Abstract
Epigenetic mechanisms, which control chromatin structure and function, mediate changes in gene expression that occur in response to diverse stimuli. Recent research has established that environmental events and behavioral experience induce epigenetic changes at particular gene loci that help shape neuronal plasticity and function, and hence behavior, and that some of these changes can be very stable and even persist for a lifetime. Increasing evidence supports the hypothesis that aberrations in chromatin remodeling and subsequent effects on gene expression within limbic brain regions contribute to the pathogenesis of depression and other stress-related disorders such as post-traumatic stress disorder and other anxiety syndromes. Likewise, the gradually developing but persistent therapeutic effects of antidepressant medications may be achieved in part via epigenetic mechanisms. This review discusses recent advances in understanding epigenetic regulation of stress-related disorders and focuses on three distinct aspects of stress-induced epigenetic pathology: the effects of stress and antidepressant treatment during adulthood, the life-long effects of early life stress on subsequent stress vulnerability, and the possible trans-generational transmission of stress-induced abnormalities.
Introduction
The importance of environmental influences in shaping brain structure and function has highlighted the profoundly plastic nature of the brain. In fact, all organisms continuously face stressful events and changes in their environment that can affect their homeostasis. Stress responses are reactions to these external challenges and include changes in the central nervous system (CNS) and in various peripheral organs that aim to reinstate the initial homeostasis (1). The perception of a stressful situation activates a large number of neuronal circuits, in particular, the hypothalamic-pituitary-adrenal (HPA) axis, the locus coeruleus, and autonomic noradrenergic centers in the brainstem. These and many other initial stress responses target numerous limbic brain regions, such as prefrontal cortex, hippocampus, amygdala, and ventral striatum (also termed nucleus accumbens or NAc) (Figure 1) (2, 3). These changes in the CNS directly affect learning and memory, alertness, arousal, and perhaps basal anxiety, and promote adaptive behavioral responses to subsequent stresses (4).
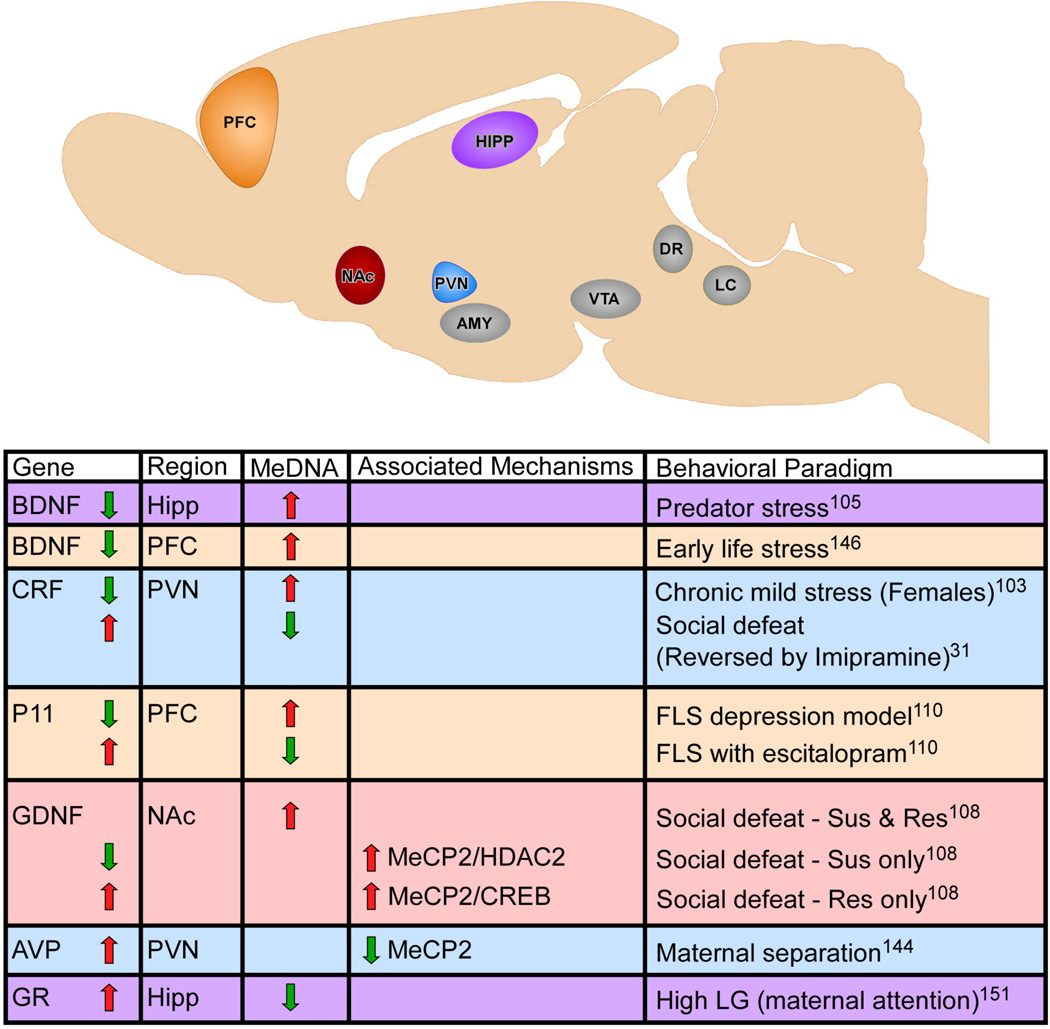
Figure 1. Effect of stress on DNA methylation and gene expression.
Cartoon of sagittal section of rodent brain depicts several inter-connected regions that have been implicated in stress responses (top). Table (bottom) lists the small number of specific genes whose methylation has been shown to date to be altered by stress, the brain regions in which this regulation occurs, and the type of stress involved. Transcriptional mechanisms associated with such altered DNA methylation are listed under Associated Mechanisms. Arrows indicate increase (red) or decrease (green) in gene expression (left) or methylation (right). References are in superscript. Hipp, hippocampus; PFC, prefrontal cortex; PVN, paraventricular nucleus of hypothalamus; NAc, nucleus accumbens; AMY, amygdala; VTA, ventral tegmental area; DR, dorsal raphe; LC, locus coeruleus; Sus, susceptible; Res, resilient.
The inability to regulate and terminate these stress responses can result in many forms of dysregulation, as illustrated by sustained elevations of glucocorticoid levels, which profoundly affect neuroendocrine systems including immune responses, metabolism, and reproduction (5). Indeed, stressful experiences have been shown to increase the prevalence of a wide array of health impairments, including coronary artery disease, chronic pulmonary disease, and certain cancers (6, 7). Stress can also have negative consequences on behavioral adaptations and predispose some individuals to depressive- or anxiety-related disorders (8). However, there is a strong inter-individual variability in susceptibility to stress, such that most individuals are resilient: they can maintain normal physiological and psychological function despite being subjected to horrendous stress (9, 10). Under the right conditions, prior exposure to moderate levels of stress might promote future resilience, a phenomenon referred to as “stress inoculation” (11). Although stress-related disorders such as depression, post-traumatic stress disorder (PTSD), and other anxiety syndromes are partly hereditary, a majority of the risk appears to be non-genetic. The central hypothesis in the field, therefore, is that exposure to a range of stressful stimuli in combination with an individual’s genetic constitution determine his or her initial coping strategy, maladaptive vs. resilient behavioral responses, and ultimate response to treatment (12, 13).
Stress-induced changes in gene expression within the brain’s limbic circuitry have long been posited to mediate this interplay between genes and environment (14). Altered expression of several candidate genes has received a great deal of attention. Induction of brain derived neurotrophic factor (BDNF) in hippocampus is important in the efficacy of antidepressant treatment and its repression may contribute to the pathophysiology of depression in animal models (15). In contrast, sustained induction of BDNF in the NAc, which persists for at least 1 month after chronic social defeat stress, mediates depression-like behavioral abnormalities, whereas reversal of this effect induces an antidepressant-like response (9, 16). Other examples include suppression of the WNT/DVL/GSK3β/β-catenin signaling cascade in several limbic brain regions in response to chronic stress, with opposite changes seen upon antidepressant treatment (17–20), and the long-lasting down-regulation of glucocorticoid receptors (GR) in hippocampus resulting from a deficiency in maternal care (21). More recently, genome-wide measures of gene expression have been used to take a more open-ended view of alterations that occur in limbic regions after chronic stress or antidepressant treatment (9, 16, 22–29). This work has also extended to the study of resilience in chronic stress models to identify specific genes whose altered expression actively opposes the development of stress-induced abnormalities (9, 30, 31).
In these various studies, the fact that chronic stress or antidepressant treatment can induce altered expression of certain genes that persist for weeks after the last exposure suggests the involvement of epigenetic mechanisms, which have been implicated in mediating highly stable changes in gene expression during development and in adult tissues. Epigenetics, in its most general sense, is the study of the regulation of a gene’s transcriptional potential in the absence of changes to the DNA sequence. The burgeoning field of epigenetics has been concerned with heritable changes in transcriptional potential as well as the role of equivalent mechanisms in controlling transcriptional regulation during the lifetime of a single organism. Numerous mechanisms underlying epigenetic regulation have been described to date, including direct modification of DNA’s transcriptional potential by methylation of cytosine bases, and indirect regulation of DNA’s accessibility through several types of chemical modification of the proteins involved in chromatin structure, including histones and many non-histone proteins (32). These mechanisms determine the accessibility of a gene to the transcriptional machinery and ultimately the ability to produce mRNA transcripts (Figure 2). An additional mode of epigenetic regulation is mediated by several types of non-coding RNAs (ncRNAs), including small noncoding RNAs (sncRNAs) and long noncoding RNAs (lncRNAs), which influence gene expression at multiple levels, for example, by modifying chromatin structure at specific genes, RNA splicing, and mRNA stability (33).
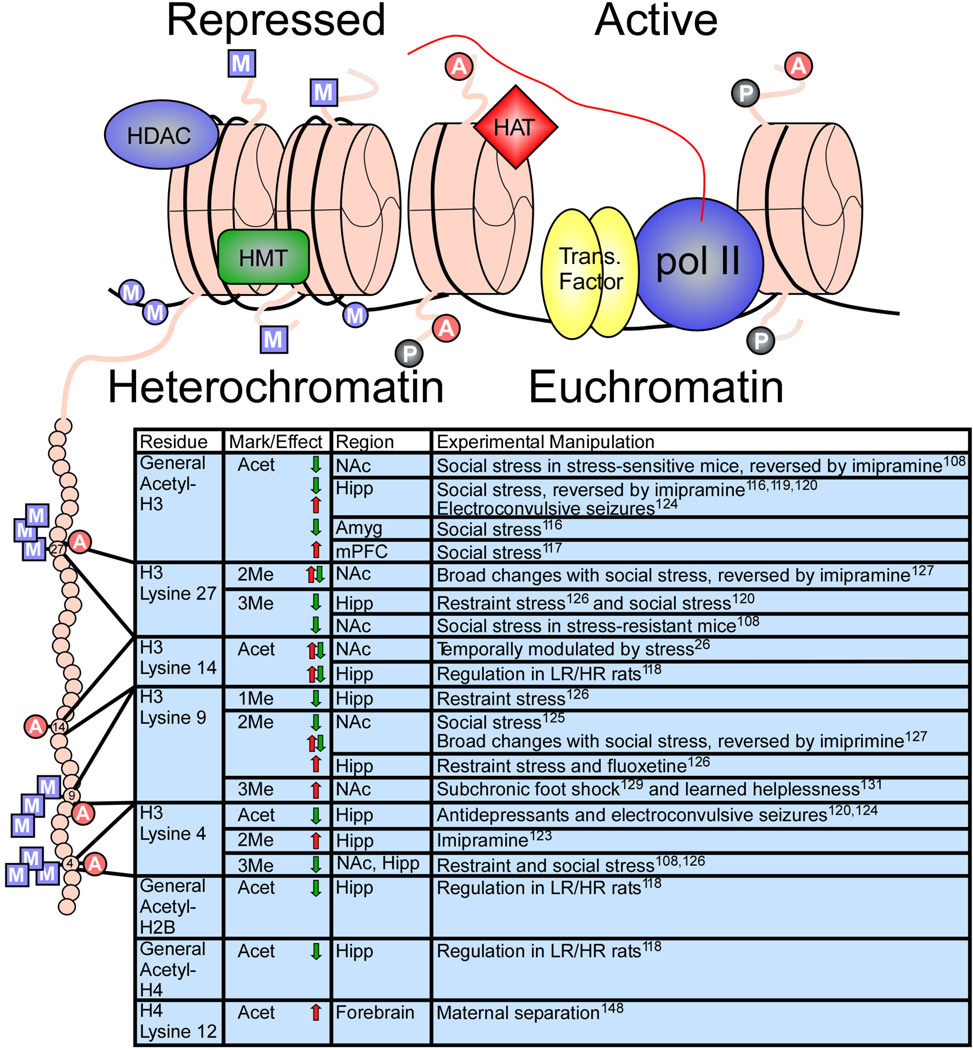
Figure 2. Chromatin modifications regulated by stress or antidepressant treatment.
