Background: SAMHD1 is an enzyme that maintains low dNTP concentrations in macrophages.
Results: Depletion of SAMHD1 decreases HIV-1 sensitivity to nucleoside reverse transcriptase inhibitors (NRTIs) in macrophages, but does not significantly alter sensitivity in T cells.
Conclusion: SAMHD1 expression levels in macrophages directly impact the efficacy of NRTIs by modulating cellular dNTP concentrations.
Significance: SAMHD1 controls HIV-1 sensitivity to NRTIs.
Keywords: DNA Polymerase, DNA Replication, HIV-1, Macrophages, Nucleoside Nucleotide Analogs
Abstract
Newly identified anti-HIV host factor, SAMHD1, restricts replication of lentiviruses such as HIV-1, HIV-2, and simian immunodeficiency virus in macrophages by enzymatically hydrolyzing and depleting cellular dNTPs, which are the substrates of viral DNA polymerases. HIV-2 and some simian immunodeficiency viruses express viral protein X (VPX), which counteracts SAMHD1 and elevates cellular dNTPs, enhancing viral replication in macrophages. Because nucleoside reverse transcriptase inhibitors (NRTIs), the most commonly used anti-HIV drugs, compete against cellular dNTPs for incorporation into proviral DNA, we tested whether SAMHD1 directly affects the efficacy of NRTIs in inhibiting HIV-1. We found that reduction of SAMHD1 levels with the use of virus-like particles expressing Vpx- and SAMHD1-specific shRNA subsequently elevates cellular dNTPs and significantly decreases HIV-1 sensitivity to various NRTIs in macrophages. However, virus-like particles +Vpx treatment of activated CD4+ T cells only minimally reduced NRTI efficacy. Furthermore, with the use of HPLC, we could not detect SAMHD1-mediated hydrolysis of NRTI-triphosphates, verifying that the reduced sensitivity of HIV-1 to NRTIs upon SAMHD1 degradation is most likely caused by the elevation in cellular dNTPs.
Introduction
Human immunodeficiency virus types 1 (HIV-1) and 2 (HIV-2) and other lentiviruses replicate in CD4+ T cells and non-dividing, terminally differentiated macrophages during the course of viral pathogenesis (1, 2). Unlike CD4+ T cells, which die upon HIV-1 infection (3), macrophages are resistant to the cytopathic effects of the virus and serve as key long-living viral reservoirs that persistently produce virus at a slow rate (2, 4–8). Also, macrophages are found at sites of primary viral transmission and in many tissues and organs such as the brain, making infected macrophages an essential cell type to target therapeutically (9). Another fundamental difference between T cells and macrophages is that T cells are rapidly dividing, whereas macrophages remain in the G0 phase of the cell cycle and do not require chromosomal DNA replication. As a consequence, macrophages contain very limited cellular dNTP pools (22–320-fold lower) when compared with activated peripheral blood mononuclear cells, primarily composed of T cells (10, 11). In addition to the lack of cell cycling, it has recently been reported that expression of sterile α motif (SAM)2 domain and HD domain-containing protein 1 (SAMHD1) also contributes to the low dNTP pools found not only in macrophages, but in other non-dividing cell types such as resting CD4+ T cells and dendritic cells (12–15). SAMHD1 acts by hydrolyzing deoxyribonucleoside triphosphates (dNTPs) into deoxyribonucleosides and triphosphates (16, 17) and acts as an antiviral host defense factor to restrict HIV-1 infection in these cell types by depleting the dNTP substrate availability for HIV-1 reverse transcriptase (RT) (12–15, 18). However, HIV-2 and several simian immunodeficiency viruses counteract SAMHD1 restriction by expression of an accessory protein, Vpx, which is able to target SAMHD1 for proteasomal degradation (13, 19–24). Degradation of SAMHD1 results in an increase in cellular dNTPs, an acceleration of proviral DNA synthesis, and ultimately enhanced viral replication in non-dividing cells such as macrophages (12, 15, 18).
Nucleoside reverse transcriptase inhibitors (NRTIs) are the most commonly used agents in highly active antiretroviral therapy (HAART), which significantly delays HIV-1 pathogenesis (25, 26). NRTIs lack the 3′-OH moiety of the ribose ring and are typically administered as their nucleoside derivatives to facilitate crossing of cellular membranes. Once inside the cell, NRTIs are phosphorylated by host nucleoside and nucleotide kinases to their triphosphate form (NRTI-TP), which competes with cellular dNTPs as substrate for viral RT. Incorporation of NRTI-TPs into HIV-1 proviral DNA halts DNA chain elongation by preventing the formation of the next phosphodiester bond with an incoming dNTP (reviewed in Ref. 27).
Since all NRTIs used for the treatment of HIV-1- and HIV-2-infected patients compete against cellular dNTPs, we hypothesized that SAMHD1 activity alters the efficacy of NRTIs in inhibiting viral replication. One mechanism by which SAMHD1 may influence NRTI efficacy toward HIV-1 is direct hydrolysis of NRTI-TPs to their inactive form, thus decreasing the active concentration of the drug in the cell. The other possible mechanism would occur upon degradation of SAMHD1, which results in an increase in cellular dNTPs (12) and would likely decrease the efficacy of NRTIs by providing more competitive dNTP substrate for RT. This would likely influence NRTI efficacy toward HIV-2. Indeed, our virological and biochemical results demonstrate that the reduction of SAMHD1 expression almost abolishes viral sensitivity to NRTIs in macrophages by elevating cellular dNTPs. This suggests that higher concentrations of NRTIs may be necessary to inhibit HIV-2 infection in macrophages. Furthermore, we could not detect SAMHD1-mediated hydrolysis of NRTI-TPs by our HPLC-based assay, suggesting that SAMHD1 does not significantly alter the active concentration of the drug. Overall, our data suggest that the anti-HIV host factor SAMHD1 directly enhances the efficacy of NRTIs in inhibiting HIV-1 infection in macrophages by hydrolyzing dNTPs much more efficiently than NRTI-TPs, thus increasing the likelihood that RT incorporates these DNA chain terminators.
EXPERIMENTAL PROCEDURES
Ethics Statement
Experiments in this study utilized primary human monocytes and CD4+ T cells isolated from human buffy coats (New York Blood Services, Long Island, NY). Human blood is publically available for purchase and is provided without indicating the identity of the subjects.
