Background: NR5A nuclear receptors are important pharmaceutical targets with poorly understood ligand regulation. Sequence divergence has potentially altered their ligand response in model organisms.
Results: Sequence divergence has differentially impacted ligand binding and protein dynamics in NR5A orthologs.
Conclusion: Mouse LRH-1 is a phospholipid-responsive receptor, whereas Drosophila NR5A2 is not.
Significance: Mice are viable therapeutic models for LRH-1-dependent diseases.
Keywords: Diabetes, Glucose Metabolism, Hormone Receptors, Mouse, Phosphatidylcholine, Phospholipid
Abstract
The members of the NR5A subfamily of nuclear receptors (NRs) are important regulators of pluripotency, lipid and glucose homeostasis, and steroidogenesis. Liver receptor homologue 1 (LRH-1; NR5A2) and steroidogenic factor 1 (SF-1; NR5A1) have therapeutic potential for the treatment of metabolic and neoplastic disease; however, a poor understanding of their ligand regulation has hampered the pursuit of these proteins as pharmaceutical targets. In this study, we dissect how sequence variation among LRH-1 orthologs affects phospholipid (PL) binding and regulation. Both human LRH-1 (hLRH-1) and mouse LRH-1 (mLRH-1) respond to newly discovered medium chain PL agonists to modulate lipid and glucose homeostasis. These PLs activate hLRH-1 by altering receptor dynamics in a newly identified alternate activation function region. Mouse and Drosophila orthologs contain divergent sequences in this region potentially altering PL-driven activation. Structural evidence suggests that these sequence differences in mLRH-1 and Drosophila FTZ-f1 (dmFTZ-f1) confer at least partial ligand independence, making them poor models for hLRH-1 studies; however, the mechanisms of ligand independence remain untested. We show using structural and biochemical methods that the recent evolutionary divergence of the mLRH-1 stabilizes the active conformation in the absence of ligand, yet does not abrogate PL-dependent activation. We also show by mass spectrometry and biochemical assays that FTZ-f1 is incapable of PL binding. This work provides a structural mechanism for the differential tuning of PL sensitivity in NR5A orthologs and supports the use of mice as viable therapeutic models for LRH-1-dependent diseases.
Introduction
The human liver receptor homologue-1 (LRH-1;3 NR5A2) is a member of the NR5A class of NRs that regulate the expression of genes central to embryonic development, cell cycle progression, reproduction, and lipid homeostasis (1) in response to activating PLs (2). This family includes steroidogenic factor-1 (SF-1; NR5A1) and Drosophila melanogaster fushi tarazu factor 1 (Ftz-F1; NR5A3). SF-1 plays a role in steroidogenesis and the proper development of the testes and adrenal glands (3, 4), and the founding member of the family, Ftz-F1, controls segmentation in flies (5).
LRH-1 plays a crucial role in early embryonic development as it is required to maintain Oct4 expression in undifferentiated embryonic stem cells (6–10), which renders LRH-1 knock-out mice unable to progress past embryonic day 6.5 (11). Beyond development, overexpression of both LRH-1 and SF-1 drives the reprogramming of murine somatic cells to pluripotent stem cells without requiring simultaneous overexpression of Oct4 (12). LRH-1 overexpression appears to drive the expression of Nanog and works synergistically with other well known factors, such as Sox2 and Klf4, to mediate cellular reprogramming (12). In fact, LRH-1 has been implicated as a new stem cell factor because it is the only protein discovered to date that can replace Oct4, which until now was considered absolutely required to manipulate cells into a pluripotent state (6).
In adults, LRH-1 is expressed predominantly in the liver, small intestine, preadipocytes, ovary, placenta, and brain (13). In the ovary, LRH-1 regulates ovarian steroidbiogenesis through control of CYP19 transcription (14). In hepatic tissues, LRH-1 regulates genes central to bile acid homeostasis, lipid and cholesterol absorption, and cholesterol reverse transport (13, 15, 16). Identifying endogenous or synthetic small molecule modulators of LRH-1 activity may lead to promising therapies to treat conditions ranging from metabolic to neoplastic diseases.
LRH-1, like most NRs, interacts with coactivators through an LXXLL motif (where X is any amino acid). Recent studies showed that apo LRH-1 interacts with widely expressed corepressors such as SMRT and NCoR (17–19) in addition to atypical NRs, which have evolved to specifically and efficiently repress LRH-1 by mimicking coactivators (15, 20).
Although the endogenous ligand for hLRH-1 is currently unknown, Lee et al. (21) recently showed that LRH-1 is specifically activated by the exogenous medium chain phosphatidylcholine isoforms, diundecanoyl (DUPC, PC 11:0/11:0) and dilauroyl (DLPC, PC 12:0/12:0) phosphatidylcholine. These medium chain PC agonists increase the ability of LRH-1 to interact with coactivators and reduce blood lipid and glucose levels in diabetic mice in an LRH-1-dependent manner (21). We have shown that DLPC is able to bind to the LRH-1 ligand binding domain (LBD) and activate the receptor by altering receptor dynamics at both an alternate activation function surface and the canonical activation function helix (AF-H) (18). The alternate activation function region in hLRH-1 is composed of residues 398–421 and makes direct contact with bound PLs. We showed that the dynamics of this region are coupled to ligand binding and that restricting motion in this region ablates receptor activation (18). Orthologs of hLRH-1, such as mLRH-1 and dmFTZ-f1, have evolved divergent sequences in this region potentially altering ligand binding and response.
Indeed, mLRH-1 showed no evidence of PL binding in crystal structures as a direct result of this late evolutionary adaptation, which resulted in six amino acid substitutions within the alternate activation function that presumably stabilizes the ligand binding pocket (LBP) in the absence of ligand (17, 18). Surprisingly, the AF-H of apo mLRH-1 was in the active conformation despite the presence of a large empty LBP (22). Subsequent mass spectroscopy analysis showed that mLRH-1 is capable of binding to PCs; however, PL binding was reduced when compared with human NR5A receptors (22). In line with these results, humanization of mLRH-1, by reversing a key sequence substitution in the alternate activation function region, increased sensitivity to PL regulation, suggesting that the mechanism for PL-driven activation has diverged in rodents (22). Placement of the derived rodent sequence in hLRH-1 slightly reduced PL binding and minimally impacted transactivation in HeLa and MCF-7 cells, yet reduced transactivation when transiently overexpressed with SRC-2 and SRC-3 when compared with wild-type hLRH-1 (23). Recent work, however, has shown that mLRH-1 is as robustly activated by DLPC as the human receptor (21). These conflicting observations highlight the need to understand the structural mechanism allowing such a divergent sequence at the alternate AF to support PL-driven regulation (22–26).
