Background: The human glucose transporter GLUT1 forms homo-oligomers but does not hetero-oligomerize with the neuronal transporter GLUT3.
Results: GLUT3 transmembrane helix 9 substitution with GLUT1 helix 9 promotes GLUT1-GLUT3 association.
Conclusion: GLUT1 and GLUT3 oligomeric states and transport activities are determined by transmembrane helix 9 sequence.
Significance: The activity of some multisubunit transporter complexes is determined by their quaternary structure.
Keywords: Erythrocyte, Glucose Transport, Membrane Proteins, Membrane Transport, Neurons
Abstract
The human blood-brain barrier glucose transport protein (GLUT1) forms homodimers and homotetramers in detergent micelles and in cell membranes, where the GLUT1 oligomeric state determines GLUT1 transport behavior. GLUT1 and the neuronal glucose transporter GLUT3 do not form heterocomplexes in human embryonic kidney 293 (HEK293) cells as judged by co-immunoprecipitation assays. Using homology-scanning mutagenesis in which GLUT1 domains are substituted with equivalent GLUT3 domains and vice versa, we show that GLUT1 transmembrane helix 9 (TM9) is necessary for optimal association of GLUT1-GLUT3 chimeras with parental GLUT1 in HEK cells. GLUT1 TMs 2, 5, 8, and 11 also contribute to a less abundant heterocomplex. Cell surface GLUT1 and GLUT3 containing GLUT1 TM9 are 4-fold more catalytically active than GLUT3 and GLUT1 containing GLUT3 TM9. GLUT1 and GLUT3 display allosteric transport behavior. Size exclusion chromatography of detergent solubilized, purified GLUT1 resolves GLUT1/lipid/detergent micelles as 6- and 10-nm Stokes radius particles, which correspond to GLUT1 dimers and tetramers, respectively. Studies with GLUTs expressed in and solubilized from HEK cells show that HEK cell GLUT1 resolves as 6- and 10-nm Stokes radius particles, whereas GLUT3 resolves as a 6-nm particle. Substitution of GLUT3 TM9 with GLUT1 TM9 causes chimeric GLUT3 to resolve as 6- and 10-nm Stokes radius particles. Substitution of GLUT1 TM9 with GLUT3 TM9 causes chimeric GLUT1 to resolve as a mixture of 6- and 4-nm particles. We discuss these findings in the context of determinants of GLUT oligomeric structure and transport function.
Introduction
Sugars are transported across cell membranes by a family of facilitative transport proteins called glucose transporters (GLUTs)3 (1). Fourteen human GLUTs have been identified (2). Where studied, each has a specific tissue distribution and substrate specificity (2), but all are members of the wider family of major facilitator superfamily proteins (3).
Erythrocyte, smooth muscle, astrocyte, and blood-tissue barrier endothelial cell glucose transport are catalyzed by GLUT1 (4–6). GLUT1 comprises 492 amino acids, contains a single exofacial, N-linked glycosylation site, and is predominantly α-helical (4, 7, 8). Hydropathy analysis, scanning glycosylation mutagenesis, limited proteolysis, antibody binding, and covalent modification studies indicate that GLUT1 contains 12 membrane-spanning α-helices (TMs), intracellular N and C termini, and a large cytoplasmic loop connecting TM6 and -7 (4, 9, 10). High resolution crystal structures of bacterial major facilitator superfamily proteins GlpT, LacY, EmrD, and XylE (11–14) are consistent with GLUT1 biochemistry and indicate that the sugar translocation pathway is formed by eight amphipathic, membrane-spanning α-helices (TM1, -2, -4, -5, -7, -8, -10, and -11) coordinated by a scaffold of four hydrophobic α-helices (TM3, -6, -9, and -12). The results of GLUT1 cysteine-scanning mutagenesis studies are largely consistent with this hypothesis (15–18).
Biophysical studies indicate that GLUT1 forms non-covalent homodimers and tetramers in the plasma membrane (19, 20). Although individual GLUT1 subunits are not disulfide-linked, GLUT1 thiol chemistry contributes to GLUT1 oligomerization (21, 22). GLUT1 cysteine residues 347 and 421 are inaccessible to alkylating reagents in membrane-resident, non-reduced, tetrameric GLUT1, and their mutagenesis or exposure to reductant disrupts GLUT1 quaternary structure (21). The ligand binding and transport inhibition behaviors of tetrameric GLUT1 indicate that the transport complex simultaneously presents at least two exofacial and two endofacial substrate/ligand binding sites (22–26). Dimeric (reduced) GLUT1 behaves as a dimer of independent alternating conformer carriers (22, 27, 28) in which each monomer presents only an exofacial or endofacial substrate binding site at any instant. These observations are consistent with the hypothesis that each GLUT1 monomer functions as a simple carrier that alternates between exofacial and endofacial orientations (29, 30). In dimeric GLUT1, subunits are structurally associated but functionally independent. In tetrameric GLUT1, cooperative interactions constrain conformational changes such that at any instant, two subunits must present exofacial substrate binding sites and two subunits present endofacial binding sites.
GLUT3 is the major glucose transporter in neurons (31). Little is known regarding GLUT3 quaternary structure. However, studies by Burant and Bell (32) suggest that if GLUT3 forms dimers or tetramers, the activity of each subunit is largely independent of its neighbor(s). GLUT1 shares 65–66% sequence identity with GLUT3 and the insulin-sensitive transporter GLUT4 but does not form heterocomplexes with these transporters when co-expressed in 3T3-L1 adipocytes or in Xenopus oocytes (32, 33).
In this study, we provide evidence consistent with the hypothesis that GLUT3 forms dimers, whereas GLUT1 forms dimers and tetramers in human embryonic kidney (HEK) cell membranes. GLUT1 and GLUT3 do not form heterocomplexes in HEK cell membranes. We exploit this behavior and the use of GLUT1-GLUT3 chimeras to systematically investigate and identify the sequence determinants of GLUT1 oligomerization and function. We identify transmembrane domain 9 (TM9) of GLUT1 as the major determinant of tetramerization. Our results further suggest that GLUT1 TM2, -5, -8, and -11 contribute to GLUT1 dimerization. Our previous studies suggested that only tetrameric GLUT1 shows cooperative ligand binding behavior. We now show that GLUT3 presents cooperative behavior, suggesting that cooperative interactions can also occur in dimeric GLUTs.
EXPERIMENTAL PROCEDURES
Materials
A 2.6-kbp BamHI/XbaI fragment of human GLUT3 cDNA packaged in the pBluescript vector was supplied by Dr. Graeme Bell (University of Chicago). Protease inhibitor was from Roche Applied Science. Protein A-SepharoseTM CL-4B was obtained from GE Healthcare. Supersignal chemiluminescence kits, EZ-Link Sulfo-NHS-SS-Biotin, and one-step 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) HRP substrate were from Pierce. Nitrocellulose and Immobilon-P were from Fisher. All other reagents were from Sigma-Aldrich.
Solutions
Hepes-KCl comprised 150 mm KCl, 5 mm HEPES, 4 mm EGTA, 5 mm MgCl2, pH 7.4. Solubilization buffer contained Hepes-KCl with 0.5% Triton X-100. Phosphate-buffered saline (PBS) comprised 140 mm NaCl, 10 mm Na2HPO4, 3.4 mm KCl, 1.84 mm KH2PO4, pH 7.3. PBS with Tween 20 (PBS-T) comprised PBS plus 0.1% Tween. Stop solution comprised ice-cold Hepes-KCl plus cytochalasin B (CB) (10 μm) and phloretin (100 μm). Sample buffer (2×) contained 0.125 m Tris-HCl (pH 6.8), 4% SDS, 20% glycerol, and 50 mm DTT. Phosphate buffer (PB) consisted of 20 mm NaH2P04 and 145 mm NaCl, pH 7.4.
Cell Culture
HEK293 and COS cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in a 37 °C humidified 5% CO2 incubator.
GLUT1 Purification
Tetrameric and dimeric GLUT1 were purified from human red blood cells as described previously (21).
Antibodies
An affinity-purified rabbit polyclonal antibody (G1-Ab) raised against a peptide corresponding to GLUT1 C-terminal residues 480–492 was used as described previously (34). Rabbit and goat antisera (G4-Ab) recognizing the C-terminal 13 and 24 amino acids of GLUT4, respectively, were obtained from Millipore (Temecula, CA). Mouse monoclonal antibody (1F8) directed against GLUT4 was from Cell Signal Technology. Donkey anti-goat IgG conjugated to horseradish peroxidase was acquired from Jackson Immunochemicals (West Grove, PA). Goat anti-rabbit and anti-mouse IgG-horseradish peroxide conjugates were purchased from Bio-Rad.
Construction of Wild-type G1c4, G3c4, and GLUT1-GLUT3 Chimeras
Two core constructs were employed: 1) G1c4, consisting of human GLUT1 in which the 13 C-terminal amino acids are replaced with the corresponding sequence of human GLUT4, and 2) G3c4, comprising human GLUT3 in which the 13 C-terminal amino acids are replaced with the corresponding sequence of human GLUT4.
To construct G1c4, a 1.5-kbp HindIII/XhoI fragment of GLUT1 cDNA was subcloned into the pcDNA3.1(+) mammalian expression vector using PCR. Two primers were used: P1, containing a HindIII restriction site immediately followed by 18 base pairs complementary to the 5′-end of GLUT1, and P2, comprising 20 base pairs complementary to GLUT1 codons 459–479 followed by sequence for the 13 terminal residues of human GLUT4 and XhoI. The same approach was used to construct G3c4. Sequences were confirmed by sequencing analysis (Davis Sequencing, Davis, CA).