Cell Cultures
THP1 cell lines expressing a scrambled shRNA (5′-CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTT-3′) or a SAMHD1-specific shRNA (5′-CCGGGCAGATGACTACATAGAGATTCTCGAGAATCTCTATGTAGTCATCTGCTTTTTTG-3′, provided by Dr. Nathaniel Landau) were prepared as described previously (28). Cells were maintained in RPMI (10% FBS with penicillin and streptomycin) with 0.5 μg/ml puromycin for selection. THP1 cells were differentiated overnight with 50 nm phorbol 12-myristate 13-acetate (Sigma-Aldrich). Primary human monocytes were isolated with MACS CD14 beads (Miltenyi Biotec), and CD4+ T cells were isolated with CD4 beads (Miltenyi Biotec) from human blood buffy coats as described previously (29). Monocytes were differentiated to macrophages with 5 ng/ml human GM-CSF (Miltenyi Biotec) in RPMI for 7 days. CD4+ T cells were activated for 5 days in the presence of 20 units/ml IL2 (Miltenyi Biotec) with 5 μg/ml phytohemagglutinin (Sigma-Aldrich) added only on day 1.
HIV-1 D3 Vector and VLP Preparation
The HIV-1 D3 plasmid encodes the genome for HIV-1 NL4-3 with eGFP in the place of nef and a deleted envelope (10). Vector and virus-like particles (VLPs) were prepared by the protocols described previously (39, 40).
Purification of RT and SAMHD1
HXB2 HIV-1 RT (p66) gene was cloned into pET28a (Novagen), containing an N-terminal hexahistidine tag. RT homodimer was expressed in BL21 (DE3) Escherichia coli and purified using three forms of chromatography: Ni2+-nitrilotriacetic acid, diethylaminoethanol-Sepharose (DEAE), and sulfopropyl (SP) anion exchange (30).
Human SAMHD1 was cloned into pGEX-6P-1 with an N-terminal GST tag (GE Healthcare, provided by Dr. Yoshio Koyanagi) and transformed into BL21 (DE3) pLysS competent cells (Invitrogen). Cells were grown at 37 °C to an A600 of 0.5, stored on ice for 2 h, and induced overnight with 0.25 mm isopropyl-α-d-1-thiogalactopyranoside at 25 °C. Cells were harvested and lysed in lysis buffer (50 mm Tris-HCl, pH 7.5, 500 mm NaCl, 2 mm EDTA, 1 mg/ml chicken egg white lysozyme, one tablet of Roche Applied Science Complete protease inhibitor mixture) for 4 h on ice. Cell debris was removed by centrifugation at 49,000 × g for 15 min, and lysate was incubated overnight at 4 °C with 1.5 ml of glutathione-Sepharose 4B bead slurry (GE Healthcare). Beads were pelleted and washed three times in wash buffer (50 mm Tris-HCl, pH 7.5, 500 mm NaCl, 1 mm dithiothreitol, 0.5% Triton X-100), equilibrated in buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 20% glycerol, 0.5% Triton X-100) and packed into a column. The column was washed two times with 10 ml of equilibration buffer, and SAMHD1 was eluted with 50 mm Tris-HCl, pH 8, 1 mm EDTA, 10% glycerol, 300 mm NaCl, 300 mm reduced glutathione. Eluted fractions were separated on 10% SDS-PAGE and stained with Coomassie Blue to determine protein concentration by comparing them with a bovine serum albumin (BSA) standard curve. Protein was dialyzed overnight in equilibration buffer.
Western Blot Analysis
THP1 cells were differentiated overnight in phorbol 12-myristate 13-acetate. Macrophages were treated with VLPs (150 ng of p27 per 1 million cells) for 24 h, and activated CD4+ T cells were treated with VLPs (375 ng of p27 per 1 million cells) for 24 h. To ensure that VLPs entered the T cells, cells were spinoculated at 1,670 × g for 5 min, resuspended, and spinoculated once more. All cells were washed in PBS and lysed in radioimmune precipitation buffer (10× PBS, 5% sodium deoxycholate, 10% SDS, nonyl phenoxypolyethoxylethanol (NP-40), Roche Applied Science Complete protease inhibitor mixture). 25 μg of total protein from THP1 cells and macrophages and 50 μg of total protein from CD4+ T cells were separated by 10% SDS-PAGE. Proteins were then transferred to a nitrocellulose membrane (Bio-Rad), and SAMHD1 along with β-actin (loading control) was detected after incubation with enzyme-specific antibodies (Abcam (1:1,000) and Sigma-Aldrich (1:3,000), respectively) followed by incubation with an anti-mouse-HRP secondary antibody (GE Healthcare). Immunoreactive protein bands were detected by chemiluminescence (Thermo Scientific SuperSignal West Femto maximum sensitivity substrate) and imaged on a Bio-Rad ChemiDoc imager. SAMHD1 expression levels were quantified by densitometry (Quantity One) and normalized to the β-actin controls.
HIV-1 RT Primer Extension Assay
Reactions (20 μl final) contained 50 nm purified RT, 10 nm 5′ 32P-labeled 17-nucleotide DNA primer (5′-CGCGCCGAATTCCCGCT-3′) annealed to a 38-nucleotide RNA template (3′-GCGCGGCUUAAGGGCGAUCGUUAUAAGACGUCGGUUCG-5′, Integrated DNA Technologies), increasing concentrations of dNTPs (10 nm, 25 nm, 50 nm, 100 nm, 250 nm, 500 nm, 1 μm, USB Products), with or without 5 μm zidovudine-triphosphate (AZT-TP) (Moravek Biochemicals), 5 μm zalcitabine-triphosphate (ddCTP) (Sigma-Aldrich), and RT reaction buffer (50 mm Tris-HCl, pH 7.5, 50 mm NaCl, 5 mm MgCl2, and 10 μm oligo(dT)). Reactions were initiated by the addition of RT, incubated for 10 min at 37 °C, terminated by the addition of 10 μl of 40 mm EDTA, 99% formamide, and denatured by incubation for 5 min at 95 °C. Denatured products (4 μl) were then separated by 16% urea PAGE (SequaGel, National Diagnostics) and imaged on a Bio-Rad personal molecular imager.
Drug Sensitivity Assays
After differentiation (THP1 cells and monocytes), activation (T cells), and VLP pretreatment (macrophages and T cells) as described above, NRTIs or nevirapine (National Institutes of Health AIDS Reagent Program) were added to the cells at the indicated concentrations. After pretreatment with NRTIs for 2 h to allow for phosphorylation, HIV-1 D3 was added. THP1 cells were collected for flow cytometry 48 h post transduction (hpt), macrophages were collected 7 days post transduction (dpt) (medium and drugs were replaced 48 hpt), and T cells were collected 40 hpt. Cells were stained with propidium iodide to monitor cell death, and the percentage of live/transduced (propidium iodide−/GFP+) cells was measured using an Accuri C6 flow cytometer, analyzed in FlowJo, and normalized to the no-drug controls.
dNTP Quantification Assay
Activated CD4+ T cells were treated with VLPs ±Vpx (375 ng of p27 per 1 million cells) for 24 h, and dNTPs were extracted and quantified using the protocol described by Diamond et al. (10). To obtain the concentration of dNTPs per cell, the volume of an activated CD4+ T cell, 320 μm3 (10), was used in the calculation.
