Background: TUT7 adds uridines to and regulates RNAs.
Results: Xenopus TUT7 (XTUT7) uridylates RNAs, possesses a basic region that binds nucleic acids, and represses translation of a polyadenylated RNA.
Conclusion: XTUT7 contains a nucleic acid-binding domain important for activity and can repress translation.
Significance: The basic region of XTUT7 may bind RNA in vivo. XTUT7 may control mRNAs by occluding poly(A).
Keywords: Enzyme Mutation, Molecular Biology, Protein-Nucleic Acid Interaction, RNA-binding Protein, Translation Control, Xenopus, RNA Uridylation, TUT7, ZCCHC6, Terminal Uridylyl Transferase
Abstract
Terminal uridylyl transferases (TUTs) catalyze the addition of uridines to the 3′ ends of RNAs and are implicated in the regulation of both messenger RNAs and microRNAs. To better understand how TUTs add uridines to RNAs, we focused on a putative TUT from Xenopus laevis, XTUT7. We determined that XTUT7 catalyzed the addition of uridines to RNAs. Mutational analysis revealed that a truncated XTUT7 enzyme, which contained solely the nucleotidyl transferase and poly(A) polymerase-associated domains, was sufficient for catalytic activity. XTUT7 activity decreased upon removal of the CCHC zinc finger domains and a short segment of basic amino acids (the basic region). This basic region bound nucleic acids in vitro. We also demonstrated that XTUT7 repressed translation of a polyadenylated RNA, to which it added a distinct number of uridines. We generated a predicted structure of the XTUT7 catalytic core that indicated histidine 1269 was likely important for uridine specificity. Indeed, mutation of histidine 1269 broadened the nucleotide specificity of XTUT7 and abolished XTUT7-dependent translational repression. Our data reveal key aspects of how XTUT7 adds uridines to RNAs, highlight the role of the basic region, illustrate that XTUT7 can repress translation, and identify an amino acid important for uridine specificity.
Introduction
The addition of nontemplated uridines to the 3′ ends of RNAs is an emerging form of RNA control that can influence RNA stability and processing (1–4). Terminal uridylation regulates both messenger RNAs (mRNAs) and small, noncoding RNAs. Uridylation of specific mRNAs in Schizosaccharomyces pombe and mammalian cells promotes removal of the 5′ cap and subsequent degradation (5–11). Uridylation of microRNA precursors (pre-miRNAs) can either block or promote processing depending on the cellular context (12–18). In nematodes, plants, and algae, uridylation destabilizes microRNAs (miRNAs)3 and/or small-interfering RNAs (siRNAs) (19–22). Mammalian miRNAs are also uridylated (23–26). Despite the apparent pervasiveness of RNA uridylation, little is known about the enzymes that add uridines to RNAs.
Terminal uridylyl transferases (TUTs; also known as poly(U) polymerases) add uridines to RNAs (27–29). TUTs are noncanonical ribonucleotidyl transferases (rNTases) of the DNA polymerase-β superfamily, which contains enzymes that add nucleotides to a variety of substrates, including RNAs (28). The nucleotide specificity of a particular rNTase is difficult to predict by amino acid sequence and must be experimentally derived as determinants for specificity remain unclear. Additionally, TUTs possess several conserved domains: the nucleotidyl transferase domain (NTD), the poly(A) polymerase-associated domain (PAPD), and the nucleotide recognition motif (NRM) (3, 28). NTDs contain the conserved catalytic motif characteristic of rNTases and the catalytic triad of acidic residues, typically aspartates (28). PAPDs encode an NRM, which mediates nucleotide specificity by contacting the base in the active site (28, 30–33).
TUT7 orthologs are poly(U)-adding enzymes implicated in the regulation of let-7 miRNA biogenesis, a family of miRNAs critical during development and oncogenesis (6, 16, 17, 29, 34). Uridylation of let-7 precursor (pre-let-7) RNAs can either block or promote processing, depending on cell type. In mammalian stem cells, the paralogs TUT7 and TUT4 add several uridines to pre-let-7 after recruitment by the RNA-binding protein LIN28 (12, 14–16). Uridylation blocks processing of pre-let-7 into mature miRNAs, as well as destabilizes pre-let-7 RNAs. In mammalian somatic cells, however, TUT7 acts independent of LIN28 and adds a single uridine to a subset of pre-let-7 RNAs (17). Monouridylation of these pre-miRNAs creates an optimal 3′ end for downstream processing into mature miRNAs.
To further understand TUT7-dependent RNA uridylation, we identified and focused on Xenopus TUT7 (XTUT7) as it may have key roles in the oocyte and/or embryo. We sought to better understand how XTUT7 adds uridines to RNAs and its potential role in mRNA regulation. We utilized Xenopus oocytes because uridylated RNAs are stable, and microinjected mRNAs are efficiently translated. With this approach, we identified XTUT7 domains important for catalytic activity, illustrated that XTUT7 can repress translation of a polyadenylated RNA, and pinpointed an important residue for uridine specificity. Our experiments also revealed a key role for a small region of basic amino acids that binds nucleic acids.
EXPERIMENTAL PROCEDURES
MS2 Fusion Protein Plasmids
The pCS2+3HA:MS2, pCS2+3HA:MS2:Xp54, and pCS2+3HA:MS2:GLD2-D242A plasmids were previously described (35). Newly constructed MS2 fusion plasmids, and the primers and restriction sites used for their construction, are listed in supplemental Table 1. All MS2 fusion proteins were designed to contain: N terminus; three hemagglutinin (3HA) tags; MS2 coat protein; protein to be tested; and C terminus. XTUT7 (also known as ZCCHC6) and XTUT4 (also known as ZCCHC11) cDNAs were cloned from both Xenopus laevis and Xenopus tropicalis stage VI oocytes. cDNAs corresponding to XTUT7-FL (X. tropicalis), XTUT7-C (X. laevis), and XTUT4-C (X. laevis) were deposited to GenBankTM with accession numbers KC493151, KC493152, and KC493153, respectively. Mutations and deletions in XTUT7 were inserted by site-directed mutagenesis using Pfu-Ultra DNA polymerase (Agilent), and mutated/deleted residues are listed in supplemental Table 1. Specific amino acids (i.e. His-1269) discussed under “Results” and “Discussion” are referenced by their location in XTUT7-FL. Domain predictions of XTUT7 proteins were completed using the InterProScan Sequence Search Tool (36, 37) and Pfam (38) on XTUT7-FL.
Multiple Sequence Alignments
XTUT7 sequence homologs were identified by reciprocal best BLAST (National Center for Biotechnology Information (NCBI)): Ciona intestinalis (GI number: 198429697), Strongylocentrotus purpuratus (115933324), Bos taurus (329664700), Canis lupus familiaris (73946401), Macaca mulatta (109112038), Mus musculus (259016375), Rattus norvegicus (293354419), Hydra magnipapillata (221116335), Monodelphis domestica (334332807), Ornithorhynchus anatinus (345314193), Danio rerio (326668285), Homo sapiens (297307111), Caenorhabditis elegans (17554128), and Amphimedon queenslandica (340382961). Sequence logos were derived from MUSCLE (39) sequence alignments of the putative XTUT7 orthologs using WebLogo (40).
Reporter RNA Plasmids
The pLG-MS2 (firefly luciferase), pLG-MS2+A39 (polyadenylated firefly luciferase), pSP65-ren (Renilla luciferase), pLGMS2-luc (RNA with three MS2-binding sites), pLGMS2+A39-luc (RNA with three MS2-binding sites and a poly(A)39 tail), pLG:FBE-ACAmut (RNA that lacked MS2 binding sites), and pLG:FBE-ACAmut+A39 (RNA with a poly(A)39 tail that lacked MS2 binding sites) plasmids have been described (41–44).
In Vitro Transcriptions
RNAs were in vitro transcribed from restriction digested plasmids using either the AmpliScribe SP6 high yield transcription or T7-Flash transcription kits (Epicentre). RNAs encoded in pCS2+3HA:MS2 (NotI, SP6), pSP65-ren (SalI, SP6), pLG-MS2+A39 (BamHI, T7), pLGMS2+A39-luc (BamHI, T7), pLG-MS2 (BglII, T7) and pLGMS2-luc (BglII, T7) plasmids were prepared with the indicated reagents. All reactions included m7G(5′)ppp(5′)G cap analog (New England Biolabs). In some cases, [α-32P]UTP was included to radiolabel the RNA.
Oocyte Injections and RNA Analysis
Oocyte injections were performed as described (41, 44). Oocytes were collected after overnight incubation (∼16 h). Total RNA was extracted from 10 oocytes using TRI reagent (Sigma). Total RNA from three oocytes was separated on denaturing 6% polyacrylamide gels and analyzed by phosphorimaging. Densitometric analyses were completed using ImageQuant software (GE Healthcare).
Luciferase Assays and Western Blotting
Dual-Luciferase assays (Promega) and Western blotting were performed as described (35, 43). Student's two-tailed t tests were used to calculate all p values.