Illustration (top) indicates histone octomers (pink) in heterochromatin (left) and euchromatin (right), along with associated proteins and histone tail/DNA modifications. A, acetylation; P, phosphorylation; square M, histone methylation; circle M, DNA methylation. Table (bottom) lists histone tail modifications of specific residues—depicted on the expanded histone tail illustration (left)—that are regulated by various stress paradigms or antidepressant treatments within the indicated brain regions. Arrows indicate an increase (red) or decrease (green) in specific modifications. References are in superscript. HDAC, histone deacetylase; HMT, histone methyltransferase; HAT, histone acetyltransferase; Pol II, RNA polymerase II; Trans. Factor, transcription factor.
Much of what we know about epigenetic mechanisms comes from developmental biology, where chromatin changes govern the acquisition and stability of cellular phenotypes throughout differentiation (34, 35). Such cellular differentiation is accomplished by complex epigenetic regulation of individual genes via cues from surrounding cells and influences from the organism’s environment. In contrast, many cell types in the adult organism no longer divide, yet their phenotype, including their transcriptional potential, changes dramatically in response to environmental challenges. In the brain, there are many thousands of cell types with distinct phenotypes that are subjected to experience-dependent phenotypic modification throughout the lifetime. There is growing evidence that epigenetic mechanisms play a key role in this plasticity, thereby mediating stable functional changes in the brain in response to environmental exposures (36–41). By stably changing diverse aspects of neuronal function, epigenetic modifications have been implicated in learning and memory (42–44), social and maternal behaviors (45, 46), and pathological behaviors, such as drug addiction (47, 48), depression (26) and PTSD (49). Such epigenetic modifications could underlie the longlasting behavioral abnormalities seen in these disorders as well as the extraordinary inter-individual variability in vulnerability to adversity. In fact, epigenetics might partly explain the difficulty in identifying the specific genetic variations that contribute to these syndromes, the significant discordance between monozygotic twins, and the differential gender susceptibility to certain psychiatric diseases (50, 51). The study of epigenetic regulation thus opens new doors to understanding normal behavioral traits as well as disease etiology in humans. Although best demonstrated in cancer and stem cell biology, pathological epigenetic marks have been observed in psychiatric disorders, for example, in postmortem tissue of depressed patients (26, 52, 53), and individuals that committed suicide (54). However, careful interpretation is required since these epigenetic marks could represent the cause of the disability or a mark of the disease state, or both.
In this review, we discuss recent progress toward understanding epigenetic regulation by stress during early life and adulthood in the brain’s limbic circuitry, and how such mechanisms might contribute to susceptibility versus resilience to stress-related disorders and to their reversal by antidepressant treatments. We also address recent insight into possible epigenetic transmission of behavioral adaptations to stress across generations.
Overview of Epigenetic Mechanisms
DNA Methylation
Cytosine bases can be covalently modified by methylation at the 5’ position, resulting in a methyl group projecting into the major groove of DNA (55). In mammals, this predominantly occurs in the palindromic sequence 5’-CpG-3’ (and occasionally 5’-CpNpG-3’) and does not interfere with normal hydrogen bonding with complementary guanine bases. Approximately 3% of all cytosines in the human genome are methylated (56), and proper cytosine methylation (5mC) is required for cell differentiation, genetic imprinting, suppression of repetitive elements, and X-chromosomal inactivation (57). The location of CpG bases in mammalian DNA is uneven, occurring at high concentrations in select regions termed CpG islands. CpG islands overlap with the promoters of 50–60% of human genes and are typically methylated to a much lower extent than CpG dinucleotides found outside of islands (58).
Although methylated cytosines can both prevent and promote the binding of various proteins to DNA (59), CpG methylation in promoter regions is generally considered to repress gene transcription (see below). DNA methylation is catalyzed by DNA methyltransferases (DNMTs), a family of enzymes that include DNMT1, DNMT2, DNMT3a, and DNMT3b (60). These enzymes play distinct roles, with DNMT1 maintaining methylation patterns during DNA replication, while DNMT3a and DNMT3b appear to catalyze de novo methylation of previously unmethylated double-strand DNA (55, 61). Several DNA demethylation pathways have been proposed, such as those mediated by methyl CpG binding domain protein 2 (MBD2) (62), growth arrest and DNA-damage-inducible protein 45 alpha (GADD45a) (63), and GADD45b (64). Recent studies indicate DNA oxidation/repair pathways may drive active DNA demethylation in nondividing neurons. Also, ten-eleven-translocation (TET) enzymes have been shown to oxidize 5mC into 5′-hydroxymethylcytosine (5hmC), which can be further oxidized into 5-formolcystosine and 5-carboxylcystosine (65–68). TET family hydroxylase-induced oxidation-deamination thus represents a new candidate for the long-sought DNA demethylase in brain (69). Interestingly, all three forms of 5mC derivatives seem to be enriched in brain, and 5hmC displays a developmentally programmed acquisition in neuronal cells (70).
DNMTs can directly interact with transcription factors, presumably allowing the methylation of promoter regions at specific locations. In a recent study, nearly 80 transcription factors were found to interact with DNMTs (71). This enables gene-specific binding of MBDs. MBDs have been shown to repress gene transcription through recruitment of corepressor complexes that can interfere with the transcriptional machinery or directly regulate chromatin structure (see below). MBDs are essential for normal development and growth, as loss of function mutations of the MBD, methyl CpG binding protein 2 (MeCP2) is associated with Rett’s Syndrome (72, 73) and the MBD Np95/ICBP90-like ring finger protein regulates the cell cycle and has been implicated in tumorogenesis (74). Although originally thought to be a very stable modification, it appears that DNA methylation is subject to dynamic regulation in the adult brain (64, 75, 76), and regulation of DNA methylation at specific genes has been demonstrated in multiple cases (77–79).
Histone Modifications
In eukaryotic cells, DNA is densely packed into chromatin through interactions with large protein complexes called nucleosomes. Individual nucleosome cores are composed of an octamer containing four histone dimers, one dimer each of histones H2A, H2B, H3, and H4, around which are wrapped 147 bp of DNA in approximately two superhelical turns (36). Variant histone proteins or post-translational modifications of histones cause changes in chromatin compaction that are correlated with more “open” (euchromatin: transcriptionally permissive) and “closed” (heterochromatin: transcriptionally repressive) states (80). The N-terminal tails of histones are exposed at the surface of the nucleosome and exhibit multiple, reversible covalent modifications that alter the accessibility of DNA to the transcriptional machinery in a regulated fashion (Figure 2). In addition, histones can be substituted with variants, such as the CENP-A variant of H3, which is found specifically in centromeric DNA, and whose tail contains a significantly different set of modifiable residues, which suggests that substitution of this histone may be essential for mitosis (81).
Histone acetylation, which negates the positive charge of lysine residues in the histone tail, is associated with transcriptional activation. This is likely due to a weakening of the interaction with the DNA strand that depends upon the negative charge of lysines (36, 82). In contrast, lack of histone acetylation correlates with gene repression. Histones are acetylated by histone acetyltransferases (HATs), which use acetyl coenzyme A as a cosubstrate, and deacetylated by histone deacetylases (HDACs). HATs comprise a large family of proteins and acetylate multiple lysine residues in the tails of both H3 and H4. HDACs are composed of multiple families of proteins divided into three classes. In the brain, Classes I and II appear to regulate histone deacetylation at most genes, while Class III enzymes (comprising the sirtuin proteins) deacetylate numerous nuclear and cytoplasmic substrates in addition to histones and are implicated in numerous cellular functions (83–86).
Unlike the clear correlation of histone acetylation with transcriptional activation, histone methylation can be associated with both transcriptional activation and repression depending on the particular residue modified and the extent of methylation. Both lysine and arginine residues can be methylated by various histone methyltransferases (HMTs), which use S-adenosylmethionine (SAM) as a cosubstrate. Methylated arginine residues are converted to citrulline by deaminases, while lysines are demethylated by lysine demethylases (87). Unlike acetylation, methylation does not substantially alter the charge of the target residues, but can dramatically change the steric profile and potential molecular interactions through multivalent addition of mono-, di-, or tri-methyl groups. Examples of the bidirectional effects of histone methylation include trimethylation of lysine 4 of H3 (H3K4me3), which is associated with transcriptional initiation, and di- and trimethylation of lysines 9 and 27 of H3 (H3K9me2/3 and H3K9me2/3), which are associated with transcriptional repression (47). In contrast, H3K36me3 is associated with transcription elongation.
Multiple additional modifications of histone tails are known, including phosphorylation, ubiquitination, sumoylation, and ADP ribosylation, among others (32, 36). For instance, phosphorylation of serine 10 of H3 is associated with increased gene transcription through a complex mechanism involving HAT recruitment and prevention of HMT activity at lysine 9 (47). Modification of one histone can also affect subsequent modification of other histones within the nucleosome, as ubiquitination of H2B appears to be a prerequisite for H3K4 methylation and the resulting initiation of transcription (88). The level of complexity in the nearly infinite possible combinations of various modifications at multiple residues of histone tails, as well as the introduction of histone variant subunits, has led to the proposal of a histone “code” whose “reading” by various histone-interacting proteins is essential for the proper regulation of gene expression (81, 89).
Noncoding RNAs
Many types of ncRNAs are well known, such as ribosomal (rRNA) and transfer (tRNA) RNA. More recently, many additional ncRNAs have been described, divided between the lncRNAs and various short varieties, including micro (miRNA), small nuclear (snoRNA), promoter associated (PASR), Piwi-interacting RNA (piRNA), and transcription initiation (tiRNA) types (33). lncRNAs are generally characterized as being longer than 200 nucleotides; they can be spliced like mRNAs to form functional secondary structures and can act as precursors for various sncRNAs (90). Although only a small number of functional lncRNAs have been characterized to date, they have been shown to control gene expression programs at every level (91). lncRNAs appear to function via multiple epigenetic mechanisms, and can modulate the status of protein-coding genes through the recruitment of chromatin remodeling complexes to specific regions of the genome, thereby regulating the chromatin structure of a single promoter region, a cluster of genes, or an entire chromosome (90). lncRNAs can also serve as an enhancer to regulate gene transcription (92).
miRNAs are the best studied sncRNAs in brain. They are post-transcriptional regulators which can bind to complementary sequences on target mRNAs and lead to translational repression or mRNA degradation (93). It appears that miRNAs can bridge with other epigenetic mechanisms to regulate neural plasticity (94). In addition, sncRNAs can influence epigenetic regulation of gene expression. For instance, sncRNAs bound to Argonaute proteins (and other factors required for inhibitory RNAs or RNAi’s, such as Dicer) have been demonstrated to generate H3K9 methylation, and have since been implicated in the process of heterochromatin formation through recruitment of lysine methyltransferases (KMTs) (95). In C. elegans, short interfering RNAs (siRNAs) and RNAi’s trigger long-term gene silencing in a dominantly heritable fashion, with multiple chromatin remodeling factors required for maintenance of the silent state (96).
Regulation of the Epigenome by Stress and Antidepressant Treatment in Adulthood
There is now considerable evidence that chromatin remodeling is a dynamic process that occurs throughout life in many organs including the brain (34, 97, 98). Patterns of DNA methylation and histone acetylation gradually change over time in monozygotic twins (99). Such drift might not occur randomly and could represent the effect of the environment and the accumulation of the individual’s experience. Chromatin remodeling processes are necessary for learning and memory, and aberrant epigenetic modification can lead to cognitive deficits (100, 101). It is within this context that an increasing number of studies have shown that exposure to stress promotes alterations in various epigenetic marks, in particular, histone acetylation and methylation as well as DNA methylation, in various limbic brain regions (Figures 1 and 2). Additionally, experimental manipulation of the epigenetic machinery within these regions has been demonstrated to potently control mood states and responses to stress and antidepressant treatments.
Regulation of DNA methylation in adulthood
Given that DNA methylation is a relatively stable epigenetic mark in post-mitotic cells, it is likely that it participates in the long-lasting change in gene expression underlying the development of stress-induced abnormalities. Recent reports show changes in CpG methylation levels at gene promoters implicated in HPA axis reactivity and antidepressant treatment. Depressive-like symptoms induced by chronic social defeat stress are accompanied by the sustained upregulation of CRF, which is not observed in mice that are resilient to the stress (31). CRF is expressed by neurons of the paraventricular nucleus (PVN) in the hypothalamus and controls HPA axis activity as well as several other stress responses in the brain. Stress induction of the Crf gene is accompanied by a decrease in DNA methylation at its promoter. Chronic imipramine treatment was sufficient to reduce Crf mRNA levels and increase DNA methylation at the Crf promoter only in socially-defeated mice. A related observation is that conditional knockout of MeCP2 in the PVN, which reduces the repression of certain methylated genes, induces an abnormal physiological stress response (102). This shows that epigenetic regulation of genes is critical for appropriate regulation of the HPA axis. Interestingly, female rats exposed to chronic mild stress show increased DNA methylation at the Crf promoter in the PVN, suggesting sex-specific alterations in HPA axis activity (103).
