Background: PI(3,5)P2 is a phosphoinositide lipid generated at the endosomal membranes.
Results: We demonstrate PI(3,5)P2-dependent transcriptional activation of the gluconeogenesis genes repressed by the general transcriptional regulator Tup1.
Conclusion: Transcriptional reprogramming from glycolysis to gluconeogenesis requires PI(3,5)P2.
Significance: Our findings illustrate, for the first time, a critical role for the endosomal PI(3,5)P2 lipid in regulating metabolic reprogramming from glycolysis to gluconeogenesis in eukaryotic cells.
Keywords: Gluconeogenesis; Glycolysis; Membrane Lipids; Metabolism; Transcription Regulation; Tup1 Conversion; Metabolic Reprogramming; Phosphoinositide PI(3,5)P2 Lipid
Abstract
Glucose/carbon metabolism is a fundamental cellular process in living cells. In response to varying environments, eukaryotic cells reprogram their glucose/carbon metabolism between aerobic or anaerobic glycolysis, oxidative phosphorylation, and/or gluconeogenesis. The distinct type of glucose/carbon metabolism that a cell carries out has significant effects on the cell's proliferation and differentiation. However, it is poorly understood how the reprogramming of glucose/carbon metabolism is regulated. Here, we report a novel endosomal PI(3,5)P2 lipid-dependent regulatory mechanism that is required for metabolic reprogramming from glycolysis to gluconeogenesis in Saccharomyces cerevisiae. Certain gluconeogenesis genes, such as FBP1 (encoding fructose-1,6-bisphosphatase 1) and ICL1 (encoding isocitrate lyase 1) are under control of the Mig1 repressor and Cyc8-Tup1 corepressor complex. We previously identified the PI(3,5)P2-dependent Tup1 conversion (PIPTC), a mechanism to convert Cyc8-Tup1 corepressor to Cti6-Cyc8-Tup1 coactivator. We demonstrate that the PIPTC plays a critical role for transcriptional activation of FBP1 and ICL1. Furthermore, without the PIPTC, the Cat8 and Sip4 transcriptional activators cannot be efficiently recruited to the promoters of FBP1 and ICL1, suggesting a key role for the PIPTC in remodulating the chromatin architecture at the promoters. Our findings expand our understanding of the regulatory mechanisms for metabolic reprogramming in eukaryotes to include key regulation steps outside the nucleus. Given that Tup1 and the metabolic enzymes that control PI(3,5)P2 are highly conserved among eukaryotes, our findings may provide important insights toward understanding glucose/carbon metabolic reprogramming in other eukaryotes, including humans.
Introduction
Glucose is the primary carbon source for many organisms, including humans. Glucose metabolism provides energy (ATP) and metabolite molecules that are utilized as a precursor for macromolecule biosynthesis. For example, glucose 6-phosphate is metabolized for biosynthesis of nucleotides and aromatic amino acids. Dihydroxyacetone phosphate is a precursor for triacylglyceride and phospholipid synthesis (Fig. 1).
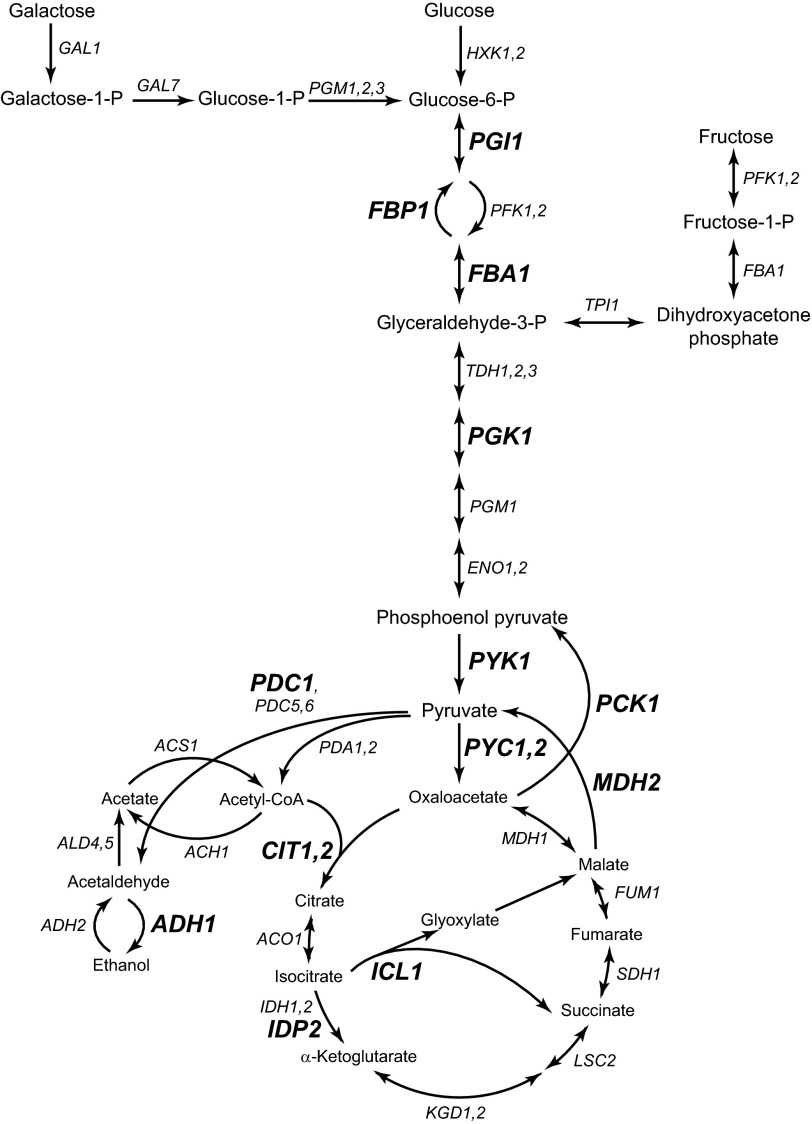
FIGURE 1.
The glycolysis and gluconeogenesis pathways. The glycolysis and gluconeogenesis pathways are presented in a schematic diagram. The TCA cycle is included. The gene encoding for the enzyme that is involved in each step is indicated. For simplicity, only certain intermediate metabolites and metabolic genes are shown in the pathways. The genes that we analyzed in this study are shown in boldface letters. Glucose 6-P, glucose 6-phosphate; Glyceraldehyde-3-P, glyceraldehyde 3-phosphate; Fructose-1-P, fructose 1-phosphate; HXK1,2, hexokinase 1,2; PGI1, phosphoglucose isomerase 1; PFK1,2, phosphofructokinase 1,2; FBP1, fructose-1,6-bisphosphatase 1; FBA1, fructose-bisphosphate aldolase 1; TPI1, triose-phosphate isomerase 1; TDH1,2,3, triose-phosphate dehydrogenase 1,2,3; PGK1, phosphoglycerate kinase; PGM1, phosphoglucomutase 1; ENO1,2, enolase 1,2; PYK1, pyruvate kinase 1; PYC1,2, pyruvate carboxylase 1,2; GAL1, galactokinase; GAL7, galactose-1-phosphate uridyltransferase; MDH1, malate dehydrogenase 1; FUM1, fumarase 1; SDH1, succinate dehydrogenase 1; LSC2, ligase of succinyl-CoA; KGD1,2, α-ketoglutarate dehydrogenase 1,2; IDP2, isocitrate dehydrogenase; NADP-specific 2; IDH1,2, isocitrate dehydrogenase 1,2; ICL1, isocitrate lyase 1; ACO1, aconitase 1; CIT1,2, citrate synthase 1,2; ACH1, acetyl-CoA hydrolase; ACS1, acetyl-CoA synthetase; ALD4,5, aldehyde dehydrogenase 4,5; ADH1,2, alcohol dehydrogenase 1,2; PDC1,5,6, pyruvate decarboxylase 1,5,6; PDA1,2, pyruvate dehydrogenase 1,2.
Glucose is metabolized to pyruvate through glycolysis (Fig. 1). In aerobic conditions, normal differentiated human cells metabolize pyruvate to CO2 through mitochondrial oxidative phosphorylation (TCA cycle) (1). Without oxygen in anaerobic conditions, normal differentiated cells metabolize pyruvate to lactate (anaerobic glycolysis) (1). In contrast, highly proliferative cancer cells metabolize pyruvate to lactate even in aerobic conditions, a phenomenon called the Warburg effect or aerobic glycolysis (1). When glucose decreases and glycolysis is no longer functional, gluconeogenesis is activated to generate metabolite precursors for macromolecule biosynthesis (Fig. 1).
Recent studies in cancer cell metabolism have provided new perspectives on the role of glucose/carbon metabolism. In addition to providing energy and biosynthetic precursors, the PKM2 isoform of pyruvate kinase possesses non-enzymatic function in regulating a critical oncogenic transcriptional factor, HIF-1α (2). A cellular energy sensor, AMP-activated protein kinase, regulates phosphorylation of p53 and histone H2B and their transcriptional regulatory functions (3, 4). In addition, certain metabolites, such as acetyl-CoA, NAD+, and β-hydroxybutyrate, directly interact with histone-modifying enzymes as a substrate, cofactor, or inhibitor and significantly affect their functions (5–7). Therefore, the type of glucose/carbon metabolic enzymes and metabolites a cell contains will significantly affect the cell's transcriptional reprogramming and physiology (8, 9). Importantly, the reprogramming of glucose/carbon metabolism determines the type of enzymes and metabolites produced by the cell. However, it is poorly understood how reprogramming of glucose/carbon metabolism is regulated.
Saccharomyces cerevisiae has been an invaluable model to study glucose/carbon metabolism and metabolic reprogramming (10, 11). Our previous study on the yeast galactose metabolism gene, GAL1, identified a novel phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2)2-dependent transcriptional regulatory mechanism (12). PI(3,5)P2 is a phosphoinositide lipid that is synthesized from phosphatidylinositol 3-phosphate by the Fab1 phosphatidylinositol-3-phosphate 5-kinase at the late endosomal/vacuolar lysosomal membranes (13, 14). In glucose-rich medium, GAL1 is repressed by the Mig1 repressor and Cyc8-Tup1 corepressor complex (15, 16). The Cyc8-Tup1 complex is a global corepressor that represses numerous genes (more than 150 genes) through interactions with sequence-specific DNA binding repressors, such as Mig1/2, Sko1, Crt1, and Rox1 (17).
