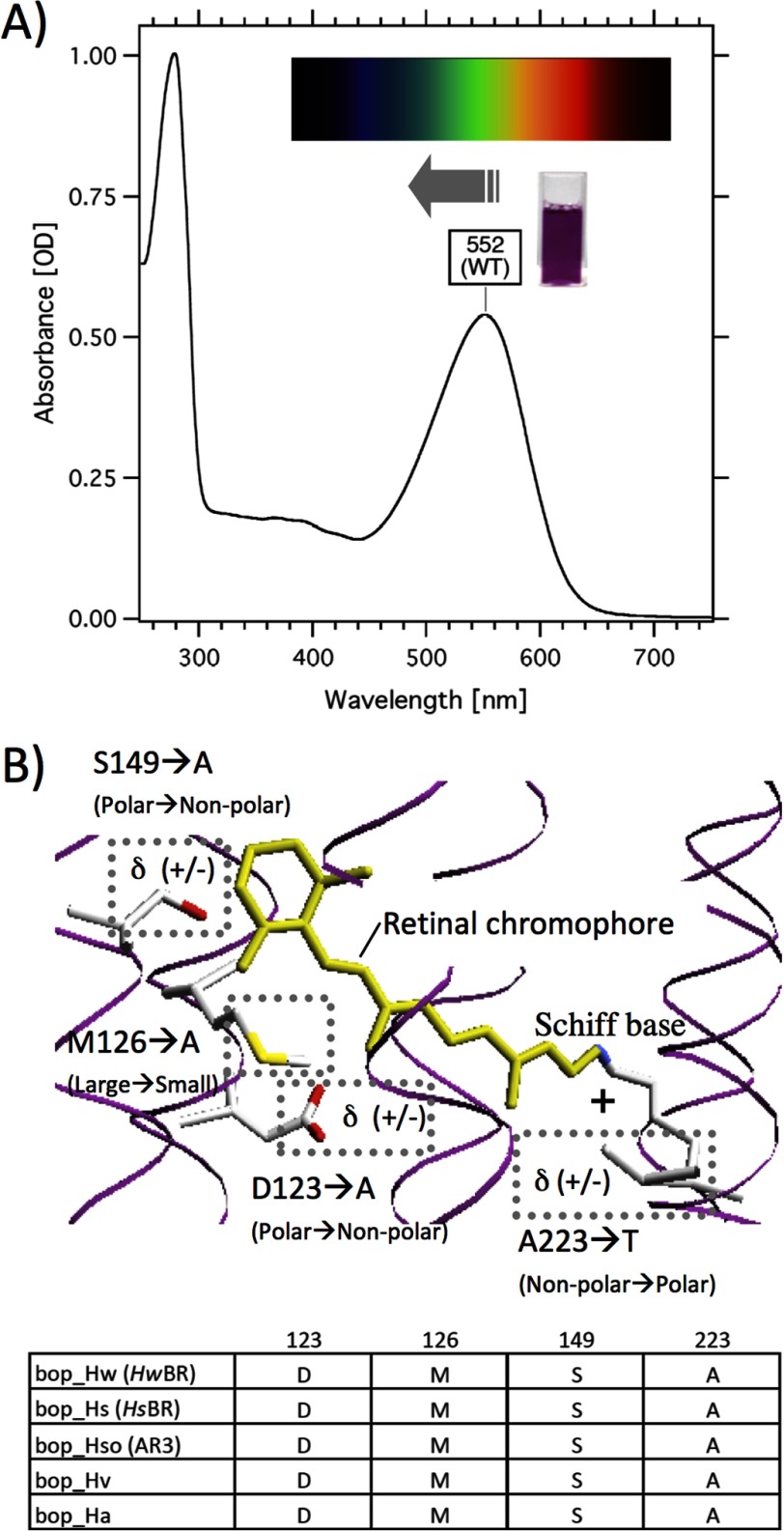
The experimental strategy. Shown are the UV-visible absorption spectrum of WT HwBR (A) and the crystal structure around the retinal chromophore of HsBR24 with the number of the amino acid residues in HwBR, which are completely conserved among archaeal proton pumping rhodopsins (B). The membrane normal is roughly in the vertical plane of this figure, and the upper and lower sides in B correspond to the cytoplasmic and extracellular sides, respectively. The bottom panel indicates the amino acid sequence alignment of BR homologs. The cuvette in A shows the purified HwBR.
EXPERIMENTAL PROCEDURES
Expression and Purification of HwBR and AR3
The expression plasmid for the wild-type HwBR was constructed as described previously (20). The mutant genes were constructed by the QuikChange site-directed mutagenesis method (Stratagene, La Jolla, CA) as described previously (21). The gene of AR3 was amplified using PCR from the genomic DNA of Halorubrum sodomense and was inserted into the arabinose-inducible expression vector as described previously (20). Consequently, the plasmid encodes AR3 with six histidines at the C terminus. The E. coli DH5a strain was used as a host for DNA manipulation. All constructed plasmids were analyzed using an automated sequencer to confirm the expected nucleotide sequences. The preparation of crude membranes and purification of proteins were performed using essentially the same method described previously (22). Briefly, proteins with a six-histidine tag at the C terminus were expressed in E. coli BL21 (DE3) cells as recombinant proteins, solubilized by n-dodecyl-β-d-maltoside, and purified with a Ni2+ affinity column. Where necessary, the samples were further purified with an anion exchange column.
UV-visible Spectroscopy, HPLC Analysis, and Light-induced pH Changes
The purified sample was concentrated and exchanged using an Amicon Ultra filter (Millipore, Bedford, MA), against a buffer containing 1 m NaCl, 50 mm Tris-Cl, and 0.05% n-dodecyl-β-d-maltoside. UV-visible spectra were recorded using a UV2450 spectrophotometer with an ISR2200 integrating sphere (Shimazu, Kyoto, Japan). The temperature was kept constant at 298 K. HPLC analysis was performed following the published procedure (23). The proton transport activity of each protein was measured by monitoring pH changes using a glass electrode. The cells of E. coli expressing HwBR were harvested by centrifugation (4,800 × g for 3 min), and then they were washed three times and resuspended in the solvent for the measurement (100 mm NaCl). 7.5 ml of cell suspension was kept in darkness and then illuminated with the output of a 100 W xenon arc lamp (LM103; Asahi Spectra, Tokyo, Japan) through a long pass glass filter for 5 min at a specific wavelength region (WT HwBR: >510 nm; M126A/S149A/A223T and D123A/M126A/S149A/A223T: >460 nm). The power of the light was controlled by ND filters (0.0625, 0.125, 0.25, 0.5, 1, 2.5, and 5 W) for measuring the power dependence of the pumping activities. For the measurement of the action spectra, interference filters (full width at half-maximum (FWHM) = 10 nm) were used to obtain the light at specific wavelengths by filtering the output of the same xenon arc lamp. The photon flux of the light was measured by a calibrated multichannel detector (IRRAD-C2000+; Ocean Optics), and the initial slopes at the specific wavelengths were scaled by these fluxes to obtain the action spectrum.