Mice serve as an important model system to study stem cell biology and both normal and aberrant hepatic biology including biliary cirrhosis, lipid dysregulation, and diabetes. Flies serve as powerful developmental models. Because both models are used to study LRH-1 biology, it is critical to determine how LRH-1 orthologs differentially interact with PLs. To address this, we used biochemical assays and mass spectrometry to show that mLRH-1 binds to PLs, whereas FTZ-f1 does not, suggesting that mLRH-1 is PL-regulated, whereas FTZ-f1 is ligand-independent. To isolate the effects of the rodent-specific sequence adaptations, we tested the ability of a variant form of LRH-1, mouse-loop LRH-1 (mlLRH-1), to interact with co-regulator peptides both in the absence and in the presence of bound PLs in vitro. Further, we determined the structure of the apo mlLRH-1 variant to 2.75 Å resolution. Finally, we used hydrogen deuterium exchange coupled with mass spectrometry (HDX-MS) to show that the mouse-loop sequence stabilizes the alternate activation function surface and AF-H in the absence of PL, whereas only minimally impacting PL binding.
EXPERIMENTAL PROCEDURES
Reagents
Chemicals were purchased from Sigma, Fisher, or Avanti PLs. pMALCH10T and the vector for His-tagged tobacco etch virus were a gift from John Tesmer (University of Texas at Austin). pLIC_MBP and pLIC_HIS were gifts from John Sondek (University of North Carolina at Chapel Hill). Peptides were synthesized by Synbiosci (Livermore, CA).
Protein Expression and Purification
The hLRH-1 and mlLRH-1 LBDs, residues 291–541, were purified as described previously (23). Pure mlLRH-1 LBD was dialyzed against 100 mm ammonium acetate (pH 7.4), 1 mm DTT, 1 mm EDTA, and 2 mm CHAPS and concentrated to 3–5 mg/ml prior to crystallization. The dmFtz-F1 LBD, residues 791–1025, and mLRH-1 LBD, residues 320–557, were cloned into the pLIC_MBP vector C-terminal to a cassette containing a His6 tag, maltose-binding protein (MBP), and a tobacco etch virus protease cleavage site. The fusion proteins were expressed in BL21(DE3) pLysS cells using standard methods and purified using affinity chromatography with tobacco etch virus cleavage of the fusion partners.
Structure Determination
Crystals of mlLRH-1 LBD were grown by hanging drop vapor diffusion at 22 °C from solutions containing 0.75 ml of protein at 6.5 mg/ml protein and 0.75 ml of the following crystallant: 9.5–15% PEG 3350, 5% glycerol, and 50 mm Bis-Tris, pH 6.4. Crystals were cryoprotected in crystallant containing 20% glycerol and flash-frozen in liquid N2. Data to 2.75 Å resolution were collected at 100 K at the South East Regional Access Team (SER-CAT) at the Advanced Photon Source (Argonne, IL) and were processed and scaled with HKL2000 (see Table 1) (27). Initial phases were determined using the structure of the mLRH-1 LBD (1PK5) as a molecular replacement search model (17). The crystals were pseudo-merohedral twins with a twinning fraction of 45%, and the data were de-twinned using the -h, l, k operator in Detwin (28). The structure was refined using the REFMAC5 within CCP4 suite of programs (28, 29), and model building was performed in COOT (30, 31). The final model contains two mlLRH-1 LBD monomers (residues 300–528) and exhibits good geometry (32). Two loops (H2-H3 residues 332–337 and H11-H12 residues 528–530) displayed poor density, presumably due to high disorder, and were omitted from the final model. The Rfactor values for the final model are 22.4% and 25.7% for R and Rfree, respectively. MolProbity was used for model validation, indicating that 95.3% of the residues fall in the most favored regions of the Ramachandran plot and none fall in disallowed regions. The overall MolProbity score was 2.51, placing mlLRH-1 in the 91st percentile for overall geometric quality among protein crystal structures of comparable resolution (33).
TABLE 1.
Data collection and refinement statistics for the mlLRH-1 crystal structure
Highest resolution shell is shown in parentheses.
| mlLRH-1 | |
|---|---|
| Data collection | |
| Wavelength (Å) | 1.00 |
| Resolution (Å) | 2.8 (2.85-2.80) |
| Space group | P212121 |
| Unit cell dimensions | |
| a, b, c (Å) | 36.3,120.0,123.8 |
| α = β = δ (°) | 90.0 |
| No. of reflections | 14,223 |
| Rsyma (%) | 7.6 (35.7) |
| Completeness (%) | 98.2 (83.7) |
| Average redundancy | 4.4 (3.6) |
| I/σ | 11.8 (3.2) |
| Refinement statistics | |
| Monomers per asymmetric unit (AU) | 2 |
| No. of protein atoms/AU | 1,874 |
| Rworkingb/Rfreec | 0.22/0.25 |
| Average B-factors, Å2 | |
| Protein | 56.5 |
| r.m.s.d deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.4 |
a Rsym = S|I − 〈I〉/S|I|, where I is the observed intensity and 〈I〉 is the average intensity of several symmetry-related observations.
b Rworking = S‖Fo| − |Fc‖/S|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively.
c Rfree = S‖Fo| − |Fc‖/S|Fo| for 7% of the data not used at any stage of the structural refinement.
d r.m.s., root mean square.
Mass Spectrometry
Samples were analyzed using electrospray ionization in the negative-ion mode to detect and identify PLs. Approximately 6 mg of wild-type or mutant forms of LRH-1 LBD and Ftz-F1 LBD was extracted with a 2:1 chloroform/methanol solution, diluted in 200 μl of chloromethylene, and analyzed by negative ion electrospray injection mass spectrometry (ESI/MS) on a Thermo LTQ Fourier transform mass spectrometer using direct injection analysis with electrospray ionization (Thermo Finnigan). All extractions were performed in duplicate. The high-resolution analyses were performed in the Fourier transform mass spectrometer at a resolution of 100,000 at 400 m/z. The MS/MS experiments were done in the ion trap portion of the instrument with a mass selection of 3 atomic mass units and a normalized collision energy of 30 V. The major PL species were identified by accurate mass measurements and MS/MS via collisional-induced dissociation, which yields product ions characteristic of the head groups and attached fatty acids. Acquisition and analyses were performed using the Analyst QS software for this instrument.
Phospholipid Quantification
Preceding PL quantification, 1 mg of protein was digested by 0.45 ml of 8.9 n sulfuric acid at >200 °C for 25 min in glass tubes. Tubes were allowed to cool before the addition of 150 μl of hydrogen peroxide. Tubes were again heated to >200 °C for 30 min. 3.9 ml of deionized water and 0.5 ml of 2.5% ammonium molybdate(VI) tetrahydrate were added, and tubes were vortexed five times each followed by the addition of 0.5 ml of 10% ascorbic acid solution and vortexing. Tubes were capped and heated at 100 °C for 7 min and then allowed to cool before determining the absorbance of each of the samples at 820 nm. All experiments were performed in triplicate and scaled to hLRH-1.