SAMHD1 Phosphohydrolysis Assay
Reactions (35 μl final) contained 1 μm purified SAMHD1, 1 mm dNTP/ddNTP/AZT-TP, ±100 μm dGTP, and reaction buffer (50 mm Tris-HCl, pH 8, 50 mm KCl, 5 mm MgCl2, and 0.1% Triton X-100). Reactions were initiated with the addition of SAMHD1, incubated for 3 h at 37 °C, and terminated by incubation for 10 min at 75 °C. Reactions were diluted in 12.5% acetonitrile containing 60 μm dCMP as a reference control and then injected into a Beckman Coulter System Gold 126 solvent module as described by Goldstone et al. (17) and White et al. (31). Nucleotide abundance was determined by integrating the area under each peak using 32 Karat 8.0 software. dNTP/ddNTP/AZT-TP levels were normalized by the amount of dCMP detected in each diluted sample and then normalized to the no-enzyme controls. Reactions conducted to test allosteric activation with ddGTP were carried out as described above, but contained 500 μm of the indicated activator and were incubated for 30 min at 37 °C.
Statistical Methods
Statistical significance was determined using an unpaired t test with a Welch's correction. All error bars represent the S.E.
RESULTS
Effect of SAMHD1 on HIV-1 Sensitivity to NRTIs in THP-1 Cells
Because SAMHD1 modulates the concentration of cellular dNTPs, which compete against NRTI-TPs as substrates for RT, we hypothesized that SAMHD1 influences HIV-1 sensitivity to NRTIs. To test this, we first utilized a THP-1 cell line, which is a human monocytic cell line that endogenously expresses SAMHD1 (12, 32). To knock down SAMHD1, a SAMHD1-specific shRNA (shSAMHD1) was stably expressed in these cells, and a scrambled shRNA (shScramble) was used as a control. Reduced SAMHD1 expression (12, 20) (Fig. 1A) and elevated dNTP levels in the shSAMHD1 cells after differentiation with phorbol 12-myristate 13-acetate were observed (12). The differentiated shScramble and shSAMHD1 THP-1 cell lines were pretreated with increasing concentrations of zidovudine (AZT) and then transduced with a vesicular stomatitis virus-G protein (VSV-G) pseudotyped HIV-1 D3 vector, which encodes the entire NL4-3 HIV-1 genome except env, and nef was replaced with eGFP (HIV-1 D3 (10)). The transduction efficiency at each AZT concentration was measured by flow cytometry for eGFP expression 48 hpt, and inhibition curves were plotted to compare relative drug efficacy. Transduction efficiencies were normalized to the no-drug controls because shSAMHD1 cells are more readily transduced than shScramble cells. As shown in Fig. 1B, there was a 12.7-fold reduction in HIV-1 sensitivity to AZT at the highest concentration tested (200 nm) in the shSAMHD1 cells when compared with the shScramble cells. This result shows that knockdown of SAMHD1 decreases HIV-1 sensitivity to NRTIs in THP-1 cells. However, when this experiment was repeated with a non-nucleoside RT inhibitor (NNRTI), nevirapine, the efficacy of nevirapine was not altered upon knockdown of SAMHD1 (Fig. 1C). This result was predicted as NNRTIs bind to an allosteric site of HIV-1 RT and therefore, unlike NRTIs, do not compete against cellular dNTPs. These inhibition curves demonstrate that SAMHD1 specifically affects the efficacy of AZT, and not nevirapine, against HIV-1 replication.
FIGURE 1.
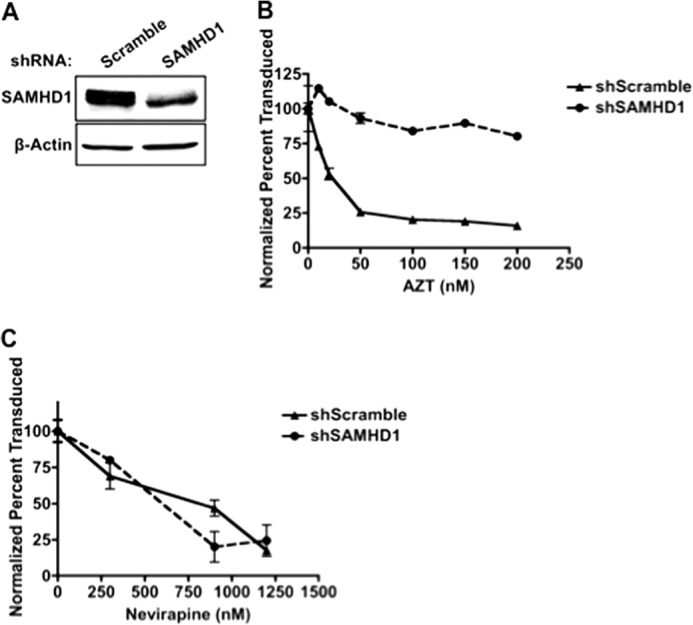
Knockdown of SAMHD1 decreases NRTI efficacy in THP1 cells. A, Western blot of SAMHD1 expression in differentiated THP1 cells with a scramble shRNA or a SAMHD1-specific shRNA. B and C, THP1 cell lines were differentiated, treated with increasing concentrations of AZT (B) or the NNRTI nevirapine (C), and transduced with HIV-1 D3. To determine transduction efficiency, cells were collected 48 hpt to measure GFP expression by flow cytometry (error bars represent the S.E., n = 3).