In galactose medium without glucose, GAL1 transcription is activated by the Gal4 activator. Gal4 recruits SAGA to the GAL1 promoter through interaction with Tra1, a crucial step to establish preinitiation complex (PIC) assembly (18–20).
In addition to its well known function as a corepressor, the Cyc8-Tup1 complex functions as a coactivator at the GAL1 promoter when associated with Cti6 (21). Cti6, a Cyc8-interacting protein, directly interacts with the SAGA complex. The Cti6-Cyc8-Tup1 coactivator contributes to the recruitment of the SAGA complex to the GAL1 promoter (21).
How the Cyc8-Tup1 corepressor undergoes conversion to the Cti6-Cyc8-Tup1 coactivator was not well understood. Our recent work showed that Tup1 and Cti6, two proteins of the three-protein coactivator complex Cti6-Cyc8-Tup1, bind PI(3,5)P2 lipid with a high specificity (12). Without PI(3,5)P2 or Cti6, GAL1 remains constitutively repressed by the repressive Cyc8-Tup1 complex in the context that the Gal4 activation pathway is compromised. We have proposed the PI(3,5)P2-dependent Tup1 conversion (PIPTC), a mechanism by which PI(3,5)P2 mediates the conversion of the Cyc8-Tup1 corepressor to the Cti6-Cyc8-Tup1 coactivator by interacting with Cti6 and Cyc8-Tup1 to facilitate the assembly of the Cti6-Cyc8-Tup1 coactivator complex. Whether the PIPTC is specific only for GAL1 transcriptional regulation or a general regulatory mechanism functioning at other Tup1-repressed genes was not clear.
Here, we show the PIPTC plays a crucial role for transcriptional reprogramming from glycolysis to gluconeogenesis. Tup1-repressed gluconeogenesis genes, in particular ICL1 and FBP1, require the PIPTC for their transcriptional activation during turning on the gluconeogenesis pathway.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
Yeast strains and plasmids were generated by standard genetic and molecular methods. The yeast strains and plasmids used in this study are summarized in Tables 1 and 2, respectively.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Reference or source |
|---|---|---|
| SEY6210 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | Robinson et al. (51) |
| fab1Δ2 | SEY6210; fab1Δ::HIS3 | Gary et al. (14) |
| BHY49 | SEY6210; tup1Δ::TRP1 | Han and Emr (12) |
| BHY130 | SEY6210; cti6Δ::TRP1 | Han and Emr (12) |
| BHY328 | SEY6210; cat8Δ::KANr | This study |
| BHY329 | SEY6210; sip4Δ::TRP1 | This study |
| BHY330 | SEY6210; cat8Δ::KANr sip4Δ::TRP1 | This study |
| BHY355 | SEY6210; CAT8-HA::HIS3 | This study |
| BHY356 | fab1Δ2; CAT8-HA::TRP1 | This study |
| BHY357 | BHY130; CAT8-HA::HIS3 | This study |
| BHY358 | SEY6210; SIP4-HA::TRP1 | This study |
| BHY359 | fab1Δ2; SIP4-HA::HIS3 | This study |
| BHY360 | BHY130; SIP4-HA::HIS3 | This study |
TABLE 2.
Plasmids used in this study
Medium and Medium Shift Experiments
Three different media (YPD, YPEtOH, and YPFructose) were used to investigate the mRNA levels of genes of interest in this study. The YP (yeast extract, peptone) medium was used as the basal medium. YPD is the standard YP medium containing 2% glucose. YPEtOH is YP medium containing 3% ethanol and 0.05% glucose. YPFructose is YP medium containing 2% fructose. Yeast cells were grown in YPD, collected at the exponential phase, washed twice with glucose-free YP medium, resuspended in YPEtOH or YPFructose, and grown for 15.5 h before collecting for mRNA analysis (indicated as YPEtOH 15.5 h or YPFruc 15.5 h). For microscopy experiments in Fig. 6, yeast cells were grown in the standard synthetic medium containing 2% glucose, collected at the exponential phase, washed twice with glucose-free YP medium, and resuspended in synthetic medium containing 3% ethanol before microscopy.
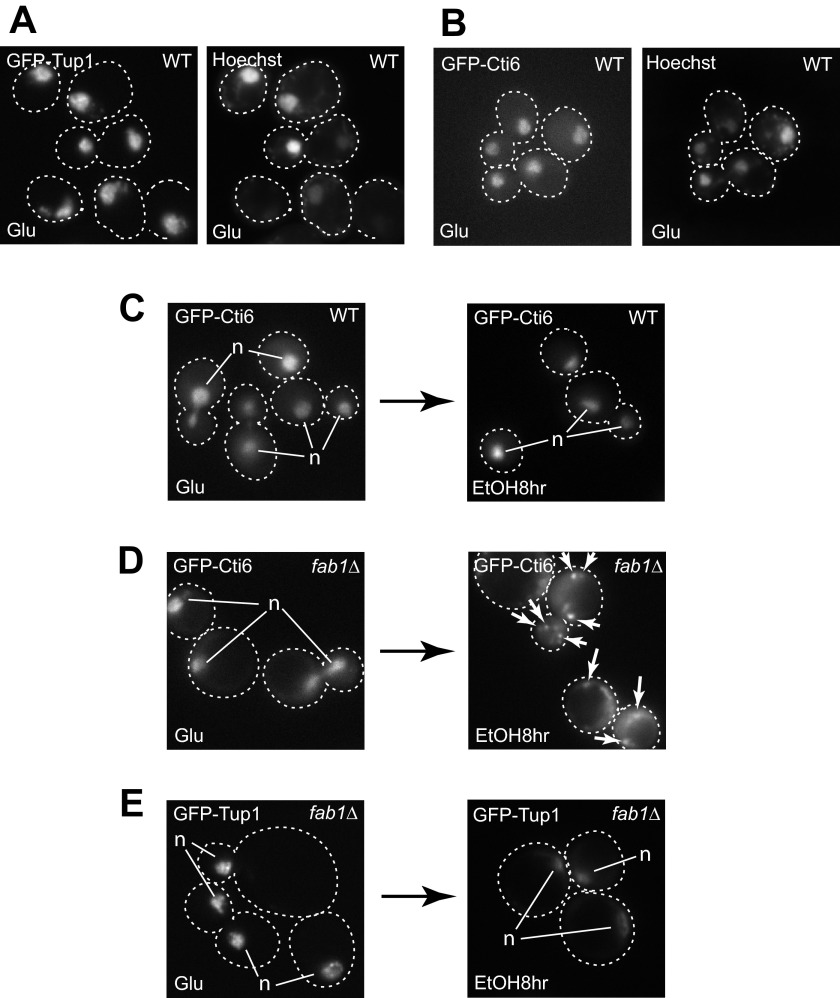
FIGURE 6.
Cti6 mislocalizes to the cytoplasm in fab1Δ cells in ethanol medium. A and B, GFP-Tup1 (A) and GFP-Cti6 (B) localizes primarily in the nucleus (stained with Hoechst 33342) in WT cells in glucose-rich medium. C, GFP-Cti6 localizes primarily in the nucleus in WT cells at 8 h after shifting from glucose to ethanol media. D, GFP-Cti6 accumulates in the cytoplasm in fab1Δ cells at 8 h after shifting from glucose to ethanol media. Some cytoplasmic Cti6 formed multiple speckles (marked by arrows). E, GFP-Tup1 localizes primarily in the nucleus in fab1Δ cells at 8 h after shifting from glucose to ethanol media. Single, bright GFP signals of Cti6 (C and D) or Tup1 (E) correspond to nuclear proteins (marked as n).
Reverse Transcription-qPCR (RT-qPCR) Analysis
Two-step RT-qPCR was performed using SYBR Green according to Han and Emr (12). Cell pellets equivalent to 3 OD were lysed by five cycles of 30-s bead beating with intermissions of 30-s cooling. Total RNA was purified using RNeasy minikit (Qiagen) and quantitated with NanoDrop 1000 (Thermo Scientific). First-strand cDNA was synthesized using the SuperScriptTMIII reverse transcriptase kit (Invitrogen). ACT1 mRNA was used as an internal reference calibrator. ΔΔCt analysis method was used to quantify mRNA. The mRNA level of a gene of interest that was transcribed in wild type (WT) cells in exponential growth phase in YPD was set as 1.0, to which the relative mRNA levels of the gene in different conditions (with different mutant cells or different medium conditions) were evaluated and indicated in Figs. 2 (A and B), 3A, and 4 (A and B). The relative mRNA levels in Figs. 3 (B–D) and 5A were evaluated by the same calculation. Three independent experiments were performed (indicated as n = 3 in the figure legends) and analyzed by standard statistical analysis for the values of average and S.D. and for t tests.
FIGURE 2.