Generation of Apo LRH-1
Apo LRH-1 and mlLRH-1 were generated using published protocols (18). Briefly, pure protein was subjected to chloroform:methanol extraction (2:1) to remove bound lipids according to the Bligh and Dyer method (34). The resulting pellet containing denatured protein was washed three times with chloroform to remove any trace lipids associated with the protein or vessel. The resulting white pellet was then dried by evaporation and resuspended in 6 m guanidinium HCl. Empty protein was then refolded by fast dilution into a buffer containing 10 mm K2HPO4, 100 mm Tris (pH 7.4), 1 mm EDTA, and 500 μm cetyl trimethylammonium bromide at 4 °C. After ∼20 h, protein was then concentrated and purified by size exclusion chromatography to ensure a homogeneous population of refolded receptors.
Cofactor Binding Assays
The polarization of fluorescein-labeled peptides derived from SHP NR box 1 (+H3N-QGAASRPAILYALLSSSLK-CO2−), PGC-1α NR box 2 (+H3N-EEPSLLKKLLLAPA-CO2−), SRC-1 NR box 2 (+H3N-SPSSHSSLTERHKILHRLLQEGSP-CO2−), SMRT (+H3N-TNM GLEAIIRKALMGKYDQW-CO2−), NCoR ID2 (+H3N-DPASNLGLEDIIRKALMGSFDDK-CO2−), or TIF2 NR box 3 (+H3N-PVSPKKKE NALLRYLLDKDDT-CO2−) was monitored with a BioTek Synergy 4 spectrophotometer with polarizers (Winooski, NJ) as a function of protein concentration. Experiments were conducted in 150 mm sodium chloride, 20 mm Tris-HCl (pH 7.4), and 5% (v/v) glycerol. All experiments were done in triplicate, and data were fit with Prism 5 (GraphPad) by the linear least-squares methods to a single site binding model.
Hydrogen-Deuterium Exchange Mass Spectroscopy
Solution-phase amide HDX was carried out with a fully automated system as described previously (35). Briefly, 4 μl of protein was diluted to 20 μl with D2O-containing HDX buffer and incubated at 25 °C for 10, 30, 60, 900, or 3,600. Following ion exchange, back exchange was minimized, and the protein was denatured by dilution to 50 μl in a low pH and low temperature buffer containing 0.1% (v/v) TFA in 5 m urea (held at 1 °C). Samples were then passed across an immobilized pepsin column (prepared in-house) at 50 μl min−1 (0.1% v/v TFA, 15 °C); the resulting peptides were trapped on a C8 trap cartridge (Hypersil Gold, Thermo Fisher). Peptides were then gradient-eluted (4% (w/v) CH3CN to 40% (w/v) CH3CN, 0.3% (w/v) formic acid over 5 min, 2 °C) across a 1 mm × 50-mm C18 HPLC column (Hypersil Gold, Thermo Fisher) and electrosprayed directly into an Orbitrap mass spectrometer (LTQ Orbitrap with electron transfer dissociation, Thermo Fisher). Data were processed with in-house software and visualized with PyMOL (Schrödinger, LLC). To measure the difference in exchange rates, we calculated the average percentage of deuterium uptake for apo mlLRH-1 LBD following 10, 30, 60, 900, and 3,600 s of on exchange. From this value, we subtracted the average percentage of deuterium uptake measured for the apo hLRH-1 LBD. Negative perturbation values indicate that exchange rates are slower for these regions within apo mlLRH-1.
RESULTS
Overall Structure
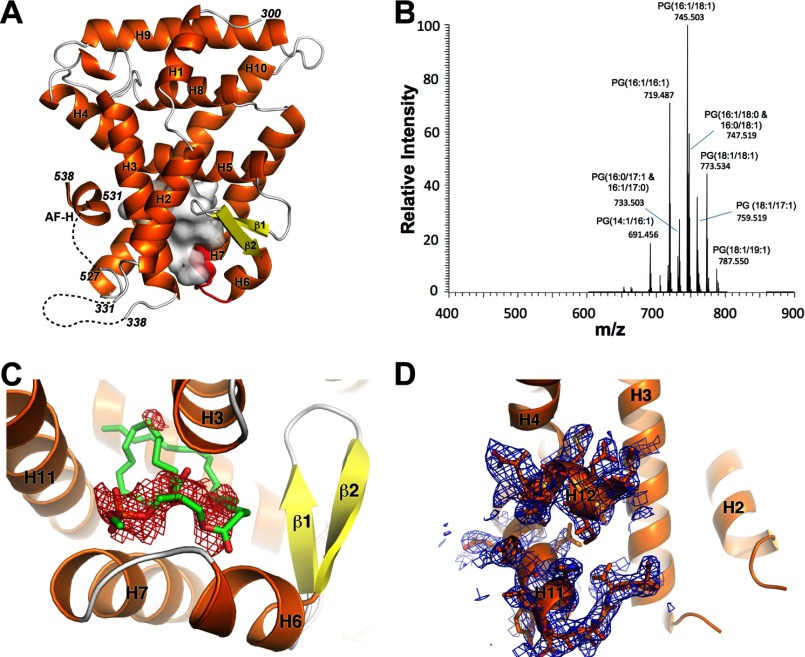
Although nearly all the residues contacting bound PLs are conserved in the LRH-1/SF-1 family, residues 419QAGATL424 in hLRH-1 are replaced by 438HTEVAF443 in mLRH-1 as a result of a late evolutionary divergence in the rodent lineage (see Fig. 3D) (22). This six-amino acid replacement creates a salt bridge between the H7 and H11 at the opening of the LBP, which presumably precludes the binding of PL as observed in hLRH-1 and SF-1 (17, 24). In line with this hypothesis, removal of the H7-H11 salt bridge, via an E440G mutation in mLRH-1, enhanced sensitivity to PL levels (17, 24). To investigate the role of these rodent-specific residues in PL binding, we crystallized the LRH-1 LBD with this six-residue replacement, termed mlLRH-1 (23), and determined its structure to 2.75 Å resolution (Table 1, Fig. 1A). The mlLRH-1 LBD crystallized with two molecules in the asymmetric unit in the P212121 space group. Previous studies showed that the mlLRH-1 LBD contains abundant phosphatidylethanolamine and phosphatidylglycerol species ranging in acyl tail length from C14 to C20, which fortuitously co-purify from Escherichia coli (23). We again confirmed the presence of PLs using mass spectrometry, which showed strong peaks for several PL isoforms (Fig. 1B). Despite the presence of bound E. coli PLs and coactivator peptide in the crystallization conditions, electron density for either bound PL or coactivator peptide was not evident. To confirm that PL was absent in the crystal structure, we modeled a C16:1-C18:1 phosphatidylethanolamine in the LBP. Clear negative Fo − Fc electron density was observed encompassing the modeled phosphatidylethanolamine backbone, confirming that the mouse-loop structure is free of bound PLs (Fig. 1C).
FIGURE 3.