TM domain chimeras were engineered as described previously (35). Using overlapping primers designed for each region of interest, fragments were amplified by Herculase polymerase PCR, purified using the MinElute gel purification kit, and joined by PCR, and the process was repeated until a full-length insert was obtained. The insert was purified using the PCR purification kit, digested with restriction enzymes, purified using the MinElute gel purification kit, and inserted into pcDNA 3.1(+) with the same restriction sites. All final constructs were subcloned into either XL1-Blue competent cells or DH5α subcloning cells, purified using a HiSpeed Maxi kit, and verified by sequence analysis. All point mutations and amino acid substitutions were engineered using QuikChange multisite-directed mutagenesis kits and verified by sequencing.
Western Blotting
Parental GLUT1 and recombinant c4-tagged GLUT1, GLUT3, and GLUT1/GLUT3 chimeras were detected by Western blot analysis as described previously (34, 35).
Transient Transfection of HEK293 Cells
Subconfluent HEK293 cells were transfected with G1c4, G3c4, and chimeric GLUT c4 cDNAs using Lipofectamine 2000 (35, 36). Cells were harvested and analyzed 48 h post-transfection.
Immunoprecipitation with C-terminal GLUT1 Antisera
Two 100-mm plates of transfected or untransfected HEK293 or COS cells were washed twice with phosphate-buffered saline containing protease inhibitor mixture. Cells were rinsed off the dish in 2 ml of solubilization buffer, incubated on ice for 30 min, and clarified by centrifugation at 100,000 × g for 1 h at 4 °C. Supernatants were assayed for total protein content and blotted with rabbit G4-Ab for semiquantitative analysis of recombinant c4 epitope-tagged protein.
HEK cell extracts containing 1–2 mg of total protein were mixed with G1-Ab (final dilution of 200-fold) and incubated overnight at 4 °C with end over end rotation. Lysates from untransfected HEK293 cells were used as negative controls. Each sample was mixed with Protein A-Sepharose CL-4B in solubilization buffer (75 μl of packed resin) and incubated for an additional 4–6 h at 4 °C. Beads were washed three times with solubilization buffer and once with Hepes-KCl and then extracted in 75 μl of 2× Laemmli sample buffer for 10 min at 50 °C. Each sample (30 μl) was loaded in duplicate onto a 10% acrylamide gel for immunoblotting.
Peptides separated by SDS-PAGE were transferred to nitrocellulose. Following blocking and four washes (5 min in PBS-T), membranes were incubated for 1 h in primary antibody (goat anti-GLUT4 C-Ab; 1:200 dilution), washed four times to remove primary antibody, and then exposed for 45 min at 37 °C to secondary antibody (donkey anti-goat-horseradish peroxidase conjugates) diluted 1:5000 in PBS-T plus blockers. After six washes with PBS-T, detection of antigen-antibody complexes was achieved using Pierce SuperSignal West Pico chemiluminescent substrate and visualized by autoradiography. Precipitated G1c4, G3c4, and GLUT1/GLUT3 chimeras were measured by densitometry.
Solid Phase ELISA
Association between endogenous HEK293 GLUT1 and chimeric protein was detected using a modified sandwich ELISA. ELISA wells were coated with rabbit G1-Ab (diluted 1:5000 in PB) and then blocked for 2 h at 37 °C in PB containing 3% gelatin and 1% milk. Following three washes in PB, plates were stored at 4 °C until use. Cell extracts were prepared as above and added (200 μl) to duplicate wells of precoated ELISA plates. Binding proceeded for 4 h at room temperature, and then wells were washed three times in PB and blocked for 2 h at 37 °C. Wells were washed three times in PB and incubated for 2 h at room temperature with 200 μl/well of goat G4-Ab diluted 1:400 in PB, 1% gelatin, 1% milk. Wells were washed three times in PB and incubated for 2 h with 200 μl of donkey anti-goat IgG HRP conjugate (1:5000) at 37 °C. Following three washes in PB, 100 μl of Pierce one-step 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) detection reagent was added to each well, and the plates were developed for 30 min at 37 °C. Complex formation was quantitated by measuring absorbance at 415 nm. Cells transfected with G1c4 were used as positive controls, whereas lysates prepared from untransfected HEK293 cells and G3c4-transfected HEK293 cells were used as negative controls.
2-Deoxy-d-glucose Uptake in HEK293 Cells
2-DG (100 μm) uptake by HEK293 cells at 37 °C was measured as described previously (34, 35, 37).
Size Exclusion Chromatography
Size exclusion chromatography was performed as described previously (21, 22). Membranes from HEK cells (transfected or untransfected) were isolated as described previously (34), and membrane proteins (50 μg) were solubilized in 50 mm phosphate buffer (pH 7.4) containing 150 mm NaCl and 60 mm cholic acid (30 min at 4 °C). Samples were centrifuged at 100,000 × g for 1 h, and the supernatant was loaded onto a Toso Haas TSK-Gel G4000-SWXL column pre-equilibrated with 50 mm phosphate buffer (pH 7.4) containing 150 mm NaCl and 20 mm cholic acid and developed at 0.25 ml/min, and fractions were collected at 0.2-ml intervals. The column was calibrated using protein standards of known Stokes radii.
ELISA
Chimeric protein in each fraction of the size exclusion chromatography assay was detected by ELISA as described previously (21, 22) using a polycolonal rabbit antibody (G4-Ab) directed against a C-terminal epitope of GLUT4.
Biotinylation and Quantitation of Cell Surface GLUTs
Cell surface protein biotinylation using 1 mm EZ-Link Sulfo-NHS-SS-Biotin was as described previously (35, 38). Biotinylated cells were harvested, resuspended in PBS-T, assayed for protein content, and then incubated with Neutravidin gel. Bound protein was eluted using DTT as per the kit instructions, and eluates from normalized loads were quantitated by Western blot analysis.
RESULTS
GLUT1 Forms Homo-oligomers in COS Cells
To assess GLUT1 self-association, we constructed a GLUT1 chimera (G1c4) in which the C-terminal 13 residues of GLUT1 were substituted by the equivalent sequence from GLUT4. This GLUT region is one the most variable within the family of GLUTs and thus permits the generation of isoform-specific, peptide-directed antibodies. Transient expression of G1c4 tripled the sugar uptake capacity of COS-7 cells, indicating that substitution of the 13 terminal residues of GLUT1 does not abrogate GLUT1 transport capacity and that the G1c4 chimeric protein is functionally expressed at the cell surface (Fig. 1). Endogenous, cellular GLUT1 was immunoprecipitated from clarified octyl glucoside extracts using a GLUT1-specific C-terminal Ab (G1-Ab), and the immunoprecipitates were probed for co-precipitation of transfected G1c4, using a GLUT4-specific antibody (G4-Ab). Transfected G1cG4 associates with endogenous GLUT1 (Fig. 1, lane 4), indicating that GLUT1 and G1c4 form oligomers in COS cell membranes. Untransfected COS-7 cells did not express G4-Ab-reactive proteins (Fig. 1, lanes 1 and 3). These blots reveal an array of G4-Ab-reactive proteins of Mr(app) 48,000–75,000. This reflects GLUT1 heterogeneous glycosylation because GLUT1 treatment with peptide:N-glycosidase F causes the array to collapse to 1–2 more mobile bands (39). The 30 kDa band is an occasionally observed, proteolytic fragment of G1c4 and is also observed in non-transfected, GLUT1-expressing cells using GLUT1-specific antibodies (34).
FIGURE 1.
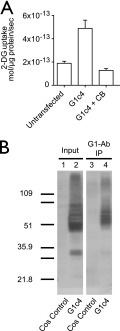
Sugar transporter activity and oligomerization in COS-7 cells. A, cells were mock-transfected or were transfected with G1-c4, and uptake of 100 μm 2-DG was measured in three separate wells at 37 °C for 5 min. Uptake was measured in the presence of 10 μm CB. Ordinate, mol of 2-DG imported/μg of total cell protein/s. Results are shown as mean ± S.E. (error bars). B, association of transfected GLUT1c4 (detected using G4-Ab) with immunoprecipitated, parental GLUT1 (immunoprecipitated (IP) using G1-Ab). Untransfected cells are shown in lanes 1 and 3. Cells transfected with G1c4 are shown in lanes 2 and 4. Lanes 1 and 2, input protein. Lanes 3 and 4, co-immunoprecipitated material. The mobility of molecular weight markers is shown at the left.
G4-Ab and G1-Ab are used to detect heterologous G1c4 and endogenous GLUT1, respectively; therefore, no direct method of comparing G1c4 and GLUT1 expression levels in COS cells is available. Assuming that all protein-mediated transport is inhibited by CB, transport is increased by 3–6-fold in G1c4-transfected COS cells versus untransfected cells. Making the further assumption that G1c4 and GLUT1 share identical catalytic properties, we conclude that transfected COS cells express 3–6-fold more surface GLUT1 than do untransfected cells. Densitometry of lanes 2 and 4 of Fig. 1B reveals that 40% of G1c4 is co-immunoprecipitated with GLUT1. Thus, for each immunoprecipitated G1c4 molecule, an additional 1.5 G1c4 molecules are not captured. Assuming that GLUT1 and G1c4 form homo- and heterotetramers; that capture of G1c4 requires 1, 2, or 3 GLUT1 molecules/heterotetramer; and that G1c4 is stochastically distributed among heterotetramers, each captured G1c4 molecule is accompanied by 1 GLUT1 molecule. Inclusion of the 1.5 G1c4 molecules that escape pull-down indicates that the GLUT1 content of transfected COS cells (GLUT1 + G1c4) is increased 3.5-fold. Because transport activity of transfected cells is increased by 3–6-fold, estimates of relative expression by transport assay and by co-immunoprecipitation are in agreement.
These initial experiments were undertaken using COS cells because we surmised that their high background GLUT1 expression (37) would facilitate co-immunoprecipitation of heterologously expressed GLUT with parental GLUT1. However, high background GLUT1 expression in COS cells results in 2-DG transport rates that are 40-fold greater than transport in HEK cells (compare Figs. 1A and 2B). This could complicate measurements of initial rates of GLUT transport function in transfected cells, so we subsequently switched to a HEK cell model in which background GLUT expression is much lower.