QM/MM Calculations
Because of the availability of x-ray crystallographic structures, we performed the molecular simulations for HsBR, which exhibits a close sequence similarity to HwBR especially in the RPSB-binding pocket. A starting simulation model of the protein embedded in a 1-palmitoyl-2-oleoylphosphatidylcholine bilayer environment was constructed with the x-ray crystallographic structure of HsBR determined by Luecke et al. (Protein Data Bank code 1C3W) (24). The simulation systems of the native protein and the triple mutant (M118A/S141A/A215T, corresponding to M126A/S149A/A223T of HwBR) with a manually rotated β-ionone ring were first equilibrated by molecular dynamics simulations with molecular mechanics force fields. Molecular structures of the active site including the RPSB and the polar groups in the vicinity of protonated Schiff base, Arg-82, Asp-85, Glu-121, and three water molecules (Wat-401, Wat-402, and Wat-406) were then refined by QM/MM reweighting free energy self-consistent field (RWFE-SCF) geometry optimizations (25). The QM/MM RWFE-SCF method optimized the molecular geometries of the active site at the QM level of theory (density functional theory with B3LYP functional) on a free energy surface constructed with MM conformational statistical samples of the surrounding protein environment obtained by the molecular dynamics simulations. The method can therefore take into account the thermal relaxation of the protein environment during the geometry optimization and thus remove artifacts of structural conflicts introduced in the mutant protein model. Conformational samples of molecular dynamics simulations for 60 and 39 ns in total for the native and mutant proteins, respectively, were used in the QM/MM RWFE-SCF geometry optimization. To examine the effect of the β-ionone ring rotation in the triple mutant on the structural stability and absorption spectral shift, we performed second order multireference Møller-Plesset perturbation (MRMP2) calculations for the RPSB moiety at the free energetically optimized geometries. The MRMP2 calculations were carried out in isolated conditions.
Locomotion Analysis of C. elegans Expressing AR3
A plasmid encoding AR3 fused with GFP at the C terminus was constructed as described previously (26). The M128A/S151A/A225T mutant gene was prepared by the site-directed mutagenesis method. The C. elegans worm strain (N2) expressing AR3 under the control of a pan-neuronal promoter F25B11p was generated and grown on nematode growth medium plates seeded with a solution of E. coli OP50 and 500 μm all-trans-retinal (Sigma-Aldrich) as described previously (26). The animals were maintained at 20 °C in the dark unless indicated otherwise. For observation of the GFP fluorescence and of the locomotion of C. elegans, we used the apparatus and procedure for the analysis described previously (26). In short, the animals were observed under a fluorescence stereo microscope (SZX12) and were illuminated with a 100 W HBO mercury lamp through an excitation bandpass filter. In most experiments, the worms were used only once for a single series of illumination experiments with varying light intensities (0–1 milliwatt/mm2) and were then discarded unless otherwise indicated. The images were acquired with a digital CCD camera Cool SNAP HQ2 (Photometrics, Tucson, AZ) using the PM Capture Pro software (Nippon Roper, Japan) and processed using the ImageJ public domain software. The intensity of 470-, 500-, and 550-nm light was measured at the object plane using an optical power meter (Model 3664, Hioki, Ueda, Japan) with an optical sensor (Model 9742, Hioki, Ueda, Japan). Various intensities of illumination were implemented by changing the magnification of the objective lens of the microscope. To calculate the intensity of illumination, we measured the radius of the illuminated area, calculated the area, and then divided the total light intensity by the area as described previously (26). In general, the worms swim well on the plate without light. The locomotion stopped completely and promptly upon photoirradiation. The stopped cells are defined as being “paralyzed” in this study.
RESULTS
Absorption Spectra of the Mutants of HwBR with Single or Multiple Substitutions
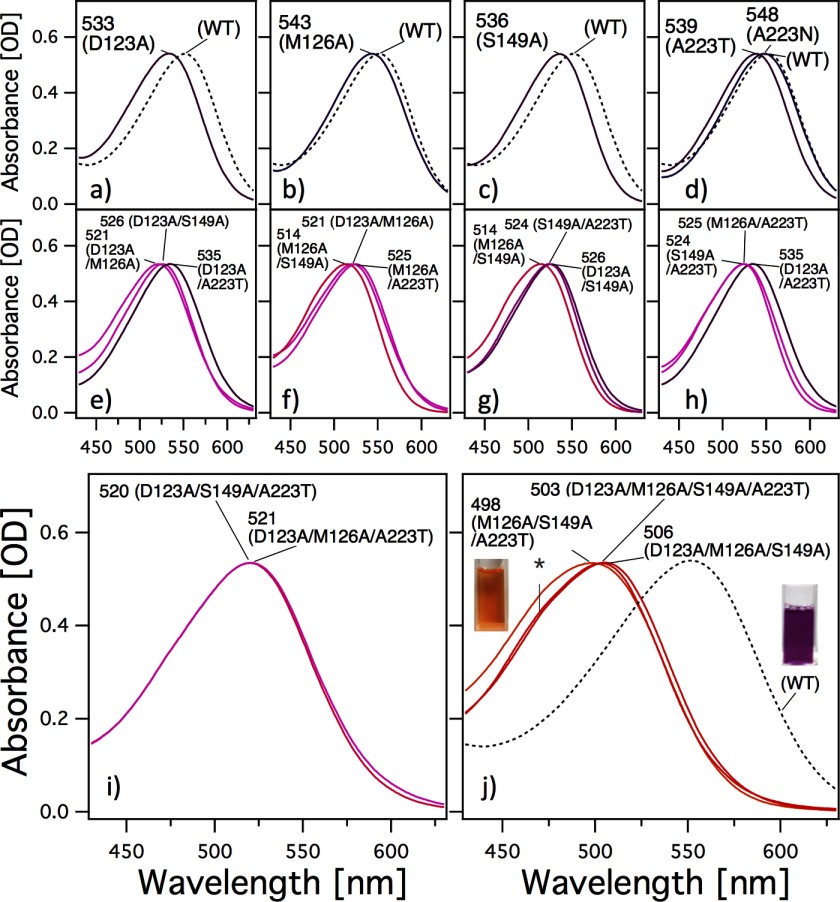