Activation of the HPA axis and the subsequent release of glucocorticoids have a profound impact on hippocampal function because hippocampal neurons express high levels of GR. Therefore, the hippocampus has received a lot of attention for its role in stress-induced behavioral and cognitive deficits and antidepressant responses. One mechanism by which stress affects hippocampal function is through repression of BDNF, as noted above (104). Chronic exposure to predator stress increases methylation of the Bdnf promoter in hippocampus (105), possibly contributing to the morphological changes and neuronal impairments reported after chronic stress in animal models and in depressed humans (106, 107).
Genetic background influences the outcome of stress exposure in animals and humans, and this could be mediated in part by DNA sequence-directed differences in epigenetic modifications in response to stress, a possibility which is just beginning to be explored. Chronic mild stress increases anxiety- and depression-like behaviors in BALB/C mice, effects not seen under the same conditions in C57BL/6 mice. Exposure to stress differentially regulates glial cell-derived neurotrophic factor (GDNF) expression in the NAc in the two strains of mice (108), with BALB/C mice displaying decreased GDNF expression and C57BL/6 mice showing increased expression. The authors report increased DNA methylation and MeCP2 binding at the Gdnf promoter in the NAc of both strains after chronic stress, changes associated with increased expression of DNMT1 and DNMT3a. However, the two strains are distinguished by the protein complexes that associate with MeCP2 at the Gdnf promoter. In BALB/C mice, MeCP2 reportedly interacts with HDAC2 to decrease H3 acetylation and concomitantly represses Gdnf transcription in NAc, whereas the association of MeCP2 with CREB, a transcriptional activator, is purported to enhance GDNF expression in the NAc of C57Bl/6 mice. The authors propose that these differences are driven by different methylation patterns at the Gdnf promoter, although much further work is needed to validate these findings as well as understand their molecular basis. Nevertheless, these observations are consistent with reports that MeCP2 can act as a repressor or activator of gene transcription (109). In the Flinders Sensitive Line (FSL) genetic rodent model of depression, elevations in DNA methylation at the P11 promoter were reduced by chronic administration of escitalopram, leading to an increase in P11 and a decrease in DNMTs in the prefrontal cortex (110). P11 interacts with serotonin and perhaps other receptors and regulates their function, and decreased levels of P11 have been observed in post-mortem depressed patients and associated with depressive-like behaviors in mice (111, 112). Finally, decreased DNA methylation at the promoter of the serum interleukin 6 (IL-6) gene in peripheral blood mononuclear cells was associated with higher levels of IL-6 in adults with a history of depression (113). IL-6 is part of the inflammatory response system that has been shown to be dysregulated in some depressed individuals (114).
As noted, induction of DNMTs occurs in NAc after chronic social defeat stress (115), and recent evidence demonstrates directly that this adaptation contributes to the development of depressive-like behaviors. Overexpression of DNMT3a in the NAc facilitates vulnerability to the deleterious effects of the stress. This mechanism appears critical for the establishment of maladaptative behaviors because intra-NAc infusion of two DNMT inhibitors, RG108 or Zebularine, reversed the behavioral deficits induced by chronic stress (108, 115). Since DNMTs generally function to repress gene transcription, these data suggest that the development of stress-induced behavioral abnormalities is accompanied in the NAc by the downregulation of genes implicated in reward and motivation. On the other hand, DNA methylation in other brain regions, such as those important for the HPA axis as discussed above, constitutively represses genes that participate in stress responses. Therefore, it is unlikely that drugs that globally affect DNA methylation would be viable therapeutic approaches, although targeting site-specific DNA methylation may be a valuable pathway for development of future therapies.
Regulation of histone acetylation in adult responses to stress
Although considered more labile, some histone post-translational modifications can contribute to the persisting abnormalities of stress-related psychopathology. After chronic social defeat in mice, there is a transient decrease in total cellular levels of H3 acetylation in NAc followed by a more persistent increase which lasts for at least 10 days after the last stress (26). This lasting increase may be mediated via the sustained repression of HDAC2 expression in this brain region. Interestingly, increased H3 acetylation and decreased HDAC2 levels are seen in the NAc of depressed humans. Nevertheless, these adaptations seem to be adaptive, since local infusion of HDAC inhibitors into the NAc, or local inhibition of HDAC2 via overexpression of a dominant negative mutant of the enzyme, exerts potent antidepressant-like actions in several behavioral assays (26, 108). Moreover, local administration of an HDAC inhibitor produced a partly similar pattern of gene expression changes in NAc as seen with systemic fluoxetine treatment (26). Chronic social defeat stress induces more transient increases in global H3 acetylation levels in other brain regions, such as hippocampus, amygdala, and medial prefrontal cortex, and these effects may also be adaptive since HDAC inhibitors are antidepressant-like when administered into these regions as well (116, 117). However, other groups have observed decreased H3 acetylation levels in hippocampus after chronic stress and the reversal of these effects by imipramine treatment (116, 118, 119).
Nevertheless, several groups have demonstrated antidepressant-like effects of HDAC inhibitors upon systemic administration (120–122), with such inhibitors having no effect on depression-like behaviors in non-stressed mice (123). These studies thus suggest that HDAC inhibitors show some potential as novel antidepressant agents. However, the lack of specificity of the inhibitors used to date, and their limited ability to penetrate the brain, are major caveats that must be considered. Given the lack of brain-specific HDAC isoforms, it seems unlikely that HDAC inhibitors of acceptable safety and specificity can be developed for treatment of depression. Still these data can buttress drug discovery efforts by helping to identify the genes important in depression models, which can then be targeted for drug development. For example, comparison of genes within a given limbic region regulated by fluoxetine versus HDAC inhibitors might allow identification of novel mechanisms that are unique to HDAC inhibition and perhaps capable of better, faster antidepressant responses (26).
Currently, little is known about the specific genes at which stress-induced changes in histone acetylation mediate such regulation of depression-like behavior. Chronic social defeat stress down-regulates Bdnf transcription in hippocampus, an effect reversed by antidepressant treatment (120). While the repression may be mediated by enhanced DNA and histone methylation (see below), antidepressant reversal of this effect is associated with increased H3 acetylation at the Bdnf promoter (120, 124). This hyperacetylation resulting from antidepressant treatment is mediated by the down-regulation of HDAC5 in hippocampus, which was shown to be necessary for the behavioral effects of chronic antidepressant administration (120). Further work is needed to identify, in a genome-wide manner, the many other genes involved in mediating the antidepressant-like effects of enhanced histone acetylation at the levels of several limbic brain regions.
Regulation of histone methylation in adult responses to stress
Chronic social defeat, coincident with the sustained increase in H3 acetylation in NAc noted above, causes a global decrease in levels of H3K9me2, a repressive mark, mediated via downregulation of the associated HMTs, G9a and G9a-like protein (GLP) (125). These latter adaptations represent stress-induced pathological changes, because conditional knockout of G9a from the NAc, or its pharmacological inhibition, promoted susceptibility to social stress (125). In contrast, G9a overexpression in NAc, which increases H3K9me2 levels, promotes stress resilience and induces antidepressant-like responses (26, 125). One gene that contributes to these effects is Ras, a small GTPase upstream of the ERK signaling cascade. Stress-induced decreases in H3K9me2 at the Ras gene in the NAc leads to an increase in its expression and to subsequent activation of the ERK signaling cascade, including activation of CREB, which then induces depression-like behavioral abnormalities (Figure 3). It is interesting that two adaptations associated with gene activation (reduced H3K9me2 and increased H3 acetylation) exert opposite behavioral effects. One explanation is that different genes are affected by these modifications, a possibility that now requires direct investigation with genome-wide methods.
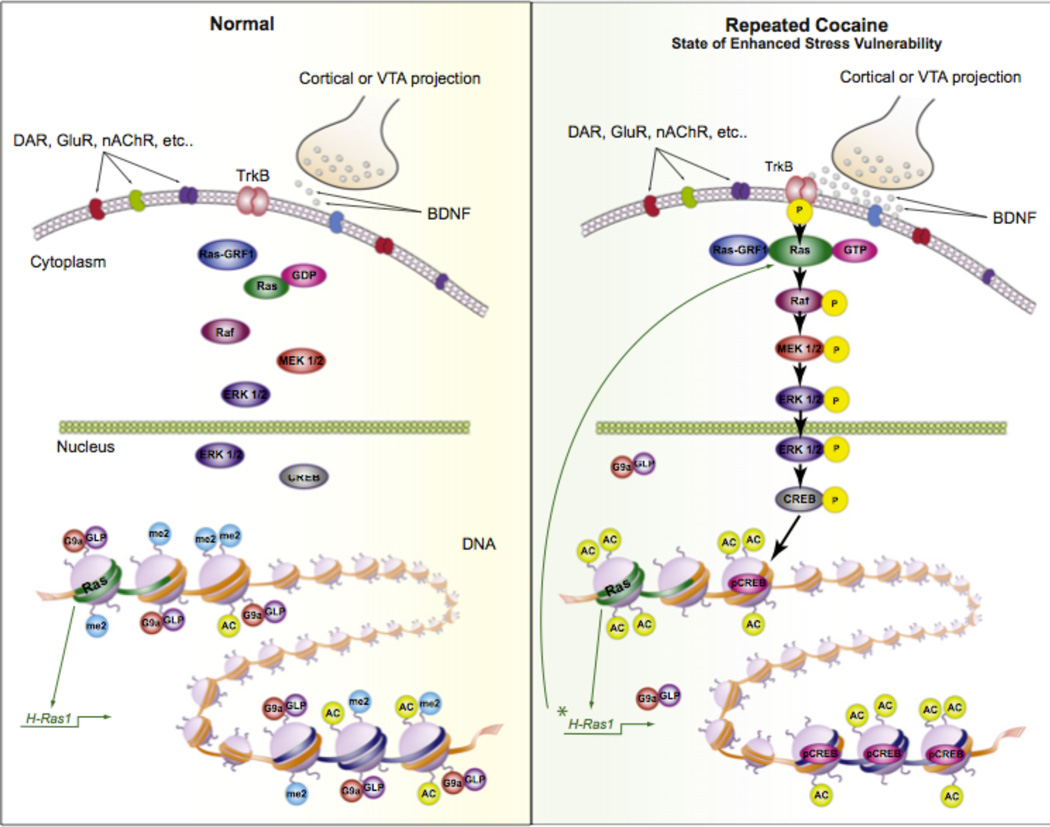
Figure 3. Enhanced Vulnerability to stress via cocaine-Induced priming of BDNF signaling in NAc.
Repeated cocaine increases vulnerability to the depressive-like effects of social defeat stress via priming BDNF signaling through Ras induction in NAc. Under control conditions (left view), BDNF activation of TrkB signaling is limited. However, after repeated cocaine (right view), BDNF-TrkB signaling is elevated in NAc, causing enhanced phosphorylation and activity of downstream-signaling mediators including CREB. This cocaine-initiated maladaptive response occurs not only through increased BDNF signaling in NAc but also through increased Ras expression as a result of decreased G9a binding, and decreased H3K9me2, at the H-Ras1 gene promoter. Chronic stress is associated with similar adaptations in this brain region. Ras also appears to be a target for CREB, creating a positive feed-forward loop, promoting CREB activation and Ras expression as well as depressive-like behavior. From 125.
Chronic stress has also been shown to alter levels of histone methylation in hippocampus. Differential effects on global levels of H3K4me3, H3K9me3, and H3K27me3 within distinct hippocampal subfields occur in response to acute versus chronic restraint stress (126). Interestingly, our group found that chronic social defeat stress increases H3K27me3 levels at the Bdnf gene promoter in hippocampus in concert with its repression (120). Histone repressive marks are often associated with increased DNA methylation, and this is consistent with increased methylation observed at this gene in response to other forms of stress (105).
The discussion above highlights the need for genome-wide assessments of chromatin changes, which will allow identification of previously unknown mechanisms involved in stress-related disorders and antidepressant action. Such studies are just now getting underway. ChIP-chip analysis (chromatin immunoprecipitation followed by genome-wide promoter arrays) for H3K9me2 identified numerous genes that display increases or decreases in this chromatin mark one month after chronic social defeat stress (Figure 4) (127). Interestingly, a similar global pattern of H3K9me2 binding was observed after prolonged adult social isolation. Although these two forms of chronic stress are ethologically distinct, they induce some similar behavioral abnormalities (9, 128). Furthermore, imipramine treatment reverses the large majority of these histone methylation changes in NAc induced by chronic social defeat stress, and the pattern of chromatin changes induced by imipramine resembles that seen in resilient mice (127). These novel findings suggest that one mechanism by which antidepressants work is to induce some of the same molecular adaptations that occur naturally in more resilient individuals, and that a new path forward in antidepressant drug discovery is to mimic such natural resilience mechanisms. For example, these data implicated the WNT/DVL/GSK3β/β-catenin pathway in the NAc in mediating susceptibility versus resilience, as well as antidepressant action, findings later validated by direct manipulation of this cascade (20).
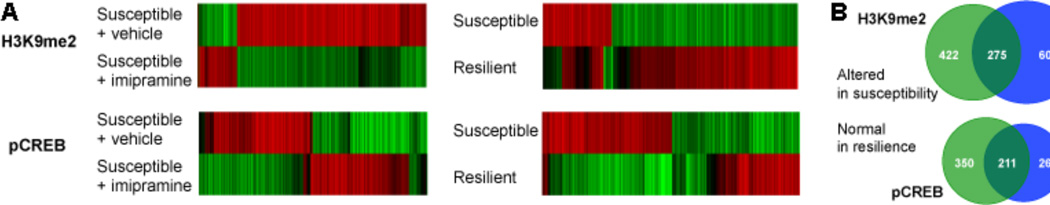
Figure 4. Regulation of H3K9me2 and phospho-CREB (pCREB) binding in NAc after chronic social defeat stress.