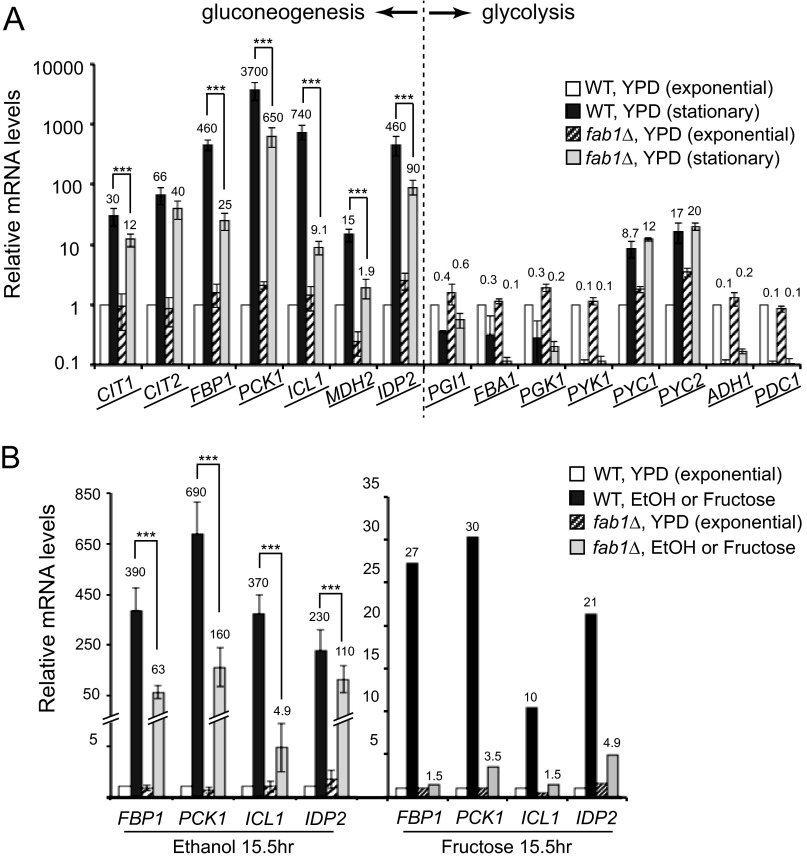
FAB1 is required for transcriptional induction of ICL1, FBP1, PCK1, and IDP2. A, mRNA levels of the genes in gluconeogenesis and glycolysis were quantified by RT-qPCR. The mRNA level of each gene in WT cells in exponential growth phase in YPD was set as 1.0. The relative mRNA levels of the gene in different conditions are presented. YPD (stationary), cultures at 48 h in YPD. The y axis is plotted on a log scale. Three independent experiments were performed and analyzed by standard statistical analysis. Error bars, S.D. ***, the two values of interest have a statistically significant difference with p values lower than 0.05. Values are shown only for the relative mRNA levels in WT and fab1Δ cells in late stationary phase in YPD. The relative mRNA levels in exponential phase are either 1.0 (WT) or close to 1.0 (fab1Δ), but for simplicity, their values are not shown. B, mRNA levels of FBP1, PCK1, ICL1, and IDP2 in WT and fab1Δ cells were quantified by RT-qPCR. mRNA levels of each gene were calculated relative to the mRNA level in WT cells in exponential phase in YPD, which was set as 1.0. Two non-glucose carbon media, ethanol (n = 3) and fructose (n = 1), were used to induce the gluconeogenesis genes. Error bars, S.D. ***, the two values of interest have a statistically significant difference with p values lower than 0.05. Values are shown only for the relative mRNA levels in WT and fab1Δ cells in YPEtOH or YPFructose. The relative mRNA levels in exponential phase in YPD are either 1.0 (WT) or close to 1.0 (fab1Δ or cti6Δ), but for simplicity, their values are not shown.
FIGURE 3.
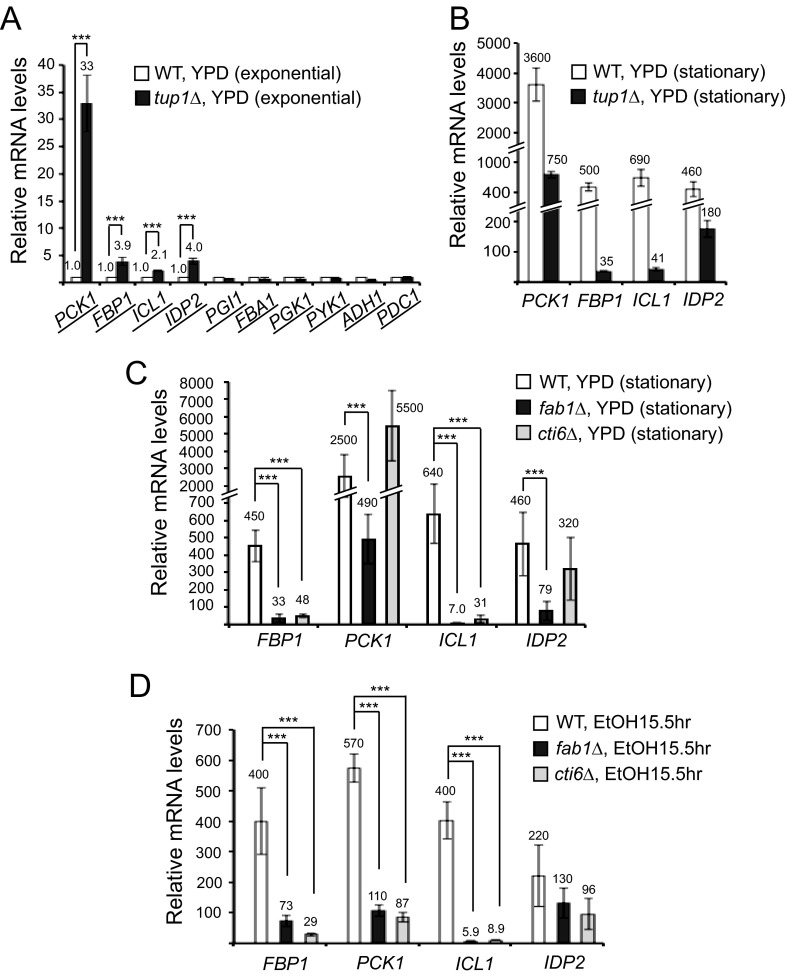
CTI6 is required for transcriptional induction of ICL1 and FBP1. A, RT-qPCR analyses showed higher transcript levels of PCK1, FBP1, ICL1, and IDP2 in tup1Δ cells compared with the levels in WT cells (set as 1.0) in the exponential growth phase in YPD (n = 3). B, relative mRNA levels of PCK1, FBP1, ICL1, and IDP2 were analyzed by RT-qPCR in WT and tup1Δ cells in late stationary phase at 48 h in YPD compared with the levels in WT cells (set as 1.0) in the exponential phase in YPD (n = 3). C, relative mRNA levels of FBP1, PCK1, ICL1, and IDP2 were analyzed by RT-qPCR in WT, fab1Δ, and cti6Δ cells in late stationary phase at 48 h in YPD compared with the levels in WT cells (set as 1.0) in the exponential phase in YPD (n = 3). Transcriptional induction of FBP1 and ICL1, but not PCK1 and IDP2, was severely defective in cti6Δ cells at 48 h in YPD. D, relative mRNA levels of FBP1, PCK1, ICL1, and IDP2 were analyzed by RT-qPCR in WT, fab1Δ, and cti6Δ cells at 15.5 h after shifting from YPD to YPEtOH compared with the levels in WT cells (set as 1.0) in the exponential phase in YPD (n = 3). Transcriptional induction of FBP1, PCK1, and ICL1, but not IDP2, was severely defective in cti6Δ cells in ethanol medium. Error bars, S.D. ***, the two values of interest have a statistically significant difference with p values lower than 0.05.
FIGURE 4.
Transcription of Cat8 and Sip4, two key transcriptional activators of gluconeogenesis genes, is under control of glucose repression. A, RT-qPCR analyses showed that the mRNA level of SIP4, but not CAT8, was higher in tup1Δ cells compared with the level in WT cells (set as 1.0) in the exponential growth phase in YPD (n = 3). Error bars, S.D. ***, the mRNA levels of SIP4 have a statistically significant difference with p values lower than 0.05 between WT and tup1Δ cells. B, relative mRNA levels of CAT8 and SIP4 were analyzed by RT-qPCR in WT, fab1Δ, and cti6Δ cells either in late stationary phase at 48 h in YPD (left) or in YPEtOH for 15.5 h after shifting from YPD to YPEtOH (right) compared with the levels in WT cells (set as 1.0) in the exponential phase in YPD (n = 3). Error bars, S.D. CAT8 was highly induced in WT, fab1Δ, and cti6Δ cells in late stationary growth phase in YPD and in ethanol medium. Transcriptional induction of SIP4 was defective in fab1Δ and cti6Δ cells. Values are shown only for the relative mRNA levels in WT and fab1Δ cells in YPEtOH or YPFructose. The relative mRNA levels in exponential phase in YPD are either 1.0 (WT) or close to 1.0 (fab1Δ or cti6Δ), but for simplicity, their values are not shown.
FIGURE 5.
The PIPTC is required for efficient recruitment of Cat8 and Sip4 activators to the gluconeogenesis gene promoters. A, relative mRNA levels of FBP1, PCK1, ICL1, and IDP2 were analyzed by RT-qPCR in WT, cat8Δ, sip4Δ, and cat8Δsip4Δ cells at 15.5 h after shifting from YPD to YPEtOH compared with the levels in WT cells (set as 1.0) in the exponential phase in YPD (n = 3). Error bars, S.D. Transcriptional induction of FBP1 and ICL1, but not PCK1 and IDP2, was significantly defective in cat8Δ cells. Deletion of SIP4 does not affect transcriptional induction of FBP1, PCK1, ICL1, and IDP2. Deletion of both CAT8 and SIP4 further reduced transcriptional induction of FBP1 and ICL1. Transcriptional induction of PCK1 was not affected in sip4Δ cells but was severely defective in cat8Δsip4Δ cells. Transcriptional induction of IDP2 was not significantly affected in either cat8Δ, sip4Δ, or cat8Δsip4Δ cells. B and C, ChIP-qPCR experiments were performed to analyze recruitment of Cat8 (B) and Sip4 (C) activators to the promoters of FBP1, ICL1, PCK1, and IDP2 in WT, fab1Δ, or cti6Δ cells grown in ethanol medium for 15.5 h after shift from YPD (n = 3). ACT1 promoter was analyzed as a negative control. Error bars, S.D. ***, the two values of interest have a statistically significant difference with p values lower than 0.05. D, the same numbers of cells in exponential growth phase in YPD were taken, washed, made in 10-fold serial dilution, and spotted on YPD and YP-ethanol media.
ChIP-qPCR Analysis
ChIP was performed based on the method of Goldfarb and Alani (22). Briefly, 45-ml cell cultures were cross-linked by treating with 0.37% formaldehyde and were quenched with 125 mm glycine. The pellets were washed twice with ice-cold TBS. Cell pellets were broken in lysis buffer by seven cycles of 30-s bead beating with intermissions of 30-s cooling. The lysates were sonicated using a Branson Sonifier 250 cell disruptor to generate chromatin fragments with an average size of 500 bp. 5% of the supernatants were saved for inputs. The rest of the supernatant was divided into two aliquots. One aliquot was immunoprecipitated with anti-HA antibody (12CA5) (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and the other was immunoprecipitated with no antibody. After reversing cross-links of the immunoprecipitates by incubation overnight at 65 °C, the DNA was isolated by phenol/chloroform extraction and quantified by qPCR. The background DNA immunoprecipitated with no antibody was subtracted from the amount of DNA immunoprecipitated with antibody. ACT1 was used as a negative control. Three independent experiments were performed and analyzed by standard statistical analysis.