FIGURE 2.
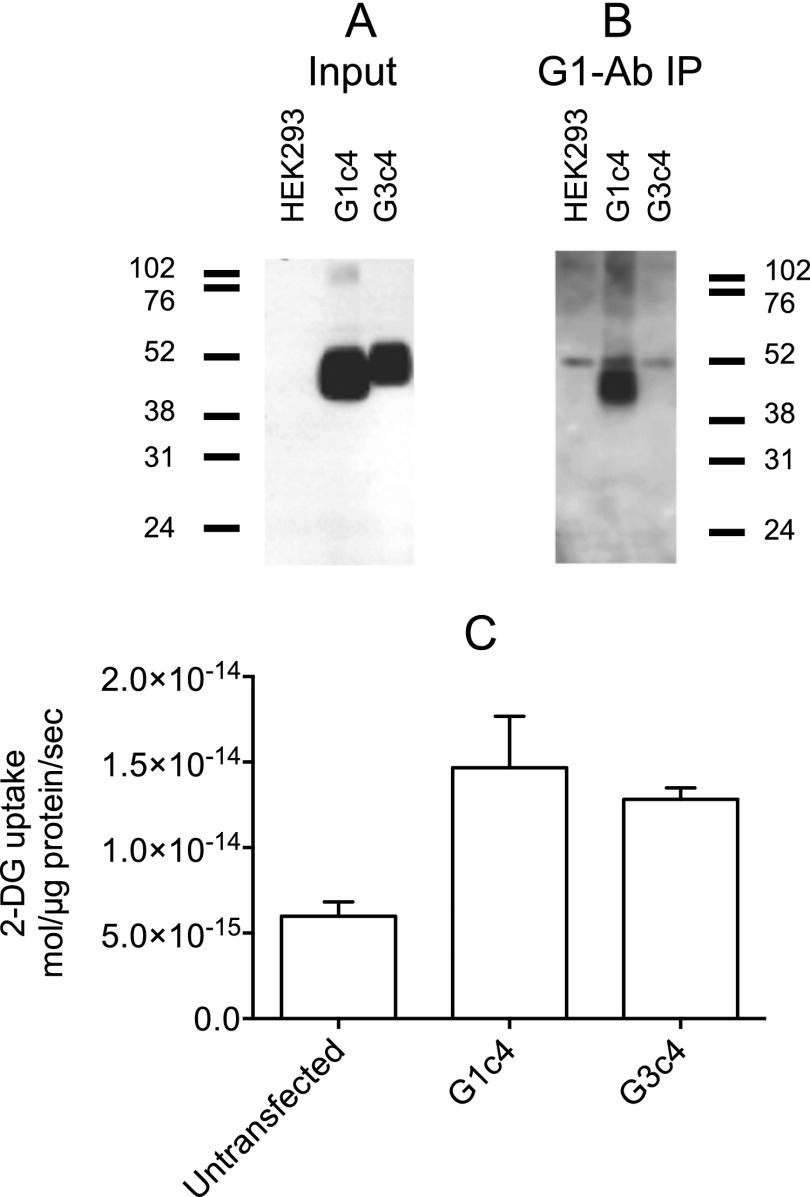
Heterologous expression of G1c4 and G3c4 in HEK cells and association with parental GLUT1. Results are shown for untransfected HEK cells (HEK293) and for cells transfected with G1c4 and G3c4. The mobility of molecular weight markers is shown at the left of each blot. A, the input blot shows detergent-solubilized cell membrane blotted with rabbit anti-G4-Ab. B, the G1-Ab immunoprecipitation (IP) blot shows goat anti-G4-Ab blots of G1-Ab immunoprecipitates of input material. C, cells were mock-transfected or were transfected with G1c4 or G3c4, and uptake of 100 μm 2-DG was measured in three separate wells at 37 °C for 5 min. Ordinate, mol of 2-DG imported/μg of total cell protein/s. Results are shown as mean ± S.E. (error bars).
GLUT1 and GLUT3 Do Not Form Heterocomplexes in HEK Cells
Human embryonic kidney cells (HEK) express low levels of endogenous GLUT1 (35). G1cG4 and G3cG4 were transiently transfected into HEK cells, membrane proteins were solubilized with Triton X-100, and association of G1cG4 and G3cG4 with GLUT1 was assessed by immunoprecipitation of parental GLUT1 using G1-Ab. Although both G1c4 and G3c4 were overexpressed in HEK cells at comparable levels 48 h post-transfection (Fig. 2A) and increased HEK cell-mediated 2-DG uptake (Fig. 2C), only G1c4 co-immunoprecipitated with parental GLUT1 (Fig. 2B). These results demonstrate that whereas G1c4 and parental GLUT1 form a heterocomplex, G3c4 and parental GLUT1 do not. This finding is consistent with earlier studies suggesting that GLUT1 and GLUT3 do not form functional hetero-oligomers (32). Similar studies also show that GLUT1 and GLUT4 do not form heterocomplexes (33). GLUT1 is only 5 amino acids shorter than GLUT3 but displays disproportionately greater electrophoretic mobility than GLUT3 upon SDS-PAGE. GLUT3 contains more acidic, fewer basic, and fewer nonpolar amino acids than GLUT1. This may result in the binding of fewer SDS molecules and a commensurate reduction in GLUT3 electrophoretic mobility (40).
The addition of Triton X-100-solubilized, purified human GLUT1 to clarified cell membrane extracts from HEK cells expressing G1c4 completely blocked G1c4 co-immunoprecipitation when using G1-Ab to immunoprecipitate parental GLUT1 (Fig. 3A). Mixing controls showed that GLUTs do not exchange between lipid/detergent/protein micelles. For example, when we combined clarified detergent extracts of cell membranes isolated from G1c4-transfected cells with extracts of cell membranes from WT GLUT1-transfected HEK cells and then immunoprecipitated using G1-Ab, we observed unchanged or even competitively reduced G1c4 pull-down with WT GLUT1. However, when HEK cells were co-transfected with G1c4 plus WT GLUT1 cDNAs, G1c4 pull-down using G1-Ab was significantly enhanced (Fig. 3B). Co-immunoprecipitation results are therefore GLUT1-specific and are not postsolubilization artifacts.
FIGURE 3.
Does exogenous GLUT1c4 associate specifically with parental GLUT1 in HEK cells? A, exogenous, purified human GLUT1 competitively inhibits G1c4/parental GLUT1 co-immunoprecipitation by G1-Ab. Shown is G1c4 pull-down in the presence and absence, respectively, of 25 μg of solubilized purified GLUT1 and immunoblot using G4-Ab. B, cells were transfected with WT GLUT1 (lane 1), G1c4 (lane 2), or WT GLUT1 and G1c4 (lane 4). 2 days later, cells were harvested, solubilized in solubilization buffer, and adjusted for total protein content. Extracts from WT GLUT1 and from G1c4-transfected cells were mixed (lane 3). All extracts were then subjected to immunoprecipitation using G1-Ab, and the precipitates were resolved by SDS-PAGE and probed using G4-Ab. The mobility of molecular weight markers is indicated to the left of each blot.
Construction and Nomenclature of GLUT1/GLUT3 Chimeric Transporters
The inability of GLUT1 to form a stable, immunoprecipitable complex with GLUT3 or GLUT4 suggests that GLUT1 oligomerization involves GLUT1-specific sequence. We exploited this behavior and GLUT1-GLUT3 homology-scanning mutagenesis to investigate the sequence determinants of GLUT1 quaternary structure. GLUT1-GLUT3 chimeras were constructed by a multistep PCR process in which sequence from one transporter was systematically substituted with the corresponding sequence from the second transporter. GLUT1 and -3 were initially divided into four quarters (TM1 to -3, TM4 to -6, TM7 to -9, and TM10 to -12), and various in-frame combinations were engineered. For instance, the “1133” chimera comprises the N-terminal half of G1c4 (TM1 to -6) followed by the C-terminal half of G3c4 (cytoplasmic loop 6-7, TM7 to -12 plus the cytoplasmic C-terminal loop). The “3311” chimera contains the N terminus of G3c4 (TM1 to -6) followed by the C terminus of G1c4 (cytoplasmic loop 6-7, TM7 to -12 plus the cytoplasmic C-terminal loop). Quarter mutants were also constructed. The “1333” chimera comprises TM1 to -3 of G1c4 followed by the nine terminal TMs of G3c4. In quarter mutants, unless otherwise indicated, the cytoplasmic C terminus and the long loop connecting TM6 and -7 correspond to sequence from the transporter from which 9 of the 12 TMs are derived. The 13 C-terminal amino acids of all chimeras, whether GLUT1 or GLUT3 sequence, were substituted with equivalent GLUT4 sequence to distinguish the protein from endogenous GLUTs. Where individual TMs are substituted, the original scaffold protein is indicated, followed by the substituted TM domain in parenthesis. For example, the GLUT1(93) chimera comprises G1c4 in which TM9 is substituted by the corresponding sequence of G3c4 TM9.
GLUT1 Oligomerization Sequence Determinants
HEK cells were transiently transfected with GLUT1/GLUT3 chimera cDNAs. Chimera expression was assayed by immunoblot analysis, transport function was assayed by 2-DG uptake assays, and chimera association with parental GLUT1 was assayed by solid phase immunoprecipitation ELISA.
Chimeric transporters were expressed at high levels in HEK293 cells and catalyze 2-DG transport. Transient expression of each chimera increased the sugar uptake capacity of HEK cells 2–3-fold, indicating that each chimera is expressed at the cell surface along with endogenous GLUT1 (Fig. 4A). Transport in transfected cells was significantly greater than transport in untransfected cells and in CB-inhibited cells in all instances (p < 0.05; Student's t test). Some variation was observed in transport rates and transport inhibition by CB. This could reflect 1) inherent variability from experiment to experiment, 2) perturbation of the CB binding site in chimeric GLUTs, or 3) variability in transient transfection and cell surface expression efficiencies.