Effect of Vpx on HIV-1 Sensitivity to NRTIs in Macrophages
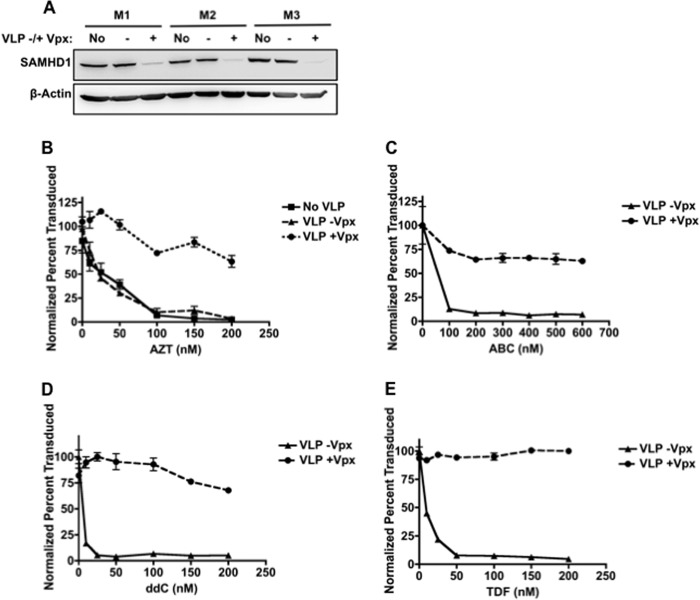
Next, our hypothesis was tested in primary human monocyte-derived macrophages. To modulate the SAMHD1 level, we treated macrophages with VLPs containing or lacking Vpx (VLPs ±Vpx) (20). As shown in Fig. 2A, the SAMHD1 expression level in macrophages was reduced on average 6-fold upon treatment with VLPs +Vpx, and we previously reported that Vpx elevates cellular dNTP levels up to 33-fold in macrophages (12, 18). After macrophages were pretreated with VLPs for 24 h, the cells were treated with NRTIs, AZT (Fig. 2B), abacavir (ABC, Fig. 2C), zalcitabine (ddC, Fig. 2D), or tenofovir (TDF, Fig. 2E) and then transduced with HIV-1 D3. Cells were collected for flow cytometry analysis 7 dpt. As shown in Fig. 2, B–E, HIV-1-transduced macrophages pretreated with VLPs +Vpx displayed significantly reduced sensitivity to the NRTIs tested when compared with cells treated with VLPs −Vpx. Furthermore, this reduction in drug efficacy could also be influenced by the level of NRTIs that become phosphorylated upon degradation of SAMHD1. It is possible that fewer NRTIs are phosphorylated by host kinases when the dNTP concentrations in the cell increase. Lahouassa et al. (12) have demonstrated that when macrophages are treated with VLPs +Vpx for 24 h prior to deoxyribonucleoside treatment, the concentration of dNTPs is 65–1,200-fold higher than when treated with VLPs +Vpx alone. This suggests that the kinases expressed in macrophages are active enough to phosphorylate a high concentration of deoxyribonucleosides into dNTPs. However, nucleoside diphosphate kinase is not as active toward NRTIs as canonical dNTPs (33). Therefore, when the concentration of dNTPs increases, the dNTPs may out-compete NRTIs for the active site of nucleoside diphosphate kinase, resulting in a decreased concentration of NRTI-TPs. Consequently, the reduced efficacy of NRTIs observed upon Vpx-mediated degradation of SAMHD1 in macrophages may be the result of direct competition of dNTPs for the active site of RT and for the active site of cellular kinases.
FIGURE 2.
Vpx-mediated degradation of SAMHD1 decreases NRTI efficacy in macrophages. A, SAMHD1 expression level in three donors of macrophages (M1, M2, and M3) treated without VLPs or with VLPs ±Vpx for 24 h. B–E, macrophages were treated with VLPs for 24 h, treated with increasing concentrations of AZT (B), ABC (C), ddC (D), or TDF (E), and then transduced with HIV-1 D3. To determine transduction efficiency, cells were collected 7 dpt to measure GFP expression by flow cytometry (error bars represent the S.E., n = 3).
Effect of Vpx on HIV-1 Sensitivity to Multiple NRTIs in Macrophages
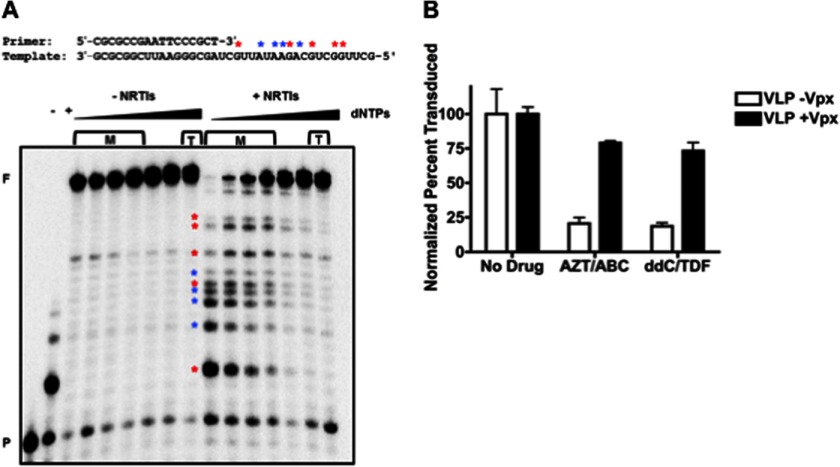
In HAART, more than one NRTI is commonly used for the treatment of infected patients (34). Therefore, we tested whether Vpx-mediated SAMHD1 degradation is also capable of diminishing the antiviral efficacy of multiple NRTIs when simultaneously applied to cells. We first biochemically tested how well combination treatment with both AZT-TP and ddCTP inhibited RT primer extension at increasing dNTP concentrations (Fig. 3A). In this assay, purified HIV-1 RT protein was incubated with a 5′ 32P-labeled 17-nucleotide DNA primer (P in Fig. 3A) annealed to a 38-nucleotide RNA template with increasing dNTP concentrations and a fixed concentration of AZT-TP and ddCTP. As shown in Fig. 3A, HIV-1 RT was able to fully extend the primer (indicated by F) at all dNTP concentrations used in the absence of drugs, but when 5 μm AZT-TP and ddCTP were added to the reactions, extension with 10–50 nm dNTPs (equivalent to dNTP levels in macrophages expressing SAMHD1, indicated by M) was significantly reduced. Inhibition under these reaction conditions is indicated by reduced levels of the full-length products and the presence of early terminated products with variable sizes (marked with asterisks) that are not present in the no-NRTI-TP controls. However, at the approximate dNTP concentrations found in activated CD4+ T cells (1–16 μm, indicated by T), the inhibitory effects of the NRTI-TPs were no longer observed, and the levels of full-length product were increased. Indeed, the dNTP concentration increase reduced the efficacy of these two NRTI-TPs to a similar extent as the addition of each drug individually (data not shown).
FIGURE 3.
Vpx-mediated degradation of SAMHD1 decreases the efficacy of combination NRTI treatment. A, purified HIV-1 RT protein was used to extend the indicated 5′ 32P-labeled primer/template with increasing dNTP concentrations (10 nm, 25 nm, 50 nm, 100 nm, 250 nm, 500 nm, and 1 μm) with or without a fixed concentration of AZT-TP and ddCTP. The dNTP concentrations found in macrophages and activated T cells are marked as M and T, respectively. The negative control (−) was without drug or dNTPs, and the positive control (+) was with drug and 1 nm dNTPs. Asterisks in blue and red indicate positions of chain termination for AZT-TP and ddCTP, respectively. F: fully extended 38-mer product; P: unextended 17-mer primer. B, VLP-treated macrophages were treated with 150 nm AZT/400 nm ABC or 150 nm ddC/200 nm TDF and transduced with HIV-1 D3. To determine transduction efficiency, cells were collected to measure GFP expression by flow cytometry (error bars represent the S.E., n = 3).