RT-PCR Assays
Total RNA was treated with 4 units of TURBO-DNase (Invitrogen) for 1 h at 37 °C and then purified using the GeneJET RNA purification kit (Fermentas). 1 μg of total RNA was reverse-transcribed using ImPromII reverse transcriptase (Promega) and 1 μm oligo(dA)18, -(dT)18, -(dC)20, or -(dG)20 for the RT primers, as indicated. cDNA was PCR-amplified using a firefly luciferase-specific forward primer (GCGTTAATCAGAGAGGCGAATTATGTG) and the corresponding RT primer. For quantitative RT-PCR assays, 100 ng of total RNA were reverse-transcribed using SuperScript III reverse transcriptase (Invitrogen) and a random hexamer primer, and 5% of the cDNA was amplified using the PerfeCTa qPCR FastMix UNG Low ROX kit (Quanta Biosciences). Firefly luciferase levels were compared with Renilla luciferase and β-actin mRNA levels.
Tail Sequencing Assays
The tail sequencing assay was performed essentially as described (45) with the following modifications. The P1 anchor primer (AATATTCACCTTGATCTGAAGC) was 5′ phosphorylated using polynucleotide kinase enzyme (Promega) and 3′-blocked with cordycepin (Sigma-Aldrich) using terminal deoxynucleotidyl transferase enzyme (New England Biolabs). 400 ng of modified P1 primer were pre-annealed with 400 ng of P1′ (GCTTCAGATCAAGGTGAATATTAAAAA) and ligated to 1–2 μg of total RNA using T4 RNA Ligase (Fermentas) at 37 °C for 1 h. Reverse transcription reactions were completed using 1 μm P1′ oligonucleotide and SuperScript III reverse transcriptase (Invitrogen). Two rounds of nested PCR amplification were performed using forward primer 1 (GCGTTAATCAGAGAGGCGAATTATGTG) and forward primer 2 (ACCTCTCTCTCTCTCTCAGGGCTGATTACTAG). P1′ was the reverse primer in both reactions. PCR products from the second PCR were TOPO-TA cloned (Invitrogen) and sequenced. 10 tails added by XTUT7-C (227 bases total) and 14 tails added by XTUT7-H1269L (751 bases) were independently cloned and sequenced.
Protein Purifications
Amino acids 453–540 of XTUT7-C (KC493152) were fused to an N-terminal maltose-binding protein (MBP) tag and a C-terminal 6-histidine (His6) tag by cloning into the XbaI restriction site of the previously described pHMTC plasmid (46). Residues of interest were mutated by site-directed mutagenesis (supplemental Table 1). Plasmids were transformed into BL21-CodonPlus(DE3)-RIL cells and grown in LB+ampicillin/chloramphenicol medium at 37 °C until A600 ∼0.6. Protein expression was induced with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside for 3 h at 37 °C. Cells were resuspended in lysis buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 0.02% (v/v) Tween 20), and lysed by incubation with 0.5 mg/ml lysozyme followed by a freeze-thaw cycle. Cleared lysates were incubated with prewashed Sepharose-amylose resin (New England Biolabs) for ∼2 h at 4 °C. The resin was washed two times in lysis buffer and one time in wash buffer (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 0.02% (v/v) Tween 20). MBP fusion proteins were eluted three times in elution buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 0.02% (v/v) Tween 20, 50 mm maltose). Eluted proteins were dialyzed overnight in 4 liters of lysis buffer and were concentrated using Vivaspin 30,000 molecular weight cut-off columns (Sartorius). Protein concentrations were measured by Bradford assays.
Electrophoretic Mobility Shift Assays
RNA that contained three MS2-binding sites (transcribed from pLGMS2-luc) was 5′ end-labeled using polynucleotide kinase enzyme (Promega) and [γ-32P]ATP. ∼3.5 fmol of end-labeled RNA were incubated with purified MBP fusion proteins (0–1 μm) in 45 mm Tris-HCl, pH 8.0, 90 mm NaCl, 0.02% (v/v) Tween 20, and 20 units of RNasin (Promega) for 30 min on ice. 3× loading buffer (6% glycerol, 0.06% bromphenol blue) was added to each reaction and then loaded onto 5% polyacrylamide/Tris-borate-EDTA gels (Bio-Rad). Gels were run for ∼1 h at 100 V at 4 °C, dried, and exposed to storage phosphor screens overnight. The percentage of nucleic acid bound was calculated using ImageQuant software (GE Healthcare). Apparent Kd values were determined using GraphPad Prism 5 software and the “One site–Specific binding with Hill slope” equation. All values were reported with associated S.E.
XTUT7 Structure Prediction
The I-TASSER server was used to generate the predicted structure of amino acids 91–422 of XTUT7-C (KC493152) (46–48). Specific amino acids discussed in the text are referred to by their position in the context of XTUT7-FL (KC493151). PyMOL was used to visualize the model, create images, and perform structural alignments.
RESULTS
XTUT7 Domain Structure
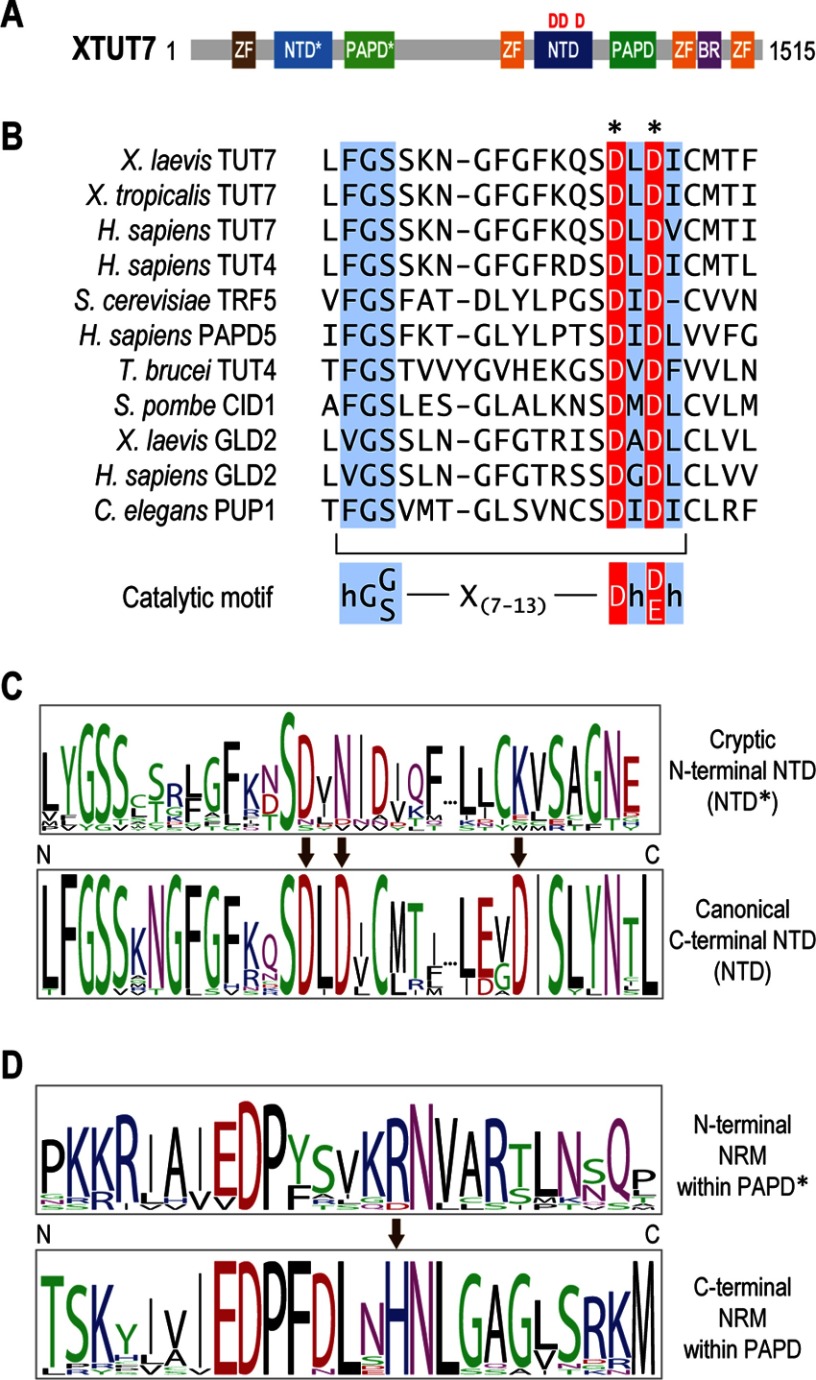
A single XTUT7 ortholog is encoded by the Xenopus genome. Putative XTUT7 cDNAs were cloned from both X. laevis and X. tropicalis oocytes and are collectively referred to as XTUT7. XTUT7 cDNA encodes conserved domains typical of rNTases (28). In particular, the domains include two NTDs, two PAPDs, and an NRM encoded in each PAPD (Fig. 1A). XTUT7 also possesses a C2H2 zinc finger domain, three CCHC zinc finger domains, and a short, arginine-rich segment of basic amino acids (the basic region or BR). The amino acids that span the C-terminal NTD and PAPD are 84% identical and 95% similar between Xenopus and H. sapiens TUT7. Overall, the proteins are 57% identical and 77% similar. XTUT7 includes the conserved motif characteristic of known rNTase active sites, which contains two of its three putative catalytic aspartates (Fig. 1B) (28).