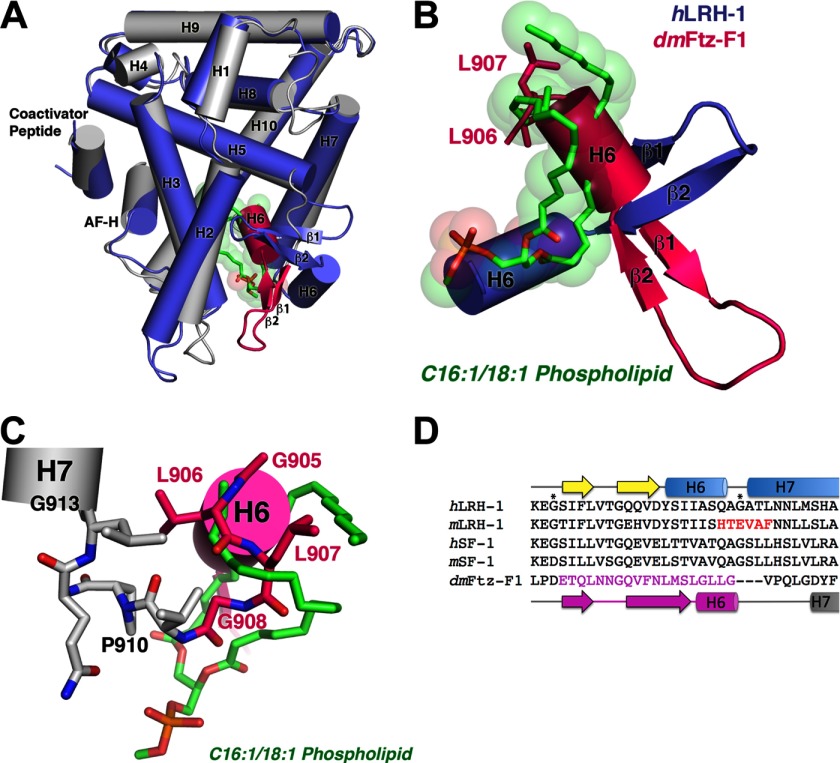
NR5A receptors have multiple modes for achieving the active conformation in the absence of ligand. A, superposition of the hLRH-1:phospholipid complex (1YUC; blue) on empty D. melanogaster Ftz-F1 (2XHS; gray/pink). Ftz-F1 residues highlighted in pink are radically repositioned with respect to all other NR5A family members and disrupt phospholipid binding. B, close-up view of the β-sheet-H6 region inside the LBP highlighting the ∼90° rotation of β-sheet-H6 region in the empty Ftz-F1. C, close-up view of the Ftz-F1 depicting the residues critical for stabilizing the empty LBP. The secondary structure is indicated, and key residues are shown as sticks. D, sequence alignment showing the β1-H7 region of the LBP in NR5A receptors. The amino acids in pink correspond with the repositioned residues in Ftz-F1. Residues in red are unique to rodent LRH-1 and were mutated to create mlLRH-1. Residues marked by asterisks are the glycines previously found to border the alternate activation function of LRH-1 (18).
FIGURE 1.
Overall structure of apo mlLRH-1. A, graphic representation of mlLRH-1 LBD with α-helixes in orange, β-strands in yellow, and the six-residue rodent-derived amino acid replacement in red. The empty LBP cavity is depicted as a white transparent surface. Disordered residues are represented by a black dashed line. B, ESI-MS of mlLRH-1. Phospholipid peaks are labeled as PG with the acyl carbon tail lengths and unsaturation characterized by collision-induced dissociation. The y-axis is scaled relative to the most abundant peak observed in the spectra. C, Fo − Fc electron density (red) contoured at −3σ for phosphatidylethanolamine modeled into the LBP of mlLRH-1. D, 2Fo − Fc electron density contoured at 1σ showing the partially active confirmation of mlLRH-1 evidenced by the discontinuous electron density between helices 11 and 12.
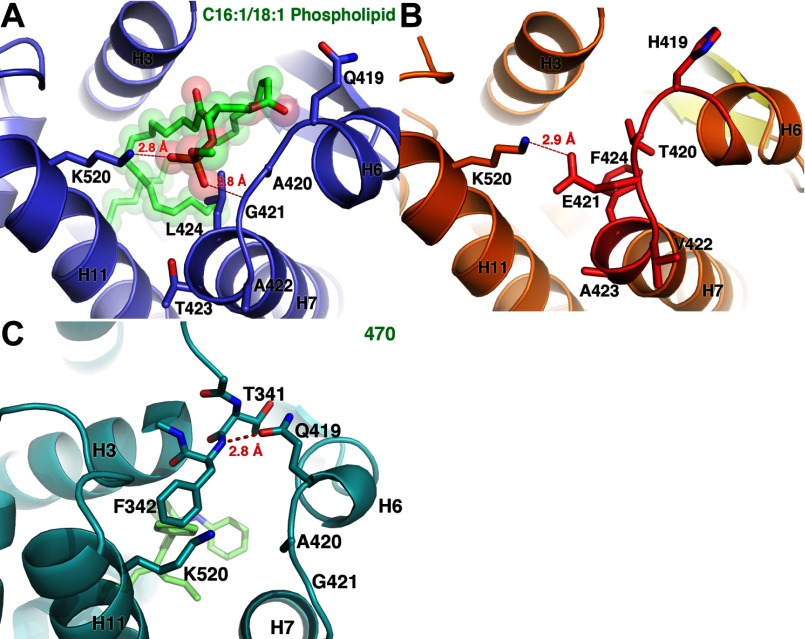
As was observed for the mLRH-1 structure (1PK5), the mlLRH-1 AF-H adopts the active orientation, and the opening of the LBP appears “closed” with a 2.9 Å ionic interaction between residues Glu-421 and Lys-520 blocking access to a well defined internal cavity with a volume of ∼941 Å3. The side chain of Glu-421 occupies the equivalent position of the lipid phosphate moiety in the hLRH-1-PL complex, and the phenyl ring of Phe-424 occupies the same space as the phosphoglycerol backbone (Fig. 2, A and B). A similar closed conformation was observed in a recent structure of the hLRH-1 LBD bound to a small molecule agonist (36), although by a completely different mechanism. Here, Gln-419 H-bonds with the backbone amide of Phe-342 to bridge H6 and H3 rather than the H6-H11 interaction facilitated by the mouse-loop substitutions (Fig. 2C). Thus, the rodent-specific substitutions appear to stabilize this closed conformation of the LRH-1 LBD without the requirement for a small molecule ligand.
FIGURE 2.
Opening to the LBP in the human LRH-1 and mlLRH-1. A, view of the opening of the LBP for hLRH-1 bound to an E. coli phospholipid (green). Blue, PDB ID: 1YUC. B and C, MlLRH-1 (orange) (B) and hLRH-1 (C) bound to a small molecule agonist (green). Orange, PDB ID: 3PLZ mlLRH-1.