Growth Assays
For growth assays on ethanol medium in Fig. 5D, yeast cells were grown in YPD to exponential phase. The same number of cells for each strain were collected, washed twice with glucose-free YP medium, and made serial diluted in YP. Dilutions were spotted on a YPD or YPEtOH medium plate.
Microscopy and Dye Staining
All imaging of yeast cells was performed using a Delta Vision RT system (Applied Precision). Hoechst 33342 (Invitrogen) was used to stain the nucleus in live yeast cells according to the manufacturer's instructions.
RESULTS
FAB1 Is Required for Transcriptional Induction of ICL1, FBP1, PCK1, and IDP2
GAL1 and many of the genes in gluconeogenesis have similar chromatin structure at their promoters. S. cerevisiae has two groups of genes, TATA box-containing and TATA-less genes (23). Each group has a distinct chromatin architecture and a distinct mechanism for the assembly of the PIC at promoters (24, 25). TATA box-containing genes, 20% of the yeast genome, are associated with highly regulated stress-responsive genes (23). In contrast, TATA-less genes are generally associated with housekeeping genes (23). TATA box-containing promoters have their +1 nucleosome positioned at the transcription start site (TSS), but the TSS at TATA-less genes is nucleosome-free (25). To establish PIC assembly during transcriptional activation, TATA box-containing genes preferentially depend on SAGA, and TATA-less genes utilize transcription factor IID (23). Recruitment of SAGA is a crucial step for TATA box-containing genes, which leads to evicting the +1 nucleosome and opening up the TSS to establish PIC assembly (18–20).
GAL1 and many of the genes in gluconeogenesis belong to the TATA box-containing gene group that utilizes SAGA rather than the transcription factor IID complex (23). Indeed, it was demonstrated for FBP1 as well as for GAL1 that SAGA plays a crucial role for establishing the activated chromatin structure at the promoter (18, 20, 26).
Importantly, like GAL1, certain gluconeogenesis genes, such as FBP1, are under control of the Mig1 repressor and Cyc8-Tup1 corepressor (27). Because GAL1 is regulated by the PIPTC, we reasoned that gluconeogenesis genes that have a similar promoter structure and utilize Tup1 repressor may also be regulated by the PIPTC.
To determine whether the transcriptional regulation of gluconeogenesis genes is dependent on PI(3,5)P2, we first analyzed how transcription of gluconeogenesis genes is affected in fab1Δ cells as compared with WT cells. As a non-glucose medium condition that induces transcription of gluconeogenesis genes, we used a very late stationary phase (at 48 h) in glucose-rich YPD medium. During the early exponential phase of growth in glucose-rich YPD medium, S. cerevisiae metabolizes glucose to pyruvate through glycolysis, and pyruvate is further metabolized primarily to ethanol through fermentation rather than to CO2 through the mitochondrial oxidative phosphorylation (TCA cycle) even in aerobic conditions (28). Later when the glucose level drops, the yeast cells switch from the fermentation pathway to the oxidative phosphorylation (TCA cycle) pathway by turning on transcription of the genes in the TCA cycle (28). Upon reaching late stationary phase, the glucose in the medium is depleted, and the yeast cells can no longer utilize glycolysis. They activate gluconeogenesis to generate glucose 6-phosphate and other glycolysis intermediate precursors for macromolecule biosynthesis. To activate gluconeogenesis, the yeast cells induce genes involved in gluconeogenesis.
Indeed, several gluconeogenesis genes were well induced in wild type cells at late stationary phase in YPD (Fig. 2A). Particularly, FBP1, PCK1, ICL1, and IDP2 were highly induced. In contrast, certain genes in the glycolysis (FBA1 and PYK1) and fermentation (ADH1 and PDC1) pathways were significantly turned off at the stationary growth phases in YPD because glycolysis and fermentation are no longer functional in the low glucose condition (Fig. 2A). The results indicate that transcriptional induction at the late stationary phase is specific for the key genes in the gluconeogenesis pathway but not for genes in the glycolysis and fermentation pathways. The results suggest transcriptional reprogramming is the primary mechanism to activate the gluconeogenesis pathway at late stationary phase in YPD.
Importantly, we found genes that are dependent on PI(3,5)P2 for their transcriptional induction, FBP1, PCK1, ICL1, MDH2, and IDP2. These gluconeogenesis genes exhibited a significant defect in transcriptional induction in fab1Δ cells (Fig. 2A). For example, mRNA levels of ICL1 (530-fold) and FBP1 (560-fold) were highly induced in WT cells in late stationary phase, whereas transcription of ICL1 (6-fold) and FBP1 (34-fold) was poorly induced in fab1Δ cells (Fig. 2A). ICL1, FBP1, and PCK1 encode three key enzymes in gluconeogenesis (Fig. 1). The defect in transcriptional induction in fab1Δ cells was specific for these gluconeogenesis pathway genes because other genes, such as PYC1 and PYC2, were induced in fab1Δ cells as much as in WT cells at the late stationary phase (Fig. 2A).
We decided to focus our remaining experiments on the four highly induced genes: PCK1, FBP1, ICL1, and IDP2. We tested if transcriptional induction of PCK1, FBP1, ICL1, and IDP2 depends on FAB1 in other non-glucose media. Signaling proteins, such as Snf1 protein kinase, are activated to induce glucose-repressed genes in non-glucose medium (29). Non-glucose carbon sources can be categorized into two groups, fermentative and non-fermentative carbon sources. Fermentative non-glucose carbon sources, such as fructose, are metabolized through part of the glycolytic pathway, but non-fermentative non-glucose carbon sources, such as ethanol, cannot utilize glycolysis (Fig. 1). Transcription of the gluconeogenesis genes can be regulated differentially between non-fermentative non-glucose medium and fermentative non-glucose medium. We decided to test both non-fermentative ethanol medium and fermentative fructose medium.
Transcription of ICL1, FBP1, PCK1, and IDP2 was all highly induced in WT cells in both ethanol and fructose media (Fig. 2B). Overall, the levels of transcriptional induction of ICL1, FBP1, PCK1, and IDP2 were higher in ethanol medium than in fructose medium (Fig. 2B). For example, FBP1 transcripts were induced about 420-fold in WT cells in ethanol medium, whereas the transcripts were induced 27-fold in fructose medium (Fig. 2B). These results indicate that glucose (or glucose-dependent Snf1 kinase) is not the sole factor that determines transcriptional induction of the genes. There may be multiple metabolite-responsive elements at their promoters that sense diverse metabolic signals in addition to glucose.
In both ethanol and fructose media, transcriptional induction of ICL1, FBP1, PCK1, and IDP2 was severely defective in fab1Δ cells (Fig. 2B). For example, in WT cells, ICL1 transcripts were induced 380-fold and 10-fold in ethanol and fructose media, respectively, but the transcripts were induced only 4.5- and 1.5-fold in fab1Δ cells in ethanol and fructose media (Fig. 2B). The results clearly demonstrate that FAB1 is required for transcriptional induction of ICL1, FBP1, PCK1, and IDP2 in all of the non-glucose conditions we tested.
ICL1, FBP1, PCK1, and IDP2 Are under Control of Tup1
Next, we asked if ICL1, FBP1, PCK1, and IDP2 are repressed by Cyc8-Tup1. FBP1 and ICL1 genes were reported to have a consensus sequence for Mig1/Mig2 binding (30, 31). It was demonstrated that FBP1 is repressed by the Mig1-Cyc8-Tup1 complex (27). We examined if Tup1 represses ICL1, FBP1, PCK1, and IDP2 in cells in the exponential phase in the glucose-rich YPD medium, a condition of glucose repression. ICL1, FBP1, PCK1, and IDP2 showed higher transcript levels in tup1Δ cells compared with WT cells (Fig. 3A), suggesting that they are repressed by Tup1, directly or indirectly. Increased transcription of ICL1, FBP1, PCK1, and IDP2 in tup1Δ cells indicates transcriptional derepression at the gene promoters without TUP1. In contrast, genes in glycolysis (PGI1, FBA1, PGK1, and PYK1) and fermentation (ADH1 and PDC1) expressed similar levels of transcripts in tup1Δ cells compared with WT cells in the exponential growth phase in YPD (Fig. 3A).
Next, we examined if the derepressed state of ICL1, FBP1, PCK1, and IDP2 in tup1Δ cells in the exponential phase in YPD can be further activated in the low glucose condition that activates gluconeogenesis. Indeed, in late stationary phase in YPD, the mRNA levels of ICL1, FBP1, PCK1, and IDP2 further increased in tup1Δ cells from the level in the exponential phase (Fig. 3, A and B). For example, PCK1 mRNA levels increased from 33- to 750-fold, whereas FBP1 mRNA levels increased from 3.9- to 35-fold. The results indicate the mRNA levels of ICL1, FBP1, PCK1 and IDP2, which are derepressed in tup1Δ cells in the glucose-rich condition at the exponential phase in YPD medium, further increase in the low glucose condition at late stationary phase. When glucose drops in late stationary phase, transcriptional activation signals are turned on to activate the transcription of gluconeogenesis genes farther from the derepressed state. Upon glucose depletion, Snf1 kinase is one of the key transcriptional activation signaling proteins. Also, transcriptional activators Cat8 and Sip4 are induced in low glucose conditions (see Fig. 4B). These signaling proteins and transcriptional activators further increase the mRNA levels of ICL1, FBP1, PCK1, and IDP2 genes in a transcriptional activated state from the levels in a derepressed state in tup1Δ cells at exponential phase in YPD medium.
However, the mRNA levels of ICL1, FBP1, PCK1, and IDP2 in tup1Δ cells at late stationary phase are significantly lower than those in WT cells (Fig. 3B). Without Tup1, transcription of the gluconeogenesis genes is not fully activated in the low glucose condition, presumably due to the lack of Tup1 coactivator function. The results here are consistent with the notion that the gluconeogenesis genes are under control of the dual functionality (corepressor and coactivator) of Tup1.