FIGURE 4.
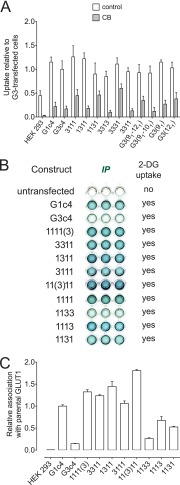
Determinants of GLUT1 oligomerization. HEK cells were transfected with GLUT1c4/GLUT3c4 chimeras and assayed for 2-DG transport and chimera association with parental GLUT1 in G1-Ab pull-downs. A, representative 2-DG uptake in untransfected cells and in cells transfected with chimeras. Ordinate, rate of 2-DG uptake from medium containing 100 μm 2-DG relative to uptake by GLUT3c4-transfected cells. This experiment summarizes uptake measurements made in triplicate in the absence (open bars) and presence (gray bars) of 10 μm CB. Analysis by Student's t test indicates that transport in transfected cells is significantly greater than transport in untransfected cells or in cells exposed to CB (p < 0.05). B, representative solid phase immunoprecipitation results (ELISA) for half and quarter G1c4/G3c4 chimeras (see “Results” for chimera nomenclature). Construct, chimera tested; IP, the solid phase immunoprecipitation readout (color development) shown as 3 wells; 2-DG update, sugar transport catalyzed by the construct (no, not greater than untransfected cell transport; yes, greater transport than observed in untransfected cells). C, results of B quantitated by ELISA. All measurements are shown as mean ± S.E. (error bars) of three or more determinations.
Solid phase ELISA applied clarified detergent extracts of HEK cells to ELISA wells containing preadsorbed immunoprecipitating rabbit G1-Ab. Following binding, goat G4-Ab was applied, and the extent of G4-Ab binding was assessed by the addition of donkey anti-goat IgG HRP conjugate followed by HRP substrate. Using this assay, we observed that G1c4 associates with parental GLUT1 in HEK cells (Fig. 4, B and C, 1111). In contrast, G3c4 and parental GLUT1 did not form heterocomplexes in HEK cells detectable by solid phase ELISA (Fig. 4, B and C, 3333). Substitution of GLUT1 TM6 to -12 with the corresponding sequence from GLUT3 produced a transport-competent chimera that did not associate with parental GLUT1 (Fig. 4, B and C, 1133). The reverse half-chimera (3311; GLUT3 TM1 to -6 plus GLUT1 TM7 to -12) did associate with parental GLUT1. The middle loops (loop 6-7) and the C termini of GLUT1 and -3 are interchangeable, indicating that isoform-specific sequence in these areas is not required for oligomerization (Fig. 4, B and C, 11(3)11 and 1111(3), respectively).
GLUT1-specific TM9 Sequence Promotes Oligomerization
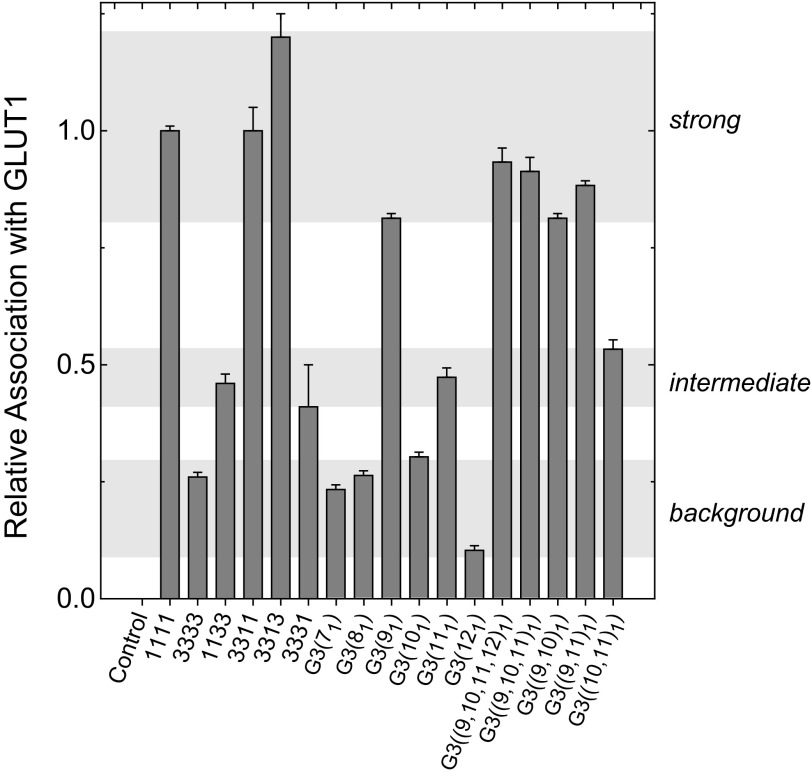
Analysis of quarter chimeras reveals that G1c4 association with parental GLUT1 does not require GLUT1-specific sequence in TM1 to -3 (Fig. 4, B and C, 3111) or TM3 to -6 (Fig. 4, B and C, 1311). When TM10 to -12 (Fig. 4, B and C, 1113) were replaced with equivalent GLUT3 sequence, chimera association with parental GLUT1 was reduced by 25%. When TM7 to -9 of GLUT1 were replaced with corresponding GLUT3 sequence, chimera association with endogenous GLUT1 was reduced by 50% (Fig. 4, B and C, 1131). These results indicate that the C-terminal half of GLUT1 is critical but the 1113 and 1131 results are less definitive in demonstrating which specific C-terminal TMs are most important. We therefore undertook analyses using single and double TM swaps in which individual GLUT1 TMs or pairs of TMs were substituted into a G3c4 scaffold (Fig. 5). Substitution of GLUT1 TM9 into G3c4 or into any of the GLUT3 chimeras induced parental GLUT1 association with the chimera. This demonstrates that TM9 sequence is sufficient to promote hetero-oligomerization between GLUT1 and GLUT3. G3c4 chimeras containing GLUT1 TM11 also associated with endogenous GLUT1 but not to the same extent as GLUT3 chimeras containing GLUT1 TM9 (Fig. 4, B and C). The effect of replacing TM9 plus TM11 of G3c4 with GLUT1 sequence was not additive to that of TM9 alone. GLUT3 constructs lacking GLUT1 TM9 but containing GLUT1 TM11 either alone or in combination with other TMs (e.g. 3331, GLUT3(111), and GLUT3(101111)) and the 1133 chimera show intermediate levels of heteroassociation with parental GLUT1 (Fig. 5).
FIGURE 5.
Determinants of GLUT1 oligomerization. HEK cells were transfected with G1c4, with G3c4, or with G3c4 in which six, four, three, two, or only one TM was substituted with equivalent GLUT1 sequence. Cells were assayed for chimera association with parental GLUT1 in G1-Ab pull-downs. Chimera/parental GLUT1 association was assayed by solid-phase immunoprecipitation (ELISA) and quantitated by absorbance measurements, and results are shown as mean ± S.E. (error bars) of three or more determinations and are expressed relative to association observed with GLUT1-transfected cells. Three levels of association of transfected constructs with parental GLUT1 are observed and are indicated by the shaded, horizontal bars: background, intermediate, and strong.
GLUT1 and GLUT3 TM9 sequence is non-homologous at nine locations. Seven of these locations map to a common surface of an α-helix (Table 1). No single point mutation (GLUT1 sequence converted to GLUT3 sequence) was sufficient to block G1c4-parental GLUT1 co-immunoprecipitation (Table 1).
TABLE 1.
GLUT1–GLUT3 TM9 sequence conservation
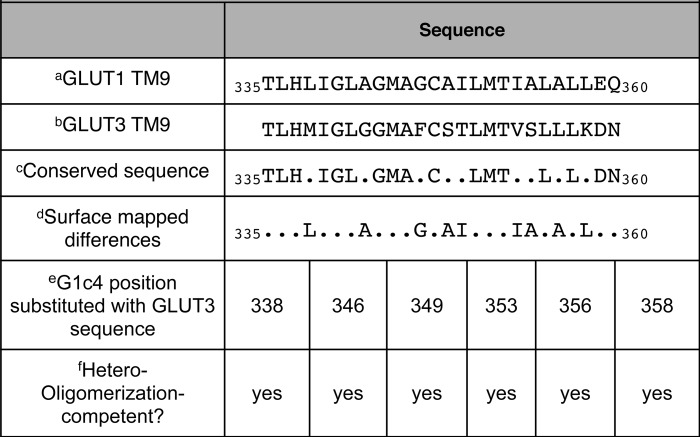
a Analysis of GLUT1 TM9 sequence.
b Analysis of GLUT3 TM9 sequence.
c TM9 sequence that is invariant or homologous.
d GLUT1 TM9 residues that map to the lipid bilayer-exposed surface of transmembrane helix 9 (41).
e Position in GLUT1 sequence mutagenized to equivalent GLUT3 sequence.
e Does G1c4 containing the indicated TM9 point mutation form a heterocomplex with HEK cell, endogenous GLUT1 to the same extent as non-mutagenized G1c4?