FIGURE 1.
XTUT7 domain structure. A, diagram representing XTUT7 and its predicted protein domains (amino acids indicated). Zinc finger domain (ZF) (brown), C2H2 zinc finger domain (227–252); NTD*, cryptic NTD (295–437); PAPD*, cryptic PAPD (526–575); NTD, nucleotidyl transferase domain (998–1146); PAPD, poly(A) polymerase-associated domain (1215–1268); zinc finger domain (orange), CCHC zinc finger domains (946–962, 1327–1343, 1432–1448); BR, basic region (1344–1361). The red D's denote putative catalytic aspartates. B, multiple sequence alignment of the characteristic catalytic motifs contained in the NTD of XTUT7 and other previously identified nucleotidyl transferases. The consensus rNTase catalytic motif, hG[G/S]X7–13Dh[D/E]h, is shown below the sequence alignment, where h indicates any hydrophobic amino acid (28). S. cerevisiae, Saccharomyces cerevisiae; T. brucei, Trypanosoma brucei. C, sequence logos representing multiple sequence alignments of the putative catalytic motifs contained in the NTD and NTD* of XTUT7 orthologs. The arrows indicate analogous positions in the NTD and NTD* and highlight amino acid substitutions of putative catalytic aspartates in the NTD*. D, sequence logos representing multiple sequence alignments of the NRMs encoded in the PAPDs and PAPD*s of XTUT7 orthologs. The arrow indicates an invariant histidine in the C-terminal NRM.
XTUT7 and its orthologs encode two distinct NTDs and PAPDs. XTUT7 orthologs have canonical NTDs in their C-terminal halves and cryptic NTDs (NTD*) in their N-terminal halves. The NTD contains the three canonical catalytic aspartates (Fig. 1C). In contrast, the NTD* contains aspartate to asparagine and aspartate to lysine substitutions of aspartates two and three, respectively. XTUT7 orthologs also have distinct NRMs encoded in their N- and C-terminal PAPDs. The NRMs encoded in their C-terminal PAPDs contain highly conserved type-2 NRMs characteristic of rNTases, which includes an invariant histidine (Fig. 1D) (28). The NRMs encoded in their cryptic, N-terminal PAPDs (PAPD*) are more divergent among species and contain an arginine in place of the histidine (Fig. 1D).
XTUT7 Is a Poly(U)-adding Enzyme
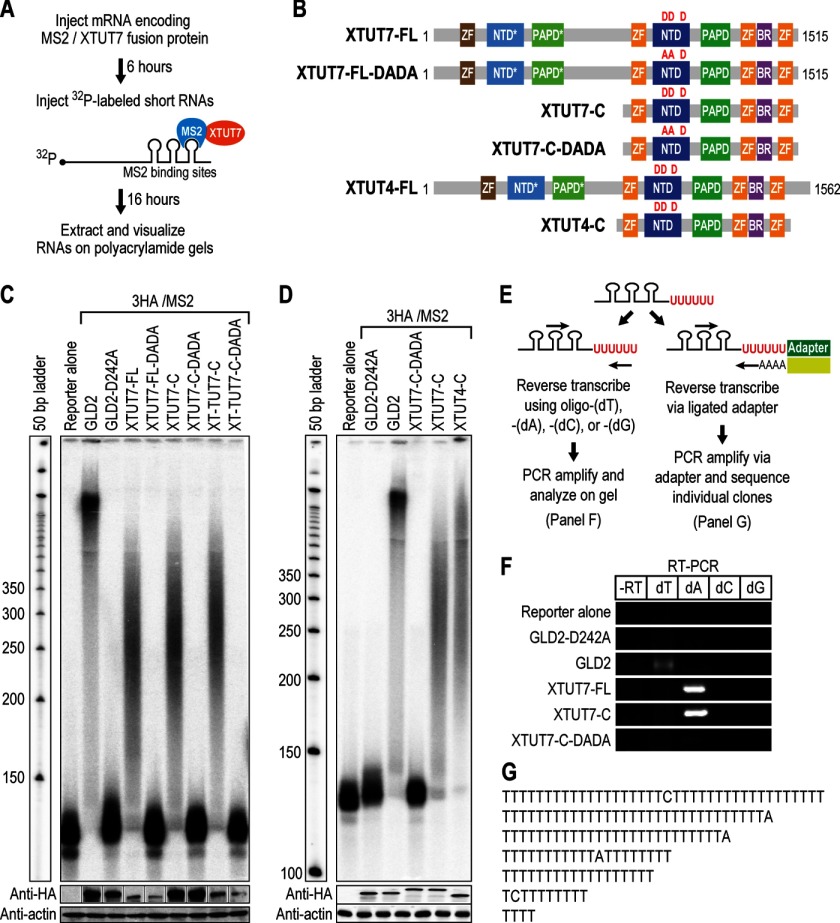
To test whether XTUT7 can catalyze rNTase activity, XTUT7 was tethered to reporter RNA using MS2 coat protein and MS2-binding sites (Fig. 2A). mRNAs encoding portions of XTUT7 fused to three HA tags and MS2 coat protein (3HA/MS2) were microinjected into X. laevis oocytes to directly translate the proteins. Radiolabeled RNA substrates that contained three MS2-binding sites were then microinjected and analyzed by polyacrylamide gel electrophoresis. Diagrams of the proteins tested are shown in Fig. 2B.
FIGURE 2.
XTUT7 is a poly(U)-adding enzyme. A, mRNAs encoding MS2 coat protein fused to XTUT7 were microinjected into oocytes. Following a 6-h incubation to allow translation of the fusion proteins, 32P-labeled RNA that contained MS2-binding sites was injected. MS2 coat protein binds the MS2-binding sites, thus tethering XTUT7 to the reporter RNA. For clarity, only one binding event is depicted. After 16 h, RNAs were extracted, analyzed on polyacrylamide gels, and visualized by phosphorimaging. B, diagrams representing proteins tested in subsequent panels. The red D's denote catalytic aspartates, whereas the red A's denote aspartate to alanine substitutions; ZF, zinc finger domain. The XTUT4-FL diagram represents the domain structure of full-length XTUT4, which is a paralog of XTUT7. XTUT4-C represents the construct tested in panel D, which is analogous to XTUT7-C. C, extracted RNAs were separated on denaturing polyacrylamide gels. All proteins assayed were 3HA/MS2 fusions. The reporter alone sample serves as a negative control for extension, whereas the GLD2 sample is a positive control for extension. GLD2-D242A is a mutant GLD2 containing an alanine in place of a catalytic aspartate. 3HA/MS2/XTUT7 fusion proteins tested are depicted in panel B. XTUT7-FL and XT-TUT7-C are derived from X. tropicalis, and XTUT7-C is derived from X. laevis. XTUT7-DADA proteins contain aspartate to alanine substitutions as indicated in panel B. The bottom panels indicate protein levels as determined by Western blotting for HA-tagged fusion proteins and actin. D, the indicated 3HA/MS2 fusion proteins were assayed as in panel C. All proteins tested here and in subsequent figures were derived from X. laevis, except when noted. The bottom panels indicate protein levels as determined by Western blotting for HA-tagged fusion proteins and actin. E, schematic of the assays used to determine the nucleotide(s) added by XTUT7. First, RNAs were assayed by RT-PCR (left half, for panel F). Second, RNAs were assayed by a tail sequencing assay (right half, for panel G). F, RT-PCR analysis for the indicated fusion proteins. All samples were analyzed on the same gel but were separated for clarity. RT primers are indicated. The dT lane indicates the addition of adenosines, the dA lane indicates addition of uridines, and so forth. G, seven representative, independently cloned sequences that illustrate the nucleotides added by XTUT7-C are shown.
XTUT7 proteins that contained a wild-type NTD added nucleotides to the end of the reporter RNA. Tethered XTUT7-FL and the control poly(A) polymerase GLD2 (47, 48) yielded a heterogeneous mix of slower migrating RNA relative to the reporter alone control (Fig. 2C). This indicated that the enzymes added long nucleotide tails to the reporter RNA. Substituting catalytic aspartates with alanines in the NTD of XTUT7 (DADA) or GLD2 (D242A) prevented extension of the reporter RNA. Mutant and wild-type enzymes were expressed comparably; therefore, differences in activity were not due to differences in expression levels. Truncated X. laevis and tropicalis XTUT7 proteins (XTUT7-C and XT-TUT7-C, respectively) that lacked the NTD* and PAPD* were as active as the full-length protein, and again inactivated by mutation of catalytic aspartates. A construct of X. laevis TUT4 (XTUT4-C) that lacked its NTD* and PAPD* extended the reporter RNA much like XTUT7-C (Fig. 2D). The C-terminal half of XTUT7 is therefore sufficient to add nucleotides to RNAs.
