Abstract
The molecular function of mTERFs (mitochondrial transcription termination factors) has so far only been described for metazoan members of the protein family and in animals they control mitochondrial replication, transcription and translation. Cells of photosynthetic eukaryotes harbour chloroplasts and mitochondria, which are in an intense cross-talk that is vital for photosynthesis. Chlamydomonas reinhardtii is a unicellular green alga widely used as a model organism for photosynthesis research and green biotechnology. Among the six nuclear C. reinhardtii mTERF genes is mTERF-like gene of Chlamydomonas (MOC1), whose inactivation alters mitorespiration and interestingly also light-acclimation processes in the chloroplast that favour the enhanced production of biohydrogen. We show here from in vitro studies that MOC1 binds specifically to a sequence within the mitochondrial rRNA-coding module S3, and that a knockout of MOC1 in the mutant stm6 increases read-through transcription at this site, indicating that MOC1 acts as a transcription terminator in vivo. Whereas the level of certain antisense RNA species is higher in stm6, the amount of unprocessed mitochondrial sense transcripts is strongly reduced, demonstrating that a loss of MOC1 causes perturbed mitochondrial DNA (mtDNA) expression. Overall, we provide evidence for the existence of mitochondrial antisense RNAs in C. reinhardtii and show that mTERF-mediated transcription termination is an evolutionary-conserved mechanism occurring in phototrophic protists and metazoans.
INTRODUCTION
Most of our knowledge about mitochondrial gene expression and its regulation results from research carried out with non-phototrophic organisms, especially mammalian and yeast cells (1,2). In the case of phototrophic eukaryotes, there is an additional level of complexity, as metabolism in the mitochondria needs to be coordinated to that in the chloroplast (3). As yet the molecular mechanisms underpinning the regulation of gene expression in the mitochondria of plant cells, including the unicellular green alga Chlamydomonas reinhardtii, which is an established model organism widely used to study the regulation of nuclear and organelle gene expression, are unclear.
Mitochondria of C. reinhardtii retain a small, but information-dense, genome (4), which contains genes encoding eight proteins, including the complex IV subunit 1, five complex I subunits, apocytochrome b of complex III, a reverse transcriptase-like protein and a strain-dependent number of introns (5). The rRNA genes encoding large (L) and small (S) ribosomal rRNAs are split into modules encoding rRNA segments (eight L and four S modules), which are interspersed with one another or protein-coding and tRNA genes (6). All the mitochondrial rRNAs are encoded by the mitochondrial genome of C. reinhardtii (4), but the majority of tRNAs have to be imported from the cytosol (7). In vitro labelling experiments and physical mapping indicated that the mitochondrial DNA (mtDNA) of C. reinhardtii is a linear monomeric molecule (8), but electron microscopy (9) demonstrated that mtDNA preparations contain a small fraction of circular molecules. The telomere structure, known to be decisive for mtDNA architecture (10), of the C. reinhardtii mtDNA is unusual and differs from structures described for other organisms (11). Both strands of the C. reinhardtii mtDNA contain transcription units (12–14), and two sequences identified in the intergenic region located between the transcription units might act as a bi-directional promoter (15). Transcription of the C. reinhardtii mtDNA generates long polycistronic transcripts (12–14), which are further processed to yield the mature mRNAs. The use of few transcription initiation sites to drive the expression of large transcription units is a feature frequently found in mitochondria of protists (16) and mammals (1), whereas higher plant chondromes represent a more complex system with multiple transcription initiation sites (17).
Recent work has identified a nuclear gene of C. reinhardtii termed mTERF-like gene of Chlamydomonas (MOC1), which plays a role in fine-tuning mitochondrial transcription on changes in illumination. Absence of MOC1 in the stm6 mutant causes a pleiotropic phenotype characterized by perturbed mitorespiration (18) and multiple effects on the physiological state of the plastid resulting in light-sensitivity (18,19) and, interestingly, the enhanced production of biohydrogen (20).
MOC1 is a member of the mTERF (mitochondrial transcription termination factor) family of transcription factors, which are found in metazoans (21) including higher plants (mono- and dicotyledonous) (22), but which are absent in fungal and prokaryotic genomes (21). Current knowledge about mTERF function was mainly obtained by the characterization of metazoan mTERF proteins (23). All vertebrate mTERF factors are targeted to the mitochondrion, where they are implicated in the regulation of transcription (24–28), translation (29) and replication (30,31). The knockout of mTERF2 (27) and tissue-specific inactivation of mTERF3 (26) in mice caused severe mitochondrial phenotypes resulting from aberrant transcription of the organellar genome, possibly at the level of transcription initiation. To date, three mTERF factors from different organisms have been shown to terminate mtDNA transcription (23). Human mTERF1 binds specifically to a sequence within the tRNALeu(UUR) gene and arrests RNA polymerase progression in vitro (24,32). Simultaneous binding of mTERF1 to the termination site and the heavy strand promoter in vitro (25) was proposed as a mechanism facilitating rapid re-initiation of transcription after termination at the tRNALeu(UUR) site, but evidence that the higher amount of rRNA relative to mRNA in vivo depends on this mechanism remains to be provided. Another well-described mTERF protein is the Drosophila homologue of human mTERF named DmTTF, which possesses two specific binding sites in the Drosophila mtDNA and terminates transcription in vitro (33) and in vivo (34).
Given the current interest in developing strains of green algae with enhanced production of biohydrogen, we decided to investigate the molecular function of MOC1. To this end, we analysed possible DNA-binding properties of recombinant MOC1 and investigated the phenotypic consequences of an MOC1 knockout in the mutant stm6. The sum of our data shows that mTERF-mediated transcription termination occurs in mitochondria of lower photosynthetic eukaryotes, for the first time demonstrating that this mechanism is not restricted to animal mitochondria. MOC1 binds specifically to an octanucleotide sequence within the mitochondrial rRNA-coding module S3, and a loss of MOC1 increases read-through transcription at the S3-binding site thereby causing elevated antisense RNA levels in the mutant stm6. Impaired transcription termination in stm6 alters the mitochondrial transcriptome leading to a growth defect under strictly heterotrophic conditions.
MATERIALS AND METHODS
Strains and culture conditions
The MOC1 knockout mutant stm6 and the complemented strain B13 were generated as described by Schönfeld et al. (18). Liquid cultures of C. reinhardtii were cultivated mixotrophically (50 µmol photons m−2 s−1) and heterotrophically (dark cultivation) in Tris–acetate phosphate medium [TAP, (35)] or phototrophically in high-salt medium (35). Dark growth as well as phototrophic low light (50 µmol photons m−2 s−1) and high-light growth (1500 µmol photons m−2 s−1) was analysed in a FMT 150 photobioreactor (Photon Systems Instruments, Brno, Czech Republic) with continuous monitoring of the optical density at 735 nm as a measure for cell density.
Plasmid construction
To construct expression vector pRSET-A(Thr)MOC1ΔN46, the MOC1 cDNA [(36); clone HCL008g04; Accession No. AV640010; Kasuza DNA Research Institute (Kisarazu, Japan)] was first amplified with primers MOC1-MTS_A fw/rev (Supplementary Table S1), and the resulting polymerase chain reaction (PCR) product was sub-cloned into vector pGem T-Easy (Promega). The MOC1-coding sequence lacking the first 46 amino acids of the N-terminus was then either cloned into a derivative (kind gift of Dr Ernesto Cota Segura, Imperial College London) of expression vector pRSET A (Invitrogen) or vector pGEX-6-P3 (GE Healthcare). For the construction of pQE-80 L-MOC1, the full coding sequence of MOC1 was amplified with primers MOC1Ex fw/rev (Supplementary Table S1) using the MOC1 cDNA clone as a template and sub-cloned into vector pGem T-Easy before cloning into the Escherichia coli expression vector pQE80L (Quiagen).
Production of recombinant MOC1 in E. coli for binding studies
Expression of recombinant MOC1 in E. coli KRX cells (Promega) transformed with plasmid pRSET-A(Thr)MOC1ΔN46 was induced by adding 0.01% (w/v) l(+)-rhamnose to cultures grown at 37°C until OD600 = 0.5. Subsequently cultures were transferred to 18°C and cultivated for 16 h before the cells were harvested by centrifugation. For the expression of glutathione S-transferase (GST)-MOC1 in KRX cells, cultures were grown at 37°C to an OD600 of 0.4 before 1 mM IPTG (isopropyl β-d-1-thiogalactopyranoside) was added. Expression was carried out at 37°C overnight. Recombinant His-MOC1 was purified by nickel-nitrilotriacetic acid (Ni-NTA) chromatography and GST-MOC1 affinity purified on glutathione–cellulose (Supplementary Methods).
Generation of an antiserum directed against MOC1
For the production of an MOC1-specific antiserum, full-length MOC1 was expressed in E. coli KRX cells (Promega) transformed with plasmid pQE-80 L-MOC1. The His-tagged protein was purified from inclusion bodies according to standard protocols for denaturing Ni-NTA chromatography and used to raise an antiserum in rabbits at Seqlab (Göttingen, Germany).
Isolation of mitochondria from C. reinhardtii cells
Mitochondria were purified from the cell wall-deficient C. reinhardtii strain cc400 as described by Atteia et al. (37) using the BioNeb cell disruption system (Glas-Col, Terre Haute, USA) with N2 (20 psi).
Sodium dodecyl sulphate–polyacrylamide gel electrophoresis and immunoblotting
Protein samples were separated on 12.5% (w/v) denaturing sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) gels containing 6 M urea and stained with Coomassie blue R-250 or electroblotted onto nitrocellulose membrane (0.2-μm pore size; GE Healthcare, Germany). Immunoblotting analyses were performed using specific primary antibodies and a horseradish peroxidase-conjugated secondary antibody (GE Healthcare, Germany). Signals were visualized using the FUSION-FX7 detection system (Peqlab, Germany) or ECL Western Blotting Substrate (Pierce, Germany) in combination with X-ray films. The following polyclonal antisera were used as primary antibodies: rabbit anti-MOC1, rabbit anti-histone H3 (Agrisera, Sweden, product AS10710), rabbit anti-LHCBM4/6 (kindly provided by M. Hippler, Institute of Plant Biology and Biotechnology, University of Muenster, Muenster, Germany) and anti-COXIIb (Agrisera, Sweden, product AS06 151).
In vitro binding studies with dsDNA–cellulose
Recombinant MOC1 (1.8 µM) or bovine serum albumin (BSA) (1.8 µM) in DNA-cellulose binding buffer (DCBB) buffer [50 mM hydroxyethyl piperazineethanesulfonic acid (HEPES), pH 8.0, 50 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA); 1 mM β-mercaptoethanol and 1 mM Pefabloc® SC protease inhibitor] were incubated for 2 h with 60 mg double-stranded DNA–cellulose resin (Sigma, D8515) at room temperature. The resin was washed using DCBB buffer, and a step gradient (100, 300, 600 and 1000 mM NaCl in DCBB) was used to elute bound protein.