Apo mlLRH-1 Adopts a Destabilized Active Orientation
The residues located in the loop preceding the AF-H are known to play a critical role in NR activation, and mutations in this region dramatically alter co-regulator recruitment (17, 23, 37). In the mlLRH-1 structure, there is no interpretable electron density for the H11-H12 loop preceding the AF-H (Fig. 1D). This is mirrored in the structure of mLRH-1, which shows discontinuous electron density in the pre-AF-H loop and roughly 2-fold higher B-factors (∼80 Å2 versus 45 Å2 for all protein atoms) in this region (17). Thus, this region is destabilized in the absence of ligand. Although this disorder does not displace the AF-H, it likely represents the structure of a “destabilized agonist” conformation previously observed in NRs when complexed with weak competitive antagonists (38–40).
Sequence Divergence Allows NR5A Receptors to Adopt Multiple Conformations to Achieve the Active State
Because the Drosophila ortholog of LRH-1, Ftz-f1, also displays sequence divergence in the alternate activation function region (Fig. 3D), we analyzed the dmFtz-F1 (NR5A3) crystal structure (Protein Data Bank (PDB) ID: 2XHS) (41). Interestingly, Ftz-F1 also crystallized in the active orientation without evidence for PLs in the LBP. Instead, residues in the β1-β2 and H6-H7 region adopt an unprecedented conformation that turns 90° inward toward the ligand pocket, filling the cavity occupied by PL in the human NR5A receptors (Fig. 3, A–C).
We have previously shown that flexibility in this region is important for hLRH-1 activation (18). Given the mobility of elements comprising this “wall” of the LBP among NR5A receptors and the fact that crystallization selected for an empty population of mlLRH-1 receptors, it is possible that crystallization also selected for apo Ftz-f1. We therefore asked whether mLRH-1 and Ftz-F1 are able to adjust their conformation to bind PLs when expressed and purified from E. coli.
Do mLRH-1 and Ftz-F1 Bind PLs despite Their Ability to Crystallize Empty?
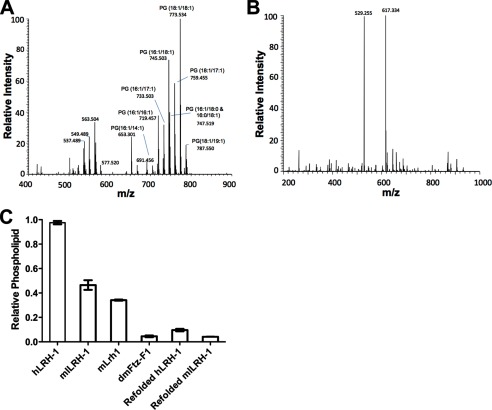
Using mass spectrometry, we discovered that indeed E. coli-expressed recombinant mLRH-1 binds to phosphatidylglycerol (PG) with the most abundant isoforms being PG(18:1/18:1) and PG(18:1/16:1) (Fig. 4A). Mass spectrometry also detects peaks in the 500 m/z range; however, these peaks do not represent PLs, and the collision-induced decomposition analysis reveals that these peaks likely represent an unknown cyclic compound. Attempts to identify this compound were unsuccessful. Recombinant Ftz-F1, however, showed no detectable PL binding (Fig. 4B). Two low abundance peaks with an m/z of 529 and 617 were present in the Ftz-F1 MS data at ∼0.5% of the signal observed for the major peaks in the mLRH-1 spectra. These peaks do not correspond to known compounds, and given their low abundance, attempts to identify these compounds were unsuccessful. To quantify the level of PL bound to Ftz-F1, mLRH-1, and mlLRH-1, we performed an assay to detect lipid phosphorous (Fig. 4C). In line with the mass spectrometry analysis, Ftz-F1 showed almost no PL binding. mLRH-1 shows a diminished ability to bind E. coli PLs versus the human receptors but is on par with the mouse-loop variant of the receptor (23).
FIGURE 4.
Phospholipid analysis of mLRH-1 and Ftz-F1. A and B, ESI-MS of mLRH-1 (A) and Ftz-F1 (B). Identified phospholipid peaks are labeled. The y-axis is scaled relative to the most abundant peak observed in the spectra. C, total phospholipid quantification of LRH-1 variants following chloroform-methanol extraction. Data are represented as mean ± S.E. from three experiments repeated in triplicate.
Testing Ligand Regulation
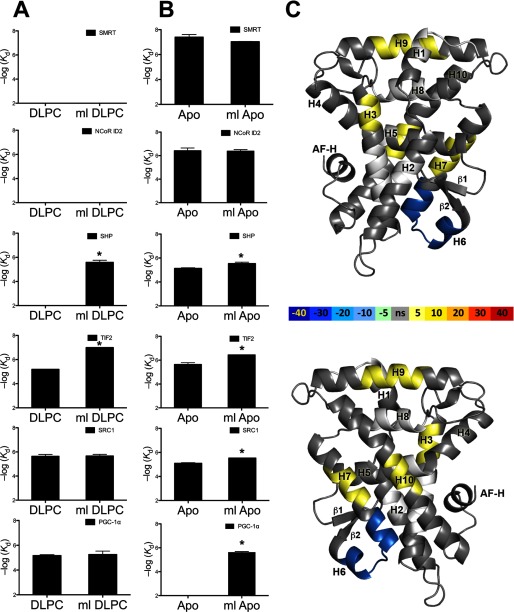
Previous cell-based studies showed that the transcriptional activity of mlLRH-1 ranged from nearly wild-type activity to half of that observed for the human receptor depending on cell type (23). Co-transfection with coactivators exaggerated this difference (23). To fully characterize the link between PL binding and receptor activation in this LRH-1 variant, we would ideally compare the coactivator preference and transactivation potential of the apo- versus PL-bound form of mlLRH-1 versus hLRH-1. However, we are currently unable to determine or control the levels of apo LRH-1 in mammalian cells. We are also unable to measure the levels or identity of bound endogenous PLs or to determine whether the receptor is fully occupied with exogenously supplied DLPC when treated with this lipid. Therefore, we tested the impact of the H6-H7 substitutions on receptor activity in vitro by measuring the ability of either apo or DLPC-bound human and mlLRH-1 LBD to interact with coactivator- and corepressor-derived peptides. Refolded receptor was verified PL-free following chloroform:methanol extraction (Fig. 4C). Specifically, we monitored the ability of protein variants to recruit coactivator peptides derived from SRC-1, TIF2, PGC-1α, and corepressor peptides derived from SHP, SMRT, and NCoR ID2 using fluorescence polarization (Fig. 5 and Table 2).
FIGURE 5.
Apo mlLRH-1 is more active than WT LRH-1 by enhancing stability in the alternate activation region. Binding affinities for various peptides derived from coactivator and corepressor proteins were expressed as Kd ± S.E. A, human and mlLRH-1 LBD bound to DLPC. B, apo human and mlLRH-1 (ml) LBD. C, differential HDX-MS between apo hLRH-1 and apo ml LRH-1 (ml) LBD mapped onto PDB 1YUC. The difference in percentage of deuterium incorporation is indicated by the colored scale bar. Figures were generated in PyMOL.
TABLE 2.