Panels a–d in Fig. 2 show the visible absorption spectra obtained for the WT and for various mutants of HwBR in which a single amino acid residue was replaced with Ala for Asp-123, Met-126, or Ser-149 or with Thr (or Asn) for Ala-223. To avoid denaturation, the spectra shown were measured immediately after purification. Their λmax values (main absorption) and retinal configurations obtained by HPLC analysis are listed in Table 1. The λmax of all of the mutants showed spectral blue shifts of 19 nm (646 cm−1) for D123A, 9 nm (300 cm−1) for M126A, 16 nm (541 cm−1) for S149A, 13 nm (437 cm−1) for A223T, and 4 nm (132 cm−1) for A223N, respectively. Furthermore, the retinal compositions of the mutants were investigated (Table 1). It is well known that one of the retinal conformations, the all-trans-isomer, is the functional form of all microbial rhodopsins studied so far. As shown in Table 1, all HwBR single mutants contain above 60% all-trans-retinal similar to WT HwBR (79%) as well as HsBR (66.4%) in n-dodecyl-β-d-maltoside micelles, indicating that the mutants maintain functional proteins. To further characterize the effects of these residues, combination mutants of HwBR were constructed, and the absorption spectra and retinal compositions were measured (Fig. 2, e–j, and Table 1). All these multiple HwBR mutants contain above 53% all-trans-retinal with a minor proportion of other isomers, such as 13-cis, 11-cis, and 9-cis, similar to the single mutants, indicating that these mutants also maintain functional proteins. Panel j in Fig. 2 shows the absorption spectra of the most blue-shifted mutants, M126A/S149A/A223T, D123A/M126A/S149A/A223T, and D123A/M126A/S149A, where the λmax values are located at 498, 503, and 506 nm, respectively. These mutants have an orange color (Fig. 2j) and show a vibrational fine structure at ∼460 nm, which has also been seen in other blue-shifted microbial rhodopsins such as SRII absorbing light of ∼500 nm (27, 28). The origin of this fine structure will be discussed later. The shift in absorption of the M126A/S149A/A223T mutant from that of the WT was 1964 cm−1, which corresponds to the difference observed between SRII and other microbial rhodopsins (BR, HR, and SRI) (∼2000 cm−1). Thus, we succeeded in the production of blue-shifted mutants of HwBR. It should also be noted that light-dark adaptation did not lead to an increase in the all-trans conformer, which was observed in the WT, except for the M126A, S149A, and A223T mutants (Table 1).
FIGURE 2.
Visible absorption spectra of WT HwBR and its mutants. Panels a–d and e–j correspond to the single and multiple residue substitutions, respectively. The spectra are scaled to be the similar intensity as to WT by multiplying 1.03–1.55 for mutants. The large blue-shifted mutants have a spectral shoulder at ∼470 nm (asterisk in panel j). The cuvette in panel j shows the purified M126A/S149A/A223T mutant of HwBR.
TABLE 1.
Absorption maximum, opsin shift, and retinal configuration of the WT and of various HwBR mutants both in the dark and light
| Opsin type | λmax | Δυ | All-trans |
|
|---|---|---|---|---|
| Dark-adapted | Light-adapted | |||
| nm | cm−1 | % | ||
| WT (HwBR) | 552 | 79 ± 2 | 90 ± 3 | |
| D123A | 533 | 646 | 61 ± 2 | 60 ± 2 |
| M126A | 543 | 300 | 60 ± 4 | 75 ± 3 |
| S149A | 536 | 541 | 69 ± 2 | 76 ± 2 |
| A223N | 548 | 132 | 77 ± 2 | 81 ± 3 |
| A223T | 539 | 437 | 78 ± 2 | 96 ± 2 |
| D123A/M126A | 521 | 1078 | 65 ± 3 | 58 ± 1 |
| D123A/S149A | 526 | 895 | 62 ± 3 | 69 ± 2 |
| D123A/A223T | 535 | 576 | 66 ± 2 | 70 ± 2 |
| M126A/S149A | 514 | 1339 | 70 ± 2 | 71 ± 3 |
| M126A/A223T | 525 | 932 | 81 ± 2 | 73 ± 2 |
| S149A/A223T | 524 | 968 | 81 ± 2 | 77 ± 3 |
| D123A/S149A/A223T | 520 | 1115 | 53 ± 1 | 48 ± 2 |
| D123A/M126A/A223T | 521 | 1078 | 71 ± 2 | 56 ± 1 |
| D123A/M126A/S149A | 506 | 1647 | 65 ± 2 | 58 ± 1 |
| M126A/S149A/A223T | 498 | 1964 | 73 ± 2 | 66 ± 2 |
| D123A/M126A/S149A/A223T | 503 | 1765 | 76 ± 1 | 72 ± 2 |
Properties of the Blue-shifted Mutants
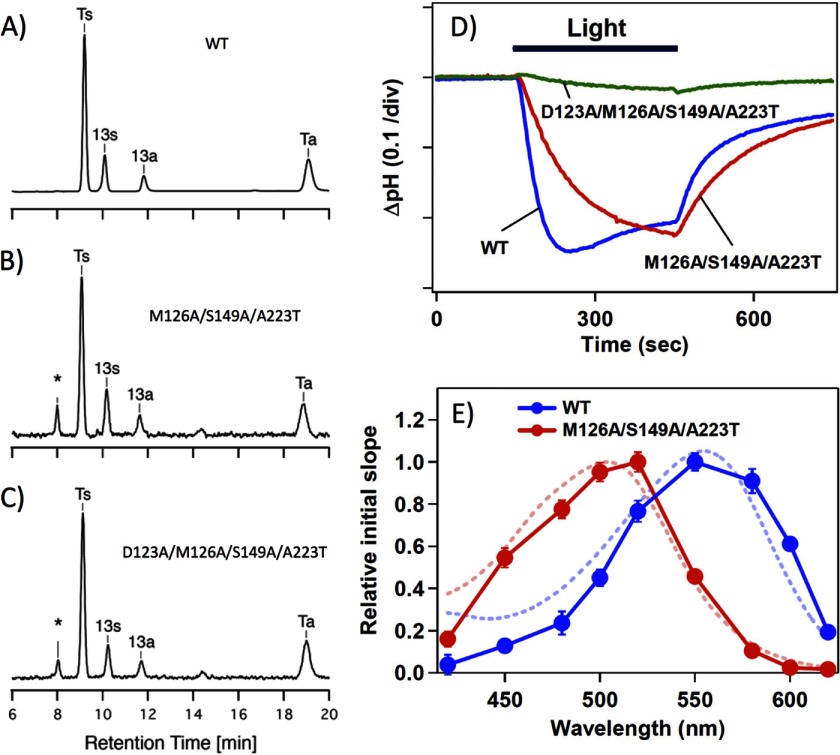
In contrast with fluorescent and luminescent proteins, color conversion is not sufficient for rhodopsins to work as optogenetic tools, but their function must also be maintained. As already mentioned, all biological functions of microbial rhodopsins studied so far are triggered by the trans-cis isomerization of the all-trans-retinal (29). The dominant proportion of the all-trans-isomer of the WT (Fig. 3A) and two of the most blue-shifted mutants of HwBR, M126A/S149A/A223T (Fig. 3B) and D123A/M126A/S149A/A223T (Fig. 3C), suggest that their functioning might be maintained upon introducing these mutations (see also Table 1). In the next step, this was tested by measuring the proton pumping activity of the mutants (M126A/S149A/A223T and D123A/M126A/S149A/A223T shown as the red and green curves in Fig. 3D, respectively) compared with that of the WT (blue curve) by probing the light-induced pH change in the E. coli cells expressing the corresponding proteins. As shown, a strong light-induced negative signal, which corresponds to a decrease in pH (outward proton transport), was observed for M126A/S149A/A223T as well as the WT, whereas only a small signal was observed for the D123A/M126A/S149A/A223T mutant, indicating its weak proton pumping activity. This poor activity of the quadruple mutant might be caused by a change(s) of some photochemical properties such as an abnormal pKa change during the photocycle. In contrast, the triple mutant (M126A/S149A/A223T) pumped, although slowly, almost the same amount of protons out of the cells.