Electrophoretic mobility shift assays
Double-stranded DNA probes (500–550 bp) for the screening of the entire mtDNA of C. reinhardtii were prepared by PCR using primers labelled with the fluorescent dye Cy-3 (Supplementary Table S1). PCR products were purified using the peqGOLD Cycle-Pure Kit (peqlab, Germany). Recombinant MOC1 (39 pmol) was incubated for 20 min with Cy3-labelled probes (0.12 pmol) in 25 µl of electrophoretic mobility shift assays (EMSA) binding buffer [10 mM HEPES, pH 7.5, 100 mM KCl, 5 mM dithiothreitol (DTT), 0.1% (w/v) Tween 20, 20% (w/v) glycerol, 12.5 mM MgCl2 and 0.2 mg/ml of BSA] containing unspecific competitor (salmon sperm DNA added in mass excess relative to probe) at room temperature. After 20 min, samples were immediately put on ice before they were loaded onto pre-cooled 1.5% (w/v) agarose or 6% (w/v) polyacrylamide gels prepared in 1 × Tris-acetate-EDTA (TAE) buffer (40 mM Tris, pH 7.5, and 2.5 mM EDTA), which were pre-run in the cold room for 30 min before sample loading. Electrophoresis (3 min/300 V followed by 45–60 min/90 V) was carried out in the cold room. For the EMSA with probes S3, S2/nd1 IGR and LHCBM6 NG (0.36 pmol each), 26 pmol recombinant MOC1 was added, and complexes were separated from free probe in a 2% (w/v) agarose gel. Small (24 bp) double-stranded oligonucleotide probes were prepared by annealing equal molarities of complementary single-stranded oligonucleotides (Supplementary Table S1), which were modified with Cy-3 at their 5′-end. Binding reactions contained 17 pmol recombinant MOC1 and 1.5 pmol Cy-3-labelled probes. Poly dI:dC (Sigma) was added as an unspecific competitor (mass excess relative to probe), and unlabelled probes were added to the labelled probe in molar excess. Cy-3 fluorescence was detected using a Typhoon scanner (GE Healthcare).
Determination of the relative mtDNA content in stm6
Total DNA samples (5 ng) extracted from stm6 and the MOC1-complemented strain B13 were used as a template for quantitative PCR (Q-PCR) using the KAPA SYBR FAST Universal reagent (peqlab, Germany) and the DNA Engine Opticon (Bio-RAD, Germany). The primer pair Chlamy nd1 fw/rev (Supplementary Table S1) was used to amplify part of the mitochondrial nd1 gene, and the primers TBP for and TBP rev (Supplementary Table S1) were applied to amplify a sequence of the nuclear TBP (TATA-box–binding protein) gene (NCBI Gene ID: 5716470). Cycle threshold (Ct) values were used to calculate ΔCt values [Ct(nd1)-Ct(TBP)] and the relative mtDNA content in each strain (2−ΔCt). DNA was extracted from two different cultivations per strain, and for each DNA sample, three technical Q-PCR replicates were used.
Extraction of nucleic acids
Total RNA was extracted according to Chomczynski and Sacchi (38), and total cellular DNA was isolated using the cetyltrimethylammonium bromide method (39).
Reverse transcriptase–Q-PCR
Real-time reverse transcriptase (RT)–PCR was performed with total RNA samples, which were subjected to DNaseI (RQ1 RNase-free DNase, Promega) digest before reverse transcription and PCR amplification using the SensiFAST™ SYBR No-ROX One-Step Kit (BIOLINE, Germany). SYBR Green I fluorescence was recorded on a DNA Engine Opticon (Bio-RAD, Germany). For the quantification of mature transcripts, 100 ng of total RNA was used, and CYN19-3 (Gene ID: 5719445) served as a housekeeping gene (primers CYN 19-3 fw/rev). The amount of unprocessed transcripts was quantified using 400 ng of total RNA, and TBP (TATA-box–binding protein) served as a reference gene. Relative expression values for mitochondrial transcripts in mutant stm6 and the complemented strain (B13) were calculated according to Pfaffl (40). For the quantification of antisense transcripts, strand-specific reverse transcription was carried out, and RT reactions in the absence of primer served as controls to exclude primer-independent cDNA synthesis. Primer sequences are given in Supplementary Table S1.
Cyclic amplification and selection of targets
The randomized DNA library was prepared as described by Blackwell (41) using a 56-nt-long oligonucleotide template (Supplementary Table S1) containing 20 internal random nucleotides, which were flanked on either side by 18 nt representing BamHI and HindIII restriction sites. A double-stranded and Cy-3-labelled library was prepared by PCR using 1.13 pmol single-stranded template and 30 pmol of primers SELEX Cy-3 fw and SELEX rev. Excess primers were removed from the library by gel purification using 14% native polyacrylamide gels. For sequence selection, recombinant MOC1 (3.3–17.5 pmol) was incubated with the double-stranded library (1.65 pmol) for 20 min in 25 µl of EMSA binding buffer [10 mM HEPES, pH 7.5, 100 mM KCl, 5 mM DTT, 0.1% (w/v) Tween 20, 20% (w/v) glycerol, 12.5 mM MgCl2 and 0.2 mg/ml of BSA] at room temperature before binding reactions were stopped on ice. Unbound library was separated from complexes by electrophoresis (5 min at 300 V then 90 min at 120 V) in 10% native polyacrylamide gels, which were run in the cold room using pre-cooled 1 × TAE buffer (40 mM Tris, pH 7.5, and 2.5 mM EDTA) as the running buffer. Complex bands were excised from the gel, and DNA was extracted according to Blackwell (41). Extracted DNA served as a PCR template for the generation of a target-enriched library used in the subsequent round of selection. During the selection process, the amount of recombinant MOC1 was gradually reduced, and unspecific competitor (poly dI:dC) was added to increase selection stringency. The progress of selection was analysed by sequencing of library sequences after cloning into vector pGem-T-Easy (Promega).
Detection of a transcript containing cob in sense and L2b in antisense orientation
First 2 µg of total RNA extracted from the wild-type strain cc400 was reverse transcribed using M-MLV reverse transcriptase (Promega) and 170 pmol of primer cob-L2b as N1 rev (Supplementary Table S1) according to the manufacturer’s instructions. In a control reaction, reverse transcriptase was replaced by nuclease-free water (−RT control). Excess primer was removed from the cDNA using the peqGOLD Cycle-Pure Kit (peqlab, Germany). Diluted (1:20 in nuclease-free water) cDNA (2 µl) from ±RT reactions was used as a template for the first PCR with primers cob-L2b as N1 fw/rev. Purified and diluted (1:10) products (2 µl ± RT) from the first PCR were then used in a second nested PCR with primers cob-L2b as N2 fw/rev. PCR products were analysed in a 1.5% (w/v) agarose gel, which was stained with SYBR Gold (Invitrogen, Germany). The dominating band migrating at 700 bp was excised, and DNA was purified using the peqGOLD gel extraction kit (peqlab, Germany) before cloning into pGem-T-Easy and sequencing.
RESULTS
MOC1 is targeted to the mitochondrion
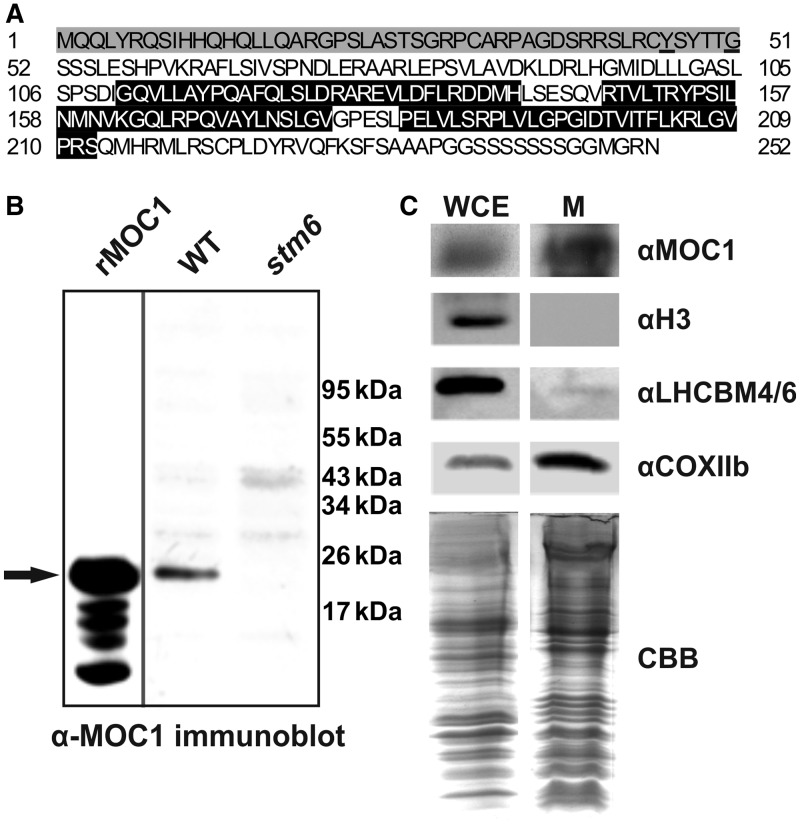
As reported before (22), the nuclear genome of C. reinhardtii encodes six distinct members of the mTERF family (Table 1). Two of the C. reinhardtii mTERF proteins (MOC5 and MOC6) contain putative transit peptides for chloroplast targeting and four mTERFs (MOC1–4) contain mitochondrial targeting sequences according to the sub-cellular prediction tool Predalgo (43), which was developed for use with proteins from C. reinhardtii or other green alga. Among the four proteins is MOC1, which is in the centre of interest in this study. In silico analyses performed with the amino acid sequence of MOC1 (Figure 1A) and the sub-cellular prediction tool MitoProtII (44) predicted that this protein localizes to the mitochondrion (probability of export to mitochondria 0.53; cleavage site 46) (Figure 1A). Cleavage site predictions (amino acid 46 MitoProtII/amino acid 51 Predalgo) are in good agreement with the observed apparent molecular weight of mature MOC1 detected in a C. reinhardtii crude protein extract by immunoblotting (Figure 1B; WT) using an antiserum raised against the full-length MOC1 protein. Processed MOC1 in the algal protein extract displayed an apparent molecular weight similar to that of a recombinant His-tagged MOC1 (Figure 1B; rMOC1) expressed in E. coli (Supplementary Figure S1) and lacking the mitochondrial targeting sequence (amino acids 1–45; Figure 1A) predicted by MitoProtII. This recombinant protein was used throughout the present study and will from here on be referred to as recombinant MOC1. In a previous study, the mitochondrial localization of MOC1 was reported (18), but the amino acid sequence (UniProtKB Q8LJS6_CHLRE) available at that time predicted an MOC1 protein consisting of 373 amino acids and containing a C-terminal part, which was not related to mTERF domains. Consequently, biochemical localization studies with purified organelles and a new antiserum (raised against full-length MOC1; UniProtKB A8IXZ5_CHLRE) were repeated to confirm that this protein indeed localizes to algal mitochondria. To confirm a mitochondrial localization of MOC1, whole-cell extracts (Figure 1C; WCE) and mitochondrial extracts (Figure 1C; M) were compared regarding their MOC1 content by immunoblot experiments (Figure 1C; αMOC1) clearly demonstrating that MOC1 becomes highly enriched during mitochondria purification. A mitochondrial marker (Figure 1C; αCOXIIb), the nucleus-encoded cytochrome c oxidase subunit IIb, showed a comparable enrichment during mitochondria purification, whereas chloroplast-targeted light-harvesting proteins (Figure 1C; αLHCBM 4/6) and the nuclear histone H3 (Figure 1C; αH3) were depleted during mitochondria isolation. In summary, in silico studies and the enrichment of MOC1 during mitochondria purification demonstrate that this protein resides in C. reinhardtii mitochondria.
Table 1.