Coactivator peptide recruitment
Kd values are μm ± S.E. NB, no binding detected.
| SMRT | NCoR ID2 | SHP | PGC-1α | SRC1 | TIF2 | |
|---|---|---|---|---|---|---|
| hLRH-1 APO | 0.04 ± 0.01 | 0.39 ± 0.03 | 7.36 ± 0.4 | NB | 8.0 ± 0.9 | 2.3 ± 0.4 |
| mlLRH-1 APO | 0.09 ± 0.002 | 0.42 ± 0.02 | 2.9 ± 0.3 | 2.4 ± 0.1 | 2.9 ± 0.1 | 0.36 ± 0.004 |
| hLRH-1 DLPC | NB | NB | NB | 6.8 ± 0.5 | 2.3 ± 0.4 | 6.5 ± 2.0 |
| mlLRH-1 DLPC | NB | NB | 2.6 ± 0.5 | 5.6 ± 0.2 | 2.2 ± 0.3 | 0.1 ± 0.01 |
As was observed for hLRH-1, apo mlLRH-1 bound to all corepressor peptides tested. The addition of DLPC prevented association between mlLRH-1 and canonical LXXXIXXX(I/L) containing corepressor-derived peptides. mlLRH-1-DLPC binds much more tightly to TIF than hLRH-1. Surprisingly, although DLPC relieves SHP binding for hLRH-1, the mlLRH-1-DLPC complex binds to SHP in vitro, which may explain why mlLRH-1 was observed to have lower basal activation in cells (23). These results are in line with the hypothesis that the rodent-specific divergence alters the dynamics of the apo LBD and suggests potential differences in SHP interaction and regulation between mice and humans. These data show that show that DLPC binding alters co-regulator preference for mlLRH and suggests that rodent divergence may have altered, but not ablated, ligand regulation.
mlLRH-1 Is Specifically Stabilized Versus hLRH-1
The co-regulator peptide recruitment data show that apo mlLRH-1 binds with higher affinity than apo hLRH-1 to all coactivators, suggesting that it is better stabilized in the active orientation. To discover the differences in protein dynamics that underlie this enhanced ability to bind coactivators, we performed HDX-MS (Fig. 5C), comparing apo mlLRH-1 and apo hLRH-1. As hypothesized, the rodent-specific sequence replacement confers increased protection in the helix 6–7 region as evidenced by nearly 40% less deuterium exchange in this region. Surprisingly, no protection was observed for the AF-H; rather, the effect of the mouse-loop replacement is to stabilize the alternate activation function region in LRH-1. Previous work showed that mobility in the alternate activation region of LRH-1 is critical to support normal activation (18).
DISCUSSION
A small number of orphan NRs have acquired the ability to act in a ligand-independent fashion by evolving diverse structural mechanisms to stabilize their overall fold in the absence of ligand (42). Still more NRs have been crystallized in the absence of ligand despite being ligand-dependent (18, 43, 44). In fact, hLRH-1 was recently crystallized without ligand, showing unexpected plasticity in the LBP (18). Despite the fact that crystallization selects for empty receptor, we show that mouse and the mouse-loop variants of LRH-1 bind PLs and that PL binding alters co-regulator selectivity in vitro. The six-amino acid mouse-loop replacement is within the LRH-1 alternate activation function surface and serves to enhance its conformational stability as evidenced by HDX. Thus, rodent LRH-1 tunes its PL sensitivity in vitro by altering receptor dynamics to slightly enhance coactivator and SHP interactions in the absence of ligand. This same sequence difference also confers binding to PGC-1α-derived peptides in the absence of ligand in vitro, which is contrasted with the PL-dependent binding of PGC-1α in hLRH-1. Importantly, the six-amino acid replacement does not over-stabilize the LBP because apo mlLRH-1 retains the ability to interact with canonical corepressors motifs in vitro. It remains unclear how mLRH-1 would coordinate activating PLs such as DLPC. Substitution of Gly-420 to alanine (Glu-440 in mLRH-1) abrogates both PL binding and transactivation (18). Gly-420 is strictly conserved in all non-rodent LRH-1 orthologs and coordinates the lipid phosphate moiety via H-bonds with its backbone amide (22). The conformational mobility of this glycine is thought to permit PL interaction, suggesting that the additional rodent-specific substitutions within the alternate activation function region were likely compensatory substitutions to tolerate this drastic G420E change (18).
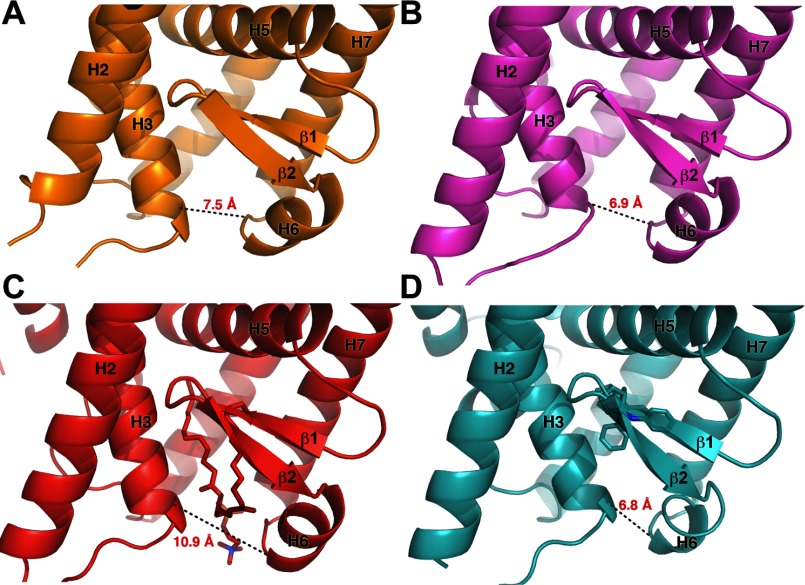
It is clear that remarkable plasticity exists in the NR5A fold in regions outside the canonical activation function surface. This is evidenced by the ability of mLRH-1 and mlLRH-1 to effectively close the entrance to the LBP and by the dramatic structural rearrangement observed in dmFtz-F1 to fill the LBP entirely. This structural plasticity is also supported by the observation that binding of synthetic agonists drives an ∼3 Å constriction of the opening of the LBP versus PL-bound receptor, which is on par with that of the mLRH-1 and mlLRH-1 crystal structures (Fig. 6) (36). The ability of synthetic agonists to induce this constriction of the pocket may be a general feature of non-PL activators.
FIGURE 6.
Coordination at the mouth of the LRH-1 LBP is important for receptor activation. A, mlLRH-1. B, mLRH-1 (PDB ID: 3PK5). C, hLRH-1 bound to an activating phosphatidylcholine (PDB ID: 4DOS). D, hLRH-1 bound to a small molecule agonist (PDB ID: 3PLZ).