CTI6 Is Required for Transcriptional Induction of ICL1 and FBP1
Cti6 is an essential player in converting the Cyc8-Tup1 corepressor to a Cti6-Cyc8-Tup1 coactivator complex. We showed previously that without Cti6, transcriptional induction of GAL1 is severely defective (12). We examined if transcriptional induction of PCK1, FBP1, ICL1, and IDP2 is dependent on Cti6. Indeed, cti6Δ cells showed a significant defect in transcriptional induction of ICL1 and FBP1 genes but not PCK1 and IDP2 (Fig. 3, C and D). For example, ICL1 transcripts were induced 640- and 400-fold in WT cells in the late stationary growth phase in YPD and in ethanol medium, respectively, compared with those in WT cells in the exponential growth phase in YPD. However, in cti6Δ cells, ICL1 transcripts were induced only 31- and 8.8-fold in the late stationary growth phase in YPD and in ethanol medium, respectively (Fig. 3, C and D). Interestingly, although PCK1 transcription is repressed by Tup1 (Fig. 3A) and its proper induction requires Cti6 in ethanol medium (Fig. 3D), PCK1 transcripts were well induced without Cti6 at the late stationary growth phase in YPD (Fig. 3C). The exact reason for this phenomenon is not clear at present. Perhaps PCK1 has multiple regulatory elements at its promoter, and they may be regulated by diverse signals that are differentially generated in different medium conditions. These results, taken together, suggest that transcriptional induction of ICL1 and FBP1, but not IDP2, is regulated by the PIPTC.
Transcription of Cat8 and Sip4, Two Key Transcriptional Activators of Gluconeogenesis Genes, Is under Control of Glucose Repression
Many of the genes of the gluconeogenesis pathway contain the carbon source-responsive element (CSRE) at their promoters, a binding site for Cat8 and Sip4 transcriptional activators (32, 33). CAT8 itself, but not SIP4, contains the URE that is recognized by Mig1/Mig2, and CAT8 is significantly derepressed in mig1Δ cells (27, 33, 34). Although SIP4 does not contain the URE for Mig1/Mig2, it was reported that transcription of SIP4, like CAT8, is significantly affected by TUP1 (27). Therefore, we examined how transcription of CAT8 and SIP4 is affected by TUP1 and FAB1.
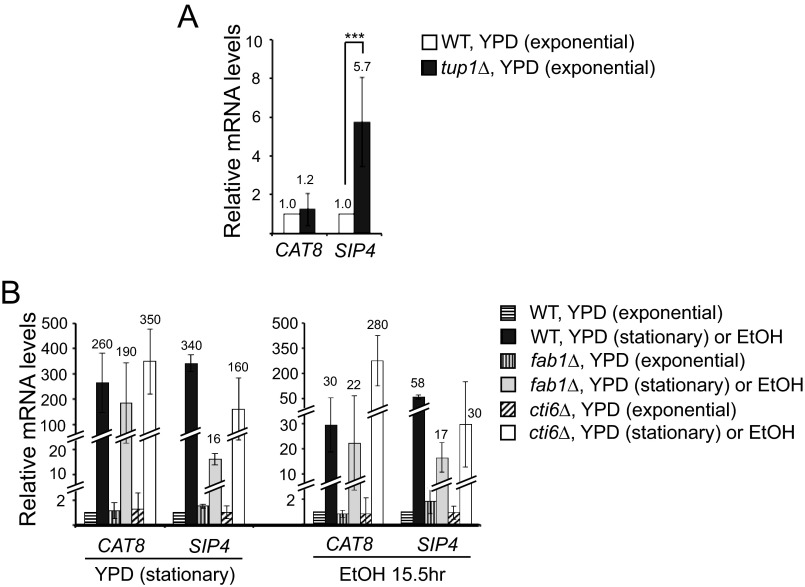
Transcription of SIP4 is significantly derepressed in the exponential growth phase in YPD medium by deletion of TUP1 (Fig. 4A), consistent with previous reports (27). Although SIP4 does not contain a URE for Mig1/Mig2 at the promoter, deletion of TUP1 results in substantial derepression of SIP4 (Fig. 4A). However, contrary to the previous report (27), CAT8 transcription was not significantly affected in tup1Δ cells (Fig. 4A).
The mRNA levels of CAT8 and SIP4 were highly induced in WT cells both in the late stationary growth phase in YPD (250-fold for CAT8; 340-fold for SIP4) and in ethanol medium (28-fold for CAT8; 58-fold for SIP4) (Fig. 4B), consistent with previous reports (33, 34). In contrast, transcriptional induction of SIP4 was defective in fab1Δ or cti6Δ cells in both medium conditions (Fig. 4B). However, CAT8 induction was modestly defective in fab1Δ cells and higher in cti6Δ cells in both medium conditions compared with WT cells (Fig. 4B). These results suggest that transcriptional induction of SIP4 but not of CAT8 is modestly influenced by the PIPTC. SIP4 may contain a URE for an unidentified repressor, which recruits a Cyc8-Tup1 corepressor complex. Alternatively, because Cyc8-Tup1 corepressors have been reported to influence chromatin architecture of a gene at a distance (35), Cyc8-Tup1 corepressors that are positioned far from the SIP4 locus may affect transcription of SIP4.
Cat8 and Sip4, Two Key Transcriptional Activators of Gluconeogenesis Genes, Function Redundantly to Activate Transcription of ICL1, FBP1, and PCK1
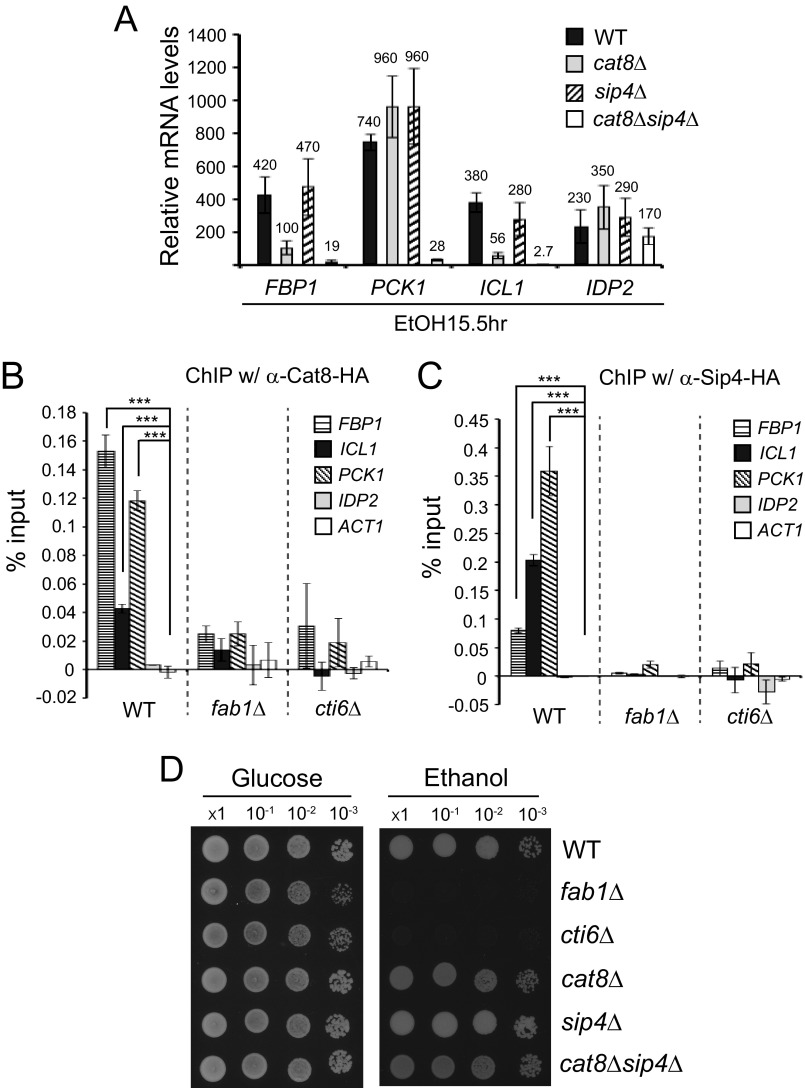
Many of the gluconeogenesis genes, including ICL1 and FBP1, contain the CSRE at their promoters (36). The transcriptional activators, Cat8 and Sip4 bind to the CSRE and activate gene transcription (33, 34, 36). ICL1, FBP1, PCK1, and IDP2 genes all contain the CSRE, and their transcriptional induction was reported to require Cat8 (34, 36, 37). It was shown that Sip4 could bind the Cat8-binding CSRE (33). The severe defect of transcriptional induction of ICL1 and FBP1 observed in fab1Δ or cti6Δ cells may be in part due to the reduced expression of Sip4. We decided to examine how significantly the Cat8 and Sip4 activators contribute to the transcriptional induction of ICL1, FBP1, PCK1, and IDP2. In ethanol medium, sip4Δ cells showed no effect on ICL1, FBP1, PCK1, and IDP2 transcription (Fig. 5A). However, cat8Δ cells showed a substantial defect in transcriptional induction of ICL1 and FBP1 but not PCK1 and IDP2 (Fig. 5A). Deletion of both CAT8 and SIP4 resulted in a severe defect in transcriptional induction of ICL1, FBP1, and PCK1 but not IDP2 (Fig. 5A). Therefore, it appears that Cat8 and Sip4 function redundantly for transcriptional activation of ICL1, FBP1, and PCK1, but Cat8 is the primary transcriptional activator for ICL1 and FBP1.
Given that Cat8 transcription is only modestly reduced in fab1Δ cells and even higher in cti6Δ cells (Fig. 4B) and Sip4 does not affect ICL1 and FBP1 transcription (Fig. 5A), the defect in transcriptional induction of ICL1 and FBP1 in fab1Δ or cti6Δ cells appears not to be related with the changed expression levels of the transcriptional activators, Cat8 and Sip4.