The six distinct mTERF factors encoded by the C. reinhardtii nuclear genome
| UniProtKB | Gene | No. of mTERF motifs | Sub-cellular localization prediction by Predalgo (score) | Closest homologue A. thaliana (max score) | Closest homologue Homo sapiens (max score) | Closest homologue Drosophila melanogaster (max score) | |
|---|---|---|---|---|---|---|---|
| MOC1 | A8IXZ5 | Cre12.g542500a | 3 | Mitochondrion (1.54) | NM_116533 (164) | mTERF1 (mTERF) (73.9) | D-mTERF3 (78.6) |
| MOC2 | E1VD14 | Cre10.g427000 | 5 | Mitochondrion (1.23) | NM_129964 (342) | mTERF1 (mTERF) (112) | D-mTERF3 (87.4) |
| MOC3 | A8JHP3/E1VD13 | Cre12.g560750 | 6 | Mitochondrion (1.54) | NM_129964 (273) | mTERF1 (mTERF) (115) | D-mTERF3 (73.6) |
| MOC4 | Cre09.g408050 | 4 | Mitochondrion (0.78) | NP_566005.1 (316) | mTERF1 (mTERF) (99) | D-mTERF3 (80.9) | |
| MOC5 | Cre14.g611200 | 2 | Chloroplast (3.51) | NM_116533 (126) | mTERF1 (mTERF) (49.3) | D-mTERF3 (52.0) | |
| MOC6 | A8IH52 | Cre07.g330650 | None | Chloroplast (0.99) | NP_566005.1 (92) | mTERF1 (mTERF) (45.4) | D-mTERF3 (35) |
For each of the mTERF genes, the locus name (C. reinhardtii v5.3; http://www.phytozome.com/) and available UniProtKB entries (http://www.uniprot.org/) are given. Predalgo (42), https://giavap-genomes.ibpc.fr/cgi-bin/predalgodb.perl?page=main), was used to predict the sub-cellular localization in silico. The number of mTERF motifs was determined using the SMART tool (http://smart.embl-heidelberg.de/). In addition the closest (highest Max score) homologue (NCBI Delta-BLAST; http://www.ncbi.nlm.nih.gov/) from H. sapiens, D. melanogaster and A. thaliana is indicated for each C. reinhardtii mTERF.
aPredicted protein truncated.
Figure 1.
In silico prediction and biochemical evidence for mitochondrial targeting of MOC1. (A) Amino acid sequence (UniProtKB A8IXZ5) of MOC1. The mitochondrial targeting sequence is highlighted in grey, and cleavage sites predicted by MitoProtII or Predalgo are underlined. The three mTERF motifs are highlighted in black. (B) Comparison of the apparent molecular weight of recombinant His-tagged MOC1 lacking the mitochondrial targeting sequence predicted by MitoProtII (rMOC1) and mature MOC1 in a C. reinhardtii wild-type protein extract (WT) by immunodetection. An arrow indicates the position of recombinant and native MOC1. The MOC1 knockout mutant stm6 served as a control to assess the specificity of the antiserum raised against MOC1. (C) Immunodetection of MOC1 (αMOC1) in whole-cell extracts (WCE) and purified mitochondria (M). Immunoblots using antibodies directed against histone H3 (αH3) and light-harvesting proteins (αLHCBM4/6) were performed to assess the purity of the mitochondrial fraction. COXIIb served as a mitochondrial marker protein. A Coomassie brilliant blue (CBB) stain of mitochondrial fractions and the whole-cell extract served as a loading control.
In vitro MOC1 binds specifically to mitochondrial DNA
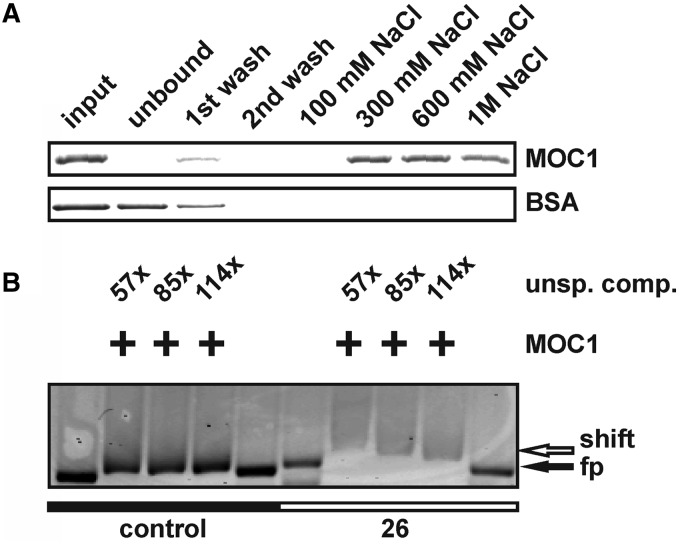
Most of the mTERF factors characterized to date bind double-stranded DNA, thereby regulating the transcription or replication of mtDNA (23), but additional functions such as a role in translational regulation have been reported (29). To investigate whether MOC1 binds to double-stranded DNA (dsDNA), as is suggested by the existence of mTERF motifs (Figure 1A), its binding to double-stranded DNA cellulose was assessed (Figure 2A). The recombinant MOC1 bound to dsDNA–cellulose (Figure 2A; input versus unbound; MOC1), whereas a non–DNA-binding control protein (Figure 2A; input versus unbound; BSA) was found in the unbound fraction. Binding of MOC1 to the DNA resin could be reversed by high ionic strength (Figure 2A; 300 mM–1 M NaCl; MOC1), typical for DNA–protein interactions (45). The wash–stable interaction of MOC1 with dsDNA–cellulose indicated a dsDNA-binding activity of the protein. Against the background that structure and gene content of mtDNAs vary enormously between eukaryotic organisms (1,2,16), the already identified binding sites for mammalian or insect mTERFs (23) cannot be used to identify respective binding sites in C. reinhardtii mtDNA. With the aim to identify genome regions containing binding sites for MOC1, the entire mtDNA of C. reinhardtii was systematically screened for putative binding sites (Supplementary Figure S2). For this, 30 overlapping and fluorescently labelled DNA probes (550–600 bp) covering the entire mitochondrial genome were used in EMSA. Among the tested probes, an mtDNA fragment No. 26 (NCBI Accession NC_001638; nt 13064–13604), containing parts of the ribosomal L8 and S3 rRNA-coding modules, showed binding to recombinant MOC1 even in presence of excess unspecific competitor (Supplementary Figure S2 and Figure 2B; +MOC1; 57–144 × unsp. comp.).
Figure 2.
In vitro studies with the recombinant MOC1 protein indicate specific binding to dsDNA. (A) Incubation of recombinant MOC1 or BSA with dsDNA–cellulose. Input samples taken before incubation with DNA cellulose and supernatant (unbound) samples after incubation were analysed by SDS–PAGE and CBB staining. Wash fractions (first and second wash) and eluted fractions (100 mM–1 M NaCl) were analysed as well. BSA served as a control to assess the stringency of the chromatographic conditions. (B) Gel shift assay with fluorescently-labelled mtDNA probe 26 (NCBI Accession NC_001638; nt 13064-13604) and a control probe representing part of the nuclear LHCBM6 gene (NCBI Accession M24072; nt 756–1289). Fluorescent probes were detected via their Cy-3 labels. Addition of recombinant MOC1 is indicated by ‘+’, and the mass excess of unspecific competitor (unsp. comp.) is given as well. The position of free probe (fp) and shifted probe (shift) is indicated by arrows.
Identification of an MOC1-binding site within the mitochondrial rRNA-encoding module S3
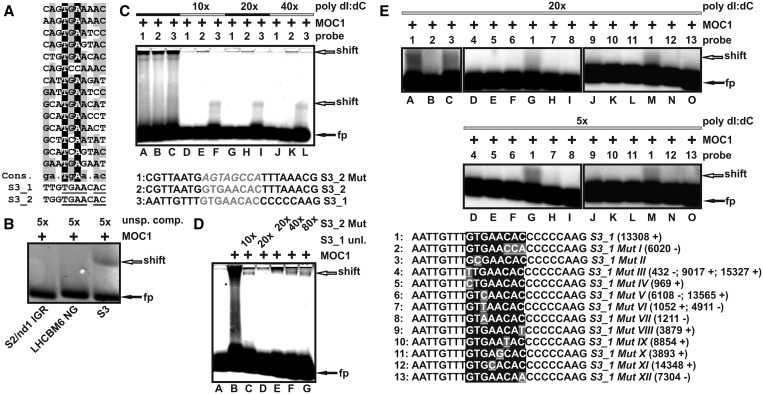
Because of the fact that the probes used within the EMSA screen of the mitochondrial genome of C. reinhardtii (Supplementary Figure S2 and Figure 2B) were 550–600-bp long and thus contained many potential binding sites, an additional strategy had to be applied to narrow down the actual binding site on probe 26 (Figure 2B). CAST [cyclic amplification and selection of targets; (46)] was conducted to derive a consensus sequence (Figure 3A and Supplementary Figure S3), which could be used to identify the binding site on the mtDNA fragment representing parts of the L8 and S3 rRNA-coding modules. After 10 rounds of selection, 14 of 29 sequences (Figure 3A) contributed to a motif (Supplementary Figure S3) identified by the software tool MEME (multiple EM for motif elicitation) as part of the MEME suite (47). Two identical octanucleotide sequences (Figure 3A; S3_1 and S3_2) showing similarity to the motif obtained by the CAST experiment were identified within the mtDNA fragment (No. 26), which showed binding to MOC1 in the EMSA screen [Supplementary Figure S2 and Figure 2B]. Both sequences GTGAACAC are located in the small-subunit rRNA-coding module S3 of the mitochondrial genome (S3_1 nt position 13308–13315 and S3_2 nt position 13406–13413; NCBI Accession NC_001638). In a first step, the part of probe 26 harbouring the octanucleotide sequences was analysed by EMSA and exclusive binding of this probe (Figure 3B; S3) but not of control probes (Figure 3B; S2/nd1 IGR and LHCBM6 NG) to recombinant MOC1 in the presence of unspecific competitor further suggested that the sequences S3_1 and S3_2 represent MOC1-binding sites. The sequences S3_1 and S3_2 were further analysed by mobility shift assays using oligonucleotide probes in which the octanucleotide GTGAACAC was flanked by eight nucleotides at either side [Figure 3C; S3_1 (probe 3) and S3_2 (probe 2)]. An additional oligonucleotide served as a control and was derived from S3_2 with the exception that the GTGAACAC sequence was disrupted by shuffling the eight nucleotides contained in the motif [Figure 3C; S3_2 Mut (probe 1)]. All three probes showed binding to recombinant MOC1 in the absence of unspecific competitor (Figure 3C, lanes A–C), but only probes having an intact GTGAACAC motif bound to MOC1 in the presence of excess unspecific competitor (Figure 3C; lanes D–L; poly dI:dC). Similar results were obtained using a GST-tagged version of recombinant MOC1 (Supplementary Figure S4). To confirm specific recognition of the identified octanucleotide motif further, specific and unspecific competition experiments were carried out with the fluorescently-labelled probe S3_1 (Figure 3D). When the unlabelled probe S3_1 (Figure 3D; S3_1 unl.) was added in excess as a specific competitor to the labelled probe S3_1, a 20-fold excess abolished complex formation almost completely (Figure 3D; lane D), whereas the same and even higher amounts of unlabelled unspecific competitor (Figure 3D; S3_2 Mut) still allowed complex formation (Figure 3D; lanes E–G). Further evidence that the motif GTGAACAC is specifically recognized by MOC1 was obtained by binding studies with mutants of the motif (Figure 3E). When motif positions 7 and 8 (Figure 3A; ultimate and penultimate positions AC) were permuted to yield probe S3_1 MutI binding to recombinant MOC1 was no longer stable in the presence of unspecific competitor (Figure 3E; lane A versus B). Replacement of thymine at position two by cytosine had a smaller effect on complex stability, but decreased complex formation compared with the probe containing the wild-type sequence (Figure 3E; lane A versus C). As single mutations introduced into the S3_1 motif did not completely abolish complex formation, additional single mutations were tested (Figure 3E; sequences 4–13). A systematic analysis of all possible octanucleotide motifs derived from S3_1 and deviating from the wild-type motif at only a single position led to the identification of 10 single mutants, which can be found in the mtDNA of C. reinhardtii. For these mutated motifs, binding to MOC1 could not be detected in the presence of unspecific competitor (Figure 3E; 20× and 5× poly dI:dC; lanes D–F, H–—L, N and O). These data allow us to conclude that the octanucleotide motif GTGAACAC represents a target binding site for MOC1, and that similar motifs found in the mitochondrial genome of C. reinhardtii are bound with less affinity in vitro.