Taken together, these results show how sequence divergence in the NR5A alternate activation function region has differentially tuned the sensitivity of NR5A receptors to PLs. It is possible that mLRH-1 evolved a tempered PL response as a result of differential co-regulator expression or limited access to activating PLs. Importantly, these data support PL-dependent regulation for mLRH-1, strengthening conclusions generated in previous studies (17, 24), and support the use of mice as viable models for studying PL-dependent LRH-1 signaling. Future studies focused on the identification and quantification of endogenous ligands and the mechanisms that govern the spatial and temporal access of LRH-1 to PLs are critically needed to understand how PL sensing via NR5A receptors regulates metabolism, lipid flux, steroid synthesis, and immunity.
Finally, we show by mass spectrometry and lipid phosphorous assays that dmFtz-F1 is devoid of PLs when expressed and purified from E. coli, supporting the claim that that dmFtz-F1 is a truly constitutive receptor (41). The selective advantage for PL-independent Ftz-F1 signaling in arthropods is unclear. The NR5A family of NRs arose before the divergence of Placozoans and Eumetazoans from a ligand-activated ancestor (42). The LRH-1 ortholog in nematodes, nhr-25, was recently shown to bind to long chain phosphatidylinositol phosphates and to directly participate in the control of enzymes required to maintain the cellular lipid pool (45). The arthropod-nematode split occurred nearly 1 billion years ago (46), suggesting that PL binding was gained deep in the metazoan lineage and that the constitutive activity of Ftz-F1 was a more recent adaptation in arthropods. Recent examples in marine invertebrates show that losing ligand regulation is possible even in complex systems such as estrogen signaling, which controls diverse gene programs in vertebrate biology (42). It is possible that Drosophila evolved ligand independence to escape the requirement for spatial/temporal ligand presentation during critical developmental processes driven by this receptor.
Although this in vitro study focuses on the isolated LRH-1 LBD, out of the context of post-translational modifications and co-regulator pools that often dictate activation, these results suggest that flexibility in key regions of the receptor should be taken into account and may be capitalized upon to aid in further drug design efforts for LRH-1. For example, stabilization of the alternate activation function region may be a novel strategy for the design of LRH-1 agonists. Conversely, disruption of this region would represent a novel approach toward antagonist design.
Acknowledgments
Data were collected at SE Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract Number W-31-109-Eng-38.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1DK095750 from the NIDDK (to E. A. O.), R01GM084041 from the NIGMS (to P. R. G.), and Emory–NIEHS Graduate and Postdoctoral Training in Toxicology Grant T32ES012870 (to P. M. M.). This work was also supported by American Heart Association (AHA) Predoctoral Grant 12PRE12060583 (to P. M. M.).
The atomic coordinates and structure factors (code 4IS8) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- LRH-1
- liver receptor homologue-1
- hLRH-1
- human LRH-1
- mLRH-1
- mouse LRH-1
- dmFTZ-f1
- Drosophila FTZ-f1
- mlLRH-1
- mouse-loop LRH-1
- NR
- nuclear receptor
- SF-1
- steroidogenic factor-1
- PL
- phospholipid
- PC
- phosphatidylcholine
- PG
- phosphatidylglycerol
- DLPC
- 1,2-didodecanoyl-sn-glycero-3-phosphocholine
- LBD
- ligand binding domain
- LBP
- ligand binding pocket
- AF-H
- activation function helix
- MBP
- maltose-binding protein
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol
- ESI/MS
- electrospray injection mass spectrometry
- HDX-MS
- hydrogen deuterium exchange coupled with mass spectrometry
- SMRT
- nuclear receptor co-repressor 2
- NCoR
- nuclear receptor co-repressor 1
- SHP
- small heterodimer partner (NR0B2)
- TIF
- nuclear receptor coactivator 2
- PGC-1α
- peroxisome proliferator-activated receptor gamma coactivator 1-α
REFERENCES
- 1. Fernandez-Marcos P. J., Auwerx J., Schoonjans K. (2011) Emerging actions of the nuclear receptor LRH-1 in the gut. Biochim. Biophys. Acta 1812, 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Musille P. M., Kohn J. A., Ortlund E. A. (2013) Phospholipid–driven gene regulation. FEBS Lett. 587, 1238–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Parker K. L., Schimmer B. P. (1997) Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr. Rev. 18, 361–377 [DOI] [PubMed] [Google Scholar]
- 4. Hammer G. D., Ingraham H. A. (1999) Steroidogenic factor-1: its role in endocrine organ development and differentiation. Front. Neuroendocrinol. 20, 199–223 [DOI] [PubMed] [Google Scholar]
- 5. Ueda H., Sonoda S., Brown J. L., Scott M. P., Wu C. (1990) A sequence-specific DNA-binding protein that activates fushi tarazu segmentation gene expression. Genes Dev. 4, 624–635 [DOI] [PubMed] [Google Scholar]
- 6. Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 7. Yu J., Vodyanik M. A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J. L., Tian S., Nie J., Jonsdottir G. A., Ruotti V., Stewart R., Slukvin I. I., Thomson J. A. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 [DOI] [PubMed] [Google Scholar]
- 8. Kelly V. R., Xu B., Kuick R., Koenig R. J., Hammer G. D. (2010) Dax1 up-regulates Oct4 expression in mouse embryonic stem cells via LRH-1 and SRA. Mol. Endocrinol. 24, 2281–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kelly V. R., Hammer G. D. (2011) LRH-1 and Nanog regulate Dax1 transcription in mouse embryonic stem cells. Mol. Cell Endocrinol. 332, 116–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wagner R. T., Xu X., Yi F., Merrill B. J., Cooney A. J. (2010) Canonical Wnt/β-catenin regulation of liver receptor homolog-1 mediates pluripotency gene expression. Stem Cells 28, 1794–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paré J. F., Malenfant D., Courtemanche C., Jacob-Wagner M., Roy S., Allard D., Bélanger L. (2004) The fetoprotein transcription factor (FTF) gene is essential to embryogenesis and cholesterol homeostasis and is regulated by a DR4 element. J. Biol. Chem. 279, 21206–21216 [DOI] [PubMed] [Google Scholar]
- 12. Heng J. C., Feng B., Han J., Jiang J., Kraus P., Ng J. H., Orlov Y. L., Huss M., Yang L., Lufkin T., Lim B., Ng H. H. (2010) The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell 6, 167–174 [DOI] [PubMed] [Google Scholar]
- 13. Fayard E., Auwerx J., Schoonjans K. (2004) LRH-1: an orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 14, 250–260 [DOI] [PubMed] [Google Scholar]