The PIPTC Is Required for Efficient Recruitment of Cat8 and Sip4 Activators to the Gluconeogenesis Gene Promoters
It was not known how the recruitment of Cat8 and Sip4 activators to the promoters of the gluconeogenesis genes is affected when the genes are constitutively repressed by the Cyc8-Tup1 corepressor without the PIPTC. Therefore, we examined recruitment of Cat8 and Sip4 to the promoters of ICL1, FBP1, PCK1, and IDP2 in WT, fab1Δ, and cti6Δ cells by ChIP-qPCR analysis. Indeed, both Cat8 and Sip4 occupied the promoters of FBP1, ICL1, and PCK1 much more compared with the ACT1 promoter (negative control) in WT cells in ethanol medium (Fig. 5, B and C). However, Cat8 and Sip4 were not recruited to the IDP2 promoter. These ChIP-qPCR results are consistent with the Fig. 5A results, in which deletion of both CAT8 and SIP4 resulted in a severe defect in transcription of FBP1, ICL1, and PCK1 but not of IDP2.
In contrast, recruitment of Cat8 and Sip4 to the promoters of FBP1, ICL1, and PCK1 was significantly reduced in fab1Δ or cti6Δ cells (Fig. 5, B and C). The reduced recruitment of Cat8 and Sip4 to the promoters in fab1Δ or cti6Δ cells is unlikely to be due to their reduced expression in the mutants. In fact, the mRNA level of CAT8 was even higher in cti6Δ cells than in WT cells in ethanol medium, but Cat8 recruitment to the promoters was strikingly reduced (Figs. 4B and 5B).
Taken together, these results suggest that without the PIPTC in fab1Δ or cti6Δ cells, the chromatin structure at the promoters of FBP1, ICL1, and PCK1 is much less accessible to Cat8 and Sip4. In addition, this suggests that the conversion of the Cyc8-Tup1 corepressor to a Cti6-Cyc8-Tup1 coactivator by the PIPTC leads to remodulation of the chromatin architecture at these promoters, which may be a prerequisite for Cat8 and Sip4 recruitment.
Multiple gluconeogenesis genes contain the CSRE for Cat8 or Sip4 binding at their promoters, and certain gluconeogenesis genes also contain the URE for Mig1/Mig2-binding. In addition, CAT8 and SIP4 themselves contain the URE and the CSRE, respectively (33, 34). Therefore, transcriptional induction of the gluconeogenesis genes is under a combinatorial regulatory mechanism by the PIPTC and the transcriptional activators, Cat8 and Sip4. It is currently not clear to what degree the PIPTC or the Cat8/Sip4 activation pathway individually contributes to transcriptional induction of the genes in gluconeogenesis and the TCA cycle. We performed growth assays for WT, fab1Δ, cti6Δ, cat8Δ, sip4Δ, and cat8Δsip4Δ cells on non-fermentative ethanol medium, in which the genes in the gluconeogenesis and the TCA cycle are highly induced. Although cat8Δ, sip4Δ, and cat8Δsip4Δ cells did not show a significant growth defect on ethanol medium, fab1Δ and cti6Δ cells showed a severe growth defect on ethanol medium (Fig. 5D). Although a more accurate estimation requires global range quantification of transcription of the genes in gluconeogenesis and the TCA cycle, the growth phenotype on ethanol medium suggests that the PIPTC may be more crucial than the Cat8/Sip4-dependent transcriptional activation for transcriptional induction of the genes of gluconeogenesis and the TCA cycle.
Without PI(3,5)P2, Cti6 Mislocalizes to the Cytoplasm in Ethanol Medium
Both GFP-Tup1 and GFP-Cti6 primarily localize in the nucleus (stained with Hoechst 33342) (Fig. 6, A and B), as shown in a previous study (12). We previously observed that Tup1 and Cti6 dynamically shuttle between the nucleus and cytoplasm when shifting from glucose to galactose media (12). Cti6 and Tup1 bind to the endosomal PI(3,5)P2 lipid with a high specificity. Without PI(3,5)P2, Cti6, but not Tup1, accumulates in the cytoplasm in galactose medium (12). Under these conditions, the cytoplasmic Cti6 that is not in the Cti6-Cyc8-Tup1 complex is more susceptible to degradation (12). PI(3,5)P2 is required for Cti6 to shuttle back to the nucleus. Presumably, this is because the form of Cti6 that shuttles back into the nucleus is the Cti6-Cyc8-Tup1 coactivator complex that is assembled at the endosomal/vacuolar membrane through the interactions of PI(3,5)P2 with Cti6 and Tup1.
Because the PIPTC functions for transcriptional induction of ICL1 and FBP1 in ethanol medium as it does for GAL1 transcriptional induction in galactose medium, we decided to investigate how PI(3,5)P2 affects the cellular localization of Cti6 in ethanol medium. Cti6 localizes primarily in the nucleus in WT cells in both glucose and ethanol media (Fig. 6C). Although Cti6 localizes primarily in the nucleus in fab1Δ cells in glucose medium, Cti6 accumulates in the cytoplasm and occasionally forms cytoplasmic punctae upon shifting from glucose to ethanol media (Fig. 6D). In contrast, Tup1 localizes primarily in the nucleus in fab1Δ cells in both glucose and ethanol media (Fig. 6E).
DISCUSSION
In response to varying environments, human cells reprogram their glucose/carbon metabolism between aerobic and anaerobic glycolysis, the TCA cycle, and/or gluconeogenesis. In aerobic conditions, normal differentiated cells metabolize glucose through the TCA cycle, but highly proliferative embryonic cells or cancer cells do not use the TCA cycle. The distinct glucose/carbon metabolism that a cell carries out significantly influences the physiology of the cell. Therefore, metabolic reprogramming to maintain a specific type of glucose/carbon metabolism has a fundamental importance, but how it is regulated is poorly understood. We showed here that metabolic reprogramming from glycolysis to gluconeogenesis is regulated by a novel PI(3,5)P2-dependent regulatory mechanism in S. cerevisiae.
The Endosomal PI(3,5)P2 Lipid-dependent Transcriptional Activation of Gluconeogenesis Genes
There are conspicuous similarities in transcriptional regulatory mechanisms between GAL1 and the two gluconeogenesis genes, ICL1 and FBP1. First, they all belong to TATA box-containing genes (23). TATA box-containing genes have the +1 nucleosome positioned at the transcription start site (TSS) and depend on SAGA rather than transcription factor IID to evict the +1 nucleosome and establish the PIC (25). Indeed, a crucial role for SAGA to establish an activated chromatin structure was observed at both the GAL1 and FBP1 promoters (18, 20, 26). Second, GAL1 as well as ICL1 and FBP1 all contain both an upstream activating sequence/CSRE and a URE at their promoters, and thus their transcriptional regulation is under control of both transcriptional activation and transcriptional repression. Third, they all are regulated by the same repressor complex (Mig1/Mig2-Cyc8-Tup1). Fourth, transcription of the activator gene(s) (CAT8 activator gene for ICL1/FBP1; GAL4 activator gene for GAL1) itself is repressed by the same transcriptional repressor complex (Mig1/Mig2-Cyc8-Tup1). Thus, relief of the Mig1/Mig2-Cyc8-Tup1 repressor function will also induce expression of the transcriptional activators.
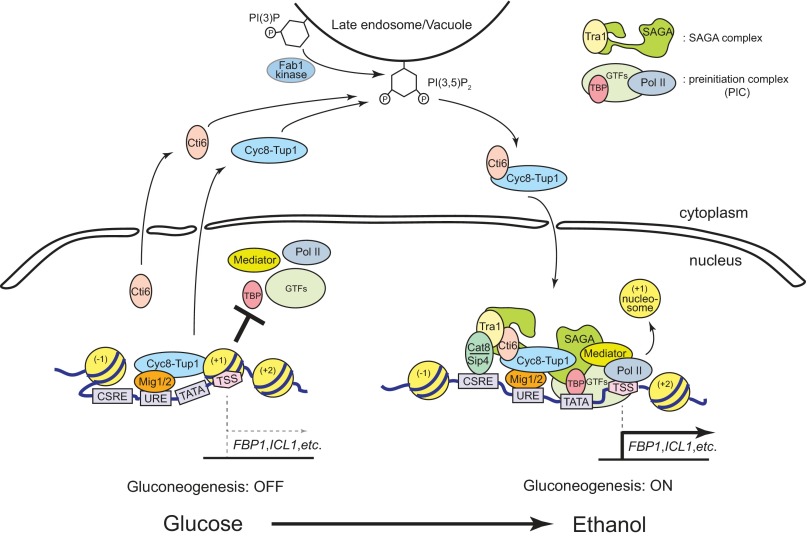
Based on our results here and the previously characterized transcriptional regulatory mechanisms at the GAL1 promoter, we propose a model for the PIPTC-mediated transcriptional activation of the gluconeogenesis genes, ICL1 and FBP1. In glucose-rich medium, transcription of ICL1 and FBP1 is repressed by a Mig1/2-Cyc8-Tup1 complex (Fig. 7). As a member of the TATA box-containing genes, we predict that the ICL1 and FBP1 promoters contain the +1 nucleosome positioned at the TSS (Fig. 7). In non-glucose carbon medium, the Cyc8-Tup1 corepressor is converted into a Cti6-Cyc8-Tup1 coactivator by the PIPTC (Fig. 7). Meanwhile, the transcription activators, Cat8 and Sip4 are expressed and bind to the CSRE in non-glucose medium (Fig. 7). Without the PIPTC, recruitment of Cat8 and Sip4 to the promoters of ICL1 and FBP1 is significantly inhibited, suggesting the Tup1-repressed chromatin structure at the promoters is much less accessible to the activators. SAGA complex is recruited to the promoters by the Cti6-Cyc8-Tup1 coactivator and Cat8/Sip4 activator proteins, presumably through the interaction between Tra1 of SAGA complex and Cti6 and/or Cat8/Sip4 proteins (Fig. 7). It has been shown that Tra1 interacts with transcription activators, Gal4, Gcn4, and Hap4 in S. cerevisiae (19). Therefore, we predict that Cti6 and/or Cat8/Sip4 activators recruit SAGA to the ICL1 and FBP1 promoters by interaction with Tra1 (Fig. 7).
FIGURE 7.