Figure 3.
Identification of two MOC1-binding sites located in the mitochondrial rRNA-coding module S3. (A) Alignment of the CAST library sequences, which contributed to the motif identified by MEME. A derived consensus (Cons.) is shown together with the two octanucleotide sequences (S3_1 and S3_2) found in the rRNA-coding S3 module. A consensus >50%, but <90% is highlighted in grey. Sequence positions with a consensus of at least 90% or higher are presented in black. Nucleotides within S3_1/S3_2, which match to the consensus sequence, are underlined. (B) EMSA with a 147-bp probe (NCBI Accession NC_001638; + strand; nt 13293–13439) representing a fragment of probe 26, which contains both octanucleotide sequences S3_1 and S3_2 (S3). Control probes (negative controls) were derived from the intergenic region between the mitochondrial S2 and nd1 genes (NCBI Accession NC_001638; + strand; nt 10269–10381; 134 bp) or the nucleus-encoded LHCBM6 gene (NCBI Accession M24072; nt 756–906; 152 bp). Probes were detected by fluorescence emission of their Cy-3 modification. Addition of recombinant MOC1 (+), the mass excess of unspecific competitor (unsp. comp.), and the position of free (fp) and shifted probe (shift) are indicated. (C) EMSA with short fluorescent oligonucleotide probes (bottom part; sequences of S3_2 Mut, S3_2 and S3_1) and recombinant MOC1 (MOC1 ‘+’) in the presence or absence of unspecific competitor (poly dI:dC) in mass excess relative to the amount of probe (10–40×). Numbers (top part; probe) indicate which probe was added to the binding reaction, and arrows are the positions of free probe (black arrow; fp) and complexes (white arrow; shift) in the gel. (D) Specific and unspecific competition experiments with probe S3_1. The fluorescently labelled probe S3_1 was added to recombinant MOC1 (MOC1 ‘+’) in the presence of its unlabelled counterpart (S3_1 unl.), which was added in molar excess relative to the labelled probe (10×; 20×). The unlabelled probe S3_2 Mut (sequence given in Figure 3C) was used as an unspecific competitor and added to the labelled probe S3_1 in molar excess (20×; 40×; 80×). (E) EMSA binding studies with probe S3_1 (sequence No. 1) and 12 probes containing mutations of the GTGAACAC recognition motif (S3_1 MutI-XII) in the presence of unspecific competitor (poly dI:dC) with mass excess indicated. For each tested octanucleotide motif, the mtDNA locations (NCBI Accession NC_001638) on the ‘+’ or ‘−’ strand is given in brackets.
Phenotypic consequences of an MOC1 knockout
The knockout mutant stm6, which is devoid of a functional MOC1 gene, and which was identified in a forward genetics screen aiming at the identification of mutants affected in short-term light acclimation mechanisms (18), was used to analyse the consequences caused by loss of MOC1. Localization studies demonstrated that this protein is targeted to the mitochondrion of C. reinhardtii (Figure 1C), and in vitro DNA-binding studies (Figures 2 and 3) revealed that MOC1 binds specifically to an octanucleotide motif, which can be found twice in the mitochondrial rRNA-encoding module S3. Consequently, replication and transcription of the mitochondrial genome were investigated in mutant stm6 (Figure 4), as mTERF proteins are known to regulate both processes (23–34).
Figure 4.
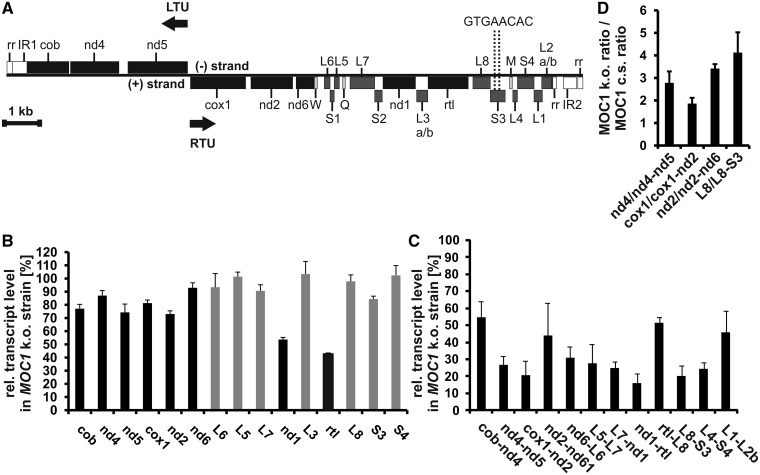
Effects on mtDNA content and transcript levels caused by a knockout of MOC1 in the mutant stm6. (A) Map of the C. reinhardtii mitochondrial genome. Genes are presented as rectangles, and black arrows indicate the directions of transcription. Protein-coding genes are shown in black, ribosomal genes in dark grey and tRNA genes in light grey. nd4, nd5, nd2, nd6 and nd1 encode subunits of the Nicotinamide adenine dinucleotide dehydrogenase complex (complex I). cob is apocytochrome b belonging to complex III. cox1 represents subunit 1 of cytochrome c oxidase (complex IV) and rtl a reverse transcriptase-like protein. W, Q and M represent tRNA genes for tryptophan, glutamine and methionine codons. The genes L1–L7 encode large-subunit rRNA-coding modules and genes S1–S4 small-subunit modules. LTU comprises the genes cob, nd4 and nd5 encoded on the (−)-strand and RTU comprises all remaining genes on the (+)-strand. Structural elements of the left and right telomeres are indicated as IR (inverted repeat region) and rr (86-bp repeat) according to Vahrenholz et al. (4). The map is drawn to scale except for the telomere region (white rectangles; rr and IR). (B) Reverse transcription–quantitative-PCR analyses with all mitochondrial genes encoding OXPHOS complex subunits, the rtl gene encoding a reverse transcriptase-like protein (black bars) and selected ribosomal RNA genes (grey bars). Transcript levels in stm6 (MOC1 k.o. strain) are given in per cent and were calculated relative to the level in the MOC1-complemented strain (set to 100%). Error bars indicate the standard error derived from two biological replicates each including three technical replicates (n = 6). (C) Analysis of bicistronic transcript levels in the MOC1 knockout strain and the complemented strain by RT–Q-PCR. The relative transcript level (MOC1-complemented strain set to 100%) in the knockout strain is plotted on the y-axis, and error bars indicate the standard error (n = 6). (D) Relative expression level ratios of processed to unprocessed transcripts were determined in the MOC1 knockout (MOC1 k.o.) and complemented strain (MOC1 c.s.) by RT–Q-PCR. Ratios in the knockout mutant (y-axis) are given relative to those found in the complemented strain (set to 1), and analysed RNA species are indicated on the x-axis. Error bars (n = 6) are derived from two biological replicates each including three technical replicates.
First the relative mtDNA content was analysed (Supplementary Figure S5) because a role of mTERF proteins in mitochondrial genome maintenance has been reported previously (31). The knockout of MOC1 in stm6 (Supplementary Figure S5; MOC1 k.o.) resulted in a 30% reduction [MOC1 k.o. 71.2% ± 9.8% (SD.)] of the relative mtDNA content compared with the MOC1-complemented strain [Supplementary Figure S5; MOC1 cs; strain B13 (18)]. In general, this reduction can be either because of a reduction in the number of mitochondria in the algal cell or because of a decreased mtDNA copy number in mitochondria of stm6.
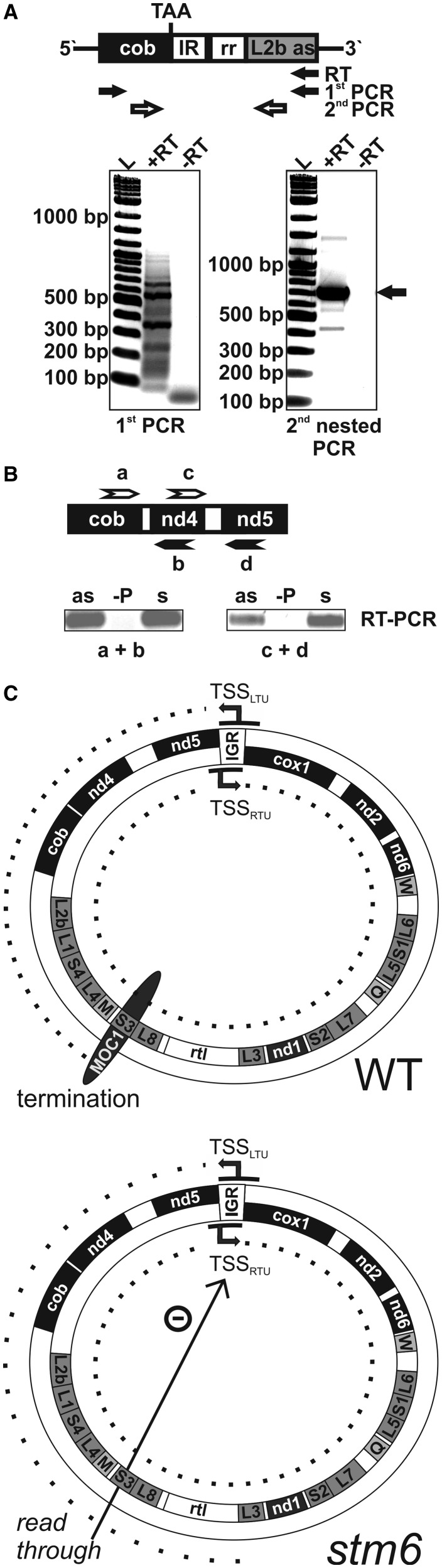
The mitochondrial genome of C. reinhardtii (Figure 4A) is expressed by the generation of long co-transcripts, which are subsequently processed by precise endonucleolytic cleavages (12–14). Two bi-directional transcriptional units exist (13), whose expression could be driven by two potential promoters (one for the leftward and one for the rightward unit) with similarity to the consensus promoter sequence found in fungal mtDNA (A/T)TA(T/G)(T/A)RR(T/G)N (48) and located in the intergenic region between cox1 and nd5. There is evidence in favour (15) and against (13) a function of these sequences serving as a bi-directional promoter, but to date, there is no reported identification of transcription initiation sites for the mtDNA of C. reinhardtii. The rightwards unit (Figure 4A) starts with the cox1 gene and the leftwards unit begins with the gene nd5. For the rightwards transcription unit (RTU), four different polycistronic transcripts were identified (13) by transcript mapping studies (cox1-nd2-nd6-trnW, L6-S1-L5, L7-S2-nd1-L3-rtl-L8-S3 and trnM-L4-S4-L1-L2). A longer precursor covering the entire rightwards unit could not be identified, but our RT–PCR data demonstrated co-transcription of nd6-L6, L5-L7 and S3-L4 providing evidence that a long precursor covering the entire rightwards unit is generated (Supplementary Figure S6). Expression of the leftwards transcription unit was shown to result in co-transcripts containing the open reading frames of nd5 and nd4 (12), and transcriptional linkage of cob and nd4 (Supplementary Figure S6) might indicate that all three genes of the leftwards unit are transcribed as a long polycistronic message.