A. The heatmaps show that, for both marks, the large majority of changes (red, up; green, down) seen in susceptible mice are not seen after chronic imipramine; as well, most changes observed in susceptible mice are not seen in resilient mice and vice versa. Note the lower bars in each heatmap are normalized to the upper bars, meaning that green would depict reversal of a red change, not a change in the opposite direction. B. The Venn diagrams show the appreciable overlap between apparent mechanisms of imipramine and of resilience. From 127.
Another ChIP-chip study mapped the genome-wide binding of SETDB1, an H3K9 HMT, in mouse prefrontal cortex and identified the NR2B NMDA glutamate receptor subunit as one key target (129). Interestingly, these authors also demonstrated that overexpression of SETDB1 in forebrain exerts antidepressant-like actions, consistent with the observations, noted above, that G9a, another H3K9 HMT, also produces antidepressant-like effects when overexpressed in the NAc (125). The identification of NR2B as one relevant substrate emphasizes the need in the field to further characterize the influence of chronic stress and antidepressant treatments on synaptic plasticity in several key limbic regions (130). Loss of KAP1, a protein implicated in transcriptional repression, also increases stress vulnerability (131), highlighting the large number of chromatin mechanisms that likely control adaptations to chronic stress.
Regulation of miRNAs in adult responses to stress
Additional forms of epigenetic regulation are likely involved in stress and antidepressant action. Chronic administration of fluoxetine reportedly increases levels of miRNA-16 (miR-16), which targets the serotonin transporter (SERT) transcript in serotoninergic neurons and decreases SERT expression (132). More surprisingly, fluoxetine was found to down-regulate miR-16 levels in noradrenergic neurons of the locus coeruleus and to thereby enable certain serotonergic phenotypes in those cells. Further work is needed to validate this adaptation and explore its contribution to the antidepressant effects of fluoxetine. In addition, constitutive and ubiquitous manipulations of SIRT1, a Class III HDAC as noted earlier, has recently been shown to affects basal levels of anxiety, with increased SIRT1 activity associated with increased anxiety-like behaviors (133). SIRT1 has also been shown to decrease the expression of miR-134 in hippocampus, thereby enhancing CREB and BDNF levels (134), which could contribute to its effects on anxiety and stress responses. These early reports highlight the need for additional studies of miRNAs in stress and antidepressant action.
Epigenetic Regulation by Early-Life Stress
During development, the brain is sensitive to environmental changes, such as parental care and stressful events. Cognitive stimulation can lead to broad effects on brain structure and function, hence influencing cognitive and affective function in children, adolescents, and young adults (6, 7). Maltreatment, neglect, and trauma during childhood are known to increase the risk of depressive, anxiety, and substance abuse disorders as well as the risk of becoming an abusive parent (135–139). In contrast, more mild forms of stress may promote resilience (10, 11). Such bidirectional regulation during early life of susceptibility to stress-related illness in adulthood is likely mediated by stable alterations in chromatin structure of specific genes in brain. Longitudinal studies revealed changes in DNA methylation in buccal epithelial cells that were associated with early life adversity (140), although how such changes relate to brain function remains unknown. Nevertheless, animal models have clearly identified epigenetic rep rogramming of gene expression as a stable mechanism by which early life experience, in particular stress and maternal care, affect the individual’s responses to stress later in life (141).
One of the most commonly used procedures for inducing early life stress in rodents is periodic maternal separation during early postnatal life. It is characterized by long-term physiological and behavioral alterations, including elevated glucocorticoid levels and hyperactivity of the HPA axis to subsequent stresses (1, 142, 143). Murgatroyd et al. reported that arginine vasopressin (AVP) undergoes increased expression in the PVN with maternal separation. In addition to its role in water homeostasis, AVP is a key regulator of the HPA axis (144). AVP is secreted by PVN neurons and, in conjunction with CRF, regulates the release of adrenocorticotropin hormone (145). Early life stress activates calcium/calmodulin-dependent protein kinase II (CaMKII) and downstream phosphorylation of MeCP2 at serine438 in the PVN, which leads to the release of MeCP2 from the Avp gene enhancer (144). This is associated with activation of the Avp gene and increased HPA axis stress reactivity, both of which are sustained into adulthood. However, the life-long Avp gene induction is not accompanied by persistent CaMKII activation and MeCP2-S438 phosphorylation. Rather, kinase activation and MeCP2 phosphorylation are followed by Avp gene enhancer demethylation. Presumably, both CaMKII activation and MeCP2 phosphorylation exert broad transcriptional effects beyond the Avp gene, which remains a focus for future investigations. Similarly, newborn rats exposed to stressed caretakers for the first postnatal week display significant increases in DNA methylation at regulatory regions (exons IV and IX) of the Bdnf gene in prefrontal cortex and hippocampus, and persistent BDNF downregulation, in prefrontal cortex but not hippocampus (146). Such early life regulation of BDNF expression might influence stress vulnerability later in life (147).
Though the causal role of DNA methylation in transcriptional regulation during postnatal brain development has been challenged (148), these studies of early life stress demonstrate that DNA methylation is an ideal mechanism, at least from a heuristic point of view, for life-long changes in transcriptional potential. Early life stress can also modify histone acetylation levels through changes in HDACs. Maternal separation in BALB/C mice decreases HDACs levels in the adult forebrain, leading to a concomitant increase in acetylated H4 lysine 12 (149). Activation of HDACs and the resulting decrease in acetylated H4 worsen the effect of stress, suggesting that the observed reduction in HDAC expression is a positive adaptive process.
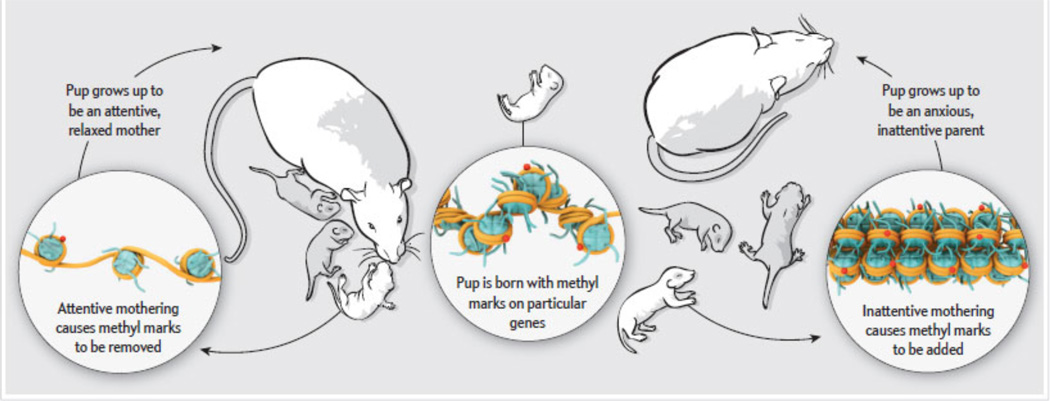
In rodents, the style of maternal care has long-term effects on the behavioral and endocrine responses to stress in offspring (142). Maternal care is measured by the frequency of pup licking and grooming (LG) behavior, which is the major source of tactile stimulation for the neonatal rat that regulates endocrine, cardiovascular, and behavioral responses (150). Variation in maternal behavior is a naturally occurring phenomenon, which can have long-term effects on neural systems that regulate learning and memory, neuroplasticity, and emotional and stress responses (150). In fact, differential levels of maternal care profoundly influence hippocampal glucocorticoid receptor (GR) levels, hence affecting the regulation of the HPA axis by stress. As adults, the offspring of high LG mothers show increased hippocampal GR expression and therefore enhanced negative feedback regulation by glucocorticoids in comparison to adult animals reared by low LG mothers (21). This range of GR expression levels has been traced inversely to levels of methylation at the GR gene promoter (Figure 5). Lower levels of methylation enhance the accessibility of the promoter to transcription factors and chromatin regulatory proteins such as CREB-binding protein (CBP) (151, 152). Increased binding of CBP, a HAT that acetylates specific sites of H3 and H4, leads to increased histone acetylation at the GR promoter in high LG offspring (153, 154). All effects of high LG rearing could be reversed in the adult offspring by intracerebroventricular infusion of L-methionine, which acts as a methyl group donor (155). Similarly, infusion of the HDAC inhibitor trichostatin A (TSA) reverses the changes induced by low LG rearing (151). These findings once again demonstrate the interplay between DNA methylation and histone modifications, and show that changes in the epigenome established by the environment during early development may be reversed by pharmacological approaches in adults– emphasizing the plasticity of chromatin regulation in the adult brain. Although low LG rats show a heightened response to stress, they show greater contextual learning under stressful conditions, suggesting that the long-term behavioral adaptations induced by maternal care have some adaptive value (156, 157), i.e., that increased stress responsiveness might prepare the individual to survive in more aversive environments. McGowan et al (158) demonstrated the clinical relevance of these observations in rats: they found increased methylation of the GR promoter and reduced GR expression in the hippocampus of suicide victims with a life history of childhood abuse.
Figure 5. Long-term epigenetic effect of maternal care.
Studies in rats have shown that epigenetics can influence maternal behavior and that this effect can be passed from one generation to the next by acting on the pup’s brain alone, without altering germ cells. When pups are born, genes involved in regulating the animals’ responses to stress are differentially methylated, which enhances sensitivity to stress. If the pups are raised by a mother that displays high levels of grooming behavior, many of these methyl marks are removed, leaving the animals less responsive to stress. When these pups mature, they, too, will be more attentive mothers. If the pups, however, are raised by a mother that displays low levels of grooming, their genes will gain methyl marks. They grow up to be more stress responsive and display lower grooming levels toward their own pups. From 212.
Other behaviors are also dependent on mother-infant interactions. Maternal LG behavior affects female offspring in their own expression of maternal care. Indeed, maternal care is a matrilineally transmitted behavior that is mediated in part by long-term alteration of oxytocin expression in the hypothalamic medial preoptic area (MPOA) and its regulation by estrogen (157). Oxytocin plays a central role in social bonding and maternal care. In female offspring of low LG mothers, there is decreased expression of estrogen receptor-alpha (ERα) through increased methylation at the ERα gene promoter. This reduces the ability of estrogen to activate oxytocin receptor expression in the MPOA (45), hence reducing the ability of oxytocin itself to promote maternal care. This epigenetic regulation of ERα expression within the MPOA emerges during the first postnatal week and is maintained into adulthood. Cross-fostering studies reveal a direct relationship between the maternal behavior and the behavioral and physiological development of the pup. In contrast, female offspring of high LG animals exhibit reduced methylation at the ERα promoter and increased ERα expression. Consequently, these females have increased estrogen sensitivity, and greater oxytocin function, which drives LG behavior. Later changes in the social environment can reverse these behavioral adaptations (159), suggesting that these epigenetic marks are subjected to modification throughout life.
A major need in the field is to extend these studies of AVP, GR, and ERα to identify the presumably many additional genes whose methylation might further contribute to these behavioral phenotypes. By using a high density tiling array, the adult rat chromosome 18, which contains the GR gene, was examined for DNA methylation, H3K9 acetylation, and transcriptional changes under differential LG mothering (160). The study demonstrated global epigenomic differences in adult brain, which span gene promoters, coding regions, and gene ends as well as intergenic regions. The data suggest that early life maternal care has a broad effect on transcription, with hundreds of genes implicated (45, 160, 161). Among these genes are those encoding several protocadherin isoforms as well as rRNA, and hypermethylation of the rRNA promoter was observed in the hippocampus of humans who died by suicide with a history of early childhood neglect/abuse (162). Further work is needed to characterize the functional consequences of these and the many other observed promoter modifications on life-long stress vulnerability.
Early life stress also encompasses environmental exposures during pregnancy. There is an extensive literature on potential effects of prenatal stress on the development of depression or other psychiatric syndromes (163–165), which would be expected to have an epigenetic basis. In humans, depressed mood during the third trimester of pregnancy induced an increased DNA methylation at the GR promoter in blood leukocytes of the offspring (166). This effect might relate to the finding that offspring of holocaust survivors with PTSD showed lower 24-hour urinary cortisol excretion than offspring of survivors without PTSD (167, 168). These early observations are intriguing and define more mechanistic experiments that are now being carried out in rodent models, as discussed further below under Possible Heritability of Behavioral Adaptations to Stress.
This discussion of epigenetic consequences of early life stress focuses on DNA methylation, which, because of its stability, is an attractive mechanism for mediating life-long behavioral adaptation. However, the notion that DNA methylation changes are subject to dynamic and bidirectional regulation in fully differentiated neurons goes against a prevailing view in the field that it represents a permanent modification during cell differentiation (169). There are thus several reports noted above that de novo DNA methylation occurs in postnatal neurons (151), and that DNA demethylation occurs throughout life (75, 76, 151). The notion that there is a wave of de novo DNA methylation in neurons early in life is supported by the finding that DNMT expression peaks in neurons during early postnatal life (170). These observations raise the interesting possibility that DNA methylation serves a crucial function in mediating the critical time windows for activity-dependent synaptic formation and refinement, and associated behavioral adaptations, long known to exist in early life (171).