Hydrodynamic Characterization of the GLUTs
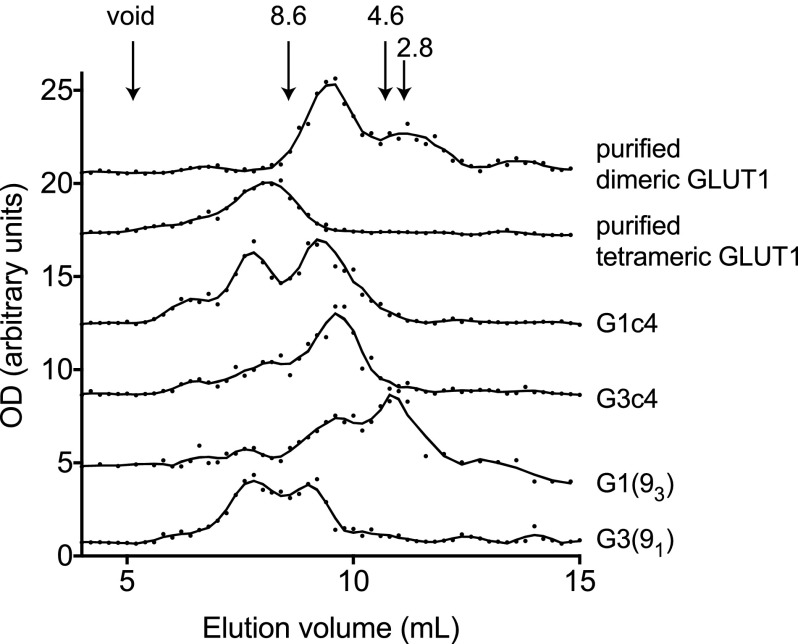
Size exclusion HPLC of cholic acid-solubilized cell membranes isolated from G1c4-transfected HEK cells shows that GLUT1 resolves in two major peaks with average Stokes radii of 10 and 6 nm (Fig. 6). These studies do not preclude the possibility that GLUT/detergent/lipid micelles also contain other protein species. However, GLUT1-containing protein/lipid/detergent micelles released from purified human GLUT1 proteoliposomes, from human red cell membranes, and from CHO cell membranes also resolve as 10- and 6-nm Stokes radius particles (Fig. 6; see also Refs. 20–22), suggesting that the hydrodynamic behavior and composition of GLUT1/lipid/detergent micelles released from cell membranes and purified GLUT1 proteoliposomes are indistinguishable. G3c4 micelles solubilized from G3c4-transfected HEK cells resolve as a major molecular species with an average stokes radius of 6 nm (Fig. 6, blue). Assuming that the lipid, detergent, and protein contents of G3c4-containing micelles are similar to those of GLUT1 micelles, this suggests that G3c4 forms dimers in HEK cell membranes.
FIGURE 6.
Size exclusion chromatography of cholic acid-solubilized, purified human red cell GLUT1 and cell membranes from HEK cells expressing GLUT1c4, GLUT3c4, and GLUT1c4/GLUT3c4 chimeras. Ordinate, relative amount of G4-Ab-reactive material (or, in the case of purified GLUT1, G1-Ab-reactive material) detected by ELISA. Abscissa, elution volume (ml). Results are shown for purified tetrameric GLUT1, purified dimeric GLUT1, G1c4, G3c4, GLUT1(93) (G1(93); G1c4 containing TM9 of GLUT3), and GLUT3(91) (G3(91); G3c4 containing TM9 of GLUT1). The column was calibrated with standards of known Stokes radius and with cholic acid-solubilized, nonreduced GLUT1, which resolves as tetrameric (10-nm Stokes radius) GLUT1 particles and with cholic acid-solubilized, reduced GLUT1, which resolves as dimeric (6-nm Stokes radius) particles (20–22).
Substitution of GLUT1 TM9 into G3c4 shifts the hydrodynamic profile of G3c4. Whereas G3c4 micelles resolved as 6-nm Stokes radius particles, GLUT3(91) resolved as a mixture of 6- and 10-nm particles (Fig. 6). This suggests that sequence within TM9 of GLUT1 promotes GLUT3 homo-tetramerization. These results are consistent with the solid phase immunoprecipitation results, which show that TM9 of GLUT1 promotes hetero-oligomerization of GLUT1 and GLUT3. Substitution of GLUT1 TM9 sequence with equivalent GLUT3 sequence to form GLUT1(93) shifted the elution profile of GLUT1 from a mixture of tetrameric (10 nm) and dimeric (6 nm) species to a largely dimeric population (Fig. 6). These observations reinforce the view that TM9 assumes a critical role in GLUT1 oligomerization.
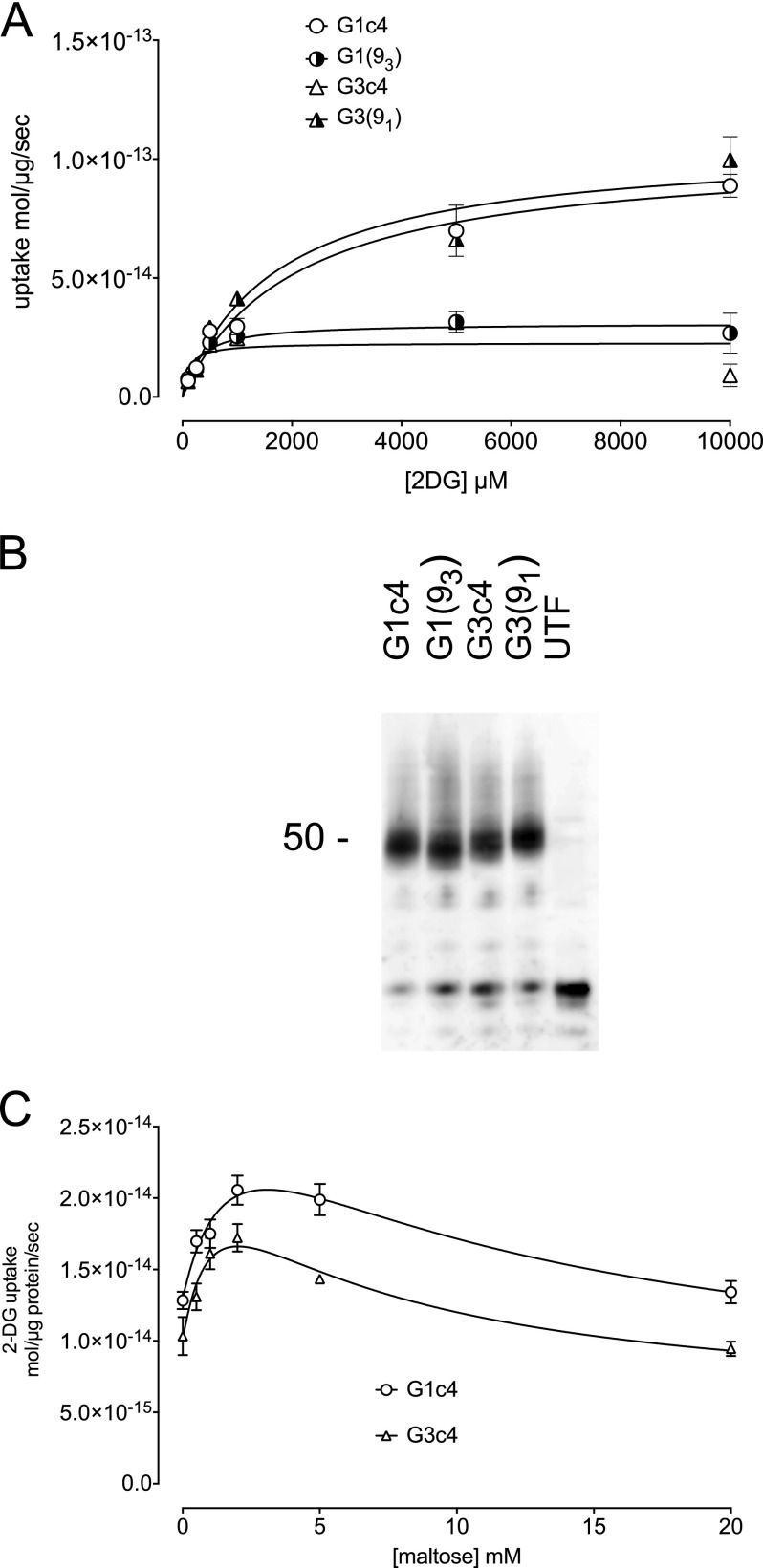
Substitution of GLUT1 TM9 Alters GLUT1 Kinetics
The concentration dependence of 2-DG uptake by HEK cells stably transfected with G1c4 or with G3c4 suggests that G1c4 is 4-fold more catalytically active than is G3c4 (Fig. 7A). Analysis of cell surface expression of G1c4 and G3c4 by cell surface protein biotinylation confirms that HEK cells express equivalent amounts of each construct at the cell surface (Fig. 7B). Substitution of GLUT1 TM9 into Gc4 to form GLUT3(91) increased HEK cell transport capacity to levels similar to those in G1c4-transfected cells, whereas GLUT1(93)-catalyzed transport resembled that of G3c4 (Fig. 7A).
FIGURE 7.
Catalytic properties of G1c4, G3c4, and G1c4/G3c4 chimeras. A, the concentration dependence of 2-DG uptake by HEK cells stably transfected with G1c4, G3c4, GLUT3(91) (G3(91)), or GLUT1(93) (G1(93)). Ordinate, rate of 2-DG uptake (mol/μg cell protein/min). Abscissa, [2-DG] (mm). Each data point represents the mean ± S.E. (error bars) of three or more separate determinations made in triplicate. The curves drawn through the points were computed by non-linear regression, assuming that transport is described by the Michaelis-Menten equation. The computed best fits are as follows: G1c4, Vmax = 103 ± 4 fmol/μg/s, Km(app) = 2.0 ± 0.5 mm; G3c4, Vmax = 23 ± 2 fmol/μg/s, Km(app) = 0.11 ± 0.11 mm; G3(91), Vmax = 106 ± 7 fmol/μg/s, Km(app) = 1.7 ± 0.6 mm; GLUT1(93), Vmax = 31 ± 2 fmol/μg/s, Km(app) = 0.27 ± 0.08 mm. B, detection of cell surface G1c4, G3c4, GLUT3(91), or GLUT1(93) by biotinylation. Cell surface proteins were biotinylated using Sulfo-NHS-SS-Biotin, membranes were solubilized, and biotinylated proteins were pulled down using streptavidin beads and then released using DTT. Released peptides were resolved by SDS-PAGE and subjected to immunoblot analysis using G4-Ab. C, G1c4 and G3c4 display cis-allostery. Shown is the concentration dependence of maltose modulation of 2-DG uptake by HEK cells stably transfected with G1c4 or with G3c4. Ordinate, rate of 2-DG uptake (mol/μg cell protein/s); abscissa, [maltose] (mm). Each data point represents the mean ± S.E. of three or more separate determinations made in triplicate. Uptake by untransfected cells was measured in parallel and subtracted from each point. Curves drawn through the points were computed assuming that uptake modulation by maltose is described as the sum of two saturable components, the first stimulatory and the second inhibitory. The computed best fits are as follows: GLUT1c4, KS = 3.1 ± 0.5 mm, KI = 6.1 ± 2.4 mm; GLUT3c4, KS = 2.6 ± 2.4 mm, KI = 2.7 ± 2.5 mm.