Analysis of mature transcript levels (Figure 4B) in stm6 (MOC1 k.o.) showed that disruption of the MOC1 gene leads to changes in the mitochondrial transcriptome, when it is compared with that of the MOC1-complemented strain (MOC1 c.s.). The mRNA amount of protein-coding genes was reduced in the MOC1-free mutant (Figure 4B; black bars) with reductions in the 10–30% range for most of the genes except for nd1 [53.6% ± 1.6% (SE)] and rtl [42.5% ± 0.9% (SE)], which showed the strongest reduction. Levels of rRNA-coding modules were less affected by a loss of MOC1 with either no change compared with the complemented strain (Figure 4B; grey bars; L5, L3, L8 and S4) or slight reductions (Figure 4B, grey bars; L6, L7 and S3). Similar to the mode of mtDNA transcription found in other protists (16), the mitochondrial genes in C. reinhardtii are co-transcribed to yield polycistronic RNAs, which are then processed to generate mature monocistronic RNAs (12–15). If RT–Q-PCR is conducted using primers binding within single-coding sequences (Figure 4B), therefore, not spanning intergenic regions, polycistronic RNA species will serve as templates for cDNA synthesis in addition to mature RNAs. Because of the far lower abundance of polycistronic species, however, analyses shown in Figure 4B will primarily detect changes in mature RNA levels. Against the background that polycistronic RNAs are the primary products of C. reinhardtii mtDNA transcription, exclusive quantification of such species seems to be more informative than a quantification of mature RNAs (Figure 4B), if changes in transcriptional activity (initiation rates) are in the focus of interest. Recent studies have shown that the relative abundance of mature RNAs encoded by distinct mitochondrial genes shows a wide variation, despite the fact that they originate from a common precursor (49–51), thus reflecting the importance of post-transcriptional regulation of mature transcript abundance in mammalian mitochondria. A similar variation of mature transcript abundance has been reported for the mitochondrial genome of C. reinhardtii with rRNAs being much more abundant than mRNAs (13).
To address effects on mtDNA transcription in stm6 more directly, additional RT–Q-PCR analyses were conducted (Figure 4C). Because of their length and extremely low abundance (49), full-length unprocessed precursor transcripts are normally not amenable to quantification by RT–Q-PCR. Therefore, primers were designed to bind in two adjacent coding sequences to exclude fully processed monocistronic mRNAs from the analysis. When RT–Q-PCR was carried out with primers spanning intergenic regions (Figure 4C), Ct (cycle threshold)-values increased by ∼10–14 cycles compared with analyses mainly detecting mature RNAs (Figure 4B), demonstrating that the abundance of mature RNAs is far higher. These analyses revealed more pronounced transcriptomic differences between stm6 (MOC1 k.o.) and the MOC1-complemented strain for both transcription units (60–80% reduction for most of the analysed genes). The subtle differences in mature RNA levels between stm6 and complemented strain (Figure 4B) along with strong reductions in the level of unprocessed transcripts in stm6 (Figure 4C) suggest that the efficiency of RNA processing could be altered in stm6. If the relative expression level ratio of mature to unprocessed (bicistronic) RNAs is determined for selected genes in total RNA preparations of mutant and complemented strain a significantly higher ratio can be observed for stm6 (Figure 4D). This is indicative of an altered processing activity in the mutant, which could act as a compensatory mechanism helping to maintain normal levels of mature RNA under conditions of impaired mitochondrial transcription control. In summary, the data clearly demonstrate that a loss of MOC1 strongly reduces the level of unprocessed RNAs derived from both transcription units without having strong effects on mature RNA abundance for most of the protein-coding genes except for nd1 and rtl (Figure 4B and C). A changed mature RNA level of nd1 is in agreement with a previous less comprehensive study (18), which mainly focused on light-induced changes of the mitochondrial transcriptome in stm6. The reduced mtDNA content (Supplementary Figure S5) in stm6 might indicate that genome maintenance is affected in this mutant. Changes in the relative abundance of mature versus unprocessed transcripts in stm6 (Figure 4D) demonstrate that transcriptomic differences cannot be simply attributed to the reduced mtDNA content. Against the background that MOC1 binds to mtDNA in a specific fashion, it seems more reasonable to assume a deregulated transcription initiation or termination as the primary cause. Growth of the MOC1 knockout mutant stm6 is severely impaired under strictly heterotrophic conditions (Supplementary Figure S7), indicating that normal mitochondrial function requires MOC1.
The absence of MOC1 causes aberrant transcription of the non-coding mtDNA strand
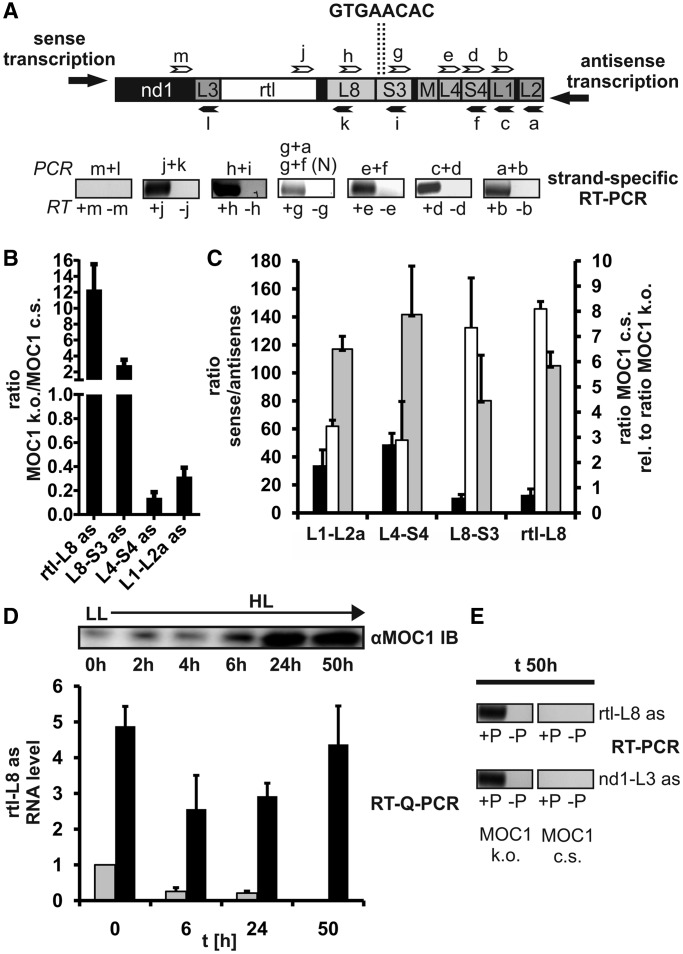
Analysis of the mitochondrial transcriptome in the MOC1-free mutant stm6 and the fact that MOC1 binds to mitochondrial DNA suggest that the transcription of both transcription units could be impaired. The two binding sites, which were identified in vitro, are both located in the rRNA-coding module S3 (Figure 4A; GTGAACAC), a region distal to the mtDNA region harbouring potential promoters for both units. A direct effect of MOC1 on transcription initiation rates, therefore, seems to be an unlikely consequence of MOC1 binding to the S3 sites. Among the described functions of mTERF factors is the termination of mtDNA transcription at specific sites (32,33), so that a potential terminator function of MOC1 was investigated (Figure 5). Impaired termination of transcription in a knockdown cell line depleted of the Drosophila mTERF protein DmTTF (34) resulted in a higher level of transcripts encoded by genes located downstream of the termination site, as a result of read-through transcription. Similar observations have recently been made for human mTERF (28), which showed that manipulation of mTERF protein levels affected read-through transcription of the antisense mtDNA strand. An investigation of sense-transcript levels in stm6 did not reveal a higher amount of transcripts encoded by genes located downstream of the S3-binding sites (Figure 4B and C), which challenges a function of MOC1 as a terminator of sense-transcription of the rightwards transcription unit. The existence of antisense (as) transcripts in the mitochondrial transcriptome of C. reinhardtii has never been reported before, but strand-specific RT–PCR demonstrated the existence of antisense transcripts derived from the non-coding strand of the right mtDNA arm (Figure 5A; strand-specific RT–PCR). The gene cluster nd1-L3-rtl-L8 is located downstream of the MOC1-binding sites, which are contained in the module S3 (Figure 5A; upper part), if transcription proceeds in the antisense direction. In contrast, parts of the S3 module and the cluster M-L4-S4-L1-L2 are located upstream of the GTGAACAC binding sites. For all rRNA modules situated upstream of S3, antisense RNA could be detected by RT–PCR in wild-type total RNA samples (Figure 5A; a + b, c + d, e + f and g + f), whereas for the cluster nd1-L3-rtl-L8, only antisense RNA derived from rtl and L8 was detectable (Figure 5A; h + i and j + k). Longer polycistronic antisense transcripts are of very low abundance, and detection by RT–PCR required two successive PCRs [Figure 5A; g + a and g + f (N)]. As shown by these RT–PCR experiments, only parts of the non-coding strand of the right mtDNA arm are transcribed to an extent allowing their detection. Furthermore, the results indicate that transcription of the non-coding strand ends in the rtl-L8 region adjacent to the S3 module harbouring the two MOC1-binding sites identified in vitro.
Figure 5.
Detection and quantification of antisense RNAs encoded by sequences upstream or downstream of the MOC1-binding site. (A) Upper panel: arrangement of genes in the genome area surrounding the MOC1-binding sites (dotted lines) identified in vitro. Binding sites of forward primers (white arrows) and reverse primers (black arrows) used for reverse transcription and PCR are indicated. Lower panel: SYBR Gold-stain of RT–PCR products separated in an agarose gel after 40 PCR cycles. RT–PCR was carried out with total RNA samples from C. reinhardtii wild-type cc400. Strand-specific reverse transcription was performed in the presence (+) or absence (−) of gene-specific forward primers (white arrows; b, d, e, g, h, j and m) to rule out primer-independent cDNA synthesis by self-priming of RNA. Reverse primers (black arrows; a, c, f, i, k and l) were subsequently added to amplify cDNA by PCR. Detection of larger antisense RNAs required a second nested (N) PCR. (B) Quantification of antisense RNA levels in stm6 (MOC1 k.o.) and the MOC1-complemented strain (MOC1 c.s.) by strand-specific RT–Q-PCR. Values are given as the ratio of stm6 levels to those of the complemented strain. Error bars indicate the standard error derived from two biological replicates each including three technical replicates (n = 6). (C) Quantification of sense-to-antisense transcript level ratios in stm6 (black bars) and the MOC1-complemented strain (grey bars) by strand-specific RT–Q-PCR. The RNA species analysed is given on the x-axis and sense-to-antisense ratios on the left y-axis. The relative difference (MOC1 c.s. versus MOC1 k.o.) in sense-to-antisense ratios (white bars) is given on the right y-axis with ratios in the knockout mutant set to 1. Error bars indicate the standard error derived from three biological replicates, each including three technical replicates (n = 9). (D) Upper panel: anti-MOC1 immunoblot showing the time course of MOC1 accumulation after shifting phototrophic cultures of the complemented strain from low light (LL; 0 h) to high light (HL; 2, 4, 6, 24 and 50 h). Lower panel: RNA levels (y-axis) of antisense transcripts derived from the gene rtl and the rRNA-encoding module L8 in the MOC1-complemented strain (grey bars) and knockout mutant (black bars) determined by RT–Q-PCR. Time points (h) used for sample collection and RNA extraction during the LL-to-HL shift experiment are indicated on the x-axis. The RNA level at time point t0 in the complemented strain was set to 1. Error bars indicate the standard error derived from four technical replicates (n = 4). (E) SYBR Gold-stain of RT–PCR products separated in an agarose gel after 44 PCR cycles. RNA extracted from complemented strain and knockout mutant after 50 h of high-light exposure was used to detect two antisense RNA species encoded by sequences located downstream of the MOC1-binding sites.