A model for the regulation of transcriptional induction of ICL1 and FBP1 by the PIPTC. A schematic model for the PIPTC-mediated regulation of chromatin architecture at the ICL1/FBP1 promoter is presented. ICL1 and FBP1 contain a positive (CSRE) and a negative (URE) transcriptional regulatory element at their promoters. The CSRE binds to Cat8 or Sip4 transcriptional activators. The URE binds to the Mig1/2 transcriptional repressor. FBP1 and ICL1 belong to TATA box-containing genes that have the +1 nucleosome positioned at the TSS and depend on SAGA rather than transcription factor IID to establish the PIC. In glucose-rich medium, glycolysis is actively functional (ON) (left). Gluconeogenesis is inactive (OFF), and transcription of ICL1 and FBP1 is repressed by the Mig1/2-Cyc8-Tup1 complex. Transcription of CAT8 and SIP4 is repressed in glucose-rich medium, and therefore transcriptional activation function from the CSRE at the ICL1/FBP1 promoter is tenuous. In non-glucose carbon medium, in which glycolysis is no longer functional (OFF), gluconeogenesis is activated (ON) (right). The Cyc8-Tup1 corepressor is converted to the Cti6-Cyc8-Tup1 coactivator by the PIPTC at the late endosomal/vacuolar membrane. The PIPTC appears to play a critical role in remodulating the chromatin architecture at the ICL1 and FBP1 promoters to a promoter structure more accessible to the Cat8 and Sip4 activators. The Cti6-Cyc8-Tup1 coactivator and the Cat8/Sip4 activators recruit the SAGA coactivator complex to the ICL1 and FBP1 promoters, presumably through the interaction between Tra1 of the SAGA complex and Cti6 or Cat8/Sip4 proteins. Inferring from the characterized features at the GAL1 promoter, we predict that SAGA may recruit the Swi/Snf nucleosome remodeler that evicts the +1 nucleosome positioning at the TSS and opens the promoter to establish the PIC. Both the PIPTC and the Cat8/Sip4-dependent activation pathway play critical roles in robustly establishing the activated state (right) from the repressed state (left) of chromatin architecture at the ICL1 and FBP1 promoters. PI(3)P, phosphatidylinositol 3-phosphate.
It was recently proposed that DNA-binding, Cyc8-Tup1 interacting repressors such as Mig1 and Sko1 function as a transcriptional activator in addition to a repressor (38). The Cyc8-Tup1 corepressor was proposed to regulate the dual functionality of DNA-binding repressor/activator proteins by masking the activation domain of repressor/activator proteins (38). This proposed mechanism does not involve Cti6. Instead, the model proposed that DNA-binding, Cyc8-Tup1-interacting repressors, such as Mig1 and Sko1, recruit coactivators, such as SAGA, by direct interaction through their activation domain. It may function at other Tup1-repressed promoters, but we believe it is the Cyc8-Tup1 complex that performs dual functionality in transcriptional regulation at the promoters of the gluconeogenesis genes, ICL1 and FBP1, and GAL1. The dual functionality of the Cyc8-Tup1 complex is modulated by Cti6 and the PIPTC as we have proposed. We clearly demonstrated a critical role played by Cti6 in activating transcription of ICL1 and FBP1 in this study and GAL1 in our previous study (12). Cti6 and Tup1 are highly specific PI(3,5)P2 interactors (12). Without Cti6 or PI(3,5)P2, the chromatin structure at the promoters of ICL1 and FBP1 is less accessible to the Cat8 and Sip4 activators as shown in Fig. 5, B and C, suggesting a role for the PIPTC in remodulating the chromatin architecture at the ICL1 and FBP1 promoters prior to binding of the Cat8 and Sip4 activators during transcriptional activation of the genes. Furthermore, the expression levels of the Cat8 and Sip4 activators are highly regulated between high glucose, ICL1/FBP1-repressing conditions and low glucose, ICL1/FBP1-activating conditions, as shown in Fig. 4B. The results of the promoter accessibility to Cat8 and Sip4 (Figs. 5, B and C) and the highly regulated expression of Cat8 and Sip4 (Fig. 4B), taken together, suggest that transcriptional induction of ICL1 and FBP1 is a dynamically regulated, multistep process that requires both conversion of the Cyc8-Tup1 corepressor to the Cti6-Cyc8-Tup1 coactivator and recruitment of the Cat8 and Sip4 activators to the promoters. Our results showed that, without either Tup1 conversion or Cat8/Sip4 activators, an activated state of chromatin architecture cannot be robustly established at the ICL1 and FBP1 promoters even in the presence of the Mig1 repressor/activator contrary to the previously proposed model (38).
The PIPTC May Be a General Mechanism Required for Transcriptional Activation of Other Tup1-repressed Genes
The Cyc8-Tup1 complex is a global repressor that regulates more than 150 genes in S. cerevisiae. From our study of GAL1, one of the best characterized Tup1-repressed genes, we identified the PIPTC to play a crucial role for the recruitment of the SAGA coactivator to the GAL1 promoter during GAL1 transcriptional activation (12). Here we show that the PIPTC also plays a critical role for metabolic reprogramming from glycolysis to gluconeogenesis. Particularly, ICL1 and FBP1, two key gluconeogenesis genes that are repressed by Tup1, require the PIPTC for their transcriptional activation. Our results suggest that the PIPTC represents a general mechanism functioning at Tup1-repressed genes. Tup1 regulates numerous genes, and Tup1-repressed genes are diverse in terms of their transcriptional regulatory mechanisms (17). In addition to the Tup1-repressed genes subject to glucose repression, there are sets of Tup1-repressed genes that are controlled by other stimuli: pheromone, salt concentration, heme concentration, etc. Therefore, to grasp a more comprehensive picture of how general the PIPTC is for the regulation of Tup1-repressed genes will require the examination of additional sets of Tup1-repressed genes under different induction conditions.
The primary role of the Cti6-Cyc8-Tup1 coactivator in GAL1 transcription is to interact with and recruit the SAGA coactivator to the GAL1 promoter (21). We believe that the Cti6-Cyc8-Tup1 coactivator plays the same role in recruiting the SAGA complex to the ICL1 and FBP1 promoters. The PIPTC illustrates that endosomal membranes provide a signaling platform for functional regulation of the general transcriptional regulator Cyc8-Tup1 complex and consequently Cti6-Cyc8-Tup1 coactivator-dependent SAGA recruitment to gene promoters. Interestingly, Ada2, a component of the SAGA complex, is capable of interacting with the lipid phosphatidylserine, and the lipid binding activity of Ada2 was suggested to regulate the function of the SAGA complex (39). In addition, it was suggested that folding of Tra1, a component of the SAGA complex, or assembly of a subcomplex of SAGA may take place on cytoplasmic membranes (40). These observations suggest that regulation of the function and/or the assembly of the SAGA complex may be closely linked with cytoplasmic membranes.
SAGA and Tup1 are highly conserved general transcriptional regulators (41, 42). The human SAGA and Tup1 homologs (TLE proteins) regulate fundamental cellular and developmental processes by interacting with crucial transcriptional factors, such as c-Myc, E2F, and/or p53 (43, 44). The PI(3,5)P2-dependent regulatory mechanism of Tup1 function and its role for SAGA recruitment that we identified in yeast may provide important insights to understand how human SAGA and TLE proteins are regulated in humans.
Glucose starvation may change vacuolar contents and properties, such as PI(3,5)P2 levels and vacuolar luminal pH, which may trigger signals for the PI(3,5)P2-dependent Tup1 conversion at the vacuolar membrane to activate transcriptional reprogramming for gluconeogenesis.
Similar to our observations, there are reports showing a role for endosomal/lysosomal membranes as a physiological/nutritional sensor and as a signaling platform in humans. The transcription factor EB (TFEB), a key transcriptional activator for lysosomal biogenesis genes, localizes at the lysosomal membranes in rich medium. When cells are starved, transcription factor EB is activated by mTORC1 at the lysosomal membranes to function in transcriptional reprogramming for lysosomal biogenesis (45).
Like our demonstration of the PI(3,5)P2-dependent regulation of Tup1 and Cti6, there are a few known cases where cells utilize membrane lipids to regulate the function of transcriptional factors. The function of the sterol-responsive element-binding protein (SREBP) transcription factor is regulated by cholesterol in the endoplasmic reticulum membrane (46). The Tubby transcription factor is regulated by interaction with PI(4,5)P2 at the plasma membrane (47). The endosomal phosphatidylinositol 3-phosphate lipid provides a signaling platform for activation of the Smad2 transcription factor (48). A lipid-mediated functional regulation of transcription regulators may have been evolved to achieve a highly specific signaling reaction in the complex intracellular milieu.
Are Human Gluconeogenesis Genes Regulated by TLE Repression and the PIPTC?
It is currently unknown if human gluconeogenesis genes are also regulated by the human Tup1 homolog TLE proteins or if there is a PI(3,5)P2-dependent mechanism that regulates transcriptional reprogramming from glycolysis to gluconeogenesis in humans.
Interestingly, it was reported that human PCK1 is regulated by hepatic nuclear factor-3β (HNF3β) and that TLE proteins interact with HNF3β (49, 50). This raises the interesting possibility that human gluconeogenesis genes also are regulated by the Tup1 repression and PIPTC that we identified in yeasts.
This work was supported by funds from a Cornell University Research Grant (to S. D. E.).
- PI(3,5)P2
- phosphatidylinositol 3,5-bisphosphate
- PIC
- preinitiation complex
- PIPTC
- PI(3,5)P2-dependent Tup1 conversion
- qPCR
- quantitative PCR
- TSS
- transcription start site
- CSRE
- carbon source-responsive element
- URE
- upstream regulatory element
- SAGA
- Spt-Ada-Gcn5-acetyltransferase
- TLE
- transducin-like enhancer.