GLUT1-mediated sugar transport in human red cells displays the phenomenon of cis-allostery, in which increasing concentrations of extracellular maltose initially stimulate and then inhibit sugar uptake (24), a behavior we had assumed was specific to tetrameric GLUT1. Subsaturating concentrations of maltose produced a modest stimulation of 2-DG uptake by G1c4-transfected HEK cells (Fig. 7C), followed by transport inhibition at higher concentrations of maltose. We were surprised to observe that sugar uptake by G3c4-transfected cells shows a similar behavior (Fig. 7C).
DISCUSSION
GLUT1 TM9 Sequence Promotes Oligomerization
Previous biophysical studies have shown that human GLUT1 forms homodimers and tetramers (20–22), whereas transport studies (32) suggest that GLUT3 is a functional monomer and that GLUT1 and GLUT3 do not form functional heterocomplexes. GLUT1-GLUT4 co-expression studies also indicate that GLUT1 and GLUT4 do not form heterocomplexes (33). The present study 1) confirms that GLUT1 forms dimers and tetramers, 2) shows for the first time that GLUT3 forms dimers, 3) physically demonstrates by co-immunoprecipitation assay that GLUT1 and GLUT3 do not form heterocomplexes, 4) systematically investigates and defines the molecular determinants of GLUT oligomerization, and 5) demonstrates that GLUT tetramerization confers a significant catalytic advantage to the transporter.
Using homology-scanning mutagenesis, co-immunoprecipitation assays, and size exclusion chromatography, we find that sequences within GLUT1 TM9 are necessary and sufficient for GLUT1 tetramerization. Replacement of GLUT3 TM9 with the corresponding sequence from GLUT1 allows heterologously expressed GLUT3/GLUT1 chimeras to associate with parental GLUT1 in HEK cells and to resolve as a mixture of 6- and 10-nm Stokes radius particles upon size exclusion chromatography of detergent-solubilized membranes. The use of purified GLUT1 to competitively inhibit parental GLUT1 immunoprecipitation prevents heterocomplex detection. This indicates that isolation of GLUT1/GLUT chimera complexes is GLUT1-specific and that the physical behavior of GLUT/lipid/detergent micelles released from HEK cells is indistinguishable from that of GLUT1/lipid/detergent micelles released from purified GLUT1 proteoliposomes.
Conversely, substitution of GLUT3 TM9 into the GLUT1 scaffold disrupts GLUT1 tetramerization. Comparison of GLUT1 and GLUT3 TM9 amino acid sequence shows that 7 of the 9 non-homologous residues are located on a common helix face exposed to the lipid bilayer in a threaded homology-modeled GLUT1 structure (Table 1 (41)). This surface may, as with glycophorin (42–44), form an interface for isoform-specific oligomerization. Systematic mutagenesis of non-conserved residues along this interface reveals that no individual amino acid contributes significantly to oligomerization; rather, oligomerization may be the result of the interaction of multiple residues or TM9 in its entirety.
Previous mutagenesis and differential alkylation studies implicate Cys-347 (which is proposed to lie in the middle of TM9 facing the interior of the GLUT1 structure (41)) and Cys-421 (which lies in loop 11–12) in GLUT1 oligomerization (21). Cys-347 is accessible to alkylation only following GLUT1 reduction, which also causes tetrameric GLUT1 to dissociate into dimers (21). Cys-347 modification may reorient TM9 to enhance or preclude TM9 interaction with adjacent subunits. Cys-347 (but not Cys-421) is conserved in GLUT1 and GLUT3, indicating that it is not sufficient for oligomerization.
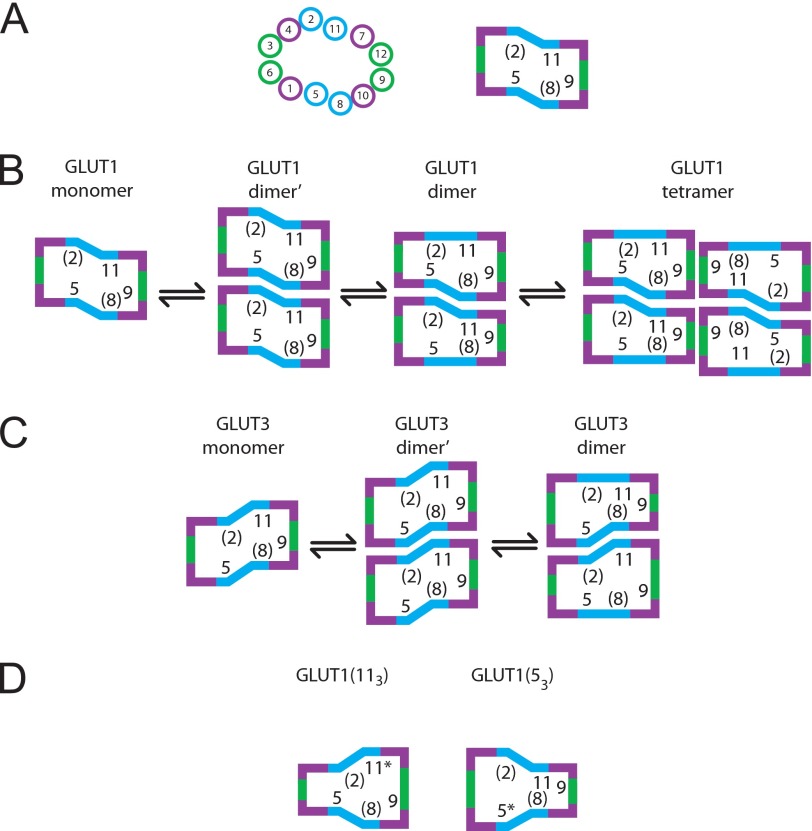
Dimerization Interface
Excluding the scaffolding TMs (TM3, -6, -9, and -12), only TM2, -5, -8, and -11 expose a membrane-spanning helical face to the bilayer hydrocarbon core in homology-modeled GLUT1. These TMs, therefore, present additional potential surfaces for GLUT oligomerization. GLUT1 and GLUT3 TM2 sequence is identical. Bilayer-exposed residues in TM8 are conserved in GLUT1 and -3. Membrane-exposed residues in TM5 and -11 are 70 and 88% conserved, respectively (Table 2). The key change in GLUT3 TM11 is cysteine substitution of GLUT1 phenylalanine 409. This permits tentative assignment of the dimerization interface to TM2 and -11 of one subunit plus TM5 and -8 of the second subunit (Fig. 8). This arrangement would permit TM9 of each subunit to serve as the interface for dimerization of dimers (i.e. tetramerization). To avoid GLUT concatemerization, dimerization of two subunits via TM2 and -11 of subunit A and TM5 and -8 of subunit B must promote conformational changes in TM5 and -8 of subunit A and in TM2 and -11 of subunit B that render these surfaces incompatible (Fig. 8B).
TABLE 2.
Conservation between GLUT1 and GLUT3 TM2, -5, -8, and -11
Analysis of the lipid bilayer exposure of GLUT1 membrane-spanning TM1, -2, -4, -5, -7, -8, -10, and -11 according to a homology-modeled GLUT1 structure (41) indicates that TM1, -4, -7, and -10 are only partially exposed at the bilayer interface.
| TM domain | Residues exposed at the external surface of GLUT1a | Conservation in GLUT3b | Nonconserved changesc |
|---|---|---|---|
| % | |||
| 2 | Leu-64, Leu-67, Ser-68, Ile-71, Gly-75, Gly-79, Val-83, Phe-86, Val-87 | 100 | |
| 5 | Gly-154, Thr-158, Leu-162, Val-166, Ile-170, Phe-174, Asp-177, Ser-178, Gly-181, Asn-182 | 70 | I170V, S178F, N182S |
| 8 | Gln-304, Tyr-308, Gly-312, Val-316, Phe-320, Val-323, Phe-326 | 100 | |
| 11 | Gly-398, Ala-402, Val-406, Phe-409, Ser-410, Ser-414, Val-418, Phe-422 | 88 | F409C |
a Those GLUT1 amino acid residues in TM2, -5, -8, and -11 that are exposed at the lipid interface.
b Conservation between exposed residues in GLUT1 and equivalent residues in GLUT3.
c Those GLUT1 residues that are not conserved and their substitution in GLUT3.
FIGURE 8.
A model for isoform-specific GLUT oligomerization. A, arrangement of GLUT1 TMs viewed from the cytoplasm (41) and simplified at the right to highlight oligomerization TMs. Bilayer-exposed residues in TM2 and -8 (in parenthesis) are conserved in GLUT1 and -3. GLUT1 TM9 drives tetramerization. B, GLUT1 oligomerization. GLUT1 TM5 and -8 interact with TM2 and -11 in an adjacent subunit to form an initial dimer (dimer′). Dimer′ undergoes conformational change at non-complexed TMs (TM2 + TM11 and TM5 + TM8), adopting the stable dimer conformation and preventing concatemerization. Dimers then associate via TM9 in each subunit to form a tetramer. C, GLUT3 oligomerization; similar to B, except that GLUT3 dimerization surfaces adopt a conformation that is incompatible with GLUT1 surfaces. Stable GLUT3 dimers do not tetramerize because they lack GLUT1 TM9. D, substituting GLUT3 TM11 into GLUT1 to form GLUT1(113) allows the TM5 + TM8 surface of GLUT1(113) to associate with TM2 + TM11 of WT GLUT1, but GLUT1(113) is unable to homodimerize due to its lack of self-compatible surfaces. Similarly, substituting GLUT3 TM5 into GLUT1 to form GLUT1(53) allows TM2 + TM11 of GLUT1(53) to associate with TM5 + TM8 of WT GLUT1, but GLUT1(53) is unable to homodimerize due to its lack of self-compatible surfaces.