FIGURE 3.
Photochemical properties of the mutants. A–C, chromophore configurations extracted from the WT and the M126A/S149A/A223T and D123A/M126A/S149A/A223T mutants of HwBR. The detection beam was set to 360 nm. Ts, Ta, 13s, and 13a stand for the all-trans 15-syn retinal oxime, the all-trans 15-anti retinal oxime, the 13-cis 15-syn retinal oxime, and the 13-cis 15-anti retinal oxime, respectively. The molar composition of each retinal isomer was calculated from the areas of the peaks in the HPLC patterns, and the estimated molar compositions are listed in Table 1. D, light-driven pH changes in cell suspensions containing the WT (blue) or the M126A/S149A/A223T (red) or D123A/M126A/S149A/A223T (green) mutants of HwBR. Light indicates the illumination period (with yellow light, >500 nm for the WT and >460 nm for the mutants), and the negative signal corresponds to a decrease in pH (outward proton transport). One division of the vertical axis corresponds to 0.1 pH units. E, action spectra for the proton pumping activity in transformants containing the WT (blue circles) or the M126A/S149A/A223T mutant (red circles) of HwBR, which were estimated from the initial slopes of the pH change. The error bars indicate the standard deviations of three identical experiments. Blue and red dashed lines represent the absorption spectra of the light-adapted WT HwBR and M126A/S149A/A223T mutant, respectively.
Thus, one of these blue-shifted mutants, the M126A/S149A/A223T mutant, produced a sufficiently robust response to permit the measurement of its action spectrum. For this, the light intensity at various wavelengths of light was adjusted and normalized by using interference filters, and the initial slope (the “proton pumping activity”) was plotted against the excitation wavelength (Fig. 3E). The action spectrum of the M126A/S149A/A223T mutant is blue-shifted by ∼50 nm compared with that of the WT and matches well with the shift in absorption between the triple mutant and the WT (dotted lines in Fig. 3E). The good agreement between the action spectra and the absorption spectra of both the WT and M126A/S149A/A223T confirms the fidelity of our quantification of the proton pumping activity.
The Origin of the Large Spectral Blue Shift in the Mutant
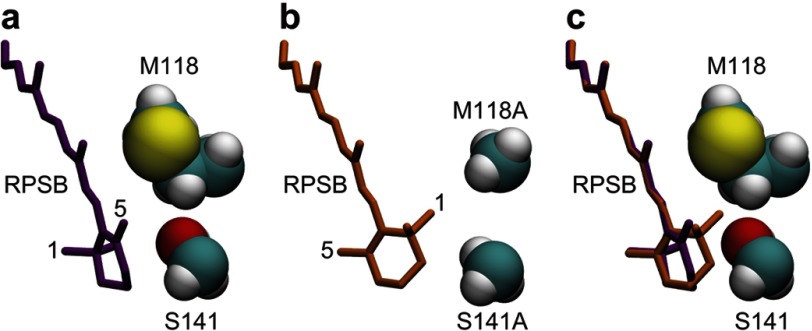
To investigate the molecular origin of the large spectral shift in the mutant, we performed molecular simulations of the native protein and the mutant. Because of the lack of a crystal structure of HwBR, a high resolution crystal structure of BR from H. salinarum (HsBR) (24) was used as a simulation model. Fig. 4 shows free energetically optimized structural models of the native protein and the M118A/S141A/A215T triple mutant corresponding to M126A/S149A/A223T in HwBR that well explain the observed large blue shift in the mutant. The polyene chain of the chromophore in the native protein of HsBR possesses a planer conformation, resulting in a long π conjugation. On the other hand, the β-ionone ring of the chromophore in the triple mutant is allowed to rotate. It should be noted that a rotation of the β-ionone ring can, despite breaking the π conjugation, easily occur in isolated condition (30) because of the steric repulsions between the methyl groups of the β-ionone ring at positions 1 and 5 and the hydrogens of the polyene chain at positions 7 and 8. In fact, MRMP2 calculations of the chromophore in optimized structures indicate that the conformation having a rotated β-ionone ring in the triple mutant is slightly energetically favorable compared with the wild type (Table 2). It is also noteworthy that the twisted conformation at the 6-S bond has also been found in the x-ray crystallographic structures of a retinal protein, the visual receptor rhodopsin (31). The rotation of the β-ionone ring is, however, prevented in the native protein with the side chains of Met-118 and Ser-141 by the steric repulsion of the methyl group at position 1 and the CH2 moiety at position 2 of the β-ionone ring (Fig. 4c). The replacement of those side chains with the smaller alanines in the triple mutant removes this steric hindrance that imposes the planer conformation in the native protein. Trp-86, Tyr-185, and Pro-186 (not shown in Fig. 4), well preserved in the triple mutant, also provide a good recognition of the methyl group of the β-ionone ring at position 5, which is placed by the rotation of the β-ionone ring into almost the same position as the methyl group at position 1 in the native protein (Fig. 4c). Therefore, the breakage of the π conjugation caused by the rotation of the β-ionone ring, in addition to electrostatic contributions of the S141A and A215T mutation (see below), is likely to contribute largely to the blue shift of the photoabsorption of the chromophore as reported previously (30, 32, 33), which is in agreement with the MRMP2 calculations (Table 2).
FIGURE 4.
Structures of the RPSB and mutated protein groups in the vicinity of the β-ionone ring of the RPSB in HsBR. Snap shots of the parts of the protein taken from conformational samples of molecular dynamics simulations at the last steps of the free energy geometry optimizations are shown. a, structure of the native HsBR. Met-118 and Ser-141 in HsBR correspond to Met-126 and Ser-149 in HwBR, respectively. Conformation of the polyene part of the RPSB is almost planar, as typically seen in x-ray crystallographic structures of microbial rhodopsins. The dihedral angle of C5=C6-C7=C8 is 174.2°. b, a structural model of the triple mutant, M118A/S141A/A215T, of HsBR. The β-ionone ring is remarkably twisted with respect to the π conjugated plane of the polyene part of the RPSB, leading to the breakage of the π conjugation. The dihedral angle of C5=C6-C7=C8 is −47.9°. c, comparison of the structures of the native protein and the triple mutant. The RPSB moiety of the triple mutant (orange) is superimposed on the structure of the native protein.