To identify the nucleotide(s) added by XTUT7, RNAs extended by XTUT7 were assayed by RT-PCR using oligo(dT), -(dA), -(dC), or -(dG) as the RT primer (Fig. 2E). XTUT7-C and XTUT7-FL samples yielded RT-PCR products solely in oligo(dA) primed reactions and only when the catalytic aspartates were present (Fig. 2F). Therefore, XTUT7 added uridines to the reporter RNA. Conversely, the control poly(A) polymerase GLD-2 yielded products only with an oligo(dT) primer, which indicated that the enzyme added adenosines to the reporter RNA (47, 48). Sequencing of cloned XTUT7-C RT-PCR products confirmed that uridines had been added (Fig. 2G). Thus, XTUT7 is a poly(U)-adding enzyme.
XTUT7 Extends RNAs Independent of MS2 Tethering
Two lines of evidence demonstrate that XTUT7 extends RNAs independent of MS2 tethering. First, XTUT7-FL and XTUT7-C extended an RNA that lacked MS2-binding sites by ∼30–50 nucleotides, and this activity was eliminated by the mutation of catalytic aspartates (Fig. 3A). Second, an XTUT7-C construct that lacked both MS2 coat protein and the 3HA tag extended a reporter RNA that contained three MS2-binding sites by up to 50 nucleotides (Fig. 3B, last lane). Tethered XTUT7-C extended the same RNA by ∼200 nucleotides on average. As expected, both tethered and untethered XTUT7-C added uridines to reporter RNA (Fig. 3C). Therefore, the C-terminal half of XTUT7 uridylates RNAs independent of MS2 tethering, and the activity of XTUT7 is increased when tethered.
FIGURE 3.
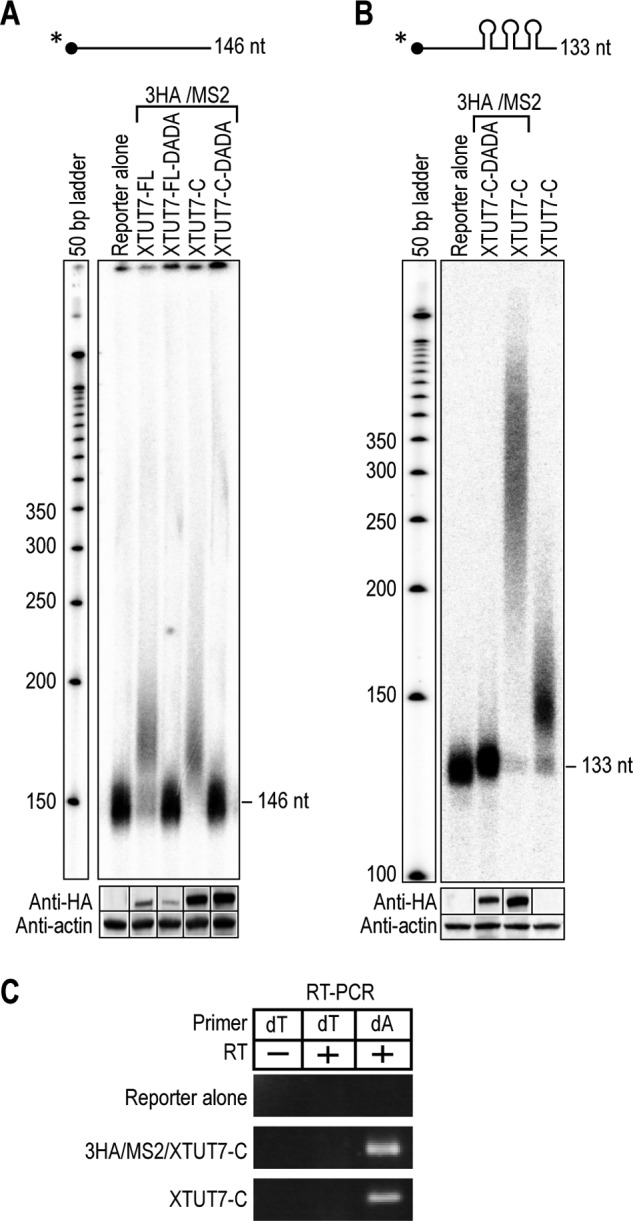
XTUT7 extends RNAs independent of MS2 tethering. A, the indicated 3HA/MS2/XTUT7 fusion proteins were assayed on a radiolabeled reporter RNA that lacked MS2-binding sites. The size of the unmodified reporter RNA is 146 nucleotides (146 nt). Diagrams of the XTUT7 proteins tested are depicted in Fig. 2B. The bottom panels indicate protein levels as determined by Western blotting for HA-tagged fusion proteins and actin. B, the indicated XTUT7 proteins were assayed on the radiolabeled reporter RNA with three MS2-binding sites. The size of the unmodified reporter RNA is 133 nucleotides (133 nt). The last XTUT7 protein tested lacks both the MS2 fusion protein and the 3HA tag. The bottom panels indicate protein levels as determined by Western blotting for HA-tagged fusion proteins and actin. C, RT-PCR assays were performed as in Fig. 2F for the indicated proteins. All samples were analyzed on the same gel but were separated for clarity. RT primers are indicated. The dT lane indicates the addition of adenosines, and the dA lane indicates the addition of uridines.
The BR and CCHC Zinc Finger Domains Mediate the Tethering-independent Activity of XTUT7
To examine the role of the BR and CCHC zinc finger domains in XTUT7, mutant enzymes were constructed in the context of the C-terminal half of the protein (Fig. 4A), which possesses the same rNTase activities as the full-length protein (Figs. 2 and 3). The mutant enzymes were first assayed on a radiolabeled reporter RNA that contained MS2-binding sites. Deletion of a single zinc finger (ΔZ1), all three zinc fingers (ΔZ123), or the BR (ΔBR) yielded nucleotide tail lengths similar to XTUT7-C (Fig. 4B). The XTUT7 enzyme that lacked both the zinc fingers and the BR (ΔZ123ΔBR) added many fewer nucleotides than wild-type XTUT7-C when expressed at comparable levels (Fig. 4D). Consequently, the BR and zinc finger domains likely act redundantly to contribute to the catalytic activity of XTUT7. However, the diminished activity also could result from a population of misfolded enzyme.
FIGURE 4.
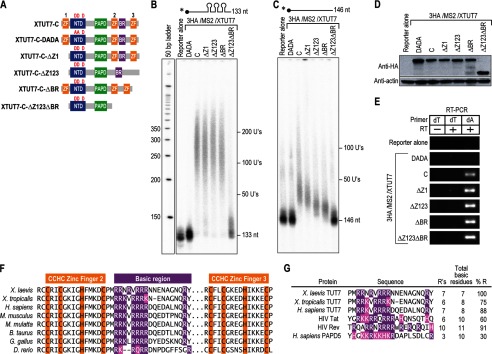
The BR and CCHC zinc finger domains mediate the tethering-independent activity of XTUT7. A, diagram of XTUT7 mutants tested. CCHC zinc fingers (ZF) 1, 2, and 3 are indicated. The red D's denote catalytic aspartates, whereas the red A's denote aspartate to alanine substitutions. B and C, indicated 3HA/MS2/XTUT7 fusion proteins were assayed in parallel on radiolabeled reporter RNA with (B) or without (C) three MS2-binding sites as in Fig. 2C. The sizes of the unmodified reporter RNAs are 133 and 146 nucleotides (nt), respectively. All proteins tested in panels B and C were assayed in parallel and analyzed on the same gel, which was subsequently split into two panels for clarity. D, protein levels for all samples tested in panels B and C as determined by Western blotting for HA fusion proteins and actin. E, RT-PCR assays were performed as in Fig. 2. All samples were analyzed on the same gel but were separated for clarity. RT primers are indicated. The dT lane indicates the addition of adenosines, and the dA lane indicates addition of uridines. F, multiple sequence alignment of the BR-containing regions from the indicated XTUT7 orthologs. Critical residues for defining CCHC zinc finger domains are highlighted in orange. The BR was defined by the above deletion experiments. Conserved arginines are highlighted in purple. The … in the alignments indicates a break in sequence. G, BRs from XTUT7 orthologs were compared with ARMs from viral RNA-binding proteins and a basic stretch from PAPD5. Purple, arginines; pink, lysines and histidines. The percentage of arginine composition of each region was also calculated as a comparison.
To further examine the role of the BR and CCHC zinc finger domains, the XTUT7 mutants were assayed on a reporter RNA that lacked MS2-binding sites. The mutant XTUT7 enzyme that lacked the zinc fingers and BR (ΔZ123ΔBR) was inactive on the RNA without binding sites (Fig. 4C). Mutant XTUT7 enzymes that lacked the BR (ΔBR) or the zinc fingers (ΔZ123) were less active than wild-type XTUT7-C when expressed at a comparable level (Fig. 4D). In addition, tethered XTUT7 mutants retained uridine specificity (Fig. 4E). Thus, the BR, as well as the CCHC zinc fingers, mediates the tethering-independent uridylation activity of XTUT7.