There are normally tens to hundreds of CpG dinucleotides in a given gene’s regulatory and coding regions, and the combination of “on” (CpG demethylation) and “off” (CpG methylation) signals at each CpG site produces a complex “methylome” for each gene. Since methylation changes at distinct regions of a gene might exert different functional effects, measures of total DNA methylation do not provide an accurate measure of that gene’s transcriptional potential. We are still learning about the functional consequences of different patterns of DNA methylation at neural genes, and much further work is needed to translate the methylome, acting in concert with the newly defined 5hmC, as well as large numbers of other forms of chromatin modifications, into understanding the gene-specific epigenetic effects of early life stress.
Possible Heritability of Behavioral Adaptations to Stress
Our discussion to this point has focused on a role for epigenetic mechanisms in mediating stable, in some cases life-long, neural and behavioral adaptations in response to environmental challenges. Far more provocative is the idea of the transmission of parental adaptations to environmental challenges to offspring when the young do not experience the challenges themselves (Figure 6). Inheritance of acquired traits might offer an advantage to the population by increasing the fitness of the species, and enabling adaptation to variations in the environment at a faster rate than natural selection would allow. Such non-genetic inheritance of acquired behavioral traits is reminiscent of the ideas put forth by Jean-Baptiste Lamarck in his 1809 book, Philosophie Zoologique, which have been widely discredited. Rather, the trans-generational transmission of behavioral traits and psychiatric disorders in humans is believed to be mediated via genetic mechanisms working in concert with parenting and culture. Nevertheless, recent advances in epigenetics have renewed interest in possible non-genetic, non-behavioral transmission of behavioral experience via epigenetic mechanisms through the germline. Such use of the term epigenetics, to be distinguished from epigenetic mechanisms occurring within a single organism, continues to cause confusion in the field (172).
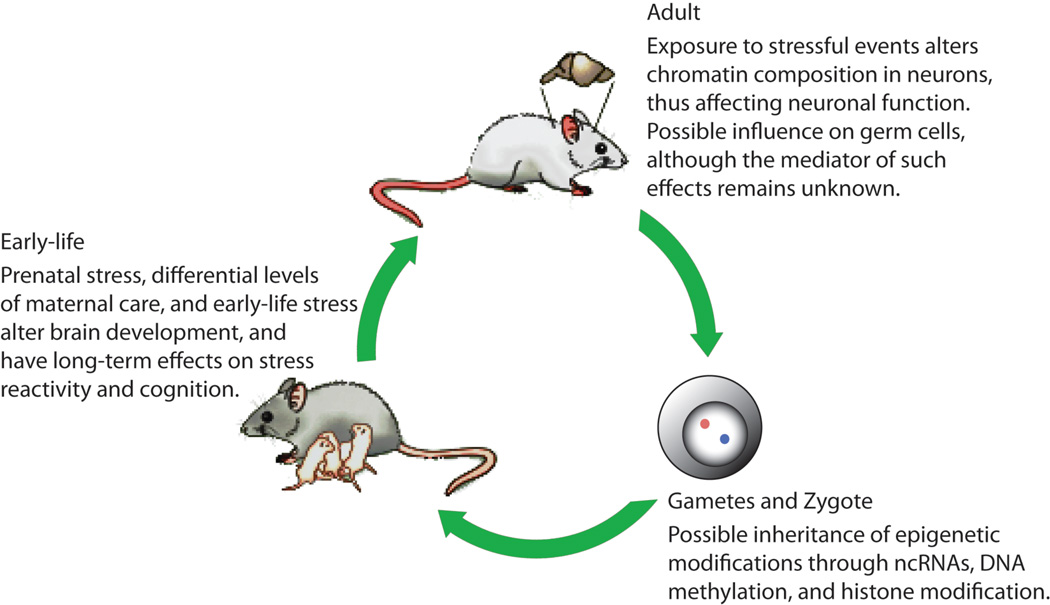
Figure 6. Contribution of epigenetic modifications throughout life cycle.
Environmental challenges, such as stress, have been associated with several short- and long-term chromatin modifications in the brain affecting neural development and function. Gametes and primordial germ cells are also sensitive to stress and might carry epigenetic transformation through subsequent generations. The development of germ cells is characterized by the genome-wide demethylation of DNA in gametes and the establishment of parental chromatin marks required for genomic imprinting. Following fertilization, there is another genome-wide DNA demethylation phase. However, some epigenetic marks escape such reprogramming (imprinted genes, DNA methylation at specific location, some histone post-translational modifications). Moreover, thousands of non-coding RNAs present in the gametes could transmit their information and potentially affect the cellular phenotype in a non-genomic manner. Modified with permission from 41.
Several groups have begun to explore the transmission of stress-related pathologies to subsequent generations via epigenetic mechanisms. Franklin et al. (173) showed that most of the behavioral alterations induced by early life maternal separation are expressed by the F1 offspring of males subjected to maternal separation. These male F1 offspring, when bred with normal females, transmitted their behavioral abnormalities almost exclusively to female offspring for 2 generations (F2 and F3). Maternal separation was reported to alter DNA methylation levels in the promoter regions of the genes encoding MeCP2, the CRF2 receptor, and the cannabinoid receptor-1 (CB1) in the germline of the separated males. Similar DNA methylation patterns were observed in the cerebral cortex of female offspring. No differences in maternal care were observed, suggesting a paternal epigenetic transmission of stress-related behavior, but in a sexually dimorphic manner. In a model of early life stress with impaired maternal care, abnormal DNA methylation and expression patterns of BDNF in the prefrontal cortex are induced for up to two generations (146). Such effects on methylation patterns are not totally reversed by cross-fostering, suggesting that some of these effects are not the result of postnatal experiences. However, females with a history of early life maltreatment show increased anxiety while pregnant, which could influence fetus development. Although prenatal and postnatal effects cannot be ruled out, transgenerational perpetuation of DNA methylation changes could be attributable to epigenetic gametic inheritance as well. Prenatal stress during the early gestation stage leads to dysmasculinized male offspring with altered stress responsiveness (174, 175). This effect was transmitted to the F2 male generation and was associated with reduction in miRNA levels in the brains of male neonates (176). These miRNAs were sensitive to testosterone levels and may play a role in organizing the brain’s sexual dimorphism.
While these studies support the robust transmission of behavioral experience through the germline, different effects were obtained in a model of adult exposure to stress. Dietz et al. (177) first showed the transmission of dramatic depression- and anxiety-like behavioral abnormalities induced in adult male mice by chronic social defeat stress to both their male and female offspring. Abnormalities in peripheral biomarkers of chronic stress (plasma levels of BDNF, VEGF, and corticosterone) were also transmitted to the offspring. However, the vast majority of these abnormalities were not observed in offspring of socially-defeated males generated by in vitro fertilization; only subtle behavioral effects were apparent. These data suggest that the primary mode of transmission of the effects of chronic stress may not be epigenetic, but behavioral, perhaps through female mice adjusting their reproductive investment depending on interactions with an impaired male, a known phenomenon (178). On the other hand, it is possible that the process of in vitro fertilization corrupted any epigenetic marks in the sperm of defeated males. The discrepancies between this study of adult mice, and those involving early life stress, raise the possibility that epigenetic mechanisms may be more important when they occur during development.
There are several reports of the trans-generational epigenetic transmission of other behavioral experiences, such as diet, drugs of abuse, and chemical toxins. Exposure to cocaine in adult male mice mated with naïve females resulted in offspring that displayed deficits in sustained attention and spatial working memory performance (179). This is associated with a decrease in DNMT1 and an increase in DNMT3 levels in spermatozoa, which might cause alterations in the DNA methylation of certain genes. Maternal high-fat diet increased body size and reduced insulin sensitivity in offspring that persisted across multiple generations (180). At the third generation, this phenotype was only transmitted to female offspring through the paternal lineage, suggesting a stable germline-based trans-generational mode of inheritance (181). This effect might be the consequence of the regulation of paternally imprinted genes that influence developmental changes in growth and body size and result in paternal transmission specifically to daughters (181). Similarly, exposure to endocrine disruptors (vinclozolin) affects DNA methylation at imprinted genes in the male germline, leading to behavioral and physiological abnormalities that are trans-generationally persistent (182–184).
What are the most plausible mechanisms for such epigenetic heritability? Epigenetic marks such as DNA methylation and histone modifications are extensively erased in primordial germ cells during gametogenesis in mice (E10.5 – E13.5) and immediately after fertilization in early embryogenesis to ensure the totipotency of cells in the early embryo. This epigenetic reprogramming limits the potential in mammals for epigenetic trans-generational inheritance. However, there are a few examples in mammals of epigenetic marks that are resistant to both phases of reprogramming: transgenes, retrotransposons, and heterochromatin at centromeres (172, 178, 185–187). During spermatogenesis, while most of the histones are replaced by protamines, some histones are retained in a locus-specific manner and could play a role in early embryo development (188). If retained after fertilization, these epigenetic modifications could participate in the trans-generational transfer of information. Another possible mechanism of trans-generational phenotypic persistence is through cytoplasmic RNAs, including mRNA, siRNA, piRNA, and miRNA (189). Indeed, RNAs from gametes allow the newly fertilized oocyte to initiate transcription and have been observed in large quantity in human sperm (190).
Only maternal-derived imprinted genes are protected from demethylation after fertilization (191). Therefore, altered DNA methylation of these imprinted genes could not persist trans-generationally unless new methylation sites are induced during critical developmental periods by environmental factors. Genomic imprinting is a form of epigenetic regulation that distinguishes the maternal and paternal genomes, leading to a bias in expression between maternally and paternally transmitted genes. Imprinted genes are silenced through DNA methylation and related chromatin modifications established in the germline (192). Proper expression of imprinted genes is essential for embryonic development (193) and numerous postnatal functions (46, 194–196). It is thus not surprising that genetic abnormalities involving imprinted genes cause developmental disorders such as Prader-Willi and Angelman’s syndromes. Besides gene imprinting and X-inactivation, whereby DNA methylation coordinates the random silencing of either X chromosome in females, there is growing evidence that allele-specific methylation occurs across the entire genome and is heterogeneous across tissues and individuals (197, 198). Most of this allele-specific methylation is determined by DNA sequence, hence showing Mendelian inheritance patterns. There is also allele-specific methylation occurring through non-cis events, which might contribute to heritability of complex diseases (199). Indeed, a recent genome-wide screen indicated that over 1,300 loci show parental-origin allelic expression bias with complex regulation throughout brain development in a cell-type and gender-specific manner (200, 201). Moreover, DNA methylation at some non-imprinted genes escapes the post-fertilization DNA methylation reprogramming (202). Therefore, allele-specific methylation could contribute significantly to the heritability of complex etiological disorders, including depression and other stress-related disorders, and explain the significant discordance of psychiatric disorders between monozygotic twins, the difference in gender susceptibility to certain psychiatric diseases, and the largely inconsistent genetic association studies of depression (50, 51).
This discussion highlights the difficulties in defining epigenetic inheritance of stressful experiences. First, it is very challenging to fully control for parental behavior. As noted, maternal behavior may be affected by the behavior of the impregnating male. As well, while cross-fostering can control for post-natal maternal care, it does not control for intra-uterine factors which might also be crucial. Likewise, in vitro fertilization is not a perfect test for germline mechanisms, since the epigenetic state of sperm may vary as a function of how and when it is harvested with respect to the stress exposure. There may also be other factors in semen that influence the mother or offspring. Ultimately, to establish epigenetic mechanisms for the trans-generational transmission of stressful experiences, it will be essential to demonstrate epigenetic modifications at specific genes in sperm or egg cells and understand how those are transmitted into alterations in neural function in adult offspring across several generations. Currently, there are no known mechanisms by which such transmission can occur, although this is a challenge for future research.
Conclusions
Epigenetic investigations promise to improve our understanding of the mechanisms by which individuals exhibit widely varying responses to adverse life events, both during development and in adulthood. Animal models of stress-related disorders are beginning to reveal specific chromatin modifications that maintain stable patterns of gene expression and thereby mediate the aberrant neuroplasticity associated with these disorders. Equivalent studies have focused on chromatin modifications that contribute to antidepressant responses. Several different types of epigenetic mechanisms have been implicated to date, including stress- or antidepressant-induced changes in DNA methylation, histone acetylation and methylation, and miRNAs. However, the modifications examined to date represent the tip of the iceberg of the diverse and complex range of epigenetic mechanisms that are likely involved in stress and antidepressant action.
Epigenetic mechanisms are regulated throughout life in neurons with some stress-sensitive time windows, such as early life, being particularly important. Although several specific genes have been identified as being permanently regulated (e.g., GR in hippocampus, CRF and AVP in PVN), many other genes probably play a role in stress vulnerability as well. Moreover, in addition to sustained alterations in steady-state mRNA levels of certain genes, it is likely that early life stress also modifies the transcriptional potential of many additional genes through their epigenetic priming or desensitization. Identifying all of the genes that are epigenetically regulated by stress, and important for controlling stress vulnerability across the life cycle, will therefore require the superimposing of several levels of analysis, involving genome-wide analyses of RNA expression along with those of DNA methylation, histone modifications, chromatin remodeling factors, and transcription factors, among many others. Such work will also require the optimization of bioinformatic tools that enable the examination of the vast datasets involved.