TABLE 2.
Comparison of S0 energy and the excitation energy (kcal/mol) of the chromophore in conformations of the native protein and the triple mutant, M118A/S141A/A215T of HsBR (M126A/S149A/A223T of HwBR), calculated at the MRMP2 level of theory in the isolated condition
a Difference in the excitation energy between the triple mutant and the native protein.
b The absolute energy is −949.60270 au.
Controlling the Locomotion of C. elegans by a Blue-shifted Color Variant
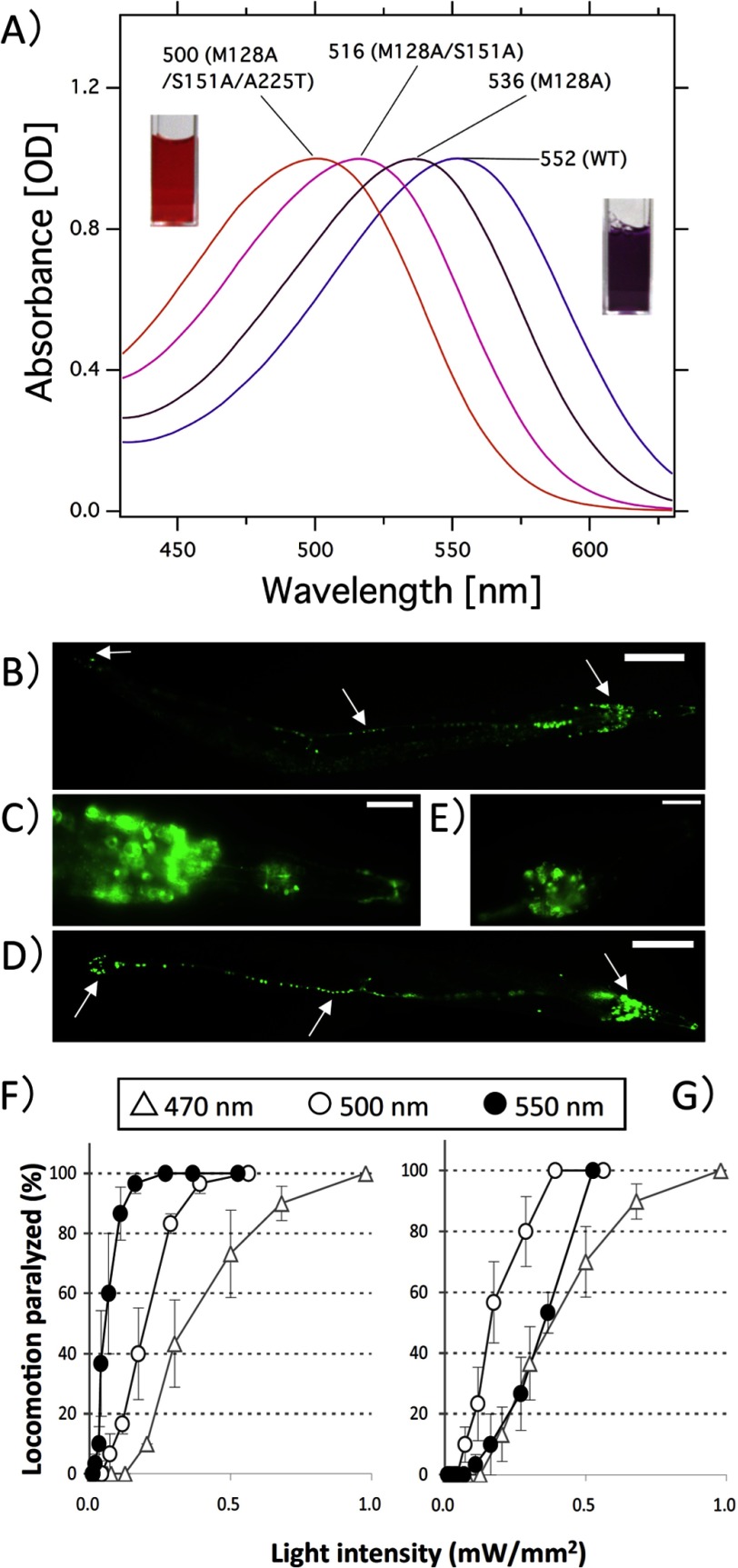
Such a blue-shifted proton pump is of great interest for optogenetic applications. Therefore, the same color-tuning mutations were introduced into archearhodopsin-3 (AR3), a protein pump that has already been successfully utilized as a light-induced neural silencer by Prof. Boyden and co-workers (5). As expected, the λmax of the triple mutant M128A/S151A/A225T (M128A, S151A, and A225T correspond to Met-126, Ser-149, and Ala-223 in HwBR, respectively) was ∼500 nm (Fig. 5A), close to that of the corresponding mutant in HwBR (498 nm), and the content of the all-trans-retinal was similar to that of the WT (Table 3). The mutation was introduced into C. elegans cells as a fusion construct with GFP under the control of a pan-neuronal promoter following a procedure described previously (26). Fig. 5 shows the fluorescent images of the cells expressing WT AR3 (Fig. 5B) and M128A/S151A/A225T (Fig. 5D). GFP signals were observed in the soma and the axon of many neurons in the head, mid-body, and tail for both the WT (Fig. 5, B and C) and the mutant AR3 (Fig. 5, D and E).
FIGURE 5.
Locomotion paralysis of C. elegans cells upon blue light illumination. A, visible absorption spectra of WT AR3 and its mutants. The spectra are scaled to be of similar intensity to the one of the WT by multiplying with 1.03 for M128A, 1.10 for M128A/S151A, and 1.08 for M128A/S151A/A225T, respectively. Met-128, Ser-151, and Ala-225 in AR3 correspond to Met-126, Ser-149, and Ala-223 in HwBR, respectively. The cuvettes colored orange and purple correspond to the purified M128A/S151A/A225T mutant of AR3 and the wild-type AR3, respectively. B and D, expression of WT AR3 (B) and the M128A/S151A/A225T mutant (D) of AR3 fused with GFP in C. elegans. GFP is expressed in the neurons (head neurons, tail neurons, and the ventral nerve cord) for both the WT and the mutant (arrow). C and E, expanded view. Scale bars, B and D = 100 μm; C and E = 10 μm. The anterior end is toward the right. F and G, dependence of the locomotion paralysis on light intensity. Animals expressing AR3::GFP were illuminated with different wavelengths of light (open triangles, 470 nm; open circles, 500 nm; closed circles, 550 nm) at varying light intensities for wild-type AR3 (F) and the M128A/S151A/A225T mutant (G). Error bars indicate ± S.E. Five animals were examined for each trial. We used the following three bandpass filters from Andover: 470 nm: FWHM = 10 nm; 500 nm: FWHM = 10 nm; 550 nm: FWHM = 10 nm.