The BR is a conserved domain that may bind nucleic acids. The BR and CCHC zinc finger domains of XTUT7 are conserved among XTUT7 orthologs, including H. sapiens TUT7 (Fig. 4F). The BR resembles arginine-rich motifs (ARMs) found in viral RNA-binding proteins, such as HIV Rev (regulator of expression of virion proteins) and Tat (transactivator of transcription), as both the BR and the ARMs are composed primarily of arginine (Fig. 4G) (49–51). In contrast, a recently identified basic stretch of amino acids in PAPD5, a poly(A) polymerase related to XTUT7, is composed primarily of lysine (52, 53). Intriguingly, the ARMs in Rev and Tat, as well as the basic stretch in PAPD5, directly bind RNA (49–52).
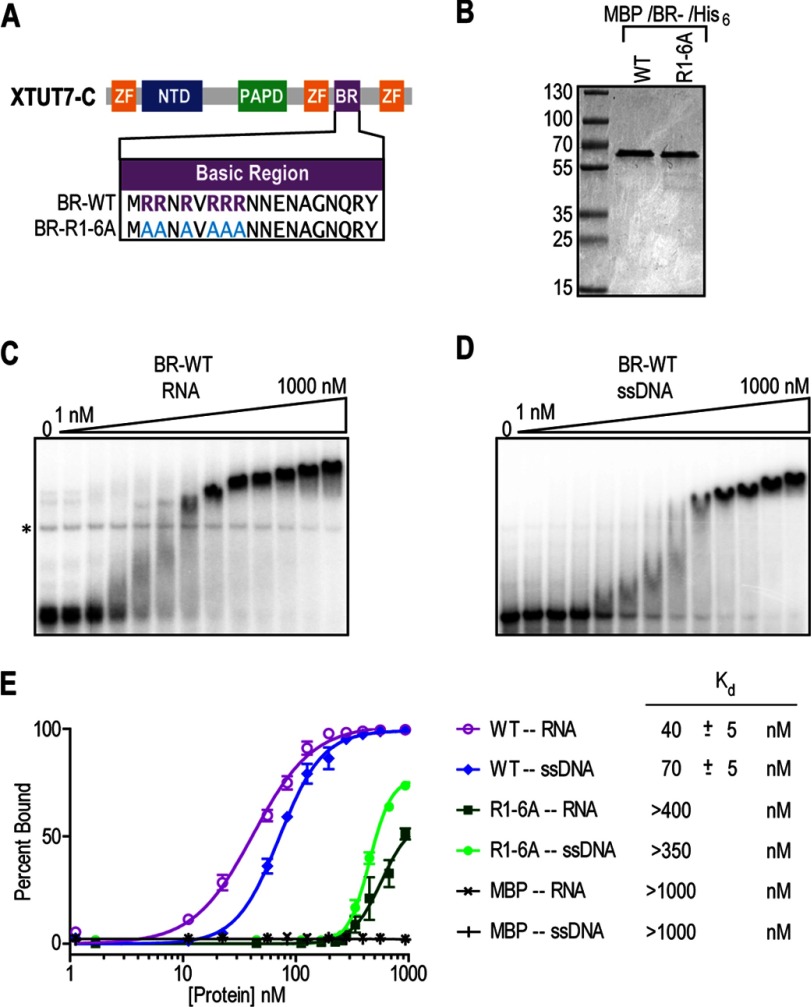
The BR Binds Nucleic Acids
To test whether the BR directly binds nucleic acids, wild-type (BR-WT) and mutant BR (BR-R1–6A) segments were fused to an MBP-His6 tag (Fig. 5A), purified (Fig. 5B), and tested using electrophoretic mobility shift assays. BR-WT bound an RNA that contained three MS2-binding sites in a concentration-dependent manner, with an apparent Kd of 40 ± 5 nm (Fig. 5, C and E). BR-WT also bound an ssDNA substrate of an equivalent sequence to the RNA substrate, with an apparent Kd of 70 ± 5 nm (Fig. 5, D and E). At the highest protein concentrations, the protein-nucleic acid complexes migrated progressively more slowly, which may indicate that multiple copies of BR-WT can bind the same nucleic acid molecule. A mutant BR in which arginines 1–6 had been changed to alanine (BR-R1–6A) bound the RNA and ssDNA substrates poorly, with estimated apparent Kd values of greater than 400 and 350 nm, respectively (Fig. 5E). Protein-RNA complexes were not observed with MBP-His6 backbone alone on either substrate (Fig. 5E). Therefore, the BR directly binds nucleic acids and requires conserved arginines for optimal binding.
FIGURE 5.
The BR binds nucleic acids in vitro. A, diagram of the XTUT7 BR. MBP/BR/His6 fusion proteins were recombinantly expressed, purified, and tested by EMSAs. The R1–6A mutant protein was created by making the indicated amino acid substitutions in the BR. ZF, zinc finger domain. B, protein gel stained with Coomassie Blue. Molecular mass ladder (in kDa) (MW) is indicated. The expected molecular mass of the proteins was ∼57 kDa. C, EMSA using the wild-type BR fusion protein and a radiolabeled RNA containing three MS2-binding sites. Protein concentrations ranged from 1 nm to 1 μm, as indicated above the gels. The * indicates an RNA artifact that is present even in the absence of proteins. D, EMSA using the wild-type BR fusion protein and a radiolabeled ssDNA of equivalent sequence to the RNA substrate. Protein concentrations ranged from 1 nm to 1 μm, as indicated above the gels. E, the average percentage of nucleic acid substrate bound at each protein concentration was calculated and plotted using nonlinear regression analysis from three experiments. The apparent Kd values for fusion protein and nucleic acid substrate pairs are indicated to the right of the plot and reported with associated S.E.
XTUT7 Represses a Polyadenylated RNA
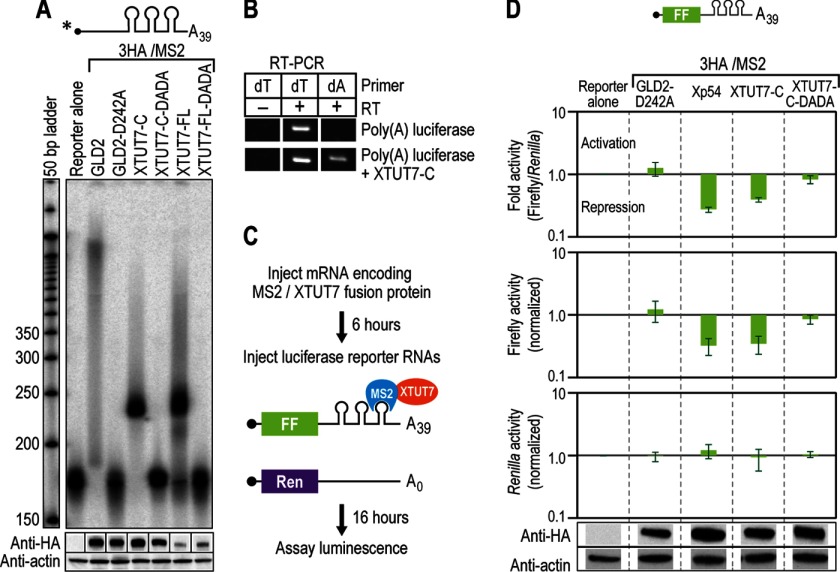
To examine the rNTase activity of XTUT7 on polyadenylated RNA, we tethered XTUT7 to an RNA with a poly(A)39 tail. XTUT7-FL and XTUT7-C extended the polyadenylated reporter RNA by a distinct number of nucleotides, which on average was 60 ± 10 nucleotides (Fig. 6A). RT-PCR of the polyadenylated RNAs extended by XTUT7-C yielded products with both oligo(dT) and oligo(dA) primers, whereas the substrate yielded a single band in the oligo(dT) lane (Fig. 6B). XTUT7 therefore added a poly(U) tail of discrete length to an RNA substrate with 39 adenosines.
FIGURE 6.
XTUT7 represses a polyadenylated RNA. A, 3HA/MS2 fusion proteins were assayed as in Fig. 2C, except that the reporter RNA contained poly(A)39 on the 3′ end. The bottom panels indicate protein levels as determined by Western blotting for HA-tagged fusion proteins and actin. B, samples were assayed by RT-PCR as in Fig. 2F, except the RNA contained poly(A)39 on the 3′ end. All samples were analyzed on the same gel but isolated for clarity. C, schematic of the assay used to determine the effect of XTUT7 on translation. RNAs were injected as in Fig. 2A, except that two nonradiolabeled reporter RNAs were co-injected. The first contained the firefly luciferase open reading frame (FF) upstream of three MS2-binding sites and a poly(A)39 tail. The second contained the Renilla luciferase open reading frame (Ren) that lacked MS2-binding sites and a poly(A) tail. After 16 h, luciferase levels were determined. D, relative luciferase levels in oocytes that express the indicated fusion proteins were determined. Luciferase levels were normalized to reporter alone samples (no fusion protein) in each panel. Error bars represent the S.D. from three experiments. The bottom and middle panels represent the average relative Renilla or firefly luciferase levels, respectively. The top panel represents the average firefly/Renilla luciferase levels. Protein levels from a representative experiment are depicted below the luciferase data, and all bands are from the same Western blot but were separated for clarity.