Next, we investigated whether Vpx-mediated SAMHD1 degradation also abolishes the efficacy of multiple NRTIs when simultaneously applied to macrophages. For this test, primary human macrophages were again exposed to VLPs ±Vpx and then simultaneously treated with AZT and ABC or ddC and TDF followed by transduction with HIV-1 D3. As shown in Fig. 3B, combination NRTI treatment in macrophages exposed to VLPs +Vpx only reduced transduction efficiency to a similar degree as single NRTI treatment. Therefore, these data support that Vpx-mediated SAMHD1 degradation, which elevates all four dNTPs, efficiently counteracts the antiviral efficacy of combination NRTI treatment commonly used in HAART.
Effect of Vpx on HIV-1 Sensitivity to NRTIs in Activated CD4+ T Cells
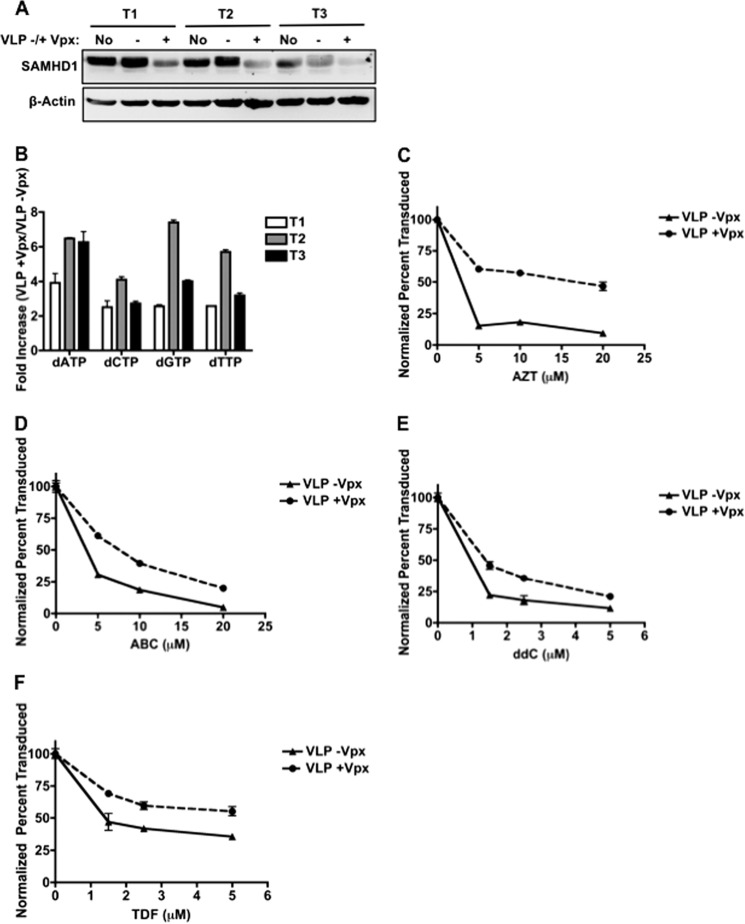
SAMHD1 expression levels directly influence cellular dNTP concentrations in non-dividing cells (12–15). Next, we tested whether degradation of SAMHD1 in primary human activated CD4+ T cells, which are a dividing cell type, affects cellular dNTP concentrations and HIV-1 sensitivity to NRTIs. After treatment with VLPs ±Vpx for 24 h, SAMHD1 expression levels decreased ∼2-fold in the VLP +Vpx-treated cells (Fig. 4A). Cellular metabolites from these donors were also extracted, and cellular dNTP concentrations were measured using a single nucleotide RT primer extension assay (10). Fig. 4B shows that activated CD4+ T cells treated with VLPs +Vpx contain 2.5–7.8 times higher dNTP levels than those treated with VLPs −Vpx. This elevation of cellular dNTPs in activated T cells is much smaller than the Vpx-mediated dNTP level elevation in macrophages (up to 33-fold (12)). Exact nucleotide concentrations averaged from the three donors are shown in supplemental Table 1. Additionally, the transduction efficiency in T cells was not significantly altered by Vpx treatment (data not shown), whereas Vpx enhanced transduction in macrophages by 20–30%. This is likely caused by the kinetic parameters of RT. The dNTPs found in macrophages are below the Km and Kd of RT (11); therefore, the increase in dNTPs upon Vpx treatment greatly increases the rate of DNA synthesis (18). However, the 2.5–7.8-fold increase in dNTPs in T cells does not alter the rate of RT because the base-line dNTP levels are sufficient to saturate RT. To determine whether NRTI efficacy is affected by SAMHD1 degradation, activated T cells were again treated with VLPs and increasing concentrations of AZT, ABC, ddC, or TDF and then transduced with HIV-1 D3. As shown in Fig. 4, C–F, Vpx treatment decreased the efficacy of all four NRTIs tested, but to a much lesser degree as VLP +Vpx-treated macrophages. These data support that SAMHD1 degradation only minimally affects HIV-1 sensitivity to NRTIs in activated CD4+ T cells.
FIGURE 4.
Vpx-mediated degradation of SAMHD1 decreases NRTI efficacy in activated T cells. A, SAMHD1 expression level in three donors of activated CD4+ T cells (T1, T2, and T3) treated without VLPs or with VLPs ±Vpx for 24 h. B, using the same three donors, dNTPs were collected and measured using a single nucleotide RT primer extension assay (10). -Fold increase of dNTPs in VLP +Vpx-treated cells when compared with VLP −Vpx-treated cells is shown (error bars represent the S.E., n = 2). C–F, activated CD4+ T cells were treated with VLPs ±Vpx for 24 h, treated with increasing concentrations of AZT (C), ABC (D), ddC (E), or TDF (F), and then transduced with HIV-1 D3. To determine transduction efficiency, cells were collected to measure GFP expression by flow cytometry (error bars represent the S.E., n = 3).