TABLE 3.
Absorption maximum, opsin shift, and retinal configuration of the WT and of various AR3 mutants both in the dark and light
| Opsin type | λmax | Δυ | All-trans |
|
|---|---|---|---|---|
| Dark-adapted | Light-adapted | |||
| nm | cm−1 | % | ||
| WT (AR3) | 552 | 53 ± 2 | 90 ± 2 | |
| M128A | 536 | 541 | 68 ± 1 | NDa |
| M128A/S151A | 516 | 1264 | 53 ± 2 | ND |
| M128A/S151A/A225T | 500 | 1884 | 53 ± 2 | ND |
a ND, not determined.
Then the locomotion of the worms carrying WT (Fig. 5F) or M128A/S151A/A225T (Fig. 5G) was measured under various wavelengths and intensities of light (470, 500, and 550 nm). As expected, the illumination with light of 550 nm was most effective for the WT (closed circles), whereas higher and much higher light intensities were required for the locomotion paralysis by light of 500 (open circles) or 470 nm (open triangles), respectively. In contrast to the WT, illuminating the sample with light of 500 nm was most effective for the M128A/S151A/A225T mutant (open circles), and higher light intensities were required for light of both 550 (closed circles) and 470 nm (open triangles). The order of the efficiency follows the magnitude of the absorption at the corresponding wavelength shown in Fig. 5A, and therefore, the wavelength dependence can be explained by the absorption spectrum. Thus, we successfully produced a blue-shifted proton pump for neural silencing in C. elegans.
DISCUSSION
Color-tuning Mechanism in Proton Pumping Rhodopsins
In this study, we identified four residues, Asp-123, Met-126, Ser-149, and Ala-223, which are involved in the color tuning of HwBR. Similar to the spectral shift of the A223T mutant of HwBR (437 cm−1), it has already been reported that an Ala to Thr or Thr to Ala mutation at position 223 causes a spectral blue shift in HsBR and SRI or a red shift in SRII, respectively, with similar shift values (Δυ = 356–545 cm−1) (15, 17, 34), confirming that this residue is one of the important color-tuning residues in microbial rhodopsins. In the case of the residues Asp-123 and Ser-149, their contributions to the color tuning have also been reported in mutants of HsBR, SRI, and SRII (15, 17, 19). For Met-126, however, the shift value caused by the M118A mutation in HsBR was 3284 cm−1 (19), which is much larger than those of both HwBR (300 cm−1), AR3 (541 cm−1), and SRII (160 cm−1) (15). In the case of HsBR, the researchers have used refolded proteins, without showing functional data of the mutants of HsBR (19). Therefore, the spectrum of the mutant might as well contain partially folded proteins, leading to a larger spectral blue shift.
As already mentioned, in the spectra of the largely blue-shifted mutants, a spectral shoulder at ∼460 nm (Fig. 2j, asterisk) becomes more visible. The deconvolution of the spectra using skewed Gaussians revealed the existence of four vibronic bands, similar to the ones observed for SRII (16, 28). Interestingly, the same is observed for the WT, in which the spectral shoulder is not visible by eyes. The first two vibronic bands (∼560 and 500 nm for the WT and ∼510 and 460 nm for the blue-shifted mutants) are commonly assigned to the S1–S0 and S2–S0 transition. This is supported by the fact that the observed blue shift, associated with a stabilization of the ground state of the chromophore, does not change the difference between those states (∼2000 cm−1). One should note that the differences in retinal composition were not taken into account, and therefore further studies are needed to obtain a better understanding of the origin of all the vibronic bands.
Do the four residues (Asp-123, Met-126, Ser-149, and Ala-223) affect each other's influence on the color tuning? To investigate the synergism of the mutations, the effects of the multiple replacements are estimated by comparing the shifts to the summations of the shifts caused by their single replacements (Table 4). A relatively large positive synergistic effect was observed for M126A/S149A/A223T (ΔΔ = +686 cm−1) and M126A/S149A (ΔΔ = +498 cm−1), which both contain the M126A and S149A mutations. These residues are located close to the β-ionone ring of the retinal chromophore, and it is likely that they both alter the conformation of the 6-S bond synergistically through a steric effect(s), leading to the breakage of the π conjugation on the polyene chain (Fig. 4). On the other hand, a relatively large negative synergistic effect was observed for D123A/S149A (ΔΔ = −292 cm−1), D123A/M126A/A223T (ΔΔ = −305 cm−1), D123A/A223T (ΔΔ = −507 cm−1), and D123A/S149A/A223T (ΔΔ = −509 cm−1). All these mutants contain the D123A and A223T mutations, except for D123A/S149A. Both replacements cause a change in polarity (Fig. 1), and therefore the observed negative synergism could be explained by both replacements causing a similar effect on the electrostatic environment in the vicinity of the retinal chromophore, such as the alteration of the strength of the electrostatic interaction or of the polarity/polarizability. Thus, the action mechanism can be assumed to be different between the positive and negative synergism. This is supported by the fact that there is almost no effect in the quadruple mutant (ΔΔ = −159 cm−1), which contains all of the replacements.
TABLE 4.