This endeavor is further complicated by the large number of brain regions involved in stress-related psychopathology. Most studies to date have focused by necessity (e.g., cost) on single brain regions, despite the knowledge that depression and anxiety disorders involve abnormal functioning of numerous brain circuits. Not only will each brain region have to be investigated independently, but several distinct neuronal and non-neuronal cell types within each region will have to be characterized separately. For example, epigenetic modifications that occur in glutmatergic pyramidal neurons in prefrontal cortex are likely very different from those that occur in several different subtypes of GABAergic interneurons, astrocytes, oligodendrocytes, and so on. Fortunately, tools are emerging that permit the cell type-specific investigation of chromatin modifications in a heterogeneous tissue such as brain (203).
To fully understand epigenetic mechanisms of stress and antidepressant action, the field must transition increasingly to genome-wide approaches, most particularly those utilizing massive parallel sequencing following ChIP (ChIP-seq), RNA isolation (RNA-seq), or selective DNA isolation (e.g., for methylcytosine). These recently developed methodologies provide whole-genome coverage as well as improved sensitivity compared to older ChIP-chip and other microarray approaches. As noted above, the combination of numerous chromatin modifications (204) will allow the delineation of the stress and antidepressant “epigenomes.” As our knowledge advances, other types of chromatin modifications and epigenetic regulation that are sensitive to the environment will be indentified, such as alterations in retrotransposons (205–207). Similar to cocaine exposure (208), stress exposure might increase genomic insertion of retrotransposable elements that regulate the transcriptome (209). A newly recognized form of DNA methylation, 5hmC, is highly abundant in neurons and predominantly localized around the coding region in association with other histone repressive marks, which suggests that it might participate in the epigenetic control of neuronal function (70, 210). These are just some examples of the new lines of investigation that have the potential of revealing fundamentally novel mechanisms controlling stress-related phenomena.
Finally, there has been interest, as noted in this review, in the possible therapeutic applications of drugs aimed at chromatin modifying enzymes (211). HDAC inhibitors exert potent antidepressant-like responses in diverse animal models, as just one example. Whether such drugs offer realistic possibilities for drug discovery remains uncertain, given that most chromatin modifying proteins are broadly expressed throughout the brain and peripheral tissues. Nevertheless, there are many hundreds of known chromatin regulatory proteins; targeting those proteins that are enriched in brain is a viable path forward. Moreover, there is no question that epigenetic characterization of stress models will reveal a far more complete view of the range of proteins and ncRNAs that mediate stress-induced pathology, the reversal of that pathology during antidepressant treatment, and the resistance against developing such pathology in resilient individuals. This knowledge will provide for the first time a comprehensive roadmap for future drug discovery efforts.
References
- 1.de Kloet ER, Joels M, Holsboer F. Stress and the brain: from adaptation to disease. Nat Rev Neurosci. 2005;6:463–475. doi: 10.1038/nrn1683. [DOI] [PubMed] [Google Scholar]
- 2.Chrousos GP. Stress and disorders of the stress system. Nat Rev Endocrinol. 2009;5:374–381. doi: 10.1038/nrendo.2009.106. [DOI] [PubMed] [Google Scholar]
- 3.Shin LM, Liberzon I. The neurocircuitry of fear, stress, and anxiety disorders. Neuropsychopharmacology. 2009;35:169–191. doi: 10.1038/npp.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McEwen BS. Central effects of stress hormones in health and disease: Understanding the protective and damaging effects of stress and stress mediators. Eur J Pharmacol. 2008;583:174–185. doi: 10.1016/j.ejphar.2007.11.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Von Holst D. The concept of stress and its relevance for animal behavior. Advances in the Study of Behavior. 1998:1–131. [Google Scholar]
- 6.Hackman DA, Farah MJ, Meaney MJ. Socioeconomic status and the brain: mechanistic insights from human and animal research. Nat Rev Neurosci. 2010;11:651–659. doi: 10.1038/nrn2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shonkoff JP, Boyce WT, McEwen BS. Neuroscience, molecular biology, and the childhood roots of health disparities: building a new framework for health promotion and disease prevention. JAMA. 2009;301:2252–2259. doi: 10.1001/jama.2009.754. [DOI] [PubMed] [Google Scholar]
- 8.Yehuda R, McFarlane AC, Shalev AY. Predicting the development of posttraumatic stress disorder from the acute response to a traumatic event. Biol Psychiatry. 1998;44:1305–1313. doi: 10.1016/s0006-3223(98)00276-5. [DOI] [PubMed] [Google Scholar]
- 9.Krishnan V, Han MH, Graham DL, Berton O, Renthal W, et al. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell. 2007;131:391–404. doi: 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 10.Feder A, Nestler EJ, Charney DS. Psychobiology and molecular genetics of resilience. Nat Rev Neurosci. 2009;10:446–457. doi: 10.1038/nrn2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyons DM, Parker KJ, Schatzberg AF. Animal models of early life stress: implications for understanding resilience. Dev Psychobiol. 2010;52:616–624. doi: 10.1002/dev.20500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uher R. Genes, environment, and individual differences in responding to treatment for depression. Harv Rev Psychiatry. 2011;19:109–124. doi: 10.3109/10673229.2011.586551. [DOI] [PubMed] [Google Scholar]
- 13.Kendler KS. The dappled nature of causes of psychiatric illness: replacing the organic-functional/hardware-software dichotomy with empirically based pluralism. Mol Psychiatry. 2012 doi: 10.1038/mp.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyman SE, Nestler EJ. Initiation and adaptation: a paradigm for understanding psychotropic drug action. Am J Psychiatry. 1996;153:151–162. doi: 10.1176/ajp.153.2.151. [DOI] [PubMed] [Google Scholar]
- 15.Duman RS, Monteggia LM. A neurotrophic model for stress-related mood disorders. Biol Psychiatry. 2006;59:1116–1127. doi: 10.1016/j.biopsych.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 16.Berton O, McClung CA, Dileone RJ, Krishnan V, Renthal W, et al. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science. 2006;311:864–868. doi: 10.1126/science.1120972. [DOI] [PubMed] [Google Scholar]
- 17.Gould TD, O'Donnell KC, Picchini AM, Dow ER, Chen G, Manji HK. Generation and behavioral characterization of beta-catenin forebrain-specific conditional knock-out mice. Behav Brain Res. 2008;189:117–125. doi: 10.1016/j.bbr.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gould TD, Einat H, O'Donnell KC, Picchini AM, Schloesser RJ, Manji HK. Beta-catenin overexpression in the mouse brain phenocopies lithium-sensitive behaviors. Neuropsychopharmacology. 2007;32:2173–2183. doi: 10.1038/sj.npp.1301338. [DOI] [PubMed] [Google Scholar]
- 19.Okamoto H, Voleti B, Banasr M, Sarhan M, Duric V, et al. Wnt2 expression and signaling is increased by different classes of antidepressant treatments. Biol Psychiatry. 2010;68:521–527. doi: 10.1016/j.biopsych.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkinson MB, Dias C, Magida J, Mazei-Robison M, Lobo M, et al. A novel role of the WNT-dishevelled-GSK3beta signaling cascade in the mouse nucleus accumbens in a social defeat model of depression. J Neurosci. 2011;31:9084–9092. doi: 10.1523/JNEUROSCI.0039-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu D, Diorio J, Tannenbaum B, Caldji C, Francis D, et al. Maternal care, hippocampal glucocorticoid receptors, and hypothalamic-pituitary-adrenal responses to stress. Science. 1997;277:1659–1662. doi: 10.1126/science.277.5332.1659. [DOI] [PubMed] [Google Scholar]
- 22.Karssen AM, Her S, Li JZ, Patel PD, Meng F, et al. Stress-induced changes in primate prefrontal profiles of gene expression. Mol Psychiatry. 2007;12:1089–1102. doi: 10.1038/sj.mp.4002095. [DOI] [PubMed] [Google Scholar]
- 23.Surget A, Wang Y, Leman S, Ibarguen-Vargas Y, Edgar N, et al. Corticolimbic transcriptome changes are state-dependent and region-specific in a rodent model of depression and of antidepressant reversal. Neuropsychopharmacology. 2009;34:1363–1380. doi: 10.1038/npp.2008.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrus BM, Blizinsky K, Vedell PT, Dennis K, Shukla PK, et al. Gene expression patterns in the hippocampus and amygdala of endogenous depression and chronic stress models. Mol Psychiatry. 2012;17:49–61. doi: 10.1038/mp.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernard R, Kerman IA, Thompson RC, Jones EG, Bunney WE, et al. Altered expression of glutamate signaling, growth factor, and glia genes in the locus coeruleus of patients with major depression. Mol Psychiatry. 2011;16:634–646. doi: 10.1038/mp.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Covington HE, 3rd, Maze I, LaPlant Q, Vialou V, Ohnishi Y, et al. Antidepressant actions of histone deacetylase inhibitors. J Neurosci. 2009;22:11451–11460. doi: 10.1523/JNEUROSCI.1758-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duric V, Banasr M, Licznerski P, Schmidt HD, Stockmeier CA, et al. A negative regulator of MAP kinase causes depressive behavior. Nat Med. 2010;16:1328–1332. doi: 10.1038/nm.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le-Niculescu H, Patel SD, Niculescu AB. Convergent integration of animal model and human studies of bipolar disorder (manic-depressive illness) Curr Opin Pharmacol. 2010;10:594–600. doi: 10.1016/j.coph.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 29.Shelton RC, Claiborne J, Sidoryk-Wegrzynowicz M, Reddy R, Aschner M, et al. Altered expression of genes involved in inflammation and apoptosis in frontal cortex in major depression. Mol Psychiatry. 2011;16:751–762. doi: 10.1038/mp.2010.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vialou V, Robison AJ, Laplant QC, Covington HE, 3rd, Dietz DM, et al. DeltaFosB in brain reward circuits mediates resilience to stress and antidepressant responses. Nat Neurosci. 2010;13:745–752. doi: 10.1038/nn.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elliott E, Ezra-Nevo G, Regev L, Neufeld-Cohen A, Chen A. Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nat Neurosci. 2010;13:1351–1353. doi: 10.1038/nn.2642. [DOI] [PubMed] [Google Scholar]
- 32.Gibney ER, Nolan CM. Epigenetics and gene expression. Heredity. 2010;105:4–13. doi: 10.1038/hdy.2010.54. [DOI] [PubMed] [Google Scholar]
- 33.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 34.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 35.Van Speybroeck L. From epigenesis to epigenetics: the case of C. H. Waddington. Ann N Y Acad Sci. 2002;981:61–81. [PubMed] [Google Scholar]
- 36.Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60:961–974. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang Y, Langley B, Lubin FD, Renthal W, Wood MA, et al. Epigenetics in the nervous system. J Neurosci. 2008;28:11753–11759. doi: 10.1523/JNEUROSCI.3797-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neves G, Cooke SF, Bliss TV. Synaptic plasticity, memory and the hippocampus: a neural network approach to causality. Nat Rev Neurosci. 2008;9:65–75. doi: 10.1038/nrn2303. [DOI] [PubMed] [Google Scholar]
- 40.Roth TL, Sweatt JD. Regulation of chromatin structure in memory formation. Curr Opin Neurobiol. 2009;19:336–342. doi: 10.1016/j.conb.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dulac C. Brain function and chromatin plasticity. Nature. 2010;465:728–735. doi: 10.1038/nature09231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 44.Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- 45.Champagne FA, Weaver IC, Diorio J, Dymov S, Szyf M, Meaney MJ. Maternal care associated with methylation of the estrogen receptor-alpha1b promoter and estrogen receptor-alpha expression in the medial preoptic area of female offspring. Endocrinology. 2006;147:2909–2915. doi: 10.1210/en.2005-1119. [DOI] [PubMed] [Google Scholar]
- 46.Garfield AS, Cowley M, Smith FM, Moorwood K, Stewart-Cox JE, et al. Distinct physiological and behavioural functions for parental alleles of imprinted Grb10. Nature. 2011;469:534–538. doi: 10.1038/nature09651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maze I, Covington HE, 3rd, Dietz DM, LaPlant Q, Renthal W, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chertkow-Deutsher Y, Cohen H, Klein E, Ben-Shachar D. DNA methylation in vulnerability to post-traumatic stress in rats: evidence for the role of the post-synaptic density protein Dlgap2. Int J Neuropsychopharmacol. 2010;13:347–359. doi: 10.1017/S146114570999071X. [DOI] [PubMed] [Google Scholar]
- 50.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 51.Mill J, Petronis A. Molecular studies of major depressive disorder: the epigenetic perspective. Mol Psychiatry. 2007;12:799–814. doi: 10.1038/sj.mp.4001992. [DOI] [PubMed] [Google Scholar]
- 52.Hobara T, Uchida S, Otsuki K, Matsubara T, Funato H, et al. Altered gene expression of histone deacetylases in mood disorder patients. J Psychiatr Res. 2009;44:263–270. doi: 10.1016/j.jpsychires.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 53.Poulter MO, Du L, Weaver IC, Palkovits M, Faludi G, et al. GABAA receptor promoter hypermethylation in suicide brain: implications for the involvement of epigenetic processes. Biol Psychiatry. 2008;64:645–652. doi: 10.1016/j.biopsych.2008.05.028. [DOI] [PubMed] [Google Scholar]