SAMHD1 Enzymatic Activity toward AZT-TP and ddNTPs
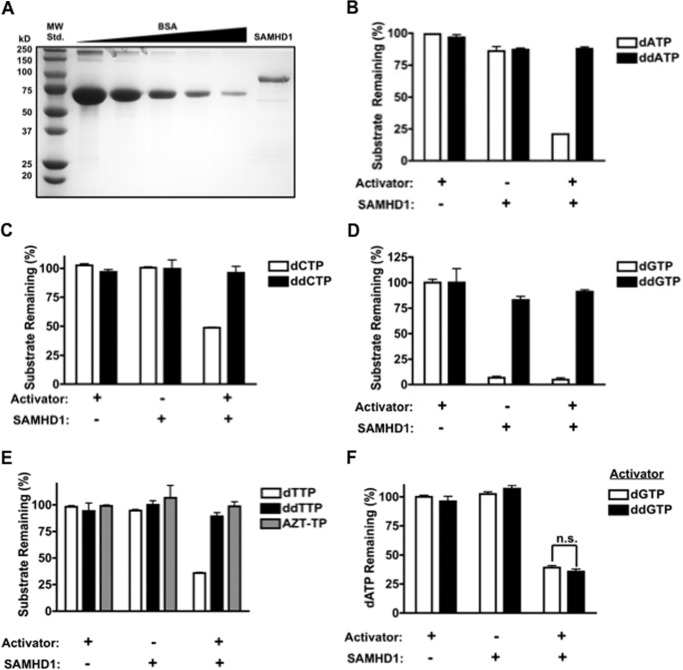
Finally, we tested the possibility that SAMHD1 directly influences viral sensitivity to NRTIs by enzymatically hydrolyzing NRTI-TPs. Although our data demonstrate that expression of SAMHD1 maintains low dNTPs and allows for NRTIs to efficiently inhibit RT, it is possible that SAMHD1 hydrolyzes NRTI-TPs at a slower rate than dNTPs. Therefore, determining whether NRTI-TPs are substrates for SAMHD1 may influence future drug design such that NRTI-TPs are at the highest possible concentration in the cell. To detect whether SAMHD1 has the ability to hydrolyze all four ddNTPs and AZT-TP, we incubated these nucleotides with purified recombinant human SAMHD1, shown in Fig. 5A. When assayed using thin layer chromatography (TLC), recombinant GST-SAMHD1 had similar specific activity as immunoprecipitated HA-SAMHD1 from 293FT cells, 50 versus 58 nm dA/ng SAMHD1/min, respectively (data not shown). Therefore, recombinant SAMHD1 was incubated separately with each substrate with or without the allosteric activator dGTP to ensure that hydrolysis is not due to contaminating phosphatases in the protein preparation. As controls, SAMHD1 was also incubated with each canonical nucleotide. The remaining non-hydrolyzed nucleotide substrates in the triphosphate form were then separated and quantified with anion exchange HPLC (17, 31). As shown in Fig. 5, B–E, hydrolysis of AZT-TP or any of the ddNTPs could not be detected by this assay, but SAMHD1-mediated hydrolysis of all dNTPs could be detected, indicating that our protein is active. We cannot rule out that SAMHD1 may hydrolyze NRTI-TPs at a very slow rate because our assay is not sensitive enough to detect very low concentrations of nucleotides. Overall, these data suggest that for efficient hydrolysis to occur, the 3′-OH moiety on the ribose may be necessary either for the SAMHD1 active site binding or for proper alignment of the α-phosphate with the catalytic residues. It also suggests that SAMHD1 does not significantly alter the concentration of the active form of NRTIs in the cell and that the effect of SAMHD1 on NRTI efficacy results primarily from modulation of cellular dNTP concentrations.
FIGURE 5.
SAMHD1 enzymatic activity toward ddNTPs and allosteric activation with ddGTP. A, recombinant SAMHD1 and a standard curve of bovine serum albumin (8 μg-0.5 μg, BSA) were run on SDS-PAGE and stained with Coomassie Blue. MW Std., molecular weight standard. B–E, purified SAMHD1 was incubated with the indicated nucleotide in the presence or absence of 100 μm dGTP as an allosteric activator. Nucleotide substrates remaining in the triphosphate form were separated and quantified using HPLC (error bars represent the S.E., n = 3). F, SAMHD1 was incubated with 1 mm dATP and 500 μm of dGTP or ddGTP. The quantity of dATP remaining after incubation was measured using HPLC and quantified as described under “Experimental Procedures” (p, 0.2599). Statistical significance was measured using an unpaired t test with a Welch's correction, n = 3.
Allosteric Activation of SAMHD1 with ddGTP
Previous studies have shown that SAMHD1 is allosterically activated by dGTP (16, 17); however, it has yet to be determined whether ddGTP is capable of activating SAMHD1. To test this, SAMHD1 was incubated with dATP as substrate in the presence of dGTP or ddGTP as activators, and the percentage of dATP remaining was quantified using HPLC. As shown in Fig. 5F, ddGTP activated SAMHD1 to the same extent as dGTP. This result provides further insight into the specificity of the allosteric site of SAMHD1 and demonstrates that an interaction with a 3′-OH moiety in the allosteric site of SAMHD1 is not necessary for activation because ddGTP is fully capable of activating SAMHD1. More importantly, these data suggest that ddGTP, and possibly other dideoxyguanosine analogues, can modulate cellular dNTP concentrations by activating SAMHD1.
DISCUSSION
Unwanted interactions of NRTIs with host molecules such as DNA polymerase γ, the mitochondrial DNA polymerase, can lead to off-target effects (35–37). Because SAMHD1, the newly identified antiviral host factor, interacts with and hydrolyzes cellular dNTPs, which chemically mimic NRTI-TPs, we tested whether SAMHD1 influences the antiviral efficacy of NRTIs by directly hydrolyzing NRTI-TPs. With our HPLC-based assay, we could not detect SAMHD1-mediated hydrolysis of ddNTPs or AZT-TP (Fig. 5); however, it is possible that there was minimal hydrolysis of NRTI-TPs that would require a more sensitive assay to be detected. These data suggest that SAMHD1 does not significantly alter the concentration of NRTI-TPs in the cell. Additionally, we show that ddGTP can activate SAMHD1, which may result in further reduction of dNTPs when dideoxyguanosine analogues are applied to cells.