This model explains how 3331, GLUT3(111), GLUT3(101111), and the 1133 chimera form intermediate levels of heteroassociation with parental GLUT1 (Table 3 and Fig. 8). Furthermore, if the GLUT1 dimer is only partially stable in detergent, this would explain why GLUT1(93), which is tetramerization-incompetent, resolves as dimers and monomers.
TABLE 3.
Which TMs contribute to dimerization between parental GLUT1 and GLUT1/GLUT3 chimeras?
Shown is an analysis of possible TM contributions to parental GLUT1 - GLUT1/GLUT3 chimera dimerization.
| Chimeraa | Forms weak complex with parental GLUT1?b | Parental GLUT1 contributions to interfacec | Chimera contributions to interfaced | Possible surface contacts eliminated by the findingse |
|---|---|---|---|---|
| 3333 | No | |||
| 1133 | Yes | TM2, TM11 | TM5, TM8 | TM2 and TM11 in one subunit with TM2 and TM11 in a second subunit |
| 3331 | Yes | TM5, TM8 | TM2, TM11 | TM5 and TM8 in one subunit with TM5 and TM8 in a second subunit |
| 1131 | Yes | TM2, TM11 or TM5, TM8 | TM5, TM8 or TM2, TM11 | |
| GLUT3(11) | Yes | TM5, TM8 | TM2, TM11 | |
| GLUT3(101111) | Yes | TM5, TM8 | TM2, TM11 |
a Chimeras considered.
b Does the chimera form a weak complex with parental GLUT1 (see Fig. 5)?
c Assuming that TM1, -4, -7, and -10 do not contribute to dimerization (see Table 2), these are the proposed parental GLUT1 TMs that contribute to dimerization with GLUT1/GLUT3 chimeras.
d These are the proposed GLUT1/GLUT3 chimera TMs which, due to their conservation with GLUT1 sequence, are assumed to contribute to dimerization with parental GLUT1.
e These TM interactions are eliminated as contributors to dimerization between parental GLUT1 and GLUT1/GLUT3 chimeras as a result of the observations in column 2.
Many transporters (e.g. aquaporin, (45), ClC channels (46), the anion transporter (47), and nucleotide-sugar antiporters (48, 49)) form multimeric, noncovalent complexes in cell membranes, some with well defined oligomerization interfaces (46) and others with less well defined contacts between subunits (47, 50). In each case, the transporter is a noncovalent complex of two or more subunits in which individual subunits constitute a functional transport pathway. A recent survey (46) concludes that no fewer than 40% of membrane transport protein families are built on this “parallel pathway principle,” in which transporter complexes comprise noncovalent dimers, trimers, or tetramers of subunits stabilized entirely by nonpolar, intersubunit contacts occurring within the bilayer phase of the membrane. A major unanswered question is what selective advantage(s) the parallel pathway principle confers.
Transport Catalyzed by Dimers and Tetramers
Cell surface G1c4 and GLUT3(91) transport capacities are almost 4-fold greater than those of G3c4 and GLUT1(93). Because each construct forms GLUT dimers, but only G1c4 and GLUT3(91) form tetramers, this substantially confirms earlier suggestions that tetrameric GLUT1 is catalytically more efficient than dimeric GLUT1 (21). We previously suggested that gains in efficiency arose from an obligate functional antiparallel arrangement of GLUT1 subunits, where, at any instant, if one subunit presents the endofacial sugar binding site, its neighbor must present the exofacial binding site and vice versa (51). This functional coupling means that sugar import by one subunit is coupled to reorientation of its neighbor from the endofacial to exofacial conformation. Hence the availability of the exofacial sugar binding sites for additional rounds of uptake is not limited by the reorientation or relaxation of the first subunit back to the exofacial state, a conformational change that can be up to 100-fold slower than sugar translocation (27).
One implication of this hypothesis is that tetrameric GLUT1 exposes two exofacial and two endofacial sugar binding sites at any instant, a hypothesis consistent with observations of cis- and trans-allostery in sugar transport (23, 24), in which low concentrations of transport inhibitors stimulate sugar uptake and higher concentrations inhibit transport. Cis- and trans-allostery describe the actions of exofacial and endofacial inhibitors on sugar uptake, respectively. Occupation of only one sugar binding site by inhibitor was proposed to accelerate transport via the adjacent subunit, which becomes inhibited only when inhibitor concentration is increased (24). The current study demonstrates that GLUT1 and GLUT3 show cis-allostery but that GLUT3 does not form tetramers. We therefore conclude either that dimeric GLUTs are capable of cis-allostery (the occupancy state of one subunit is communicated to its neighbor) or that each GLUT1 molecule contains multiple, interacting ligand binding sites. Future studies with dimerization-incompetent GLUTs (e.g. GLUT1(93,113)) may help to answer this question.
Conclusions
GLUT1 and GLUT3 homology-scanning mutagenesis studies show that GLUT scaffolding membrane-spanning helix TM9 is essential for GLUT tetramerization, whereas translocation pathway TMs (TM2, -5, -8, and -11) are involved in GLUT dimerization. GLUT1 TM9 favors GLUT tetramerization, whereas GLUT3 TM9 does not. GLUT tetramers and dimers display allosteric transport behavior that may reflect subunit interactions. GLUT tetramers are at least 4-fold more catalytically active than GLUT dimers. This may explain why GLUT1 but not GLUT3 is expressed at blood tissue barriers and in primate red cells, where rapid glucose transfer between compartments is essential for organismal carbohydrate homeostasis, and in rapidly dividing cancer cells, where glucose delivery to the glycolytic machinery is essential for cellular survival under hypoxic conditions.
This work was supported, in whole or in part, by National Institutes of Health Grants DK 36081 and DK 44888.
- GLUT
- glucose transporter
- 2-DG
- 2-deoxy-d-glucose
- CB
- cytochalasin B
- TM
- membrane-spanning α helix
- Sulfo-NHS-SS-Biotin
- sulfosuccinimidyl 2-(biotinamido)-ethyl-1,3-dithiopropionate
- HEK
- human embryonic kidney
- PB
- phosphate buffer
- Ab
- antibody.
REFERENCES
- 1. Cura A. J., Carruthers A. (2012) Role of monosaccharide transport proteins in carbohydrate assimilation, distribution, metabolism, and homeostasis. Compr. Physiol. 2, 863–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Joost H. G., Bell G. I., Best J. D., Birnbaum M. J., Charron M. J., Chen Y. T., Doege H., James D. E., Lodish H. F., Moley K. H., Moley J. F., Mueckler M., Rogers S., Schürmann A., Seino S., Thorens B. (2002) Nomenclature of the GLUT/SLC2A family of sugar/polyol transport facilitators. Am. J. Physiol. Endocrinol. Metab. 282, E974–E976 [DOI] [PubMed] [Google Scholar]
- 3. Saier M. H., Jr., Beatty J. T., Goffeau A., Harley K. T., Heijne W. H., Huang S. C., Jack D. L., Jähn P. S., Lew K., Liu J., Pao S. S., Paulsen I. T., Tseng T. T., Virk P. S. (1999) The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1, 257–279 [PubMed] [Google Scholar]
- 4. Mueckler M., Caruso C., Baldwin S. A., Panico M., Blench I., Morris H. R., Allard W. J., Lienhard G. E., Lodish H. F. (1985) Sequence and structure of a human glucose transporter. Science 229, 941–945 [DOI] [PubMed] [Google Scholar]
- 5. Mann G. E., Yudilevich D. L., Sobrevia L. (2003) Regulation of amino acid and glucose transporters in endothelial and smooth muscle cells. Physiol. Rev. 83, 183–252 [DOI] [PubMed] [Google Scholar]
- 6. Simpson I. A., Carruthers A., Vannucci S. J. (2007) Supply and demand in cerebral energy metabolism. The role of nutrient transporters. J. Cereb. Blood Flow Metab. 27, 1766–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chin J. J., Jung E. K., Chen V., Jung C. Y. (1987) Structural basis of human erythrocyte glucose transporter function in proteoliposome vesicles. Circular dichroism measurements. Proc. Natl. Acad. Sci. U.S.A. 84, 4113–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alvarez J., Lee D. C., Baldwin S. A., Chapman D. (1987) Fourier transform infrared spectroscopic study of the structure and conformational changes of the human erythrocyte glucose transporter. J. Biol. Chem. 262, 3502–3509 [PubMed] [Google Scholar]
- 9. Hresko R. C., Kruse M., Strube M., Mueckler M. (1994) Topology of the Glut 1 glucose transporter deduced from glycosylation scanning mutagenesis. J. Biol. Chem. 269, 20482–20488 [PubMed] [Google Scholar]
- 10. Blodgett D. M., Graybill C., Carruthers A. (2008) Analysis of glucose transporter topology and structural dynamics. J. Biol. Chem. 283, 36416–36424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang Y., Lemieux M. J., Song J., Auer M., Wang D. N. (2003) Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science 301, 616–620 [DOI] [PubMed] [Google Scholar]
- 12. Abramson J., Smirnova I., Kasho V., Verner G., Kaback H. R., Iwata S. (2003) Structure and mechanism of the lactose permease of Escherichia coli. Science 301, 610–615 [DOI] [PubMed] [Google Scholar]