- 54.Keller S, Sarchiapone M, Zarrilli F, Videtic A, Ferraro A, et al. Increased BDNF promoter methylation in the Wernicke area of suicide subjects. Arch Gen Psychiatry. 2010;67:258–267. doi: 10.1001/archgenpsychiatry.2010.9. [DOI] [PubMed] [Google Scholar]
- 55.Newell-Price J, Clark AJ, King P. DNA methylation and silencing of gene expression. Trends Endocrinol Metab. 2000;11:142–148. doi: 10.1016/s1043-2760(00)00248-4. [DOI] [PubMed] [Google Scholar]
- 56.Nafee TM, Farrell WE, Carroll WD, Fryer AA, Ismail KM. Epigenetic control of fetal gene expression. BJOG. 2008;115:158–168. doi: 10.1111/j.1471-0528.2007.01528.x. [DOI] [PubMed] [Google Scholar]
- 57.Bird A. The methyl-CpG-binding protein MeCP2 and neurological disease. Biochem Soc Trans. 2008;36:575–583. doi: 10.1042/BST0360575. [DOI] [PubMed] [Google Scholar]
- 58.Wang Y, Leung FC. An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics. 2004;20:1170–1177. doi: 10.1093/bioinformatics/bth059. [DOI] [PubMed] [Google Scholar]
- 59.Prokhortchouk E, Defossez PA. The cell biology of DNA methylation in mammals. Biochim Biophys Acta. 2008;1783:2167–2173. doi: 10.1016/j.bbamcr.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 60.Weber M, Schubeler D. Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Curr Opin Cell Biol. 2007;19:273–280. doi: 10.1016/j.ceb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 61.Kim JK, Samaranayake M, Pradhan S. Epigenetic mechanisms in mammals. Cell Mol Life Sci. 2009;66:596–612. doi: 10.1007/s00018-008-8432-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 63.Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 64.Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ito S, Shen L, Dai Q, Wu SC, Collins LB, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He YF, Li BZ, Li Z, Liu P, Wang Y, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Szulwach KE, Li X, Li Y, Song CX, Wu H, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hervouet E, Vallette FM, Cartron PF. Dnmt3/transcription factor interactions as crucial players in targeted DNA methylation. Epigenetics. 2009;4:487–499. doi: 10.4161/epi.4.7.9883. [DOI] [PubMed] [Google Scholar]
- 72.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 73.Nan X, Hou J, Maclean A, Nasir J, Lafuente MJ, et al. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc Natl Acad Sci U S A. 2007;104:2709–2714. doi: 10.1073/pnas.0608056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mori T, Li Y, Hata H, Ono K, Kochi H. NIRF, a novel RING finger protein, is involved in cell-cycle regulation. Biochem Biophys Res Commun. 2002;296:530–536. doi: 10.1016/s0006-291x(02)00890-2. [DOI] [PubMed] [Google Scholar]
- 75.Feng J, Zhou Y, Campbell SL, Le T, Li E, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 77.Sharma RP, Gavin DP, Grayson DR. CpG methylation in neurons: message, memory, or mask? Neuropsychopharmacology. 2010;35:2009–2020. doi: 10.1038/npp.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Feng J, Fan G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int Rev Neurobiol. 2009;89:67–84. doi: 10.1016/S0074-7742(09)89004-1. [DOI] [PubMed] [Google Scholar]
- 79.Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron. 2011;70:813–829. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 81.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 82.Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 83.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sassone-Corsi P. Minireview: NAD+, a circadian metabolite with an epigenetic twist. Endocrinology. 2012;153:1–5. doi: 10.1210/en.2011-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bellet MM, Sassone-Corsi P. Mammalian circadian clock and metabolism - the epigenetic link. J Cell Sci. 2010;123:3837–3848. doi: 10.1242/jcs.051649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- 88.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 89.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 90.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 93.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Im HI, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat Neurosci. 2010;13:1120–1127. doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Djupedal I, Ekwall K. Epigenetics: heterochromatin meets RNAi. Cell Res. 2009;19:282–295. doi: 10.1038/cr.2009.13. [DOI] [PubMed] [Google Scholar]
- 96.Vastenhouw NL, Brunschwig K, Okihara KL, Muller F, Tijsterman M, Plasterk RH. Gene expression: long-term gene silencing by RNAi. Nature. 2006;442:882. doi: 10.1038/442882a. [DOI] [PubMed] [Google Scholar]
- 97.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Meaney MJ, Szyf M. Environmental programming of stress responses through DNA methylation: life at the interface between a dynamic environment and a fixed genome. Dialogues Clin Neurosci. 2005;7:103–123. doi: 10.31887/DCNS.2005.7.2/mmeaney. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 101.Michan S, Li Y, Chou MM, Parrella E, Ge H, et al. SIRT1 is essential for normal cognitive function and synaptic plasticity. J Neurosci. 2010;30:9695–9707. doi: 10.1523/JNEUROSCI.0027-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fyffe SL, Neul JL, Samaco RC, Chao HT, Ben-Shachar S, et al. Deletion of Mecp2 in Sim1-expressing neurons reveals a critical role for MeCP2 in feeding behavior, aggression, and the response to stress. Neuron. 2008;59:947–958. doi: 10.1016/j.neuron.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sterrenburg L, Gaszner B, Boerrigter J, Santbergen L, Bramini M, et al. Chronic stress induces sex-specific alterations in methylation and expression of corticotropin-releasing factor gene in the rat. PLoS One. 2011;6:e28128. doi: 10.1371/journal.pone.0028128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schmidt HD, Duman RS. The role of neurotrophic factors in adult hippocampal neurogenesis, antidepressant treatments and animal models of depressive-like behavior. Behav Pharmacol. 2007;18:391–418. doi: 10.1097/FBP.0b013e3282ee2aa8. [DOI] [PubMed] [Google Scholar]
- 105.Roth TL, Zoladz PR, Sweatt JD, Diamond DM. Epigenetic modification of hippocampal Bdnf DNA in adult rats in an animal model of post-traumatic stress disorder. J Psychiatr Res. 2011;45:919–926. doi: 10.1016/j.jpsychires.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sapolsky RM. Stress, Glucocorticoids, and Damage to the Nervous System: The Current State of Confusion. Stress. 1996;1:1–19. doi: 10.3109/10253899609001092. [DOI] [PubMed] [Google Scholar]
- 107.Gould E, McEwen BS, Tanapat P, Galea LA, Fuchs E. Neurogenesis in the dentate gyrus of the adult tree shrew is regulated by psychosocial stress and NMDA receptor activation. J Neurosci. 1997;17:2492–2498. doi: 10.1523/JNEUROSCI.17-07-02492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Uchida S, Hara K, Kobayashi A, Otsuki K, Yamagata H, et al. Epigenetic status of Gdnf in the ventral striatum determines susceptibility and adaptation to daily stressful events. Neuron. 2011;69:359–372. doi: 10.1016/j.neuron.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 109.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Melas PA, Rogdaki M, Lennartsson A, Bjork K, Qi H, et al. Antidepressant treatment is associated with epigenetic alterations in the promoter of P11 in a genetic model of depression. Int J Neuropsychopharmacol. 2011:1–11. doi: 10.1017/S1461145711000940. [DOI] [PubMed] [Google Scholar]
- 111.Svenningsson P, Chergui K, Rachleff I, Flajolet M, Zhang X, et al. Alterations in 5-HT1B receptor function by p11 in depression-like states. Science. 2006;311:77–80. doi: 10.1126/science.1117571. [DOI] [PubMed] [Google Scholar]
- 112.Anisman H, Du L, Palkovits M, Faludi G, Kovacs GG, et al. Serotonin receptor subtype and p11 mRNA expression in stress-relevant brain regions of suicide and control subjects. J Psychiatry Neurosci. 2008;33:131–141. [PMC free article] [PubMed] [Google Scholar]
- 113.Uddin M, Koenen KC, Aiello AE, Wildman DE, de los Santos R, Galea S. Epigenetic and inflammatory marker profiles associated with depression in a community-based epidemiologic sample. Psychol Med. 2011;41:997–1007. doi: 10.1017/S0033291710001674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schmidt HD, Shelton RC, Duman RS. Functional biomarkers of depression: diagnosis, treatment, and pathophysiology. Neuropsychopharmacology. 2011;36:2375–2394. doi: 10.1038/npp.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.LaPlant Q, Vialou V, Covington HE, 3rd, Dumitriu D, Feng J, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Covington HE, 3rd, Vialou VF, LaPlant Q, Ohnishi YN, Nestler EJ. Hippocampal-dependent antidepressant-like activity of histone deacetylase inhibition. Neurosci Lett. 2011;493:122–126. doi: 10.1016/j.neulet.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hinwood M, Tynan RJ, Day TA, Walker FR. Repeated social defeat selectively increases deltaFosB expression and histone H3 acetylation in the infralimbic medial prefrontal cortex. Cereb Cortex. 2011;21:262–271. doi: 10.1093/cercor/bhq080. [DOI] [PubMed] [Google Scholar]
- 118.Hollis F, Duclot F, Gunjan A, Kabbaj M. Individual differences in the effect of social defeat on anhedonia and histone acetylation in the rat hippocampus. Horm Behav. 2011;59:331–337. doi: 10.1016/j.yhbeh.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hollis F, Wang H, Dietz D, Gunjan A, Kabbaj M. The effects of repeated social defeat on long-term depressive-like behavior and short-term histone modifications in the hippocampus in male Sprague-Dawley rats. Psychopharmacology (Berl) 2010;211:69–77. doi: 10.1007/s00213-010-1869-9. [DOI] [PubMed] [Google Scholar]
- 120.Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 121.Schroeder FA, Lin CL, Crusio WE, Akbarian S. Antidepressant-like effects of the histone deacetylase inhibitor, sodium butyrate, in the mouse. Biol Psychiatry. 2007;62:55–64. doi: 10.1016/j.biopsych.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 122.Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- 123.Gundersen BB, Blendy JA. Effects of the histone deacetylase inhibitor sodium butyrate in models of depression and anxiety. Neuropharmacology. 2009;57:67–74. doi: 10.1016/j.neuropharm.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tsankova NM, Kumar A, Nestler EJ. Histone modifications at gene promoter regions in rat hippocampus after acute and chronic electroconvulsive seizures. J Neurosci. 2004;24:5603–5610. doi: 10.1523/JNEUROSCI.0589-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Covington HE, 3rd, Maze I, Sun H, Bomze HM, DeMaio KD, et al. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron. 2011;71:656–670. doi: 10.1016/j.neuron.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Natl Acad Sci U S A. 2009;106:20912–20917. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wilkinson MB, Xiao G, Kumar A, LaPlant Q, Renthal W, et al. Imipramine treatment and resiliency exhibit similar chromatin regulation in the mouse nucleus accumbens in depression models. J Neurosci. 2009;29:7820–7832. doi: 10.1523/JNEUROSCI.0932-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wallace DL, Han MH, Graham DL, Green TA, Vialou V, et al. CREB regulation of nucleus accumbens excitability mediates social isolation-induced behavioral deficits. Nat Neurosci. 2009;12:200–209. doi: 10.1038/nn.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jiang Y, Jakovcevski M, Bharadwaj R, Connor C, Schroeder FA, et al. Setdb1 histone methyltransferase regulates mood-related behaviors and expression of the NMDA receptor subunit NR2B. J Neurosci. 2010;30:7152–7167. doi: 10.1523/JNEUROSCI.1314-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Belujon P, Grace AA. Hippocampus, amygdala, and stress: interacting systems that affect susceptibility to addiction. Ann N Y Acad Sci. 2011;1216:114–121. doi: 10.1111/j.1749-6632.2010.05896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jakobsson J, Cordero MI, Bisaz R, Groner AC, Busskamp V, et al. KAP1-mediated epigenetic repression in the forebrain modulates behavioral vulnerability to stress. Neuron. 2008;60:818–831. doi: 10.1016/j.neuron.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 132.Baudry A, Mouillet-Richard S, Schneider B, Launay JM, Kellermann O. miR-16 targets the serotonin transporter: a new facet for adaptive responses to antidepressants. Science. 2010;329:1537–1541. doi: 10.1126/science.1193692. [DOI] [PubMed] [Google Scholar]
- 133.Libert S, Pointer K, Bell EL, Das A, Cohen DE, et al. SIRT1 Activates MAO-A in the Brain to Mediate Anxiety and Exploratory Drive. Cell. 2011 doi: 10.1016/j.cell.2011.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gao J, Wang WY, Mao YW, Graff J, Guan JS, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kaufman J, Charney D. Effects of early stress on brain structure and function: implications for understanding the relationship between child maltreatment and depression. Dev Psychopathol. 2001;13:451–471. doi: 10.1017/s0954579401003030. [DOI] [PubMed] [Google Scholar]
- 136.Lupien SJ, McEwen BS, Gunnar MR, Heim C. Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat Rev Neurosci. 2009;10:434–445. doi: 10.1038/nrn2639. [DOI] [PubMed] [Google Scholar]
- 137.Kaufman J, Zigler E. Do abused children become abusive parents? Am J Orthopsychiatry. 1987;57:186–192. doi: 10.1111/j.1939-0025.1987.tb03528.x. [DOI] [PubMed] [Google Scholar]