More detailed analyses of antisense RNA levels in stm6 (MOC1 k.o.) and complemented strain (MOC1 c.s.) by RT–Q-PCR (Figure 5B) revealed dramatic differences between both strains. For antisense transcripts containing rtl and L8 sequences, a 12-fold higher level was found in stm6 [Figure 5B; rtl-L8 as; 12.3 ± 3.2 (SE)] compared with the complemented strain. The amount of antisense transcripts derived from L8 and S3 was also higher in the MOC1-free mutant [Figure 5B; L8-S3 as; 2.8 ± 0.7 (SE)], whereas quantification of antisense transcripts encoded by genes located upstream of the S3-binding sites (Figure 5B; L4-S4 as; L1-L2a as) showed that their amount in stm6 was far lower than in the complemented strain [L4-S4 as 0.14 ± 0.05 (SE); L1-L2a as 0.31 ± 0.08 (SE)]. In addition, the relative expression level ratio of sense to antisense transcripts was determined in an independent experiment for both strains (Figure 5C). The relative expression level of sense RNAs was in both strains higher than the level of corresponding antisense RNAs, but stm6 (Figure 5C; black bars; 11- to 49-fold) showed in general a lower sense-to-antisense ratio than the complemented strain (grey bars; 80- to 140- fold). Importantly, the relative difference between stm6 and complemented strain ratios (Figure 5C; white bars) was greater for L8-S3 and rtl-L8 RNA species (7- to 8-fold higher ratios in the complemented strain), which is in agreement with the strong accumulation of these antisense RNAs in the mutant (Figure 5B). To correlate the amount of MOC1 in the complemented strain with the level of antisense RNA, cell cultures were shifted from low to high light under phototrophic conditions (Figure 5D). In previous studies (18,19), a light-sensitive phenotype was reported for the MOC1 knockout mutant and interestingly MOC1 protein accumulates when cell cultures are transferred from low-light to high-light conditions (Figure 5D; αMOC1 IB). In the complemented strain, MOC1 expression is driven by the endogenous promoter (18), and after 6 h of high-light exposure, MOC1 started accumulating to reach a plateau about 24 h after the onset of high-light stress. Accumulation of MOC1 in the complemented strain was accompanied by a reduction in the level of rtl-L8 antisense RNA (Figure 5D; grey bars; 1 at t0 versus 0.21 ± 0.05 at t24h), and after 50 h of high-light exposure, this RNA species was no longer detectable. When the MOC1-free mutant (Figure 5D; black bars) was cultivated under these conditions, rtl-L8 levels first declined within the first 24 h (4.8 ± 0.5 at t0 versus 2.9 ± 0.4 at t24h) and re-accumulated again to reach the pre-stress level (4.4 ± 1.1 at t50h). Although antisense RNAs derived from regions downstream of the MOC1-binding site could be detected in the mutant (Figure 5E; RT–PCR; MOC1 k.o.; rtl-L8 as and nd1-L3 as) after 50 h of high-light exposure, the amount of these transcripts dropped below detection limits in the complemented strain (Figure 5E; MOC1 c.s.). A higher amount of transcripts corresponding to genes located downstream of the S3-binding sites in the MOC1-free mutant under distinct growth conditions (Figure 5B, C and D) indicates that absence of MOC1 increases read-through transcription at this site, which provides evidence that MOC1 functions as a terminator of mtDNA transcription in vivo. The lower level of transcripts encoded by genes located upstream of the termination site (Figure 5B) is in agreement with a previous study reporting in vivo termination of mtDNA transcription by dmTTF in Drosophila (34). Furthermore, the negative correlation between MOC1 protein levels and the level of antisense transcripts (Figure 5D and E) is in support of the view that MOC1 is needed to terminate transcription of the non-coding strand belonging to the right mtDNA arm. In summary, quantification of transcripts resulting from antisense transcription downstream or upstream of the S3-binding sites in an MOC1-containing and an MOC1-free cell line suggests that MOC1 acts as a terminator at the S3 sites in vivo. Transcription of the non-coding strand of the right arm of the mitochondrial genome is a novel finding and raises the question of how such transcripts might be generated.
Transcriptional linkage of the cob and L2b gene indicates that the leftwards transcription unit extents to L2b of the right mtDNA arm
One possible explanation for the finding that the non-coding (antisense) strand of the right mtDNA arm is transcribed (Figure 5A) is the existence of promoter sequences at the right end of the mitochondrial genome (Figure 4A). Transcription initiation at these sites would generate mainly non-coding RNA, so that another potential explanation was investigated (Figure 6). Most of the in vitro studies conducted on the configuration (linear versus circular molecules) of C. reinhardtii mtDNA supported the view that the vast majority of mtDNA molecules in this green alga are linear (8,9). However, the existence of a small fraction of circular molecules in preparations for electron microscope studies has been reported (9), and successful PCR amplification of a sequence containing the right and left end of the linear C. reinhardtii mitochondrial genome further indicated that mtDNA populations contain at least small amounts of circular molecules (15,52). The sequence [GenBank Accession No. M83998, (52)] obtained by PCR amplification of mtDNA preparations from C. reinhardtii using primers binding to the 3′-ends of cob and L2a and published by Ma et al. (52), suggested that in a fraction of mtDNA molecules, the cob-coding sequence is fused to L2b in antisense orientation. The part connecting both genes contained only one of the inverted repeat regions present in each of the two linear molecule ends and was proposed to result from homologous recombination events (52). Transcription of such molecules starting upstream of nd5 (Figure 4A) could result in a transcript extending to L2b in antisense orientation (Figure 6A; upper part) instead of terminating downstream of cob (Figure 4A). To detect such transcripts in total RNA preparations of C. reinhardtii RT–PCR was applied and confirmed that transcripts comprising cob and L2b do indeed exist (Figure 6A; upper part). The product obtained after the second nested PCR amplification (Figure 6A; lower part; second nested PCR; +RT) of the cDNA resulting from reverse transcription using a primer specifically binding to the L2b part of the transcript was sequenced. Sequencing revealed fusion of cob and L2b, and the sequence corresponded to the one published by Ma et al. (52). In agreement with a fusion of both genome arms, antisense transcripts derived from the left arm could also be detected by strand-specific RT–PCR (Figure 6B; RT–PCR; as).
Figure 6.
Evidence for co-transcription of cob and L2b and a functional model for MOC1. (A) Upper panel: Structure of a transcript containing the coding sequence of cob and L2b in antisense (as) orientation, which results from the linkage of the left and right arm of the C. reinhardtii mitochondrial genome. The cob sequence and L2b as flank an inverted repeat region (IR) and the 86-bp region (rr) according to Vahrenholz et al. (4). TAA indicates the stop codon of the cob open reading frame. Detection of the transcript by RT–PCR involved RT with a gene-specific primer binding to the L2b as part of the transcript in the first step and two successive PCR reactions with nested primers (white arrows) used in the second PCR step. Lower panel: SYBR Gold stain of an agarose gel used to separate the PCR products of the first and second PCR reaction. Reverse transcription was carried in the absence (−RT) or presence (+RT) of reverse transcriptase. The main PCR product of the second PCR amplification (black arrow) was gel extracted and sub-cloned for sequencing. (B) Detection of antisense RNA derived from the non-coding strand of the left mtDNA arm by RT–PCR. RNA was converted to cDNA by strand-specific reverse transcription to detect antisense RNA (primers a and c; upper part) or sense RNA (primers b and d). PCR (primers a + b or c + d) was used to amplify cDNA before gel separation of products and staining (lower part). As a control, reverse transcription was performed in the absence of primers (−P). (C) A model depicting the function of MOC1 as it is suggested by the sum of data. Upper part: termination of LTU (left part of the circle) transcription by MOC1 at the S3-binding sites in a wild-type (WT) mitochondrion containing MOC1. The mtDNA of C. reinhardtii is presented as a circle. Black arrows indicate the direction of transcription, and dotted lines indicate the length of LTU/RTU-derived transcripts. Abbreviations: TSSLTU/RTU: transcription start sites of the leftward/rightward transcription units; IGR: intergenic region between nd5 and cox1; gene names as given in Figure 4A. Lower part: Lack of MOC1-mediated transcription termination in stm6. Read-through at the S3 site potentially reduces transcription of both units.
DISCUSSION
The sum of data obtained during the characterization of MOC1 function indicates that this protein acts as a factor required for terminating transcription of the leftwards mtDNA unit in wild-type C. reinhardtii mitochondria (Figure 6C; WT). In vitro binding studies led to the identification of two octanucleotide sequences in the rRNA module S3, which bound specifically to recombinant MOC1 (Figure 3). At least in vitro, the two GTGAACAC motifs are bound by MOC1 with a strong preference (Figure 3E), suggesting that the S3 motifs represent prime targets for recognition in vivo. Analyses performed to investigate potential differences between the mitochondrial transcriptome of wild-type cells and that of mutant cells devoid of MOC1 (Figure 4) revealed a general reduction of unprocessed transcript amounts (Figure 4C) in the MOC1-free mutant with both transcription units of the mitochondrial genome being affected. Although levels of unprocessed transcripts are reduced in stm6, the effects on mature RNA abundance are for most of the genes small (Figure 4B), which indicates an altered processing activity in the mutant (Figure 4D). A lower amount of unprocessed transcripts in the mutant could be caused by an increased processing activity alone, or might represent a combination of pleiotropic effects on RNA processing and reduced transcription initiation rates as a consequence of impaired termination at the S3 site. Considering that the S3 module is located in a promoter-distal area of the genome, direct modulation of transcription initiation rates seemed to be unlikely as a function of MOC1. As reported before (28,34), a consequence of impaired mTERF-mediated transcription termination is read-through transcription at respective sites. Inspection of sense transcripts levels in stm6 and comparison with the levels found in the complemented strain (Figure 4C) did not indicate that read-through occurs at the S3 site as far as coding strand transcription is concerned. Interestingly, there is to date no in vivo evidence for the function of human mTERF1 as a terminator acting on the heavy strand, which encodes the majority of sense transcripts in human mitochondria. In contrast, termination of light-strand transcription yielding mainly non-coding RNAs has recently been supported by in vivo data (28) and is in good agreement with a preferential termination of light-strand transcription in vitro (53,54).
Unexpectedly, stable antisense transcripts derived from the right arm of the mitochondrial genome could be detected by RT–PCR (Figure 5A) so that a more detailed analysis of antisense transcript levels encoded by mtDNA sequences directly upstream or downstream of the MOC1-binding site was performed (Figure 5B and C). The higher level of antisense transcripts in stm6 (Figure 5B and C), which are encoded by genes downstream of the S3-binding site (antisense orientation of transcription) can be explained by read-through at the S3 site in the absence of MOC1 (Figure 6C; lower part; stm6). In the complemented strain, the extent of read-through at this site is negatively correlated with the amount of MOC1 (Figure 5D and E). The existence of antisense RNAs in the mitochondrial transcriptome of C. reinhardtii is a novel finding, and an intriguing question raised by the detection of such transcripts was how they are generated. If transcription initiation sites at the right end of the mitochondrial genome in its linear configuration exist, the use of such promoters would generate transcripts mainly containing non-coding information. An alternative explanation considers circular templates formed by end-to-end fusion of the linear genome, thus connecting the coding strand of the leftwards transcription unit (LTU) to the non-coding strand of the RTU. This idea was supported by the detection of a transcript that comprised cob in sense and L2b in antisense orientation, demonstrating that previous models (13) regarding the transcription of C. reinhardtii mtDNA need to be refined. The most reasonable explanation for the observed transcriptional linkage of the LTU and non-coding RNA derived from the RTU is that circular mtDNA molecules (Figure 6C) serve as a template for mtDNA transcription in C. reinhardtii, and that transcription of the leftwards transcription unit extends beyond cob (Figure 6C; outmost dotted line). Restriction digests of purified mtDNA from C. reinhardtii indicated that it is a linear-mapping genome (8), and the identification of telomere-like structures with single-stranded overhangs and terminal inverted repeats was interpreted as a further confirmation of the linear organization of this mitochondrial genome (4). Mitochondrial genome architecture shows a great variability even among strains within species, where the mitochondrial genomes of different strains show an identical gene arrangement but a different DNA topology (42,55). Analyses performed with species displaying this intra-species variability (42,55) revealed features that are frequently found along with ‘true’ linear monomeric mitochondrial genomes. Preparations of mitochondrial DNA from strains or species that possess a linear mitochondrial genome often contain t-circles (56), which are thought to be derived from t-loop (57) structures resembling those found at the telomeres of chromosomes in the nucleus (58). These t-loops form by base pairing of single-stranded overhangs at the extremities of linear molecules and internal complementary sequences in the terminal inverted repeats (57). It was proposed that in analogy to their nuclear counterparts, they protect linear mtDNA from exonucleolytic attack. Both ends of the C. reinhardtii mitochondrial genome contain terminal inverted repeats and single-stranded protrusions, but only for the single-stranded overhang of the right mitochondrial genome arm, a complementary sequence within an 86-bp internal repeat exists (4), which would allow t-loop formation at the right end of the mtDNA. The left arm of the genome lacks an internal repeat precluding t-loop formation. Interestingly, the 86-bp internal repeat of the right mtDNA arm partly overlaps with the gene L2b, so that, although the overhangs found at both sides are not cohesive, (t)-loop formation involving the left single-stranded overhang and the internal repeat within and immediately downstream of L2b would lead to circularization. The structure of the detected transcript (Figure 6A) further suggests that such a circular template indeed exists because the left terminal inverted repeat (Figure 4A; IR1), including the 86-bp region (Figure 4A; rr) downstream of cob, is directly fused to L2b in antisense orientation omitting the right terminal 86-bp region (Figure 4A; right telomere; rr) and the right inverted repeat (Figure 4A; IR2). In further support for the existence of circular end-to-end fused mtDNA molecules, antisense transcripts could also be detected for the non-coding part of the left genome arm (Figure 6B). Transcripts, whose synthesis is initiated in the nd5-cox1 region and comprises the nd4-nd5-cob cluster, might be terminated at the S3-binding sites by MOC1 (Figure 6C; WT; MOC1), and this view is supported by the differential expression of non-coding information in stm6 encoded by sequences located upstream and downstream of the S3 region (Figure 5B).