Positive and negative synergistic effects of the combination mutations of HwBR on the opsin shift
| Opsin type | Actual value | Estimated value from the summation | Difference |
|---|---|---|---|
| cm−1 | cm−1 | cm−1 | |
| M126A/S149A/A223T | 1964 | 1278 | +686 |
| M126A/S149A | 1339 | 841 | +498 |
| M126A/A223T | 932 | 737 | +195 |
| D123A/M126A/S149A | 1647 | 1487 | +160 |
| D123A/M126A | 1078 | 946 | +132 |
| S149S/A223T | 968 | 978 | −10 |
| D123A/M126A/S149A/A223T | 1765 | 1924 | −159 |
| D123A/S149A | 895 | 1187 | −292 |
| D123A/M126A/A223T | 1078 | 1383 | −305 |
| D123A/A223T | 576 | 1083 | −507 |
| D123A/S149A/A223T | 1115 | 1624 | −509 |
Potential of the Blue-shifted Mutant as an Optogenetical Tool
Using fluorescent and luminescent proteins, which have the advantage over added chemicals of being genetically encoded probes, biological phenomena have been visualized in a variety of cell types. From 2005 there have been attempts to control some biological phenomena by visible light, using photo-functional proteins such as rhodopsins or flavin-containing proteins (4, 35, 36). Especially since the discoveries of channel rhodopsins, type I microbial rhodopsins have been the focus as useful research tools in neural science (4). Within this, in the past few years, some of the BR-like proton pumping or other ion pumping rhodopsins have also been found to mediate a strong silencing of the neural activity in various organisms such as mice or C. elegans (5, 26). As already mentioned, AR3 and HR absorb light of longer wavelengths (∼550–580 nm) and can function as neural silencers, whereas ChR2 absorbs light of shorter wavelengths (∼460 nm) and functions as a neural activator (4). Utilizing the difference in absorption maxima, Prof. Deisseroth and co-workers have succeeded in the activation and inactivation of neurons by light of different color (37). Currently, many researchers are trying to modify the photochemical properties of ChR2 including its photo-reaction, absorption maximum, etc. Recently, Profs. Peter Hegemann and Karl Deisseroth and co-workers have reported a novel variant of ChR2 (E123T/T159C), which exhibits both increased photocurrents and faster kinetics compared with the WT (38). E123T/T159C has been called the “perfected ChR2” (39). In this study, we have developed triple mutants of HwBR and AR3, which have shorter absorption maxima (∼500 nm) and which therefore overlap to an extent with that of WT ChR2 and coincide with that of E123T/T159C (λmax values for WT and E123T/T159C are ∼500 and ∼460 nm, respectively). Therefore, now we can control both neural activation and silencing with the same wavelength of light at the same time. In future, the communication between excitable and suppressed cells will be analyzed by using E123T/T159C of ChR2 and M128A/S151A/A225T of AR3 expressed in different types of neurons. In addition to pointing out the value of such a blue-shifted pump to optogenetics, for potential neuroscientist users, its neural silencing efficiency has to be compared with that of the wild type in the C. elegans measurements. The efficiency of the M128A/S151A/A225T AR3 mutant is ∼40% compared with the wild type, which means higher light intensities are required for neuron silencing (Fig. 5). In future, a larger concentration of retinal added to C. elegans may increase the mutant AR3 efficiency, because the mutational modification to the retinal binding site may have altered its binding affinity.
In conclusion, we have succeeded in producing a blue-shifted rhodopsin absorbing light of ∼500 nm without losing its proton pumping function. QM/MM calculation suggested that the breakage of the π conjugation of the polyene chain caused by the rotation of the β-ionone ring contributes largely to the spectral shift. The color variant was shown to be functional in C. elegans cells. The strong overlap of the absorption spectra with that of channelrhodopsins makes it possible to control both neural silencing and activation with blue light (∼500 nm) at the same time. Thus, the developed variant could be a useful research tool in a number of scientific fields.
Acknowledgments
We thank Yukie Kawase for technical assistance in sample preparation and Dr. Satoshi Tsunoda for giving us the spectral data of channelrhodopsin-2 and its mutant.
This work was financially supported by Grants 22018010, 23687019, and 23657100 (to Y. S.), 24115508 and 24655009 (to K. I.), and 23107717 and 23700580 (to S. H.) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology and by funds from the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry (to S. H.).
- AR3
- archearhodopsin-3
- BR
- bacteriorhodopsin
- HsBR
- BR from H. salinarum
- HwBR
- a BR homolog from H. walsbyi
- RPSB
- retinal protonated Schiff base
- QM/MM
- quantum mechanics/molecular mechanics
- ChR
- channelrhodopsin
- HR
- halorhodopsin
- SR
- sensory rhodopsin
- MRMP2
- second order multireference Møller-Plesset perturbation
- FWHM
- full width at half-maximum
- RWFE-SCF
- reweighting free energy self-consistent field.
REFERENCES
- 1. Nielsen M. B. (2009) Model systems for understanding absorption tuning by opsin proteins. Chem. Soc. Rev. 38, 913–924 [DOI] [PubMed] [Google Scholar]
- 2. Sudo Y. (2012) Transport and sensory rhodopsins in microorganisms. CRC Handbook of Organic Photochemistry and Photobiology, 3rd ed., pp. 1173–1193, CRC Press, Boca Raton, FL [Google Scholar]
- 3. Zhang F., Aravanis A. M., Adamantidis A., de Lecea L., Deisseroth K. (2007) Circuit-breakers. Optical technologies for probing neural signals and systems. Nat. Rev. Neurosci. 8, 577–581 [DOI] [PubMed] [Google Scholar]
- 4. Zhang F., Vierock J., Yizhar O., Fenno L. E., Tsunoda S., Kianianmomeni A., Prigge M., Berndt A., Cushman J., Polle J., Magnuson J., Hegemann P., Deisseroth K. (2011) The microbial opsin family of optogenetic tools. Cell 147, 1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chow B. Y., Han X., Dobry A. S., Qian X., Chuong A. S., Li M., Henninger M. A., Belfort G. M., Lin Y., Monahan P. E., Boyden E. S. (2010) High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature 463, 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blatz P. E., Mohler J. H., Navangul H. V. (1972) Anion-induced wavelength regulation of absorption maxima of Schiff bases of retinal. Biochemistry 11, 848–855 [DOI] [PubMed] [Google Scholar]
- 7. Honig B., Dinur U., Nakanishi K., Balogh-Nair V., Gawinowicz M. A., Arnaboldi M., Motto M. G. (1979) An external point-charge model for wavelength regulation in visual pigments. J. Am. Chem. Soc. 101, 7084–7086 [Google Scholar]
- 8. Hu J., Griffin R. G., Herzfeld J. (1994) Synergy in the spectral tuning of retinal pigments. Complete accounting of the opsin shift in bacteriorhodopsin. Proc. Natl. Acad. Sci. U.S.A. 91, 8880–8884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yan B., Spudich J. L., Mazur P., Vunnam S., Derguini F., Nakanishi K. (1995) Spectral tuning in bacteriorhodopsin in the absence of counterion and coplanarization effects. J. Biol. Chem. 270, 29668–29670 [DOI] [PubMed] [Google Scholar]