- 14. Bookout A. L., Jeong Y., Downes M., Yu R. T., Evans R. M., Mangelsdorf D. J. (2006) Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 126, 789–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goodwin B., Jones S. A., Price R. R., Watson M. A., McKee D. D., Moore L. B., Galardi C., Wilson J. G., Lewis M. C., Roth M. E., Maloney P. R., Willson T. M., Kliewer S. A. (2000) A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6, 517–526 [DOI] [PubMed] [Google Scholar]
- 16. Clyne C. D., Speed C. J., Zhou J., Simpson E. R. (2002) Liver receptor homologue-1 (LRH-1) regulates expression of aromatase in preadipocytes. J. Biol. Chem. 277, 20591–20597 [DOI] [PubMed] [Google Scholar]
- 17. Sablin E. P., Krylova I. N., Fletterick R. J., Ingraham H. A. (2003) Structural basis for ligand-independent activation of the orphan nuclear receptor LRH-1. Mol. cell 11, 1575–1585 [DOI] [PubMed] [Google Scholar]
- 18. Musille P. M., Pathak M. C., Lauer J. L., Hudson W. H., Griffin P. R., Ortlund E. A. (2012) Antidiabetic phospholipid-nuclear receptor complex reveals the mechanism for phospholipid-driven gene regulation. Nat. Struct. Mol. Biol. 19, 532–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu P. L., Kong Y. Y., Xie Y. H., Wang Y. (2003) Corepressor SMRT specifically represses the transcriptional activity of orphan nuclear receptor hB1F/hLRH-1. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 35, 897–903 [PubMed] [Google Scholar]
- 20. Clyne C. D., Kovacic A., Speed C. J., Zhou J., Pezzi V., Simpson E. R. (2004) Regulation of aromatase expression by the nuclear receptor LRH-1 in adipose tissue. Mol. Cell Endocrinol. 215, 39–44 [DOI] [PubMed] [Google Scholar]
- 21. Lee J. M., Lee Y. K., Mamrosh J. L., Busby S. A., Griffin P. R., Pathak M. C., Ortlund E. A., Moore D. D. (2011) A nuclear-receptor-dependent phosphatidylcholine pathway with antidiabetic effects. Nature 474, 506–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krylova I. N., Sablin E. P., Moore J., Xu R. X., Waitt G. M., MacKay J. A., Juzumiene D., Bynum J. M., Madauss K., Montana V., Lebedeva L., Suzawa M., Williams J. D., Williams S. P., Guy R. K., Thornton J. W., Fletterick R. J., Willson T. M., Ingraham H. A. (2005) Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell 120, 343–355 [DOI] [PubMed] [Google Scholar]
- 23. Ortlund E. A., Lee Y., Solomon I. H., Hager J. M., Safi R., Choi Y., Guan Z., Tripathy A., Raetz C. R. H., McDonnell D. P., Moore D. D., Redinbo M. R. (2005) Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP. Nat. Struct. Mol. Biol. 12, 357–363 [DOI] [PubMed] [Google Scholar]
- 24. Li Y., Choi M., Cavey G., Daugherty J., Suino K., Kovach A., Bingham N. C., Kliewer S. A., Xu H. E. (2005) Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Mol. Cell 17, 491–502 [DOI] [PubMed] [Google Scholar]
- 25. Sablin E. P., Blind R. D., Krylova I. N., Ingraham J. G., Cai F., Williams J. D., Fletterick R. J., Ingraham H. A. (2009) Structure of SF-1 bound by different phospholipids: evidence for regulatory ligands. Mol. Endocrinol. 23, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Urs A. N., Dammer E., Kelly S., Wang E., Merrill A. H., Jr., Sewer M. B. (2007) Steroidogenic factor-1 is a sphingolipid binding protein. Mol. Cell. Endocrinol. 265–266, 174–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 28. Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 29. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 30. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laskowski R. A., McArthur M.W., Moss D.S., Thornton J. M. (1993) Procheck: a program to check the stereochemical quality of protein structure. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 33. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bligh E. G., Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 35. Chalmers M. J., Busby S. A., Pascal B. D., He Y., Hendrickson C. L., Marshall A. G., Griffin P. R. (2006) Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 78, 1005–1014 [DOI] [PubMed] [Google Scholar]
- 36. Whitby R. J., Stec J., Blind R. D., Dixon S., Leesnitzer L. M., Orband-Miller L. A., Williams S. P., Willson T. M., Xu R., Zuercher W. J., Cai F., Ingraham H. A. (2011) Small molecule agonists of the orphan nuclear receptors steroidogenic factor-1 (SF-1, NR5A1) and liver receptor homologue-1 (LRH-1, NR5A2). J. Med. Chem. 54, 2266–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ortlund E. A., Bridgham J. T., Redinbo M. R., Thornton J. W. (2007) Crystal structure of an ancient protein: Evolution by conformational epistasis. Science 317, 1544–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schoch G. A., D'Arcy B., Stihle M., Burger D., Bär D., Benz J., Thoma R., Ruf A. (2010) Molecular switch in the glucocorticoid receptor: active and passive antagonist conformations. J. Mol. Biol. 395, 568–577 [DOI] [PubMed] [Google Scholar]
- 39. Raaijmakers H. C., Versteegh J. E., Uitdehaag J. C. (2009) The x-ray structure of RU486 bound to the progesterone receptor in a destabilized agonistic conformation. J. Biol. Chem. 284, 19572–19579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bledsoe R. K., Madauss K. P., Holt J. A., Apolito C. J., Lambert M. H., Pearce K. H., Stanley T. B., Stewart E. L., Trump R. P., Willson T. M., Williams S. P. (2005) A ligand-mediated hydrogen bond network required for the activation of the mineralocorticoid receptor. J. Biol. Chem. 280, 31283–31293 [DOI] [PubMed] [Google Scholar]
- 41. Yoo J., Ko S., Kim H., Sampson H., Yun J. H., Choe K. M., Chang I., Arrowsmith C. H., Krause H. M., Cho H. S., Lee W. (2011) Crystal structure of Fushi tarazu factor 1 ligand binding domain/Fushi tarazu peptide complex identifies new class of nuclear receptors. J. Biol. Chem. 286, 31225–31231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bridgham J. T., Eick G. N., Larroux C., Deshpande K., Harms M. J., Gauthier M. E., Ortlund E. A., Degnan B. M., Thornton J. W. (2010) Protein evolution by molecular tinkering: diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. PLoS Biol. 8, e1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Watkins R. E., Wisely G. B., Moore L. B., Collins J. L., Lambert M. H., Williams S. P., Willson T. M., Kliewer S. A., Redinbo M. R. (2001) The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science 292, 2329–2333 [DOI] [PubMed] [Google Scholar]
- 44. Nolte R. T., Wisely G. B., Westin S., Cobb J. E., Lambert M. H., Kurokawa R., Rosenfeld M. G., Willson T. M., Glass C. K., Milburn M. V. (1998) Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature 395, 137–143 [DOI] [PubMed] [Google Scholar]
- 45. Mullaney B. C., Blind R. D., Lemieux G. A., Perez C. L., Elle I. C., Faergeman N. J., Van Gilst M. R., Ingraham H. A., Ashrafi K. (2010) Regulation of C. elegans fat uptake and storage by acyl-CoA synthase-3 is dependent on NR5A family nuclear hormone receptor nhr-25. Cell Metab. 12, 398–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang D. Y., Kumar S., Hedges S. B. (1999) Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi. Proc. Biol. Sci. 266, 163–171 [DOI] [PMC free article] [PubMed] [Google Scholar]