Besides the strong effect seen for antisense RNA transcription in stm6, expression of coding information was also found to be affected in stm6 (Figure 4 and 6C; stm6; black arrow). A potential explanation for this finding is that impaired termination of LTU transcription has an overall repressive effect on transcription initiation of both LTU and RTU. Similar observations were made after manipulating the levels of dmTTF in Drosophila (34) and were explained by a reduced availability of transcription machinery components at initiation sites because of their engagement in read-through transcription at unoccupied termination sites. It should, however, be noted that post-transcriptional mechanisms have a major impact on mitochondrial RNA abundance (59), and many direct effects on mtDNA transcription might be obscured by pleiotropic effects on RNA processing, stability or translation efficiency. Although the level of certain antisense RNAs is higher in the MOC1-free mutant (Figure 5B), their abundance is still low in comparison with sense transcripts (Figure 5C). It, therefore, seems unlikely that the phenotype of stm6 is directly caused by detrimental effects of antisense RNA on mitochondrial gene expression (e.g. by dsRNA formation with sense counterparts). The low accumulation of these RNA species can probably be explained by the fact that RNA surveillance mechanisms like those described for higher plant organelles (60,61) are still active in the mutant. It is currently difficult to depict all the consequences of impaired transcription termination for mitochondrial gene expression, but a loss of MOC1 clearly impedes mitochondrial function (18) and heterotrophic growth (Supplementary Figure S7).
With MOC1, the first mitochondrial mTERF protein from a phototrophic organism has been characterized regarding its molecular function. Besides MOC1, only four other mTERFs from a phototrophic organism, namely, Arabidopsis thaliana, have been characterized (62–66), but for all four factors, the precise molecular function remains to be elucidated. The mTERFs SOLDAT10 (64) and MDA1 (62) are both targeted to the chloroplast, whereas SHOT1 (66) was found to reside in mitochondria, and RUG2 (65) represents an example of dual targeting to chloroplast and mitochondrion. Aberrant expression of chloroplast genes caused by the BELAYA SMERT/RUGOSA2 (63) and SOLDAT10 (64) mutations together with proteomic analyses of higher plant chloroplast nucleoids (67,68) suggest that mTERFs are crucial components of the plastid transcriptional machinery and required for unperturbed expression of organelle genomes. For the recently identified SHOT1 protein, the phenotype caused by the shot1–2 mutation indicated that higher plant mTERFs also regulate mitochondrial gene expression (66). Changes in organelle gene expression are known to be a source for retrograde signals (69) communicating the physiological state of an organelle to the nucleus where most of the mitochondrial/chloroplast proteins are encoded. mTERF proteins might, therefore, be implicated in the modulation of retrograde signals, and a changed expression of nuclear genes in A. thaliana mTERF mutants supports this idea (64). The loss of a functional MOC1 gene also results in a changed expression of nucleus-encoded genes. Among them are genes encoding subunits of mitochondrial respiratory complexes (18) or stress-related light-harvesting proteins targeted to the chloroplast (19). Perturbed mitochondrial function in stm6 affects the physiological state of the plastid causing an increased light-sensitivity of the mutant (18,19) and enhanced hydrogen production capacity in the chloroplast under anaerobic conditions (20). Against this background, stm6 represents a valuable tool for the analysis of the complex interplay between the chloroplast, the mitochondria and the nucleus.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–7 and Supplementary Methods.
FUNDING
Deutsche Forschungsgemeinschaft [WO 1591/1-1 to L.W.]; European Union [FP7-No245070 KBBE SUNBIOPATH to L.W.]. Funding for open access charge: Imperial College open access publication fund.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the Center for Biotechnology (CeBiTec) at Bielefeld University for access to the Technology Platforms. Furthermore, they are grateful to A. Uhmeyer (Bielefeld University) for help with immunoblotting and RNA extraction.
REFERENCES
- 1.Asin-Cayuela J, Gustafsson CM. Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem. Sci. 2007;32:111–117. doi: 10.1016/j.tibs.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Lipinski KA, Kaniak-Golik A, Golik P. Maintenance and expression of the S. cerevisiae mitochondrial genome—from genetics to evolution and systems biology. Biochim. Biophys. Acta. 2010;1797:6–7. doi: 10.1016/j.bbabio.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 3.Cardol P, Gloire G, Havaux M, Remacle C, Matagne R, Franck F. Photosynthesis and state transitions in mitochondrial mutants of Chlamydomonas reinhardtii affected in respiration. Plant Physiol. 2003;133:2010–2020. doi: 10.1104/pp.103.028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vahrenholz C, Riemen G, Pratje E, Dujon B, Michaelis G. Mitochondrial DNA of Chlamydomonas reinhardtii: the structure of the ends of the linear 15.8-kb genome suggests mechanisms for DNA replication. Curr. Genet. 1993;24:241–247. doi: 10.1007/BF00351798. [DOI] [PubMed] [Google Scholar]
- 5.Smith DR, Lee RW. Nucleotide diversity in the mitochondrial and nuclear compartments of Chlamydomonas reinhardtii: investigating the origins of genome architecture. BMC Evol. Biol. 2008;8:11. doi: 10.1186/1471-2148-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boer PH, Gray MW. Scrambled ribosomal-RNA gene pieces in Chlamydomonas reinhardtii mitochondrial-DNA. Cell. 1988;55:399–411. doi: 10.1016/0092-8674(88)90026-8. [DOI] [PubMed] [Google Scholar]
- 7.Vinogradova E, Salinas T, Cognat V, Remacle C, Marechal-Drouard L. Steady-state levels of imported tRNAs in Chlamydomonas mitochondria are correlated with both cytosolic and mitochondrial codon usages. Nucleic Acids Res. 2009;37:1521–1528. doi: 10.1093/nar/gkn1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grant D, Chiang KS. Physical mapping and characterization of Chlamydomonas mitochondrial DNA molecules: their unique ends, sequence homogeneity, and conservation. Plasmid. 1980;4:82–96. doi: 10.1016/0147-619x(80)90085-2. [DOI] [PubMed] [Google Scholar]
- 9.Ryan R, Grant D, Chiang KS, Swift H. Isolation and characterization of mitochondrial DNA from Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA. 1978;75:3268–3272. doi: 10.1073/pnas.75.7.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valach M, Farkas Z, Fricova D, Kovac J, Brejova B, Vinar T, Pfeiffer I, Kucsera J, Tomaska L, Lang BF, et al. Evolution of linear chromosomes and multipartite genomes in yeast mitochondria. Nucleic Acids Res. 39:4202–4219. doi: 10.1093/nar/gkq1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nosek J, Tomaska L, Fukuhara H, Suyama Y, Kovac L. Linear mitochondrial genomes: 30 years down the line. Trends Genet. 1998;14:184–188. doi: 10.1016/s0168-9525(98)01443-7. [DOI] [PubMed] [Google Scholar]
- 12.Boer PH, Gray MW. The urf-5 gene of Chlamydomonas reinhardtii mitochondria - DNA-sequence and mode of transcription. EMBO J. 1986;5:21–28. doi: 10.1002/j.1460-2075.1986.tb04172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray MW, Boer PH. Organization and expression of algal (Chlamydomonas reinhardtii) mitochondrial-DNA. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1988;319:135–147. doi: 10.1098/rstb.1988.0038. [DOI] [PubMed] [Google Scholar]
- 14.Boer PH, Gray MW. Genes encoding a subunit of respiratory nadh dehydrogenase (nd1) and a reverse transcriptase-like protein (rtl) are linked to ribosomal-RNA gene pieces in Chlamydomonas reinhardtii mitochondrial-DNA. EMBO J. 1988;7:3501–3508. doi: 10.1002/j.1460-2075.1988.tb03226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duby F, Cardol P, Matagne RF, Remacle C. Structure of the telomeric ends of mtDNA, transcriptional analysis and complex I assembly in the dum24 mitochondrial mutant of Chlamydomonas reinhardtii. Mol. Genet. Genomics. 2001;266:109–114. doi: 10.1007/s004380100529. [DOI] [PubMed] [Google Scholar]
- 16.Barbrook AC, Howe CJ, Kurniawan DP, Tarr SJ. Organization and expression of organellar genomes. Phil. Trans. R. Soc. B. 2010;365:785–797. doi: 10.1098/rstb.2009.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liere K, Weihe A, Börner T. The transcription machineries of plant mitochondria and chloroplasts: Composition, function, and regulation. J. Plant Physiol. 2011;168:1345–1360. doi: 10.1016/j.jplph.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Schönfeld C, Wobbe L, Borgstädt R, Kienast A, Nixon PJ, Kruse O. The nucleus-encoded protein MOC1 is essential for mitochondrial light acclimation in Chlamydomonas reinhardtii. J. Biol. Chem. 2004;279:50366–50374. doi: 10.1074/jbc.M408477200. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen AV, Toepel J, Burgess S, Uhmeyer A, Blifernez O, Doebbe A, Hankamer B, Nixon P, Wobbe L, Kruse O. Time-course global expression profiles of Chlamydomonas reinhardtii during photo-biological H2 production. PLoS One. 2011;6:15. doi: 10.1371/journal.pone.0029364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruse O, Rupprecht J, Bader KP, Thomas-Hall S, Schenk PM, Finazzi G, Hankamer B. Improved photobiological H2 production in engineered green algal cells. J. Biol. Chem. 2005;280:34170–34177. doi: 10.1074/jbc.M503840200. [DOI] [PubMed] [Google Scholar]
- 21.Linder T, Park CB, Asin-Cayuela J, Pellegrini M, Larsson NG, Falkenberg M, Samuelsson T, Gustafsson CM. A family of putative transcription termination factors shared amongst metazoans and plants. Curr. Genet. 2005;48:265–269. doi: 10.1007/s00294-005-0022-5. [DOI] [PubMed] [Google Scholar]
- 22.Kleine T. Arabidopsis thaliana mTERF proteins: evolution and functional classification. Front. Plant Sci. 2012;3:233. doi: 10.3389/fpls.2012.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberti M, Polosa Paola L, Bruni F, Deceglie S, Gadaleta Maria N, Cantatore P. mTERF factors: a multifunction protein family. Biomol. Concepts. 2010;1:215. doi: 10.1515/bmc.2010.015. [DOI] [PubMed] [Google Scholar]