- 13. Yin Y., He X., Szewczyk P., Nguyen T., Chang G. (2006) Structure of the multidrug transporter EmrD from Escherichia coli. Science 312, 741–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun L., Zeng X., Yan C., Sun X., Gong X., Rao Y., Yan N. (2012) Crystal structure of a bacterial homologue of glucose transporters GLUT1–4. Nature 490, 361–366 [DOI] [PubMed] [Google Scholar]
- 15. Mueckler M., Makepeace C. (2009) Model of the exofacial substrate-binding site and helical folding of the human Glut1 glucose transporter based on scanning mutagenesis. Biochemistry 48, 5934–5942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mueckler M., Makepeace C. (2008) Transmembrane segment 6 of the Glut1 glucose transporter is an outer helix and contains amino acid side chains essential for transport activity. J. Biol. Chem. 283, 11550–11555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mueckler M., Makepeace C. (2006) Transmembrane segment 12 of the Glut1 glucose transporter is an outer helix and is not directly involved in the transport mechanism. J. Biol. Chem. 281, 36993–36998 [DOI] [PubMed] [Google Scholar]
- 18. Mueckler M., Roach W., Makepeace C. (2004) Transmembrane segment 3 of the Glut1 glucose transporter is an outer helix. J. Biol. Chem. 279, 46876–46881 [DOI] [PubMed] [Google Scholar]
- 19. Jung C. Y., Hsu T. L., Hah J. S., Cha C., Haas M. N. (1980) Glucose transport carrier of human erythrocytes. Radiation-target size of glucose-sensitive cytochalasin B binding protein. J. Biol. Chem. 255, 361–364 [PubMed] [Google Scholar]
- 20. Hebert D. N., Carruthers A. (1991) Cholate-solubilized erythrocyte glucose transporters exist as a mixture of homodimers and homotetramers. Biochemistry 30, 4654–4658 [DOI] [PubMed] [Google Scholar]
- 21. Zottola R. J., Cloherty E. K., Coderre P. E., Hansen A., Hebert D. N., Carruthers A. (1995) Glucose transporter function is controlled by transporter oligomeric structure. A single, intramolecular disulfide promotes GLUT1 tetramerization. Biochemistry 34, 9734–9747 [DOI] [PubMed] [Google Scholar]
- 22. Hebert D. N., Carruthers A. (1992) Glucose transporter oligomeric structure determines transporter function. Reversible redox-dependent interconversions of tetrameric and dimeric GLUT1. J. Biol. Chem. 267, 23829–23838 [PubMed] [Google Scholar]
- 23. Cloherty E. K., Levine K. B., Carruthers A. (2001) The red blood cell glucose transporter presents multiple, nucleotide-sensitive sugar exit sites. Biochemistry 40, 15549–15561 [DOI] [PubMed] [Google Scholar]
- 24. Hamill S., Cloherty E. K., Carruthers A. (1999) The human erythrocyte sugar transporter presents two sugar import sites. Biochemistry 38, 16974–16983 [DOI] [PubMed] [Google Scholar]
- 25. Carruthers A., Helgerson A. L. (1991) Inhibitions of sugar transport produced by ligands binding at opposite sides of the membrane. Evidence for simultaneous occupation of the carrier by maltose and cytochalasin B. Biochemistry 30, 3907–3915 [DOI] [PubMed] [Google Scholar]
- 26. Helgerson A. L., Carruthers A. (1987) Equilibrium ligand binding to the human erythrocyte sugar transporter. Evidence for two sugar-binding sites per carrier. J. Biol. Chem. 262, 5464–5475 [PubMed] [Google Scholar]
- 27. Appleman J. R., Lienhard G. E. (1985) Rapid kinetics of the glucose transporter from human erythrocytes. Detection and measurement of a half-turnover of the purified transporter. J. Biol. Chem. 260, 4575–4578 [PubMed] [Google Scholar]
- 28. Gorga F. R., Lienhard G. E. (1981) Equilibria and kinetics of ligand binding to the human erythrocyte glucose transporter. Evidence for an alternating conformation model for transport. Biochemistry 20, 5108–5113 [DOI] [PubMed] [Google Scholar]
- 29. Jardetzky O. (1966) Simple allosteric model for membrane pumps. Nature 211, 969–970 [DOI] [PubMed] [Google Scholar]
- 30. Lieb W. R., Stein W. D. (1974) Testing and characterizing the simple carrier. Biochim. Biophys. Acta 373, 178–196 [DOI] [PubMed] [Google Scholar]
- 31. Vannucci S. J., Maher F., Simpson I. A. (1997) Glucose transporter proteins in brain. Delivery of glucose to neurons and glia. Glia 21, 2–21 [DOI] [PubMed] [Google Scholar]
- 32. Burant C. F., Bell G. I. (1992) Mammalian facilitative glucose transporters. Evidence for similar substrate recognition sites in functionally monomeric proteins. Biochemistry 31, 10414–10420 [DOI] [PubMed] [Google Scholar]
- 33. Pessino A., Hebert D. N., Woon C. W., Harrison S. A., Clancy B. M., Buxton J. M., Carruthers A., Czech M. P. (1991) Evidence that functional erythrocyte-type glucose transporters are oligomers. J. Biol. Chem. 266, 20213–20217 [PubMed] [Google Scholar]
- 34. Blodgett D. M., De Zutter J. K., Levine K. B., Karim P., Carruthers A. (2007) Structural Basis of GLUT1 Inhibition by Cytoplasmic ATP. J. Gen. Physiol. 130, 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vollers S. S., Carruthers A. (2012) Sequence determinants of GLUT1-mediated accelerated-exchange transport. Analysis by homology-scanning mutagenesis. J. Biol. Chem. 287, 42533–42544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Levine K. B., Robichaud T. K., Hamill S., Sultzman L. A., Carruthers A. (2005) Properties of the human erythrocyte glucose transport protein are determined by cellular context. Biochemistry 44, 5606–5616 [DOI] [PubMed] [Google Scholar]
- 37. Levine K. B., Cloherty E. K., Hamill S., Carruthers A. (2002) Molecular determinants of sugar transport regulation by ATP. Biochemistry 41, 12629–12638 [DOI] [PubMed] [Google Scholar]
- 38. Cura A. J., Carruthers A. (2012) AMP kinase regulation of sugar transport in brain capillary endothelial cells during acute metabolic stress. Am. J. Physiol. Cell Physiol. 303, C806–C814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cura A. J., Carruthers A. (2010) Acute modulation of sugar transport in brain capillary endothelial cell cultures during activation of the metabolic stress pathway. J. Biol. Chem. 285, 15430–15439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shi Y., Mowery R. A., Ashley J., Hentz M., Ramirez A. J., Bilgicer B., Slunt-Brown H., Borchelt D. R., Shaw B. F. (2012) Abnormal SDS-PAGE migration of cytosolic proteins can identify domains and mechanisms that control surfactant binding. Protein Sci. 21, 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Salas-Burgos A., Iserovich P., Zuniga F., Vera J. C., Fischbarg J. (2004) Predicting the three-dimensional structure of the human facilitative glucose transporter glut1 by a novel evolutionary homology strategy. Insights on the molecular mechanism of substrate migration, and binding sites for glucose and inhibitory molecules. Biophys. J. 87, 2990–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lis M., Blumenthal K. (2006) A modified, dual reporter TOXCAT system for monitoring homodimerization of transmembrane segments of proteins. Biochem. Biophys. Res. Commun. 339, 321–324 [DOI] [PubMed] [Google Scholar]
- 43. Leeds J. A., Boyd D., Huber D. R., Sonoda G. K., Luu H. T., Engelman D. M., Beckwith J. (2001) Genetic selection for and molecular dynamic modeling of a protein transmembrane domain multimerization motif from a random Escherichia coli genomic library. J. Mol. Biol. 313, 181–195 [DOI] [PubMed] [Google Scholar]
- 44. Russ W. P., Engelman D. M. (2000) The GxxxG motif. A framework for transmembrane helix-helix association. J. Mol. Biol. 296, 911–919 [DOI] [PubMed] [Google Scholar]
- 45. Furman C. S., Gorelick-Feldman D. A., Davidson K. G., Yasumura T., Neely J. D., Agre P., Rash J. E. (2003) Aquaporin-4 square array assembly. Opposing actions of M1 and M23 isoforms. Proc. Natl. Acad. Sci. U.S.A. 100, 13609–13614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Robertson J. L., Kolmakova-Partensky L., Miller C. (2010) Design, function and structure of a monomeric ClC transporter. Nature 468, 844–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Casey J. R., Reithmeier R. A. (1991) Analysis of the oligomeric state of Band 3, the anion transport protein of the human erythrocyte membrane, by size exclusion high performance liquid chromatography. Oligomeric stability and origin of heterogeneity. J. Biol. Chem. 266, 15726–15737 [PubMed] [Google Scholar]
- 48. Olczak M., Guillen E. (2006) Characterization of a mutation and an alternative splicing of UDP-galactose transporter in MDCK-RCAr cell line. Biochim. Biophys. Acta 1763, 82–92 [DOI] [PubMed] [Google Scholar]
- 49. Hirschberg C. B., Robbins P. W., Abeijon C. (1998) Transporters of nucleotide sugars, ATP, and nucleotide sulfate in the endoplasmic reticulum and Golgi apparatus. Annu. Rev. Biochem. 67, 49–69 [DOI] [PubMed] [Google Scholar]
- 50. Jiang J., Magilnick N., Tsirulnikov K., Abuladze N., Atanasov I., Ge P., Narla M., Pushkin A., Zhou Z. H., Kurtz I. (2013) Single particle electron microscopy analysis of the bovine anion exchanger 1 reveals a flexible linker connecting the cytoplasmic and membrane domains. PLoS One 8, e55408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Carruthers A., Zottola R. J. (1996) in Transport Processes in Eukaryotic and Prokaryotic Organisms. (Konings W. N., Kaback H. R., Lolkema J. S., eds) pp. 311–342, Elsevier, Amsterdam [Google Scholar]