To assess the effect of XTUT7 on translation, XTUT7 was tethered to a poly(A)39 firefly luciferase mRNA (Fig. 6C). As a control, we co-injected Renilla luciferase mRNA that lacked MS2-binding sites and a poly(A) tail. Tethered XTUT7 specifically reduced firefly luciferase activity nearly 3-fold (p value < 0.001), much like the characterized translational repressor Xp54 (Fig. 6D, top and middle panels) (54). XTUT7-dependent repression was specific to firefly luciferase as all proteins tested had no significant effect on Renilla luciferase luminescence (Fig. 6D, bottom panel). XTUT7-C-DADA, which lacks catalytic activity, yielded firefly luciferase levels similar to that of the control GLD2-D242A and was expressed similarly to XTUT7-C. All proteins tested had no significant effect on firefly or Renilla luciferase mRNA levels as determined by quantitative PCR. Thus, XTUT7 represses translation of polyadenylated reporter mRNA by adding uridines to the RNA.
Histidine 1269 Is Important for the Uridine Specificity and Repression Activity of XTUT7
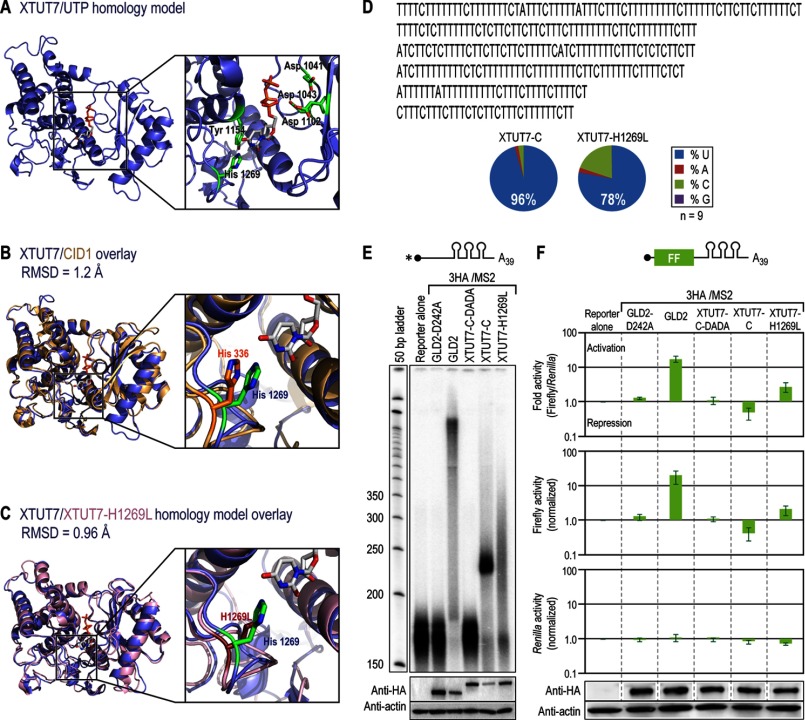
To visualize the likely structure of the active site region, the three-dimensional structure of the XTUT7 catalytic core was predicted using the I-TASSER server (55–57). This analysis yielded ligand-free and ligand-bound homology models of XTUT7 (C-score = 1.02, expected r.m.s.d. = 4.3 ± 2.9 Å) (Fig. 7A). In the model, putative catalytic aspartates 1041, 1043, and 1102 are adjacent to the triphosphate moiety of UTP (Fig. 7A). Tyrosine 1154 appears to participate in a stacking interaction with uracil. Histidine 1269, contained in the NRM of XTUT7, is predicted to contact a carbonyl oxygen in UTP. As expected, the predicted XTUT7 structure aligns well to a structure of the S. pombe poly(U)-adding enzyme CID1 (alignment r.m.s.d. = 1.2 Å, sequence identity to XTUT7 is 32%) (Fig. 7B) (30). Intriguingly, a hydrogen bond that is observed in CID1 between His-336 and UTP is predicted in XTUT7 (His-1269).
FIGURE 7.
Histidine 1269 is important for the uridine specificity and repression activity of XTUT7. A, The predicted structure of the XTUT7 core catalytic domains bound to UTP (XTUT7/UTP) as generated by the I-TASSER server (55–57). The XTUT7 homology model had a C-score of 1.02, an expected TM-score of 0.85 ± 0.08, and an expected r.m.s.d. of 4.3 ± 2.9 Å. Asp-1041, Asp-1043, Asp-1102, Tyr-1154, and His-1269 are highlighted in green. The UTP molecule is gray. B, alignment of the predicted XTUT7 structure (blue) with a described S. pombe CID1 structure (gold/orange, Protein Data Bank (PDB) ID 4E8F) (r.m.s.d. = 1.2 Å) (30). Analogous histidines and their position relative to UTP are indicated. C, alignment of the predicted wild-type XTUT7 structure (blue) to the predicted XTUT7-H1269L mutant structure (pink) (r.m.s.d. = 0.96 Å). The XTUT7-H1269L homology model had a C-score of 0.99, an expected TM-score of 0.85 ± 0.08, and an expected r.m.s.d. of 4.4 ± 2.9 Å. The position of the histidine and the H1269L substitution relative to UTP are indicated. D, the nucleotides added by XTUT7-H1269L were determined as in Fig. 2G. Six representative, independently cloned sequences that illustrate the nucleotides added by XTUT7-H1269L are shown. The average percentages of nucleotide composition of nine representative cloned tails added by XTUT7-C and XTUT7-H1269L are indicated. E, indicated proteins were assayed using the poly(A)39 radiolabeled reporter RNA as in Fig. 6A. The bottom panels indicate protein levels as determined by Western blotting for HA fusion proteins and actin. F, relative luciferase levels were determined as in Fig. 6D. Error bars represent the S.D. from three experiments. The bottom and middle panels represent the average relative Renilla or firefly luciferase levels, respectively. The top panel represents the average firefly/Renilla luciferase levels. Protein levels from a representative experiment are depicted below the luciferase data. All bands are from the same Western blot but were separated for clarity. FF, firefly luciferase open reading frame.
We reasoned that His-1269 might be important for nucleotide specificity due to its proximity to UTP. Therefore, we substituted His-1269 with leucine in the context of the XTUT7 C-terminal half (XTUT7-H1269L) because this substitution ablates a potential hydrogen bond to UTP (Fig. 7C). Indeed, tethered XTUT7-H1269L added cytosines, as well as uridines, to RNA (Fig. 7D). XTUT7-H1269L-dependent tails were ∼20% cytosine as compared with ∼3% with the wild-type enzyme (p values < 0.005). Both XTUT7-H1269L and XTUT7-C rarely added guanosines or adenosines to RNA (< 3 and 2%, respectively). Thus, His-1269 is important for the uridine specificity of XTUT7.
To test whether altered uridine specificity affected the catalytic activity of XTUT7, XTUT7-H1269L was tethered to a polyadenylated reporter RNA. XTUT7-H1269L added a heterogeneous length tail to an RNA with a poly(A)39 tail, rather than the discrete ∼60-nucleotide tail added by XTUT7-C (Fig. 7E). The tail added by XTUT7-H1269L was between ∼50 and 150 nucleotides in length, which was shorter than the tail added by the wild-type enzyme to RNA that lacked a poly(A) tail. Accordingly, incorporation of non-uridine residues by XTUT7 prevents formation of the discrete length tail on the poly(A)39 reporter RNA.
To determine the effect of XTUT7-H1269L on translational repression, XTUT7-H1269L was assayed using poly(A)39 firefly luciferase mRNA. XTUT7-H1269L not only prevented translational repression, but instead activated it ∼3-fold (p value < 0.05) (Fig. 7F). This increase in firefly luciferase activity was less than the increase yielded by the poly(A) polymerase GLD2. Firefly and Renilla luciferase mRNA levels were not significantly affected by any protein tested. Thus, the H1269L substitution alleviated XTUT7-dependent translational repression, which could result from either the relaxed nucleotide specificity of the mutant enzyme or its addition of a heterogeneous length tail to poly(A)39 firefly luciferase mRNA.
DISCUSSION
The C-terminal Half of XTUT7 Is Sufficient for rNTase Activity
Our studies demonstrate that XTUT7 is a poly(U)-adding enzyme and identify key domains and residues important for activity. The NTD and PAPD of XTUT7 were sufficient for activity, and mutations in the NTD of full-length XTUT7 abolished rNTase activity. Therefore, the NTD and PAPD are the core catalytic domains. The BR and CCHC zinc finger domains flanking the catalytic core likely enable efficient XTUT7-dependent uridylation as their removal decreased XTUT7 rNTase activity. In contrast, the conserved NTD* and PAPD* of XTUT7 lacked activity and were dispensable for it in the context of the full-length protein. The analogous domains in XTUT4 were also dispensable for catalytic activity. Thus, the C-terminal half of XTUT7 is sufficient for rNTase activity.