- 24.Kruse B, Narasimhan N, Attardi G. Termination of transcription in human mitochondria: Identification and purification of a DNA binding protein factor that promotes termination. Cell. 1989;58:391–397. doi: 10.1016/0092-8674(89)90853-2. [DOI] [PubMed] [Google Scholar]
- 25.Martin M, Cho J, Cesare AJ, Griffith JD, Attardi G. Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell. 2005;123:1227–1240. doi: 10.1016/j.cell.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 26.Park CB, Asin-Cayuela J, Camara Y, Shi YH, Pellegrini M, Gaspari M, Wibom R, Hultenby K, Erdjument-Bromage H, Tempst P, et al. mTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 27.Wenz T, Luca C, Torraco A, Moraes CT. mTERF2 regulates oxidative phosphorylation by modulating mtDNA transcription. Cell Metab. 2009;9:499–511. doi: 10.1016/j.cmet.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Hyvärinen AK, Kumanto MK, Marjavaara SK, Jacobs HT. Effects on mitochondrial transcription of manipulating mTERF protein levels in cultured human HEK293 cells. BMC Mol. Biol. 2010;11:72. doi: 10.1186/1471-2199-11-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cámara Y, Asin-Cayuela J, Park CB, Metodiev MB, Shi YH, Ruzzenente B, Kukat C, Habermann B, Wibom R, Hultenby K, et al. mTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011;13:527–539. doi: 10.1016/j.cmet.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Hyvärinen AK, Pohjoismäki JL, Reyes A, Wanrooij S, Yasukawa T, Karhunen PJ, Spelbrink JN, Holt IJ, Jacobs HT. The mitochondrial transcription termination factor mTERF modulates replication pausing in human mitochondrial DNA. Nucleic Acids Res. 2007;35:6458–6474. doi: 10.1093/nar/gkm676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hyvärinen A, Pohjoismäki J, Holt I, Jacobs H. Overexpression of mTERFD1 or mTERFD3 impairs the completion of mitochondrial DNA replication. Mol. Biol. Rep. 2011;38:1321–1328. doi: 10.1007/s11033-010-0233-9. [DOI] [PubMed] [Google Scholar]
- 32.Fernandez-Silva P, Martinez-Azorin F, Micol V, Attardi G. The human mitochondrial transcription termination factor (mTERF) is a multizipper protein but binds to DNA as a monomer, with evidence pointing to intramolecular leucine zipper interactions. EMBO J. 1997;16:1066–1079. doi: 10.1093/emboj/16.5.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberti M, Fernandez-Silva P, Polosa PL, Fernandez-Vizarra E, Bruni F, Deceglie S, Montoya J, Gadaleta MN, Cantatore P. In vitro transcription termination activity of the Drosophila mitochondrial DNA-binding protein DmTTF. Biochem. Biophys. Res. Commun. 2005;331:357–362. doi: 10.1016/j.bbrc.2005.03.173. [DOI] [PubMed] [Google Scholar]
- 34.Roberti M, Bruni F, Polosa PL, Gadaleta MN, Cantatore P. The Drosophila termination factor DmTTF regulates in vivo mitochondrial transcription. Nucleic Acids Res. 2006;34:2109–2116. doi: 10.1093/nar/gkl181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris EH. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. San Diego: Academic Press; 1989. [DOI] [PubMed] [Google Scholar]
- 36.Asamizu E, Nakamura Y, Sato S, Fukuzawa H, Tabata S. A large scale structural analysis of cDNAs in a unicellular green Alga, Chlamydomonas reinhardtii. I. Generation of 3433 non-redundant expressed sequence tags. DNA Res. 1999;6:369–373. doi: 10.1093/dnares/6.6.369. [DOI] [PubMed] [Google Scholar]
- 37.Atteia A, Adrait A, Brugiere S, Tardif M, van Lis R, Deusch O, Dagan T, Kuhn L, Gontero B, Martin W, et al. A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the alpha-proteobacterial mitochondrial ancestor. Mol. Biol. Evol. 2009;26:1533–1548. doi: 10.1093/molbev/msp068. [DOI] [PubMed] [Google Scholar]
- 38.Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat. Protoc. 2006;1:581–585. doi: 10.1038/nprot.2006.83. [DOI] [PubMed] [Google Scholar]
- 39.Murray MG, Thompson WF. Rapid isolation of high molecular-weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blackwell TK. Selection of protein-binding sites from random nucleic-acid sequences. Methods Enzymol. 1995;254:604–618. doi: 10.1016/0076-6879(95)54043-1. [DOI] [PubMed] [Google Scholar]
- 42.Kosa P, Valach M, Tomaska L, Wolfe KH, Nosek J. Complete DNA sequences of the mitochondrial genomes of the pathogenic yeasts Candida orthopsilosis and Candida metapsilosis: insight into the evolution of linear DNA genomes from mitochondrial telomere mutants. Nucleic Acids Res. 2006;34:2472–2481. doi: 10.1093/nar/gkl327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tardif M, Atteia A, Specht M, Cogne G, Rolland N, Brugière S, Hippler M, Ferro M, Bruley C, Peltier G, et al. PredAlgo, a new subcellular localization prediction tool dedicated to green algae. Mol. Biol. Evol. 2012;26:3625–3639. doi: 10.1093/molbev/mss178. [DOI] [PubMed] [Google Scholar]
- 44.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 45.Alberts B, Herrick G. DNA-cellulose chromatography. Methods Enzymol. 1971;21:198–217. [Google Scholar]
- 46.Wright WE, Binder M, Funk W. Cyclic amplification and selection of targets (CASTing) for the myogenin consensus binding site. Mol. Cell. Biol. 1991;11:4104–4110. doi: 10.1128/mcb.11.8.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tracy RL, Stern DB. Mitochondrial transcription initiation: promoter structures and RNA polymerases. Curr. Genet. 1995;28:205–216. doi: 10.1007/BF00309779. [DOI] [PubMed] [Google Scholar]
- 49.Temperley RJ, Wydro M, Lightowlers RN, Chrzanowska-Lightowlers ZM. Human mitochondrial mRNAs-like members of all families, similar but different. Biochim. Biophys. Acta. 2010;1797:1081–1085. doi: 10.1016/j.bbabio.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation and degradation of human mitochondrial RNA: the prokaryotic past leaves its mark. Mol. Cell. Biol. 2005;25:6427–6435. doi: 10.1128/MCB.25.15.6427-6435.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood A, Marie J, Haugen E, Bracken CP, Rackham O, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma DP, King YT, Kim Y, Luckett WS, Jr, Boyle JA, Chang YF. Amplification and characterization of an inverted repeat from the Chlamydomonas reinhardtii mitochondrial genome. Gene. 1992;119:253–257. doi: 10.1016/0378-1119(92)90279-x. [DOI] [PubMed] [Google Scholar]
- 53.Yakubovskaya E, Mejia E, Byrnes J, Hambardjieva E, Garcia-Diaz M. Helix unwinding and base flipping enable human mTERF1 to terminate mitochondrial transcription. Cell. 2010;141:982–993. doi: 10.1016/j.cell.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Asin-Cayuela J, Schwend T, Farge G, Gustafsson CM. The human mitochondrial transcription termination factor (mTERF) is fully active in vitro in the nonphosphorylated form. J. Biol. Chem. 2005;280:25499–25505. doi: 10.1074/jbc.M501145200. [DOI] [PubMed] [Google Scholar]
- 55.Rycovska A, Valach M, Tomaska L, Bolotin-Fukuhara M, Nosek J. Linear versus circular mitochondrial genomes: intraspecies variability of mitochondrial genome architecture in Candida parapsilosis. Microbiology. 2004;150:1571–1580. doi: 10.1099/mic.0.26988-0. [DOI] [PubMed] [Google Scholar]
- 56.Tomaska L, Nosek J, Kramara J, Griffith JD. Telomeric circles: universal players in telomere maintenance? Nat. Struct. Mol. Biol. 2009;16:1010–1015. doi: 10.1038/nsmb.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tomaska L, Makhov AM, Griffith JD, Nosek J. t-loops in yeast mitochondria. Mitochondrion. 2002;1:455–459. doi: 10.1016/s1567-7249(02)00009-0. [DOI] [PubMed] [Google Scholar]
- 58.de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rorbach J, Minczuk M. The post-transcriptional life of mammalian mitochondrial RNA. Biochem. J. 2012;444:357–373. doi: 10.1042/BJ20112208. [DOI] [PubMed] [Google Scholar]
- 60.Holec S, Lange H, Kühn K, Alioua M, Börner T, Gagliardi D. Relaxed transcription in Arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Mol. Cell. Biol. 2006;26:2869–2876. doi: 10.1128/MCB.26.7.2869-2876.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharwood RE, Halpert M, Luro S, Schuster G, Stern DB. Chloroplast RNase J compensates for inefficient transcription termination by removal of antisense RNA. RNA. 2011;17:2165–2176. doi: 10.1261/rna.028043.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robles P, Micol JL, Quesada V. Arabidopsis MDA1, a nuclear-encoded protein, functions in chloroplast development and abiotic stress responses. PLoS One. 2012;7:14. doi: 10.1371/journal.pone.0042924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Babiychuk E, Vandepoele K, Wissing J, Garcia-Diaz M, De Rycke R, Akbari H, Joubès J, Beeckman T, Jänsch L, Frentzen M, et al. Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc. Natl Acad. Sci. USA. 2011;108:6674–6679. doi: 10.1073/pnas.1103442108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meskauskiene R, Würsch M, Laloi C, Vidi PA, Coll NS, Kessler F, Baruah A, Kim C, Apel K. A mutation in the Arabidopsis mTERF-related plastid protein SOLDAT10 activates retrograde signaling and suppresses 1O2-induced cell death. Plant J. 2009;60:399–410. doi: 10.1111/j.1365-313X.2009.03965.x. [DOI] [PubMed] [Google Scholar]
- 65.Quesada V, Sarmiento-Mañús R, González-Bayón R, Hricová A, Pérez-Marcos R, Graciá-Martínez E, Medina-Ruiz L, Leyva-Díaz E, Ponce MR, Micol JL. Arabidopsis RUGOSA2 encodes an mTERF family member required for mitochondrion, chloroplast and leaf development. Plant J. 2011;68:738–753. doi: 10.1111/j.1365-313X.2011.04726.x. [DOI] [PubMed] [Google Scholar]
- 66.Kim M, Lee U, Small I, des Francs-Small CC, Vierling E. Mutations in an Arabidopsis mitochondrial transcription termination factor-related protein enhance thermotolerance in the absence of the major molecular chaperone HSP101. Plant Cell. 2012;24:3349–3365. doi: 10.1105/tpc.112.101006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmuller R. PTAC2,-6, and-12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell. 2006;18:176–197. doi: 10.1105/tpc.105.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Majeran W, Friso G, Asakura Y, Qu X, Huang M, Ponnala L, Watkins KP, Barkan A, van Wijk KJ. Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol. 2012;158:156–189. doi: 10.1104/pp.111.188474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pesaresi P, Masiero S, Eubel H, Braun HP, Bhushan S, Glaser E, Salamini F, Leister D. Nuclear photosynthetic gene expression is synergistically modulated by rates of protein synthesis in chloroplasts and mitochondria. Plant Cell. 2006;18:970–991. doi: 10.1105/tpc.105.039073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.