- 10. Yokoyama S. (2000) Molecular evolution of vertebrate visual pigments. Prog. Retin. Eye Res. 19, 385–419 [DOI] [PubMed] [Google Scholar]
- 11. Hayashi S., Tajkhorshid E., Pebay-Peyroula E., Royant A., Landau E. M., Navarro J., Schulten K. (2001) Structural determinants of spectral tuning in retinal proteins. Bacteriorhodopsin vs. sensory rhodopsin II. J. Phys. Chem. B. 105, 10124–10131 [Google Scholar]
- 12. Rajamani R., Gao J. (2002) Combined QM/MM study of the opsin shift in bacteriorhodopsin. J. Comput. Chem. 23, 96–105 [DOI] [PubMed] [Google Scholar]
- 13. Kouyama T., Murakami M. (2010) Structural divergence and functional versatility of the rhodopsin superfamily. Photochem. Photobiol. Sci. 9, 1458–1465 [DOI] [PubMed] [Google Scholar]
- 14. Sasaki J., Spudich J. L. (2008) Signal transfer in haloarchaeal sensory rhodopsin-transducer complexes. Photochem. Photobiol. 84, 863–868 [DOI] [PubMed] [Google Scholar]
- 15. Shimono K., Ikeura Y., Sudo Y., Iwamoto M., Kamo N. (2001) Environment around the chromophore in pharaonis phoborhodopsin. Mutation analysis of the retinal binding site. Biochim. Biophys. Acta 1515, 92–100 [DOI] [PubMed] [Google Scholar]
- 16. Shimono K., Hayashi T., Ikeura Y., Sudo Y., Iwamoto M., Kamo N. (2003) Importance of the broad regional interaction for spectral tuning in Natronobacterium pharaonis phoborhodopsin (sensory rhodopsin II). J. Biol. Chem. 278, 23882–23889 [DOI] [PubMed] [Google Scholar]
- 17. Sudo Y., Yuasa Y., Shibata J., Suzuki D., Homma M. (2011) Spectral tuning in sensory rhodopsin I from Salinibacter ruber. J. Biol. Chem. 286, 11328–11336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Suzuki D., Furutani Y., Inoue K., Kikukawa T., Sakai M., Fujii M., Kandori H., Homma M., Sudo Y. (2009) Effects of chloride ion binding on the photochemical properties of Salinibacter sensory rhodopsin I. J. Mol. Biol. 392, 48–62 [DOI] [PubMed] [Google Scholar]
- 19. Greenhalgh D. A., Farrens D. L., Subramaniam S., Khorana H. G. (1993) Hydrophobic amino acids in the retinal-binding pocket of bacteriorhodopsin. J. Biol. Chem. 268, 20305–20311 [PubMed] [Google Scholar]
- 20. Sudo Y., Ihara K., Kobayashi S., Suzuki D., Irieda H., Kikukawa T., Kandori H., Homma M. (2011) A microbial rhodopsin with a unique retinal composition shows both sensory rhodopsin II and bacteriorhodopsin-like properties. J. Biol. Chem. 286, 5967–5976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sudo Y., Iwamoto M., Shimono K., Kamo N. (2002) Tyr-199 and charged residues of pharaonis phoborhodopsin are important for the interaction with its transducer. Biophys. J. 83, 427–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sudo Y., Okada A., Suzuki D., Inoue K., Irieda H., Sakai M., Fujii M., Furutani Y., Kandori H., Homma M. (2009) Characterization of a signaling complex composed of sensory rhodopsin I and its cognate transducer protein from the eubacterium Salinibacter ruber. Biochemistry 48, 10136–10145 [DOI] [PubMed] [Google Scholar]
- 23. Kitajima-Ihara T., Furutani Y., Suzuki D., Ihara K., Kandori H., Homma M., Sudo Y. (2008) Salinibacter sensory rhodopsin. Sensory rhodopsin I-like protein from a eubacterium. J. Biol. Chem. 283, 23533–23541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luecke H., Schobert B., Richter H. T., Cartailler J. P., Lanyi J. K. (1999) Structure of bacteriorhodopsin at 1.55 A resolution. J. Mol. Biol. 291, 899–911 [DOI] [PubMed] [Google Scholar]
- 25. Kosugi T., Hayashi S. (2012) QM/MM reweighting free energy SCF for geometry optimization on extensive free energy surface of enzymatic reaction. J. Chem. Theory Comput. 8, 322–334 [DOI] [PubMed] [Google Scholar]
- 26. Okazaki A., Sudo Y., Takagi S. (2012) Optical silencing of C. elegans cells with arch proton pump. PLoS One 7, e35370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sudo Y., Iwamoto M., Shimono K., Kamo N. (2001) Pharaonis phoborhodopsin binds to its cognate truncated transducer even in the presence of a detergent with a 1:1 stoichiometry. Photochem. Photobiol. 74, 489–494 [DOI] [PubMed] [Google Scholar]
- 28. Chizhov I., Schmies G., Seidel R., Sydor J. R., Lüttenberg B., Engelhard M. (1998) The photophobic receptor from Natronobacterium pharaonis. Temperature and pH dependencies of the photocycle of sensory rhodopsin II. Biophys. J. 75, 999–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Spudich J. L., Yang C. S., Jung K. H., Spudich E. N. (2000) Retinylidene proteins. Structures and functions from archaea to humans. Annu. Rev. Cell Dev. Biol. 16, 365–392 [DOI] [PubMed] [Google Scholar]
- 30. Cembran A., Gonzalez-Luque R., Altoè P., Merchan M., Bernardi F., Olivucci M., Garavelli M. (2005) Structure, spectroscopy, and spectral tuning of the gas-phase retinal chromophore. The beta-ionone “handle” and alkyl group effect. J. Phys. Chem. A. 109, 6597–6605 [DOI] [PubMed] [Google Scholar]
- 31. Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Crystal structure of rhodopsin. A G protein-coupled receptor. Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 32. Wanko M., Hoffmann M., Strodel P., Koslowski A., Thiel W., Neese F., Frauenheim T., Elstner M. (2005) Calculating absorption shifts for retinal proteins. Computational challenges. J. Phys. Chem. B. 109, 3606–3615 [DOI] [PubMed] [Google Scholar]
- 33. Fujimoto K., Hayashi S., Hasegawa J., Nakatsuji H. (2007) Theoretical studies on color tuning mechanism in retinal proteins. J. Chem. Theory Comput. 3, 605–618 [DOI] [PubMed] [Google Scholar]
- 34. Sudo Y., Spudich J. L. (2006) Three strategically placed hydrogen-bonding residues convert a proton pump into a sensory receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 16129–16134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boyden E. S., Zhang F., Bamberg E., Nagel G., Deisseroth K. (2005) Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268 [DOI] [PubMed] [Google Scholar]
- 36. Losi A., Gärtner W. (2012) The evolution of flavin-binding photoreceptors. An ancient chromophore serving trendy blue-light sensors. Annu. Rev. Plant Biol. 63, 49–72 [DOI] [PubMed] [Google Scholar]
- 37. Zhang F., Wang L. P., Brauner M., Liewald J. F., Kay K., Watzke N., Wood P. G., Bamberg E., Nagel G., Gottschalk A., Deisseroth K. (2007) Multimodal fast optical interrogation of neural circuitry. Nature 446, 633–639 [DOI] [PubMed] [Google Scholar]
- 38. Berndt A., Schoenenberger P., Mattis J., Tye K. M., Deisseroth K., Hegemann P., Oertner T. G. (2011) High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc. Natl. Acad. Sci. U.S.A. 108, 7595–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pastrana E. (2011) Perfecting ChR2. Nat. Methods 8, 447. [DOI] [PubMed] [Google Scholar]