REFERENCES
- 1. Vander Heiden M. G., Cantley L. C., Thompson C. B. (2009) Understanding the Warburg effect. The metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Luo W., Hu H., Chang R., Zhong J., Knabel M., O'Meally R., Cole R. N., Pandey A., Semenza G. L. (2011) Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 145, 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bungard D., Fuerth B. J., Zeng P. Y., Faubert B., Maas N. L., Viollet B., Carling D., Thompson C. B., Jones R. G., Berger S. L. (2010) Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science 329, 1201–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jones R. G., Plas D. R., Kubek S., Buzzai M., Mu J., Xu Y., Birnbaum M. J., Thompson C. B. (2005) AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell 18, 283–293 [DOI] [PubMed] [Google Scholar]
- 5. Imai S., Guarente L. (2010) Ten years of NAD-dependent SIR2 family deacetylases. Implications for metabolic diseases. Trends Pharmacol. Sci. 31, 212–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shimazu T., Hirschey M. D., Newman J., He W., Shirakawa K., Le Moan N., Grueter C. A., Lim H., Saunders L. R., Stevens R. D., Newgard C. B., Farese R. V., Jr., de Cabo R., Ulrich S., Akassoglou K., Verdin E. (2013) Suppression of oxidative stress by β-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science 339, 211–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Takahashi H., McCaffery J. M., Irizarry R. A., Boeke J. D. (2006) Nucleocytosolic acetyl-coenzyme A synthetase is required for histone acetylation and global transcription. Mol. Cell 23, 207–217 [DOI] [PubMed] [Google Scholar]
- 8. Kim J., DeBerardinis R. J. (2013) Cancer. Silencing a metabolic oncogene. Science 340, 558–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu C., Ward P. S., Kapoor G. S., Rohle D., Turcan S., Abdel-Wahab O., Edwards C. R., Khanin R., Figueroa M. E., Melnick A., Wellen K. E., O'Rourke D. M., Berger S. L., Chan T. A., Levine R. L., Mellinghoff I. K., Thompson C. B. (2012) IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483, 474–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fraenkel D. G. (2011) Yeast Intermediary Metabolism, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 11. Racker E. (1974) History of the Pasteur effect and its pathobiology. Mol. Cell. Biochem. 5, 17–23 [DOI] [PubMed] [Google Scholar]
- 12. Han B. K., Emr S. D. (2011) Phosphoinositide [PI(3,5)P2] lipid-dependent regulation of the general transcriptional regulator Tup1. Genes Dev. 25, 984–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dove S. K., Cooke F. T., Douglas M. R., Sayers L. G., Parker P. J., Michell R. H. (1997) Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature 390, 187–192 [DOI] [PubMed] [Google Scholar]
- 14. Gary J. D., Wurmser A. E., Bonangelino C. J., Weisman L. S., Emr S. D. (1998) Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell Biol. 143, 65–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Malavé T. M., Dent S. Y. (2006) Transcriptional repression by Tup1-Ssn6. Biochem. Cell Biol. 84, 437–443 [DOI] [PubMed] [Google Scholar]
- 16. Nehlin J. O., Carlberg M., Ronne H. (1991) Control of yeast GAL genes by MIG1 repressor. A transcriptional cascade in the glucose response. EMBO J. 10, 3373–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smith R. L., Johnson A. D. (2000) Turning genes off by Ssn6-Tup1. A conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 25, 325–330 [DOI] [PubMed] [Google Scholar]
- 18. Bhaumik S. R., Green M. R. (2001) SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15, 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brown C. E., Howe L., Sousa K., Alley S. C., Carrozza M. J., Tan S., Workman J. L. (2001) Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292, 2333–2337 [DOI] [PubMed] [Google Scholar]
- 20. Larschan E., Winston F. (2001) The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15, 1946–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Papamichos-Chronakis M., Petrakis T., Ktistaki E., Topalidou I., Tzamarias D. (2002) Cti6, a PHD domain protein, bridges the Cyc8-Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol. Cell 9, 1297–1305 [DOI] [PubMed] [Google Scholar]
- 22. Goldfarb T., Alani E. (2004) Chromatin immunoprecipitation to investigate protein-DNA interactions during genetic recombination. Methods Mol. Biol. 262, 223–237 [DOI] [PubMed] [Google Scholar]
- 23. Basehoar A. D., Zanton S. J., Pugh B. F. (2004) Identification and distinct regulation of yeast TATA box-containing genes. Cell 116, 699–709 [DOI] [PubMed] [Google Scholar]
- 24. Huisinga K. L., Pugh B. F. (2004) A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 13, 573–585 [DOI] [PubMed] [Google Scholar]
- 25. Rhee H. S., Pugh B. F. (2012) Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483, 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Biddick R. K., Law G. L., Chin K. K., Young E. T. (2008) The transcriptional coactivators SAGA, SWI/SNF, and mediator make distinct contributions to activation of glucose-repressed genes. J. Biol. Chem. 283, 33101–33109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zaragoza O., Vincent O., Gancedo J. M. (2001) Regulatory elements in the FBP1 promoter respond differently to glucose-dependent signals in Saccharomyces cerevisiae. Biochem. J. 359, 193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Entian K. D., Barnett J. A. (1992) Regulation of sugar utilization by Saccharomyces cerevisiae. Trends Biochem. Sci. 17, 506–510 [DOI] [PubMed] [Google Scholar]
- 29. Hedbacker K., Carlson M. (2008) SNF1/AMPK pathways in yeast. Front. Biosci. 13, 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lundin M., Nehlin J. O., Ronne H. (1994) Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1. Mol. Cell Biol. 14, 1979–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schöler A., Schüller H. J. (1993) Structure and regulation of the isocitrate lyase gene ICL1 from the yeast Saccharomyces cerevisiae. Curr. Genet. 23, 375–381 [DOI] [PubMed] [Google Scholar]
- 32. Rahner A., Hiesinger M., Schüller H. J. (1999) Deregulation of gluconeogenic structural genes by variants of the transcriptional activator Cat8p of the yeast Saccharomyces cerevisiae. Mol. Microbiol. 34, 146–156 [DOI] [PubMed] [Google Scholar]
- 33. Vincent O., Carlson M. (1998) Sip4, a Snf1 kinase-dependent transcriptional activator, binds to the carbon source-responsive element of gluconeogenic genes. EMBO J. 17, 7002–7008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hedges D., Proft M., Entian K. D. (1995) CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 15, 1915–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fleming A. B., Pennings S. (2001) Antagonistic remodelling by Swi-Snf and Tup1-Ssn6 of an extensive chromatin region forms the background for FLO1 gene regulation. EMBO J. 20, 5219–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tachibana C., Yoo J. Y., Tagne J. B., Kacherovsky N., Lee T. I., Young E. T. (2005) Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol. Cell Biol. 25, 2138–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bojunga N., Entian K. D. (1999) Cat8p, the activator of gluconeogenic genes in Saccharomyces cerevisiae, regulates carbon source-dependent expression of NADP-dependent cytosolic isocitrate dehydrogenase (Idp2p) and lactate permease (Jen1p). Mol. Gen. Genet. 262, 869–875 [DOI] [PubMed] [Google Scholar]
- 38. Wong K. H., Struhl K. (2011) The Cyc8-Tup1 complex inhibits transcription primarily by masking the activation domain of the recruiting protein. Genes Dev. 25, 2525–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hoke S. M., Genereaux J., Liang G., Brandl C. J. (2008) A conserved central region of yeast Ada2 regulates the histone acetyltransferase activity of Gcn5 and interacts with phospholipids. J. Mol. Biol. 384, 743–755 [DOI] [PubMed] [Google Scholar]
- 40. Genereaux J., Kvas S., Dobransky D., Karagiannis J., Gloor G. B., Brandl C. J. (2012) Genetic evidence links the ASTRA protein chaperone component Tti2 to the SAGA transcription factor Tra1. Genetics 191, 765–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Buscarlet M., Stifani S. (2007) The “Marx” of Groucho on development and disease. Trends Cell Biol. 17, 353–361 [DOI] [PubMed] [Google Scholar]
- 42. Spedale G., Timmers H. T., Pijnappel W. W. (2012) ATAC-king the complexity of SAGA during evolution. Genes Dev. 26, 527–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McMahon S. B., Van Buskirk H. A., Dugan K. A., Copeland T. D., Cole M. D. (1998) The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94, 363–374 [DOI] [PubMed] [Google Scholar]
- 44. Orian A., Delrow J. J., Rosales Nieves A. E., Abed M., Metzger D., Paroush Z., Eisenman R. N., Parkhurst S. M. (2007) A Myc-Groucho complex integrates EGF and Notch signaling to regulate neural development. Proc. Natl. Acad. Sci. U.S.A. 104, 15771–15776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Settembre C., Zoncu R., Medina D. L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M. C., Facchinetti V., Sabatini D. M., Ballabio A. (2012) A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang X., Sato R., Brown M. S., Hua X., Goldstein J. L. (1994) SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell 77, 53–62 [DOI] [PubMed] [Google Scholar]
- 47. Santagata S., Boggon T. J., Baird C. L., Gomez C. A., Zhao J., Shan W. S., Myszka D. G., Shapiro L. (2001) G-protein signaling through Tubby proteins. Science 292, 2041–2050 [DOI] [PubMed] [Google Scholar]
- 48. Tsukazaki T., Chiang T. A., Davison A. F., Attisano L., Wrana J. L. (1998) SARA, a FYVE domain protein that recruits Smad2 to the TGFβ receptor. Cell 95, 779–791 [DOI] [PubMed] [Google Scholar]
- 49. Wang J. C., Strömstedt P. E., Sugiyama T., Granner D. K. (1999) The phosphoenolpyruvate carboxykinase gene glucocorticoid response unit. Identification of the functional domains of accessory factors HNF3β (hepatic nuclear factor-3β) and HNF4 and the necessity of proper alignment of their cognate binding sites. Mol. Endocrinol. 13, 604–618 [DOI] [PubMed] [Google Scholar]
- 50. Wang J. C., Waltner-Law M., Yamada K., Osawa H., Stifani S., Granner D. K. (2000) Transducin-like enhancer of split proteins, the human homologs of Drosophila groucho, interact with hepatic nuclear factor 3β. J. Biol. Chem. 275, 18418–18423 [DOI] [PubMed] [Google Scholar]
- 51. Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. (1988) Protein sorting in Saccharomyces cerevisiae. Isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell Biol. 8, 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rue S. M., Mattei S., Saksena S., Emr S. D. (2008) Novel Ist1-Did2 complex functions at a late step in multivesicular body sorting. Mol. Biol. Cell 19, 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]