Recent work on the mammalian TUT7 ortholog, TUT4, suggested that its NTD* was necessary for rNTase activity and therefore contrasts with our finding that the analogous domain of XTUT7 was dispensable (16). The NTD* of TUT4 was required for the enzyme to uridylate synthetic pre-let-7 RNA both in the presence and in the absence of LIN28, which is thought to recruit TUT4 to its pre-let-7 RNA target. Despite the high degree of similarity between Xenopus and human TUTs (77% similar), their individual domains could function differently. However, we note that in our assays, XTUT7 is tethered to RNA, which makes the assay more sensitive. Our assays are independent of LIN28 as nearly all of the XTUT7 proteins we tested lacked the C2H2 zinc finger domain needed for LIN28-dependent uridylation, likely through protein-protein contacts (16). Although the NTD* and PAPD* of XTUT7 orthologs are dispensable for catalytic activity, they nonetheless may have critical roles in vivo. For example, they may mediate protein-protein interactions, as suggested by LIN28-pre-let-7-TUT4 experiments (16). Indeed, TUT4 segments that contain its NTD* and PAPD* promote cell proliferation independent of catalytic activity (58).
The XTUT7 Basic Region
XTUT7 contains nucleic acid binding-domains, including the arginine-rich BR. XTUT7 possesses tethering-independent rNTase activity redundantly mediated by its BR and CCHC zinc finger domains. These domains are likely required for efficient catalytic activity when XTUT7 is tethered to RNA. Together, these findings suggest that the BR and at least one of the CCHC zinc finger domains bind RNA. Indeed, we show that the BR binds both RNA and ssDNA in vitro. Given the modest preference of the BR for binding RNA and that rNTases lack catalytic activity on DNA substrates (59), we suggest that the BR binds RNA in vivo.
The XTUT7 BR resembles RNA-binding domains present in certain viral proteins, such as the ARM found in HIV Rev (49, 60). ARMs are flexible RNA-binding domains that typically confer specificity for particular RNAs by recognizing RNA sequences and/or structures (61, 62). For example, the ARM in Rev specifically recognizes and binds its RNA target (63–66). Critical arginines in the ARM make base-specific contacts with the RNA and are necessary for binding. Similarly, the XTUT7 BR requires highly conserved arginines for optimal RNA binding activity.
BRs are present in other rNTases. Human PAPD5, a noncanonical poly(A) polymerase, binds a subset of RNAs likely through a small, lysine-rich stretch of amino acids (52). The basic stretch of PAPD5 is also required for efficient catalytic activity, much like the BR in XTUT7. A search for similar BRs in human rNTases reveals that five of the seven enzymes contain characterized or putative BRs, including TUT7, TUT4, and PAPD5. Thus, BRs appear to be a common feature of rNTases that are likely utilized to bind RNA substrates and/or facilitate catalytic activity.
We speculate that the BR and CCHC zinc fingers facilitate TUT7 binding to particular RNAs in vivo. TUT7 orthologs uridylate pre-let-7 in the absence of LIN28 both in vitro and in vivo, and this activity requires the pre-let-7 stem (16, 17, 67). We therefore propose that the BR of TUT7, likely in cooperation with the zinc fingers, binds the accessible region of the pre-let-7 stem.
XTUT7 Homology Model and Nucleotide Specificity
We generated a homology model of the three-dimensional structure of the XTUT7 catalytic core that identified an amino acid important for uridine specificity. Not surprisingly, the predicted structure of XTUT7 is similar to those of other poly(U)-adding enzymes, particularly CID1, and predicted that a histidine would be important for uridine specificity (30). The analogous histidine in CID1 is required for optimal uridine specificity in vitro (30–32). Indeed, substituting His-1269 with leucine broadened the nucleotide specificity of XTUT7 in vivo so that it added both uridines and cytosines. These data suggest that a histidine-UTP contact is a critical determinant for XTUT7 uridine specificity and likely represents a common mechanism of uridine recognition among XTUT7 orthologs. Direct determination of the XTUT7 structure is needed to test this rigorously.
XTUT7 and Translational Control
XTUT7 can repress translation and may represent a new class of translational repressor proteins. XTUT7 repressed translation of a polyadenylated reporter mRNA without affecting mRNA stability. We propose that the U-tail added by XTUT7 binds poly(A). The poly(A)-poly(U) hybrid may block recognition of the poly(A) tail by poly(A)-dependent factors, such as poly(A)-binding protein. This would mask the effects of the poly(A) tail, including its ability to stimulate translation. The presence of the A-U duplex is consistent with the observation that an XTUT7 mutant that added cytosines no longer repressed translation. Furthermore, the mutant enzyme also produced a heterogeneous length tail, whereas the wild-type enzyme added a discrete number of uridines to an RNA with 39 adenosines. We infer that the newly formed A-U hybrid prevents further catalysis, implying a novel mechanism that terminates poly(U) synthesis. Although RNAs with repressive A-U hybrid tails have yet to be discovered en masse in cells, they may well exist. For instance, the mammalian poly(U)-adding enzymes TUT7 and TUT4 associate with polyadenylated RNAs, and polyadenylated mRNAs in S. pombe are uridylated in vivo (6, 8, 68, 69). Thus, XTUT7 may have unanticipated roles in the regulation of mRNAs, in addition to its activities in miRNA control.
Supplementary Material
Acknowledgments
We thank members of the Wickens laboratory for helpful discussions, suggestions, and critical reading of the manuscript. We are grateful to Phil Anderson and Scott Kennedy for advice on experiments. We also thank Laura Vanderploeg and the Biochemistry Media Laboratory for help with figures.
This work was supported, in whole or in part, by National Institutes of Health Grants GM31892 and GM50942 (to M. W.).

This article contains supplemental Table 1.
- miRNA
- microRNA
- TUT
- terminal uridylyl transferase
- TUT7
- either mammalian (species indicated) or generic TUT7 enzymes
- XTUT7
- Xenopus TUT7
- rNTase
- ribonucleotidyl transferase
- NTD
- nucleotidyl transferase domain
- PAPD
- poly(A) polymerase-associated domain
- NTD*
- cryptic nucleotidyl transferase domain
- PAPD*
- cryptic poly(A) polymerase-associated domain
- NRM
- nucleotide recognition motif
- BR
- basic region
- ARM
- arginine-rich motif
- MBP
- maltose-binding protein
- r.m.s.d.
- root mean square deviation
- pre-let-7
- let-7 precursor.