One challenge with competitive inhibitors is that the drug needs to be at a high enough concentration in the cell to out-compete the canonical cellular substrates. HIV-1 and HIV-2 primarily infect CD4+ T cells and macrophages, which have a large disparity in dNTP concentrations (10, 11). Activated T cells are rapidly expanding and replicating their genomic DNA, whereas macrophages are non-dividing, terminally differentiated cells, which do not undergo chromosomal DNA replication. As a consequence, dNTP concentrations in activated peripheral blood mononuclear cells range from 1 to 16 μm, whereas dNTP concentrations in macrophages range from 20 to 70 nm (10, 11). However, the dNTP concentrations in macrophages increase from 5- to 33-fold upon Vpx-mediated degradation of SAMHD1 (12), which is likely similar to what occurs during HIV-2 infection. Aquaro et al. (38) measured the EC50 of NRTIs for inhibition of HIV-1 infection in macrophages and peripheral blood mononuclear cells, and as expected, peripheral blood mononuclear cells require a higher concentration of NRTIs to inhibit viral replication (reviewed in Ref. 34). This led us to test whether degradation of SAMHD1 impacts the efficacy of NRTIs in both macrophages and activated CD4+ T cells. Indeed, our data show that degradation of SAMHD1 by treatment with VLPs +Vpx significantly reduces HIV-1 sensitivity to NRTIs in macrophages (Fig. 2). This result was anticipated based on the previous study (38) and based on our biochemical data showing that increased dNTPs reduce AZT-TP and ddCTP inhibition of RT by active site competition. The reduction in drug efficacy observed in the cell culture experiments may also be influenced by competition for the active site of cellular kinases because NRTIs are not as efficiently phosphorylated as deoxyribonucleosides (33). Additionally, when VLPs +Vpx were added to activated T cells, NRTI efficacy was decreased to a much lesser extent than VLP +Vpx-treated macrophages (Fig. 4). This likely results from the fact that the dNTP level is only minimally elevated by Vpx in activated T cells when compared with the Vpx-treated macrophages.
NRTIs are a key component in HAART, which has been designed to treat patients with HIV-1. However, individuals infected with HIV-2 or dually infected with HIV-1 and HIV-2 also rely on HAART to maintain low viral loads. Therefore, it is essential to understand how differences in these viruses impact the efficacy of these inhibitors. This study focuses on the impact of SAMHD1 on the efficacy of NRTIs. SAMHD1 remains expressed in cells infected by HIV-1; however, HIV-2 expresses the accessory protein Vpx to target SAMHD1 for proteasomal degradation (13, 19–24). Degradation of SAMHD1 upon VLP +Vpx treatment increases dNTP concentrations in macrophages (12) and activated CD4+ T cells (Fig. 4). Overall, this study suggests that degradation of SAMHD1 directly decreases the antiviral efficacy of NRTIs in macrophages by elevating cellular dNTPs, suggesting that these drugs are not as effective at inhibiting HIV-2 infection in macrophages. Finally, this study further supports that the lack of Vpx in HIV-1 and the active site specificity of SAMHD1 to hydrolyze dNTPs more efficiently than NRTI-TPs synergistically contribute to the high efficacy of the currently available NRTIs against HIV-1.
Supplementary Material
Acknowledgments
We thank Dr. Edward Kennedy for assistance with the HPLC and Peter Gee for SAMHD1 purification protocol.
This work was supported, in whole or in part, by National Institutes of Health Grants AI077401 (to B. K.), AI049781 (to B. K.), GM049573 (to R. A. B.), Cellular, Biochemical, and Molecular Sciences Training Grant GM068411 (to S. M. A.), and Oral Cellular and Molecular Biology Training Grant DE007202 (to E. N.). This study was additionally funded by National Institutes of Health Grant 5P30-AI-50409 Centers for AIDS Research (CFAR) and the Department of Veterans Affairs.

This article contains supplemental Table 1.
- SAM
- sterile α motif
- SAMHD1
- SAM domain and HD domain containing protein 1
- RT
- reverse transcriptase
- NRTI
- nucleoside reverse transcriptase inhibitor
- NRTI-TP
- NRTI-triphosphate
- NNRTI
- non-nucleoside reverse transcriptase inhibitor
- VLP
- virus-like particle
- HAART
- highly active antiretroviral therapy
- AZT
- zidovudine
- AZT-TP
- zidovudine-triphosphate
- ddC
- zalcitabine
- ddCTP
- zalcitabine-triphosphate
- ABC
- abacavir
- TDF
- tenofovir
- ddNTP
- dideoxyribonucleoside triphosphate
- ddGTP
- dideoxy-GTP
- hpt
- hours post transduction
- dpt
- days post transduction
- HIV-1 D3
- HIV-1 NL4-3 VSV-G pseudotyped vector
- VSV-G
- vesicular stomatitis virus-G protein
- VPX
- viral protein X.
REFERENCES
- 1. Ho D. D., Neumann A. U., Perelson A. S., Chen W., Leonard J. M., Markowitz M. (1995) Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373, 123–126 [DOI] [PubMed] [Google Scholar]
- 2. Clements J. E., Zink M. C., Narayan O., Gabuzda D. H. (1994) Lentivirus infection of macrophages. Immunol. Ser. 60, 589–600 [PubMed] [Google Scholar]
- 3. Roshal M., Kim B., Zhu Y., Nghiem P., Planelles V. (2003) Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J. Biol. Chem. 278, 25879–25886 [DOI] [PubMed] [Google Scholar]
- 4. Fassati A. (2006) HIV infection of non-dividing cells: a divisive problem. Retrovirology 3, 74–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Singh D. K., Chebloune Y., Mselli-Lakhal L., Karr B. M., Narayan O. (1999) Ovine lentivirus-infected macrophages mediate productive infection in cell types that are not susceptible to infection with cell-free virus. J. Gen. Virol. 80, 1437–1444 [DOI] [PubMed] [Google Scholar]
- 6. Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. (1988) Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science 242, 899–907 [DOI] [PubMed] [Google Scholar]
- 7. Aquaro S., Bagnarelli P., Guenci T., De Luca A., Clementi M., Balestra E., Caliò R., Perno C. F. (2002) Long-term survival and virus production in human primary macrophages infected by human immunodeficiency virus. J. Med. Virol. 68, 479–488 [DOI] [PubMed] [Google Scholar]
- 8. Marchant D., Neil S. J., McKnight A. (2006) Human immunodeficiency virus types 1 and 2 have different replication kinetics in human primary macrophage culture. J. Gen. Virol. 87, 411–418 [DOI] [PubMed] [Google Scholar]