- 138.Moffitt TE, Caspi A, Harrington H, Milne BJ, Melchior M, et al. Generalized anxiety disorder and depression: childhood risk factors in a birth cohort followed to age 32. Psychol Med. 2007;37:441–452. doi: 10.1017/S0033291706009640. [DOI] [PubMed] [Google Scholar]
- 139.Neigh GN, Gillespie CF, Nemeroff CB. The neurobiological toll of child abuse and neglect. Trauma Violence Abuse. 2009;10:389–410. doi: 10.1177/1524838009339758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Essex MJ, Thomas Boyce W, Hertzman C, Lam LL, Armstrong JM, et al. Epigenetic Vestiges of Early Developmental Adversity: Childhood Stress Exposure and DNA Methylation in Adolescence. Child Dev. 2011 doi: 10.1111/j.1467-8624.2011.01641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bale TL, Baram TZ, Brown AS, Goldstein JM, Insel TR, et al. Early life programming and neurodevelopmental disorders. Biol Psychiatry. 2010;68:314–319. doi: 10.1016/j.biopsych.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Meaney MJ. Maternal care, gene expression, and the transmission of individual differences in stress reactivity across generations. Annu Rev Neurosci. 2001;24:1161–1192. doi: 10.1146/annurev.neuro.24.1.1161. [DOI] [PubMed] [Google Scholar]
- 143.Millstein RA, Holmes A. Effects of repeated maternal separation on anxiety- and depression-related phenotypes in different mouse strains. Neurosci Biobehav Rev. 2007;31:3–17. doi: 10.1016/j.neubiorev.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 144.Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- 145.Lolait SJ, Stewart LQ, Jessop DS, Young WS, 3rd, O'Carroll AM. The hypothalamic-pituitary-adrenal axis response to stress in mice lacking functional vasopressin V1b receptors. Endocrinology. 2007;148:849–856. doi: 10.1210/en.2006-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Roth TL, Sweatt JD. Epigenetic marking of the BDNF gene by early-life adverse experiences. Horm Behav. 2011;59:315–320. doi: 10.1016/j.yhbeh.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Edwards JR, O'Donnell AH, Rollins RA, Peckham HE, Lee C, et al. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome Res. 2010;20:972–980. doi: 10.1101/gr.101535.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Levine A, Worrell TR, Zimnisky R, Schmauss C. Early life stress triggers sustained changes in histone deacetylase expression and histone H4 modifications that alter responsiveness to adolescent antidepressant treatment. Neurobiol Dis. 2012;45:488–498. doi: 10.1016/j.nbd.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang TY, Meaney MJ. Epigenetics and the environmental regulation of the genome and its function. Annu Rev Psychol. 2010;61:439–466. C1–C3. doi: 10.1146/annurev.psych.60.110707.163625. [DOI] [PubMed] [Google Scholar]
- 151.Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 152.Weaver IC, D'Alessio AC, Brown SE, Hellstrom IC, Dymov S, et al. The transcription factor nerve growth factor-inducible protein a mediates epigenetic programming: altering epigenetic marks by immediate-early genes. J Neurosci. 2007;27:1756–1768. doi: 10.1523/JNEUROSCI.4164-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Conway-Campbell BL, George CL, Pooley JR, Knight DM, Norman MR, et al. The HSP90 Molecular Chaperone Cycle Regulates Cyclical Transcriptional Dynamics of the Glucocorticoid Receptor and Its Coregulatory Molecules CBP/p300 During Ultradian Ligand Treatment. Mol Endocrinol. 2011;25:944–954. doi: 10.1210/me.2010-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zeng L, Zhang Q, Gerona-Navarro G, Moshkina N, Zhou MM. Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300. Structure. 2008;16:643–652. doi: 10.1016/j.str.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Weaver IC, Champagne FA, Brown SE, Dymov S, Sharma S, et al. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bagot RC, van Hasselt FN, Champagne DL, Meaney MJ, Krugers HJ, Joels M. Maternal care determines rapid effects of stress mediators on synaptic plasticity in adult rat hippocampal dentate gyrus. Neurobiol Learn Mem. 2009;92:292–300. doi: 10.1016/j.nlm.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 157.Champagne FA. Epigenetic mechanisms and the transgenerational effects of maternal care. Front Neuroendocrinol. 2008;29:386–397. doi: 10.1016/j.yfrne.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonte B, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Champagne FA, Meaney MJ. Transgenerational effects of social environment on variations in maternal care and behavioral response to novelty. Behav Neurosci. 2007;121:1353–1363. doi: 10.1037/0735-7044.121.6.1353. [DOI] [PubMed] [Google Scholar]
- 160.McGowan PO, Suderman M, Sasaki A, Huang TC, Hallett M, et al. Broad epigenetic signature of maternal care in the brain of adult rats. PLoS One. 2011;6:e14739. doi: 10.1371/journal.pone.0014739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Weaver IC, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci U S A. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.McGowan PO, Sasaki A, Huang TC, Unterberger A, Suderman M, et al. Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PLoS One. 2008;3:e2085. doi: 10.1371/journal.pone.0002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Maccari S, Morley-Fletcher S. Effects of prenatal restraint stress on the hypothalamus-pituitary-adrenal axis and related behavioural and neurobiological alterations. Psychoneuroendocrinology. 2007;32(Suppl 1):S10–S15. doi: 10.1016/j.psyneuen.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 164.Weinstock M. The long-term behavioural consequences of prenatal stress. Neurosci Biobehav Rev. 2008;32:1073–1086. doi: 10.1016/j.neubiorev.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 165.Weinstock M. Intrauterine factors as determinants of depressive disorder. Isr J Psychiatry Relat Sci. 2010;47:36–45. [PubMed] [Google Scholar]
- 166.Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3:97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- 167.Yehuda R, Bierer LM. Transgenerational transmission of cortisol and PTSD risk. Prog Brain Res. 2008;167:121–135. doi: 10.1016/S0079-6123(07)67009-5. [DOI] [PubMed] [Google Scholar]
- 168.Yehuda R, Teicher MH, Seckl JR, Grossman RA, Morris A, Bierer LM. Parental posttraumatic stress disorder as a vulnerability factor for low cortisol trait in offspring of holocaust survivors. Arch Gen Psychiatry. 2007;64:1040–1048. doi: 10.1001/archpsyc.64.9.1040. [DOI] [PubMed] [Google Scholar]
- 169.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 170.Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- 171.Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- 172.Youngson NA, Whitelaw E. Transgenerational epigenetic effects. Annu Rev Genomics Hum Genet. 2008;9:233–257. doi: 10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
- 173.Franklin TB, Russig H, Weiss IC, Graff J, Linder N, et al. Epigenetic transmission of the impact of early stress across generations. Biol Psychiatry. 2010;68:408–415. doi: 10.1016/j.biopsych.2010.05.036. [DOI] [PubMed] [Google Scholar]
- 174.Mueller BR, Bale TL. Sex-specific programming of offspring emotionality after stress early in pregnancy. J Neurosci. 2008;28:9055–9065. doi: 10.1523/JNEUROSCI.1424-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mueller BR, Bale TL. Early prenatal stress impact on coping strategies and learning performance is sex dependent. Physiol Behav. 2007;91:55–65. doi: 10.1016/j.physbeh.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 176.Morgan CP, Bale TL. Early prenatal stress epigenetically programs dysmasculinization in second-generation offspring via the paternal lineage. J Neurosci. 2011;31:11748–11755. doi: 10.1523/JNEUROSCI.1887-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Dietz DM, Laplant Q, Watts EL, Hodes GE, Russo SJ, et al. Paternal transmission of stress-induced pathologies. Biol Psychiatry. 2011;70:408–414. doi: 10.1016/j.biopsych.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Curley JP, Mashoodh R, Champagne FA. Epigenetics and the origins of paternal effects. Horm Behav. 2011;59:306–314. doi: 10.1016/j.yhbeh.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.He F, Lidow IA, Lidow MS. Consequences of paternal cocaine exposure in mice. Neurotoxicol Teratol. 2006;28:198–209. doi: 10.1016/j.ntt.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 180.Dunn GA, Bale TL. Maternal high-fat diet promotes body length increases and insulin insensitivity in second-generation mice. Endocrinology. 2009;150:4999–5009. doi: 10.1210/en.2009-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dunn GA, Bale TL. Maternal High-Fat Diet Effects on Third-Generation Female Body Size via the Paternal Lineage. Endocrinology. 2011;152:2228–2236. doi: 10.1210/en.2010-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Skinner MK, Anway MD, Savenkova MI, Gore AC, Crews D. Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior. PLoS One. 2008;3:e3745. doi: 10.1371/journal.pone.0003745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Stouder C, Paoloni-Giacobino A. Transgenerational effects of the endocrine disruptor vinclozolin on the methylation pattern of imprinted genes in the mouse sperm. Reproduction. 2010;139:373–379. doi: 10.1530/REP-09-0340. [DOI] [PubMed] [Google Scholar]
- 185.Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- 186.Daxinger L, Whitelaw E. Transgenerational epigenetic inheritance: more questions than answers. Genome Res. 2010;20:1623–1628. doi: 10.1101/gr.106138.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Franklin TB, Mansuy IM. Epigenetic inheritance in mammals: evidence for the impact of adverse environmental effects. Neurobiol Dis. 2010;39:61–65. doi: 10.1016/j.nbd.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 188.Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Cuzin F, Rassoulzadegan M. Non-Mendelian epigenetic heredity: gametic RNAs as epigenetic regulators and transgenerational signals. Essays Biochem. 2010;48:101–106. doi: 10.1042/bse0480101. [DOI] [PubMed] [Google Scholar]
- 190.Rassoulzadegan M, Cuzin F. The making of an organ: RNA mediated developmental controls in mice. Organogenesis. 2010;6:33–36. doi: 10.4161/org.6.1.11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Delaval K, Feil R. Epigenetic regulation of mammalian genomic imprinting. Curr Opin Genet Dev. 2004;14:188–195. doi: 10.1016/j.gde.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 193.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 194.Davies W, Isles AR, Humby T, Wilkinson LS. What are imprinted genes doing in the brain? Epigenetics. 2007;2:201–206. doi: 10.4161/epi.2.4.5379. [DOI] [PubMed] [Google Scholar]
- 195.Isles AR, Davies W, Wilkinson LS. Genomic imprinting and the social brain. Philos Trans R Soc Lond B Biol Sci. 2006;361:2229–2237. doi: 10.1098/rstb.2006.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Smith FM, Garfield AS, Ward A. Regulation of growth and metabolism by imprinted genes. Cytogenet Genome Res. 2006;113:279–291. doi: 10.1159/000090843. [DOI] [PubMed] [Google Scholar]
- 197.Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 198.Schalkwyk LC, Meaburn EL, Smith R, Dempster EL, Jeffries AR, et al. Allelic skewing of DNA methylation is widespread across the genome. Am J Hum Genet. 2010;86:196–212. doi: 10.1016/j.ajhg.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Meaburn EL, Schalkwyk LC, Mill J. Allele-specific methylation in the human genome: implications for genetic studies of complex disease. Epigenetics. 2010;5:578–582. doi: 10.4161/epi.5.7.12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Gregg C, Zhang J, Butler JE, Haig D, Dulac C. Sex-specific parent-of-origin allelic expression in the mouse brain. Science. 2010;329:682–685. doi: 10.1126/science.1190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, et al. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010;329:643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Borgel J, Guibert S, Li Y, Chiba H, Schubeler D, et al. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet. 2010;42:1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 203.Cheung I, Shulha HP, Jiang Y, Matevossian A, Wang J, et al. Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc Natl Acad Sci U S A. 2010;107:8824–8829. doi: 10.1073/pnas.1001702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Muotri AR, Zhao C, Marchetto MC, Gage FH. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus. 2009;19:1002–1007. doi: 10.1002/hipo.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kuwabara T, Hsieh J, Muotri A, Yeo G, Warashina M, et al. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat Neurosci. 2009;12:1097–1105. doi: 10.1038/nn.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Maze I, Feng J, Wilkinson MB, Sun H, Shen L, Nestler EJ. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc Natl Acad Sci U S A. 2011;108:3035–3040. doi: 10.1073/pnas.1015483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci. 2010;33:345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Covington HI, Berton O. Chromatin-based treatments for affective disorders - Insight or utopia. In: CJaL BE, editor. Depression: From Psychopathology to Pharmacotherapy. Basel: Karger; 2010. pp. 182–198. [Google Scholar]
- 212.Nestler EJ. Hidden switches in the mind. Sci Am Dec. 2011:77–83. doi: 10.1038/scientificamerican1211-76. [DOI] [PMC free article] [PubMed] [Google Scholar]