REFERENCES
- 1. Wickens M., Kwak J. E. (2008) Molecular biology. A tail tale for U. Science 319, 1344–1345 [DOI] [PubMed] [Google Scholar]
- 2. Norbury C. J. (2010) 3′ Uridylation and the regulation of RNA function in the cytoplasm. Biochem. Soc. Trans. 38, 1150–1153 [DOI] [PubMed] [Google Scholar]
- 3. Schmidt M. J., Norbury C. J. (2010) Polyadenylation and beyond: emerging roles for noncanonical poly(A) polymerases. Wiley Interdiscip. Rev. RNA 1, 142–151 [DOI] [PubMed] [Google Scholar]
- 4. Kim Y. K., Heo I., Kim V. N. (2010) Modifications of small RNAs and their associated proteins. Cell 143, 703–709 [DOI] [PubMed] [Google Scholar]
- 5. Song M. G., Kiledjian M. (2007) 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA 13, 2356–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rissland O. S., Mikulasova A., Norbury C. J. (2007) Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol. Cell. Biol. 27, 3612–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mullen T. E., Marzluff W. F. (2008) Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 22, 50–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rissland O. S., Norbury C. J. (2009) Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat. Struct. Mol. Biol. 16, 616–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schmidt M. J., West S., Norbury C. J. (2011) The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA 17, 39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Su W., Slepenkov S. V., Slevin M. K., Lyons S. M., Ziemniak M., Kowalska J., Darzynkiewicz E., Jemielity J., Marzluff W. F., Rhoads R. E. (2013) mRNAs containing the histone 3′ stem-loop are degraded primarily by decapping mediated by oligouridylation of the 3′ end. RNA 19, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoefig K. P., Rath N., Heinz G. A., Wolf C., Dameris J., Schepers A., Kremmer E., Ansel K. M., Heissmeyer V. (2013) Eri1 degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat. Struct. Mol. Biol. 20, 73–81 [DOI] [PubMed] [Google Scholar]
- 12. Heo I., Joo C., Cho J., Ha M., Han J., Kim V. N. (2008) Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol. Cell 32, 276–284 [DOI] [PubMed] [Google Scholar]
- 13. Lehrbach N. J., Armisen J., Lightfoot H. L., Murfitt K. J., Bugaut A., Balasubramanian S., Miska E. A. (2009) LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 16, 1016–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hagan J. P., Piskounova E., Gregory R. I. (2009) Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 16, 1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heo I., Joo C., Kim Y. K., Ha M., Yoon M. J., Cho J., Yeom K. H., Han J., Kim V. N. (2009) TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138, 696–708 [DOI] [PubMed] [Google Scholar]
- 16. Thornton J. E., Chang H. M., Piskounova E., Gregory R. I. (2012) Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA 18, 1875–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heo I., Ha M., Lim J., Yoon M. J., Park J. E., Kwon S. C., Chang H., Kim V. N. (2012) Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell 151, 521–532 [DOI] [PubMed] [Google Scholar]
- 18. Newman M. A., Mani V., Hammond S. M. (2011) Deep sequencing of microRNA precursors reveals extensive 3′ end modification. RNA 17, 1795–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van Wolfswinkel J. C., Claycomb J. M., Batista P. J., Mello C. C., Berezikov E., Ketting R. F. (2009) CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 139, 135–148 [DOI] [PubMed] [Google Scholar]
- 20. Ibrahim F., Rymarquis L. A., Kim E. J., Becker J., Balassa E., Green P. J., Cerutti H. (2010) Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 107, 3906–3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ren G., Chen X., Yu B. (2012) Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Curr. Biol. 22, 695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao Y., Yu Y., Zhai J., Ramachandran V., Dinh T. T., Meyers B. C., Mo B., Chen X. (2012) The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr. Biol. 22, 689–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones M. R., Quinton L. J., Blahna M. T., Neilson J. R., Fu S., Ivanov A. R., Wolf D. A., Mizgerd J. P. (2009) Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat. Cell Biol. 11, 1157–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burroughs A. M., Ando Y., de Hoon M. J., Tomaru Y., Nishibu T., Ukekawa R., Funakoshi T., Kurokawa T., Suzuki H., Hayashizaki Y., Daub C. O. (2010) A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 20, 1398–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wyman S. K., Knouf E. C., Parkin R. K., Fritz B. R., Lin D. W., Dennis L. M., Krouse M. A., Webster P. J., Tewari M. (2011) Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res. 21, 1450–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jones M. R., Blahna M. T., Kozlowski E., Matsuura K. Y., Ferrari J. D., Morris S. A., Powers J. T., Daley G. Q., Quinton L. J., Mizgerd J. P. (2012) Zcchc11 Uridylates Mature miRNAs to Enhance Neonatal IGF-1 Expression, Growth, and Survival. PLoS Genet. 8, e1003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aphasizhev R. (2005) RNA uridylyltransferases. Cell. Mol. Life Sci. 62, 2194–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martin G., Keller W. (2007) RNA-specific ribonucleotidyl transferases. RNA 13, 1834–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kwak J. E., Wickens M. (2007) A family of poly(U) polymerases. RNA 13, 860–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yates L. A., Fleurdépine S., Rissland O. S., De Colibus L., Harlos K., Norbury C. J., Gilbert R. J. (2012) Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase. Nat. Struct. Mol. Biol. 19, 782–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lunde B. M., Magler I., Meinhart A. (2012) Crystal structures of the Cid1 poly(U) polymerase reveal the mechanism for UTP selectivity. Nucleic Acids Res. 40, 9815–9824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Munoz-Tello P., Gabus C., Thore S. (2012) Functional implications from the Cid1 poly(U) polymerase crystal structure. Structure 20, 977–986 [DOI] [PubMed] [Google Scholar]
- 33. Martin G., Doublié S., Keller W. (2008) Determinants of substrate specificity in RNA-dependent nucleotidyl transferases. Biochim. Biophys Acta 1779, 206–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roush S., Slack F. J. (2008) The let-7 family of microRNAs. Trends Cell Biol. 18, 505–516 [DOI] [PubMed] [Google Scholar]
- 35. Cooke A., Prigge A., Wickens M. (2010) Translational repression by deadenylases. J. Biol. Chem. 285, 28506–28513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zdobnov E. M., Apweiler R. (2001) InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17, 847–848 [DOI] [PubMed] [Google Scholar]
- 37. Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Finn R. D., Mistry J., Tate J., Coggill P., Heger A., Pollington J. E., Gavin O. L., Gunasekaran P., Ceric G., Forslund K., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A. (2010) The Pfam protein families database. Nucleic Acids Res. 38, D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gray N. K., Coller J. M., Dickson K. S., Wickens M. (2000) Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 19, 4723–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chritton J. J., Wickens M. (2010) Translational repression by PUF proteins in vitro. RNA 16, 1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cooke A., Prigge A., Opperman L., Wickens M. (2011) Targeted translational regulation using the PUF protein family scaffold. Proc. Natl. Acad. Sci. U.S.A. 108, 15870–15875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kwak J. E., Wang L., Ballantyne S., Kimble J., Wickens M. (2004) Mammalian GLD-2 homologs are poly(A) polymerases. Proc. Natl. Acad. Sci. U.S.A. 101, 4407–4412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Charlesworth A., Cox L. L., MacNicol A. M. (2004) Cytoplasmic polyadenylation element (CPE)- and CPE-binding protein (CPEB)-independent mechanisms regulate early class maternal mRNA translational activation in Xenopus oocytes. J. Biol. Chem. 279, 17650–17659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hook B. A., Goldstrohm A. C., Seay D. J., Wickens M. (2007) Two yeast PUF proteins negatively regulate a single mRNA. J. Biol. Chem. 282, 15430–15438 [DOI] [PubMed] [Google Scholar]
- 47. Wang L., Eckmann C. R., Kadyk L. C., Wickens M., Kimble J. (2002) A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419, 312–316 [DOI] [PubMed] [Google Scholar]
- 48. Rouhana L., Wang L., Buter N., Kwak J. E., Schiltz C. A., Gonzalez T., Kelley A. E., Landry C. F., Wickens M. (2005) Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA 11, 1117–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lazinski D., Grzadzielska E., Das A. (1989) Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell 59, 207–218 [DOI] [PubMed] [Google Scholar]
- 50. Weeks K. M., Ampe C., Schultz S. C., Steitz T. A., Crothers D. M. (1990) Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science 249, 1281–1285 [DOI] [PubMed] [Google Scholar]
- 51. Calnan B. J., Tidor B., Biancalana S., Hudson D., Frankel A. D. (1991) Arginine-mediated RNA recognition: the arginine fork. Science 252, 1167–1171 [DOI] [PubMed] [Google Scholar]
- 52. Rammelt C., Bilen B., Zavolan M., Keller W. (2011) PAPD5, a noncanonical poly(A) polymerase with an unusual RNA-binding motif. RNA 17, 1737–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Berndt H., Harnisch C., Rammelt C., Stöhr N., Zirkel A., Dohm J. C., Himmelbauer H., Tavanez J. P., Hüttelmaier S., Wahle E. (2012) Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. RNA 18, 958–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Minshall N., Thom G., Standart N. (2001) A conserved role of a DEAD box helicase in mRNA masking. RNA 7, 1728–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roy A., Yang J., Zhang Y. (2012) COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res. 40, W471–W477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Blahna M. T., Jones M. R., Quinton L. J., Matsuura K. Y., Mizgerd J. P. (2011) Terminal uridyltransferase enzyme Zcchc11 promotes cell proliferation independent of its uridyltransferase activity. J. Biol. Chem. 286, 42381–42389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Read R. L., Martinho R. G., Wang S. W., Carr A. M., Norbury C. J. (2002) Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc. Natl. Acad. Sci. U.S.A. 99, 12079–12084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tan R., Chen L., Buettner J. A., Hudson D., Frankel A. D. (1993) RNA recognition by an isolated α helix. Cell 73, 1031–1040 [DOI] [PubMed] [Google Scholar]
- 61. Weiss M. A., Narayana N. (1998) RNA recognition by arginine-rich peptide motifs. Biopolymers 48, 167–180 [DOI] [PubMed] [Google Scholar]
- 62. Bayer T. S., Booth L. N., Knudsen S. M., Ellington A. D. (2005) Arginine-rich motifs present multiple interfaces for specific binding by RNA. RNA 11, 1848–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Daugherty M. D., Liu B., Frankel A. D. (2010) Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat. Struct. Mol. Biol. 17, 1337–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Olsen H. S., Nelbock P., Cochrane A. W., Rosen C. A. (1990) Secondary structure is the major determinant for interaction of HIV rev protein with RNA. Science 247, 845–848 [DOI] [PubMed] [Google Scholar]
- 65. Daly T. J., Cook K. S., Gray G. S., Maione T. E., Rusche J. R. (1989) Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature 342, 816–819 [DOI] [PubMed] [Google Scholar]
- 66. Battiste J. L., Mao H., Rao N. S., Tan R., Muhandiram D. R., Kay L. E., Frankel A. D., Williamson J. R. (1996) α helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex. Science 273, 1547–1551 [DOI] [PubMed] [Google Scholar]
- 67. Yeom K. H., Heo I., Lee J., Hohng S., Kim V. N., Joo C. (2011) Single-molecule approach to immunoprecipitated protein complexes: insights into miRNA uridylation. EMBO Rep 12, 690–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Baltz A. G., Munschauer M., Schwanhäusser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M., Wyler E., Bonneau R., Selbach M., Dieterich C., Landthaler M. (2012) The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690 [DOI] [PubMed] [Google Scholar]
- 69. Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B. M., Strein C., Davey N. E., Humphreys D. T., Preiss T., Steinmetz L. M., Krijgsveld J., Hentze M. W. (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.