- 9. Murray P. J., Wynn T. A. (2011) Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 11, 723–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Diamond T. L., Roshal M., Jamburuthugoda V. K., Reynolds H. M., Merriam A. R., Lee K. Y., Balakrishnan M., Bambara R. A., Planelles V., Dewhurst S., Kim B. (2004) Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J. Biol. Chem. 279, 51545–51553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kennedy E. M., Gavegnano C., Nguyen L., Slater R., Lucas A., Fromentin E., Schinazi R. F., Kim B. (2010) Ribonucleoside triphosphates as substrate of human immunodeficiency virus type 1 reverse transcriptase in human macrophages. J. Biol. Chem. 285, 39380–39391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lahouassa H., Daddacha W., Hofmann H., Ayinde D., Logue E. C., Dragin L., Bloch N., Maudet C., Bertrand M., Gramberg T., Pancino G., Priet S., Canard B., Laguette N., Benkirane M., Transy C., Landau N. R., Kim B., Margottin-Goguet F. (2012) SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13, 223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baldauf H. M., Pan X., Erikson E., Schmidt S., Daddacha W., Burggraf M., Schenkova K., Ambiel I., Wabnitz G., Gramberg T., Panitz S., Flory E., Landau N. R., Sertel S., Rutsch F., Lasitschka F., Kim B., König R., Fackler O. T., Keppler O. T. (2012) SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nat. Med. 18, 1682–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Descours B., Cribier A., Chable-Bessia C., Ayinde D., Rice G., Crow Y., Yatim A., Schwartz O., Laguette N., Benkirane M. (2012) SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4+ T-cells. Retrovirology 9, 87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. St Gelais C., de Silva S., Amie S. M., Coleman C. M., Hoy H., Hollenbaugh J. A., Kim B., Wu L. (2012) SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology 9, 105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Powell R. D., Holland P. J., Hollis T., Perrino F. W. (2011) Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J. Biol. Chem. 286, 43596–43600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goldstone D. C., Ennis-Adeniran V., Hedden J. J., Groom H. C., Rice G. I., Christodoulou E., Walker P. A., Kelly G., Haire L. F., Yap M. W., de Carvalho L. P., Stoye J. P., Crow Y. J., Taylor I. A., Webb M. (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 [DOI] [PubMed] [Google Scholar]
- 18. Kim B., Nguyen L. A., Daddacha W., Hollenbaugh J. A. (2012) Tight interplay among SAMHD1 level, cellular dNTP levels and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J. Biol. Chem. 287, 21570–21574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hrecka K., Hao C., Gierszewska M., Swanson S. K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M. P., Skowronski J. (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., Benkirane M. (2011) SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laguette N., Rahm N., Sobhian B., Chable-Bessia C., Münch J., Snoeck J., Sauter D., Switzer W. M., Heneine W., Kirchhoff F., Delsuc F., Telenti A., Benkirane M. (2012) Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Cell Host Microbe 11, 205–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lim E. S., Fregoso O. I., McCoy C. O., Matsen F. A., Malik H. S., Emerman M. (2012) The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11, 194–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ahn J., Hao C., Yan J., DeLucia M., Mehrens J., Wang C., Gronenborn A. M., Skowronski J. (2012) HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1. J. Biol. Chem. 287, 12550–12558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goujon C., Rivière L., Jarrosson-Wuilleme L., Bernaud J., Rigal D., Darlix J. L., Cimarelli A. (2007) SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 4, 2–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Erb P., Battegay M., Zimmerli W., Rickenbach M., Egger M. (2000) Effect of antiretroviral therapy on viral load, CD4 cell count, and progression to acquired immunodeficiency syndrome in a community human immunodeficiency virus-infected cohort. Swiss HIV Cohort Study. Arch. Intern Med. 160, 1134–1140 [DOI] [PubMed] [Google Scholar]
- 26. Bangsberg D. R., Perry S., Charlebois E. D., Clark R. A., Roberston M., Zolopa A. R., Moss A. (2001) Non-adherence to highly active antiretroviral therapy predicts progression to AIDS. AIDS 15, 1181–1183 [DOI] [PubMed] [Google Scholar]
- 27. De Clercq E. (2009) Anti-HIV drugs: 25 compounds approved within 25 years after the discovery of HIV. Int. J. Antimicrob. Agents 33, 307–320 [DOI] [PubMed] [Google Scholar]
- 28. Hofmann H., Logue E. C., Bloch N., Daddacha W., Polsky S. B., Schultz M. L., Kim B., Landau N. R. (2012) The Vpx lentiviral accessory protein targets SAMHD1 for degradation in the nucleus. J. Virol. 86, 12552–12560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chugh P., Bradel-Tretheway B., Monteiro-Filho C. M., Planelles V., Maggirwar S. B., Dewhurst S., Kim B. (2008) Akt inhibitors as an HIV-1 infected macrophage-specific anti-viral therapy. Retrovirology 5, 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim B. (1997) Genetic selection in Escherichia coli for active human immunodeficiency virus reverse transcriptase mutants. Methods 12, 318–324 [DOI] [PubMed] [Google Scholar]
- 31. White T. E., Brandariz-Nuñez A., Valle-Casuso J. C., Amie S., Nguyen L., Kim B., Brojatsch J., Diaz-Griffero F. (2013) Contribution of SAM and HD domains to retroviral restriction mediated by human SAMHD1. Virology 436, 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tsuchiya S., Yamabe M., Yamaguchi Y., Kobayashi Y., Konno T., Tada K. (1980) Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer 26, 171–176 [DOI] [PubMed] [Google Scholar]
- 33. Schneider B., Xu Y. W., Sellam O., Sarfati R., Janin J., Veron M., Deville-Bonne D. (1998) Pre-steady state of reaction of nucleoside diphosphate kinase with anti-HIV nucleotides. J. Biol. Chem. 273, 11491–11497 [DOI] [PubMed] [Google Scholar]
- 34. Gavegnano C., Schinazi R. F. (2009) Antiretroviral therapy in macrophages: implication for HIV eradication. Antivir. Chem. Chemother. 20, 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Feng J. Y., Johnson A. A., Johnson K. A., Anderson K. S. (2001) Insights into the molecular mechanism of mitochondrial toxicity by AIDS drugs. J. Biol. Chem. 276, 23832–23837 [DOI] [PubMed] [Google Scholar]
- 36. Feng J. Y., Murakami E., Zorca S. M., Johnson A. A., Johnson K. A., Schinazi R. F., Furman P. A., Anderson K. S. (2004) Relationship between antiviral activity and host toxicity: comparison of the incorporation efficiencies of 2′,3′-dideoxy-5-fluoro-3′-thiacytidine-triphosphate analogs by human immunodeficiency virus type 1 reverse transcriptase and human mitochondrial DNA polymerase. Antimicrob. Agents Chemother. 48, 1300–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee H., Hanes J., Johnson K. A. (2003) Toxicity of nucleoside analogues used to treat AIDS and the selectivity of the mitochondrial DNA polymerase. Biochemistry 42, 14711–14719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Aquaro S., Perno C. F., Balestra E., Balzarini J., Cenci A., Francesconi M., Panti S., Serra F., Villani N., Caliò R. (1997) Inhibition of replication of HIV in primary monocyte/macrophages by different antiviral drugs and comparative efficacy in lymphocytes. J. Leukoc. Biol. 62, 138–143 [DOI] [PubMed] [Google Scholar]
- 39. Berger G., Durand S., Goujon C., Nguyen X. N., Cordeil S., Darlix J. L., Cimarelli A. (2011) A simple, versatile, and efficient method to genetically modify human monocyte-derived dendritic cells with HIV-1-derived lentiviral vectors. Nat. Protoc. 6, 806–816 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.